Abstract
Tunicates occupy the evolutionary position at the boundary of invertebrates and vertebrates. It exhibits adaptation to broad environmental conditions and is distributed globally. Despite hundreds of years of embryogenesis studies, the genetic basis of the invasive habits of ascidians remains largely unknown. The leathery sea squirt, Styela clava, is an important invasive species. We used the chromosomal‐level genome and transcriptome of S. clava to explore its genomic‐ and molecular‐network‐based mechanisms of adaptation to environments. Compared with Ciona intestinalis type A (C. robusta), the size of the S. clava genome was expanded by 2‐fold, although the gene number was comparable. An increase in transposon number and variation in dominant types were identified as potential expansion mechanisms. In the S. clava genome, the number of genes encoding the heat‐shock protein 70 family and members of the complement system was expanded significantly, and cold‐shock protein genes were transferred horizontally into the S. clava genome from bacteria. The expanded gene families potentially play roles in the adaptation of S. clava to its environments. The loss of key genes in the galactan synthesis pathway might explain the distinct tunic structure and hardness compared with the ascidian Ciona species. We demonstrated further that the integrated thyroid hormone pathway participated in the regulation of larval metamorphosis that provides S. clava with two opportunities for adapting to their environment. Thus, our report of the chromosomal‐level leathery sea squirt genome provides a comprehensive genomic basis for the understanding of environmental adaptation in tunicates.
Keywords: ascidian, environmental adaptation, genome sequencing, invasive species, leathery sea squirt, Styela clava
1. INTRODUCTION
Tunicates occupy the phylogenetic position at the boundary of invertebrates and vertebrates. They display diverse developmental processes and adaptive strategies, and serve as suitable models for exploring the origins of vertebrates and the molecular mechanisms of development. Tunicates are divided into three classes, including Ascidiacea, Appendicularia, and Thaliacea. Several ascidian genomes have been sequenced, such as those of Ciona intestinalis type A (Ciona robusta) (Dehal et al., 2002; Satou et al., 2019), C. savignyi (Small, Brudno, Hill, & Sidow, 2007), Botryllus schlosseri (Voskoboynik et al., 2013), Molgula spp. (Stolfi et al., 2014), and Botrylloides leachii (Blanchoud, Rutherford, Zondag, Gemmell, & Wilson, 2018). The genome resources of these organisms and some gene annotations of Phallusia, Corella and Halocynthia are now available in ANISEED, which is the main database for the worldwide community of scientists working on tunicates (Dardaillon et al., 2020). The genome data revealed that most of the vertebrate gene families could be identified in ascidians, suggesting that ascidians contain most of the ancestral genes involved in chordate development. The majority of the sequenced solitary ascidian genomes are relatively compact with sizes in the range of 100–200 Mb. Recently, a nearly complete genome assembly of C. intestinalis type A (C. robusta) was also constructed at the chromosome level (Satou et al., 2019). The sequenced ascidian genome larger than 200 Mb occurred in colonial species, which is B. schlosseri, with a genome size of about 600 Mb (Voskoboynik et al., 2013). Except for ascidians, Oikopleura dioica, an Appendicularia species, has a quite compact (only 70 Mb) and faster evolving genome compared to other tunicates (Denoeud et al., 2010; Seo et al., 2001). Salpa thompsoni, a Thaliacea species, has a relatively larger genome (>600 Mb) than most of the sequenced solitary ascidians (Jue et al., 2016).
Ascidians exhibit adaptations to diverse environmental conditions and are distributed in shallow seawaters globally (Sharyn J. Goldstien et al., 2011). Moreover, many are considered as invasive species (Lambert, 2007). Despite the great impact of invasive tunicates on biodiversity and agriculture over the past decades, the genetic adaptations and evolution of invading populations remain poorly understood (Zhan, Briski, Bock, Ghabooli, & MacIsaac, 2015). Among the recognized mechanisms underlying the rapid and adaptable responses to stress, heat‐shock proteins (hsps), a group of evolutionarily conserved protein families, are one of the crucial molecules that facilitate coping with environmental fluctuations in organisms through the correction of protein folding or the maintenance of protein homeostasis (Feder & Hofmann, 1999). Genome‐level studies could greatly facilitate the deciphering of the molecular mechanisms underpinning the invasion process (Rius, Bourne, Hornsby, & Chapman, 2015) and provide the genomic data for application in population genetic studies (Chown et al., 2015).
Ascidians, like most metazoans, have a biphasic lifespan. The fertilized eggs first develop into swimming larvae, which then attach to a substrate and metamorphose into adult‐like juveniles. Considerable changes also occur during the metamorphosis stages in ascidians (Karaiskou, Swalla, Sasakura, & Chambon, 2015), including the regression of their notochord. Such abrupt change in body structure during development provides metazoans with two opportunities for adapting to their environment. The cellular basis of larval metamorphosis has been investigated and summarized based on observations from several ascidian species (Cloney, 1982). At the molecular level, signaling pathways, such as the mitogen‐activated protein kinase pathway, the c‐Jun N‐terminal kinase pathway, and the extracellular regulated kinase (ERK) pathway, are involved in Ciona larval metamorphosis (Chambon, Nakayama, Takamura, McDougall, & Satoh, 2007). Moreover, the phosphorylated form of ERK transduces the apoptosis‐activating signal for tail regression in tail tissues of Ciona (Chambon et al., 2007). However, the signaling pathways and regulators of larval metamorphosis in ascidians still need to be further studied.
Styela has a morphology stereotypic of adult solitary ascidians, which was apparently established at least 500 Mya. The Cheungkongella ancestralis, a member of the Chengjiang fauna in Southern China has been reported to be a kind of tunicate (Shu, Chen, Han, & Zhang, 2001). The fossil tunicate, Shankouclava, which was found in South China, was also reported as the first tunicate from the Early Cambrian and is approximately 520 million years old (J. Y. Chen et al., 2003). The fossil, representing the earliest reported tunicate fossil specimen, resembles the ascidian morphologically. In addition, Styela has also long been used as a developmental model. Edwin G. Conklin made a milestone contribution to embryonic cell‐lineage tracking using Styela partita 100 years ago (Conklin, 1905).
Over the course of several decades, Styela, especially the leathery sea squirt S. clava, has also been shown to be an important ecological species because of its invasive potential and ability to adapt to new environments (Dupont, Viard, Dowell, Wood, & Bishop, 2009; S. J. Goldstien, Schiel, & Gemmell, 2010; Locke, Hanson, Ellis, Thompson, & Rochette, 2007). S. clava is native to the northwestern region of the Pacific (Sharyn J. Goldstien et al., 2011), but currently occurs globally, because of anthropogenic transport. Its global spread has had a major impact on the regional marine biodiversity and on aquaculture industries because of fouling. However, appropriate genomic resources remain scarce for this species and the genetic basis of its invasive characteristics are largely unknown.
In the present study, we sequenced the whole genome of S. clava using Pacific Bioscience (PacBio) sequencing platforms and, by combining it with a HiC approach, we achieved a chromosomal‐level assembly of 340 Mb (scaffold N50 of 20.77 Mb) with 17,480 protein‐coding genes. Using both genomic and transcriptomic data, in addition to in situ hybridization and chemical drug inhibition experiments, we expect to aid our understanding of the molecular basis of their broad environmental adaptations of S. clava.
2. MATERIALS AND METHODS
2.1. Animal collection, species identification, fertilization, and embryogenesis
The adults of S. clava (Herdman, 1881) were collected from Weihai City, China, and acclimated in sea water at 18°C in the laboratory. The 18S rRNA and actin genes were amplified, and the PCR products were sequenced for species identification. The whole body of a single adult was used for DNA extraction and genome sequencing. S. clava is a hermaphrodite. The adult animals were dissected, and the mature eggs and sperm were collected from different individuals for fertilization at room temperature. The morphology of different‐staged embryos or larvae were identified and recorded by via Nikon DIC microscopy.
2.2. Genome sequencing, assembly, and annotation
Genomic DNA was extracted from one intact starved adult using the modified phenol/chloroform method. Briefly, the procedures were as follows: the tunic was removed from one starved adult. Its tissues were cut into 1 mm3 pieces and then quickly transferred into six centrifugal tubes. lysis buffer (700 μl: 100 mM NaCl, 10 mM Tris‐HCl, 100 mM EDTA, and 1% SDS, pH8.0) and proteinase K (Merck: final concentration, 200 μg/ml) were added into each tube. The mixtures were incubated at 58°C for 3 hr. Genomic DNA was then extracted using the routine phenol/chloroform method (Engelman et al., 1995). The quality of the extracted DNA was evaluated by agarose gel. DNA with a size ≥15 Kb size and no degradation was observed. Samples were then stored at −20°C for PacBio (Pacific Biosciences) and Illumina sequencing.
Pacific biosciences RS II was used to obtain long reads for assembly. Illumina HiSeq X Ten was used to obtain paired‐end reads for genome survey and error correction of PacBio data. The genome size was estimated based on a k‐mer distribution analysis (Sambrook, 2001). FALCON was used for long‐read assembly (Marcais & Kingsford, 2011) (https://github.com/PacificBiosciences/FALCON) (length_cutoff = 5,000; pa_HPCdaligner_option: ‐k = 14, ‐h = 128, ‐l = 2000, ‐w = 8, ‐T = 8, ‐s = 700, ‐M = 32). The assembled genome completeness was validated by checking the BUSCO database (Chin et al., 2016).
Repetitive sequences in the genome were identified using RepeatModeler (version 1.0.10) and RepeatMasker (version 3.3.0). The kimura substitution rates of different types of transposable elements were calculated using the Perl script calcDivergenceFromAlign.pl from RepeatMasker. Three approaches were used to predict protein‐coding genes: homology‐based predictions, de novo predictions, and transcriptome‐based predictions. Three de novo gene prediction tools, Augustus (http://bioinf.uni‐greifswald.de/augustus/) (Simao et al., 2015), GlimmerHMM (http://ccb.jhu.edu/software/glimmerhmm/) (Stanke et al., 2006), and SNAP (http://homepage.mac.com/iankorf/) (Majoros, Pertea, & Salzberg, 2004), were used to predict genes in the repeat‐masked genome sequences. For homology‐based gene prediction, protein sequences from C. intestinalis type A (C. robusta), C. savignyi, M. oculata, B. schlosseri, O. dioica, Drosophila melanogaster, Caenorhabditis elegans, Branchiostoma floridae, Mus musculus and Homo sapiens were aligned to the genome using tblastn (Korf, 2004). The RNA‐Seq reads from different developmental stages and adult tissues were aligned to the genome using Tophat (Altschul, Gish, Miller, Myers, & Lipman, 1990). Gene predictions from the de novo approach, homology‐based approach and RNA‐Seq‐based evidence were merged to form a comprehensive consensus gene set using the EVidenceModeler software (http://evidencemodeler.github.io/) (Trapnell, Pachter, & Salzberg, 2009). Functional annotation of the obtained genes was conducted using blast (version 2.2.26) (Haas et al., 2008) against the NR, SwissProt and KEGG databases. The noncoding RNAs were also predicted in the genome. The tRNAs were searched using tRNAscan‐SE (version 1.3.1) (Altschul et al., 1990). The rRNAs were annotated by blast search. The miRNAs and snRNAs were predicted using Infernal (http://eddylab.org/infernal/) (Lowe & Eddy, 1997).
2.3. Hi‐C analysis and chromosome assembly
The muscle collected from a single adult S. clava was fixed using formaldehyde. The fixed tissue was then homogenized with tissue lysis buffer, digested with a restriction enzyme, in situ labeled with a biotinylated residue, and end‐repaired. Finally, the DNA was extracted and used for Hi‐C library preparation. The qualified library was sequenced using an Illumina HiSeq platform and used for the subsequent Hi‐C analysis. The reads were aligned to the S. clava genome using the BWA aligner (Nawrocki & Eddy, 2013). Only the valid interaction read pairs were used for the chromosome assembly.
2.4. Gene family and phylogenetic analysis
The gene families were determined via an all‐to‐all blastp search against the full protein‐coding genes from 16 selected deuterostomes including human (Homo sapines), mice (Mus musculus), frog (Xenopus tropicalis), coelacanth (Latimeria chalumnae), zebrafish (Danio rerio), flatfish (Paralichthys olivaceus), lamprey (Petromyzon marinus), sea squirts (M. oculata, C. savignyi, C. intestinalis type A (C. robusta) and O. dioica), amphioxus (Branchiostoma floridae), acorn worm (Saccoglossus kowalevskii), sea cucumber (Apostichopus japonicus), sea urchin (Strongylocentrotus purpuratus) and sea star (Acanthaster planci). Orthologous relationships between genes were inferred through all‐against‐all protein sequence similarity searches using OthoMCL (http://orthomcl.org/orthomcl/) and retained only the longest predicted transcript was retained per locus. A Venn diagram showed shared orthologous groups (duplicated genes were counted as one) on the basis of the presence of a representative gene in at least one of the grouped species. The expansion and contraction of gene families were estimated using CAFE (version 1.6) (Li & Durbin, 2009). The phylogenetic tree was constructed based on one‐to‐one orthologs, which were clustered among protein‐coding genes of the selected 17 genomes. The multiple alignments were performed by MUSCLE (version 3.7) (De Bie, Cristianini, Demuth, & Hahn, 2006) with its default setting. The ambiguously aligned positions were trimmed using Gblocks (http://molevol.cmima.csic.es/castresana/Gblocks.html). The jModelTest was used to select the best‐fit model for amino acid replacement. RAxML (http://sco.h‐its.org/exelixis/software.html) was then used to construct a maximum likelihood tree. The robustness of the maximum likelihood tree was assessed using the bootstrap method. The divergence time was estimated by mcmctree (http://abacus.gene.ucl.ac.uk/software/paml.html) in PAML (version 4.5) (Edgar, 2004). Tracer (http://beast.community/tracer) (Yang, 2007) was used to assess convergence. PAML was also used to assess the contribution of natural selection to the genes of S. clava, and the likelihood ratio tests were applied to test the significant differences.
2.5. Temperature stress experiments and quantitative real‐time PCR
The adult S. clava were acclimated to experimental conditions over the span of 1 week. The temperature controller was set at 2, 12, 22, and 32°C to maintain the sea water temperature. The animals were placed in the water at the different temperatures for 6 hr. There were three adults in each group. After the experiments, each adult was sampled in liquid nitrogen and stored at −80°C. RNA extraction and reverse transcription were conducted individually.
Quantitative real‐time PCR (qPCR) analysis was used to detect the relative expression level of genes (the sequences of primers are provided in Table S16). SYBRGreen was used as the DNA‐binding fluorescent dye. Actin gene was used as an internal standard and relative gene expression levels were calculated using the comparative Ct method with the formula 2−∆∆Ct (Rambaut, Drummond, Xie, Baele, & Suchard, 2018).
2.6. Compositional analysis and observation of the tunic structure
The monosaccharide composition of the tunic structure was determined by high‐performance liquid chromatography (Livak & Schmittgen, 2001). Two microgram of tunic tissue were hydrolyzed by 1 ml of 2 m trifluoroacetic acid at 110°C for 8 hr. The hydrolysate was labeled by 1‐phenyl‐3‐methyl‐5‐pyrazolone (Sigma) after neutralization and analyzed on an HPLC system (Agilent 1200). The composition and content of monosaccharides were determined in reference to the retention time and ultraviolet absorbance response of monosaccharide standards. For scanning electron microscopy studies of the tunic structure, the samples were fixed with 1% glutaraldehyde in 80% seawater, followed by dehydration and observation using an S‐3400N scanning electron microscope (Hitachi).
2.7. Transcriptome sequencing, gene identification, and in situ hybridization
Seven different developmental stages including 2–8‐cell embryos, gastrula embryos, neurula embryos, tailbud‐stage embryos, hatched swimming larvae, tail‐regressing larvae, and metamorphic juveniles were collected (2–8 cells, gast, neu, tb, hsl, trl, and mj, respectively). The total RNA was extracted using the RNAiso plus reagent (Takara, Japan), according to the manufacturer's instructions. RNA‐seq was performed using the Illumina HiSeq 2500 platform. Sequencing reads were aligned to the S. clava genome using HISAT (Strydom, 1994) with its default parameters. Gene expression levels were estimated by HTSeq (Kim, Langmead, & Salzberg, 2015), to acquire the fragments Per Kilobase of gene per million mapped reads (FPKM) value of each gene. The co‐expression gene network for 21 transcriptomic datasets was constructed using the R package WGCNA (Anders, Pyl, & Huber, 2015), with the parameters of softPower = 12, minimum module size = 300, cutting height = 0.99, and deepSplit = F. The genes that were expressed in at least one developmental stage were used for network construction and were assigned into nine gene modules. The intramodular connectivity (Kwithin) represents the hubness of a gene in one module (i.e., a higher Kwithin value indicates a stronger connection between one gene and the other genes in the specified module). GO enrichment analysis of the gene modules was performed using the EnrichPipeline (Langfelder & Horvath, 2008).
The complement genes were identified in the genomes of five tunicates using BLAST with an e‐value threshold of 1e‐5 against complement gene sequences from NCBI (https://www.ncbi.nlm.nih.gov/). They were further confirmed and filtered according to their NR annotation results. The remaining genes were also identified in the same way. The gene expression heatmaps were drawn according to their FPKM values. Collected embryos and larvae were fixed with 4% paraformaldehyde at 25°C for 2 hr. The probes for the THR, C3, and CR1 genes were synthesized using the primers listed in Table S17). Whole‐mount in situ hybridization was carried out essentially as published previously (S. Chen et al., 2010), using a hybridization temperature of 50–54°C.
2.8. Inhibitor treatment experiments
The fertilized eggs were reared in petri dishes with sea water at room temperature. The untreated embryos were collected and divided into several groups for experiments. Each group included 200 embryos. The treatment groups received MMI (Sigma; 301507, 1 g/L), BMS493 (Sigma; B6688, 1 μm), and UVI3003 (Sigma; SML1950, 1 μm), respectively, at the hatched swimming larval stage (15 hpf), while the control group received either the filtered fresh sea water (control for MMI) or DMSO (control for BMS493 and UVI3003) at the same developmental stage. For the dexamethasone (complement inhibitor) treatment experiments, the fertilized eggs were treated with 10 and 20 µm methanol (control) or dexamethasone (Sigma; D1756, 10 μm), respectively. The number of metamorphic juveniles and total embryos were counted at 22 hpf. The experiments were repeated three times, and the analysis of significant differences in the statistical data was performed on SPSS 17.0 via the chi‐squared test. p value <.01.
3. RESULTS AND DISCUSSION
3.1. S. clava has a larger genome size and increased gene length compared with C. intestinalis type A (C. robusta)
3.1.1. Genome sequencing and annotation
The genome of S. clava was sequenced using the PacBio RSII and Illumina Hiseq2500 platforms. A total of SMRT sequencing subreads representing over 40.73 Gb (approximately 100 × coverage) were obtained after quality filtering and were used for genome assembly. An additional Illumina pair‐end sequencing reads covering 31.74 Gb were obtained after filtering and were used for sequence correction (Table S1). The genome size was estimated at 406.87 Mb based on a k‐mer analysis. The FALCON strategy (Chin et al., 2016) was used for contig assembly, and a reference genome with 340.42 Mb and 710 contigs was constructed. The assembled genome size was smaller than the one by k‐mer analysis. This could be caused by the removement of the highly overlapped contigs during assembly. The assembled contigs displayed high continuity, with an N50 length of 758 Kb (Table S2), which is one of the longest N50 length among the sequenced genomes of ascidians and other marine invertebrates listed in this article (Table S3). Over 93% of the clean reads from the Illumina short‐insert library could be successfully mapped to the reference genome. Among the 978 Benchmarking Universal Single Copy Orthologs (BUSCOs) (Simao, Waterhouse, Ioannidis, Kriventseva, & Zdobnov, 2015) used for assessing genome completeness, 90% of the complete BUSCOs could be covered by the reference genome (Table S4). The data showed that the assembled genome of S. clava had high integrity and accuracy.
We used a Hi‐C approach to acquire a chromosome‐level assembly. The contigs were successfully clustered into 16 groups (Figure 1a, Table S5), which was consistent with previous karyotype analyses of S. clava and S. plicata (Dario Colombera, 1971; D. Colombera, 1974; Taylor, 1967). The clustered contigs corresponded to a length of 317.55 Mb (93.28% of the length of the contigs). The final assembled genome had 210 scaffolds with an N50 length of 20.77 Mb, providing a chromosomal‐level reference genome based on Hi‐C data.
FIGURE 1.
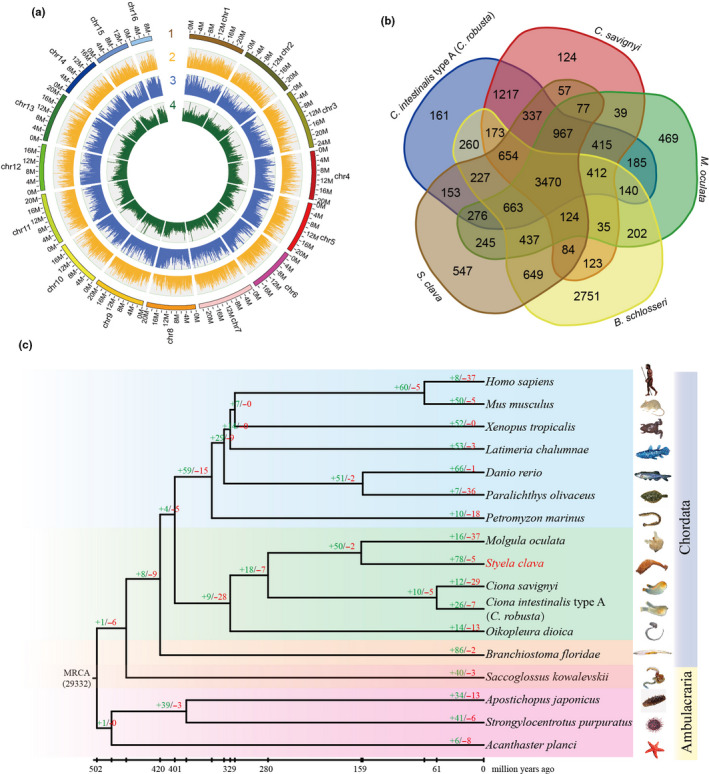
Genome map, gene family and phylogenetic position of Styela clava. a, Chromosome‐level genome map of S. clava. From outer to inner circles: 1, 16 chromosomes at megabase scales; 2, gene density across the genome (brown); 3, abundance of repetitive sequences across the genome (blue); 4, GC content across the genome drawn in 0.1 Mb nonoverlapping windows (green). b, Venn diagram of common and unique gene families among five ascidian species, i.e., C. intestinalis type A (C. robusta), C. savignyi, Molgula oculata, B. schlosseri, and S. clava. 3,470 gene families were shared among the five ascidians and 547 gene families existed exclusively in the S. clava genome. c, The phylogenetic position, divergence time estimation and gene family analysis of S. clava. The green and red colored numbers indicate the expanded gene families and the contracted gene families, respectively. The divergence years are displayed below the phylogenetic tree
The genome of S. clava showed approximately 1.72% heterozygosity based on the k‐mer analysis of short‐insert library reads. Repeat sequences made up 46.69% of the assembled genome (Table S6). A total of 17,428 protein‐coding genes were identified, 15,734 of which could be annotated in databases including NR, Swiss‐Prot, KEGG, GO, and Pfam (Table S7). Various noncoding small RNAs were also found in the genome, including 120 micro‐RNAs, 2,680 tRNAs, and 837 rRNAs (Table S6).
3.1.2. Gene family and phylogenetic analysis
The gene families among five ascidians were determined via an all‐to‐all blastp search against the full protein‐coding genes. In general, 15,673 gene families were detected among the five ascidian genomes (Figure 1b). 3,470 of them were shared by five species, and 11,621 of them were identified in at least two species. In a previous study, 8,716 families of homologous genes were present in the deuterostome ancestor (Simakov et al., 2015).
The gene families were also determined via an all‐to‐all blastp search against the full protein‐coding genes among 17 metazoans. We classified the gene families into single‐copy, multiple‐copy, unique, and other orthologs. The single‐copy family included one‐to‐one orthologous genes, the multiple‐copy family included orthologous genes with multiple copies in all the species, the unique family included genes that only appeared in one species, and other orthologs included the other orthologous genes present in at least two species. To determine the phylogenetic position of S. clava, a genome‐wide phylogenetic tree was constructed based on single‐copy genes from 17 genomes (Figure 1c). The results showed that S. clava was closest to M. oculata and that the estimated divergence time was 159 Mya. S. clava and M. oculata belong to the order Enterogona while Ciona to the order of Pleurogona. This result was consistent with that of a previous study (Delsuc et al., 2018). In total, 78 gene families were expanded in S. clava, whereas five gene families were contracted. The expanded gene families included the pulmonary surfactant‐associated protein collectin, cytochrome P450 2U1, and ficolin solute carrier family 22 (Table S8). Positively selected genes in S. clava were also identified by comparison with C. intestinalis type A (C. robusta), C. savignyi, M. oculate, and O. dioica (Table S9). The top two genes (aladin‐like and DNA mismatch repair protein Mlh1‐like) were both related to DNA repair, which indicated the adaptation of S. clava to environment.
3.1.3. Genome structure characteristics
Compared with the reported reference genomes of Ciona and Molgula, S. clava has a similar gene number but a larger genome size (Table S10). A statistical analysis revealed that S. clava had a larger average gene length and gene distance length (Table S10; Figure 2a). Moreover, the gene length and gene distance distribution revealed that S. clava had more genes with a larger intron size (>10 Kb) and a greater number of large intergenic regions (>10 Kb) compared with those from C. intestinalis type A (C. robusta) (Figure 2a). A gene structure comparison analysis revealed that the difference in average intron length was the major reason for the variation in gene length among tunicates. Further analysis also showed that the intron elongation inside each gene was not homogeneous (Figure S1). S. clava tended to have one large intron inside each gene. There were also less intron‐less genes in S. clava (Figure S2; Table S11).
FIGURE 2.
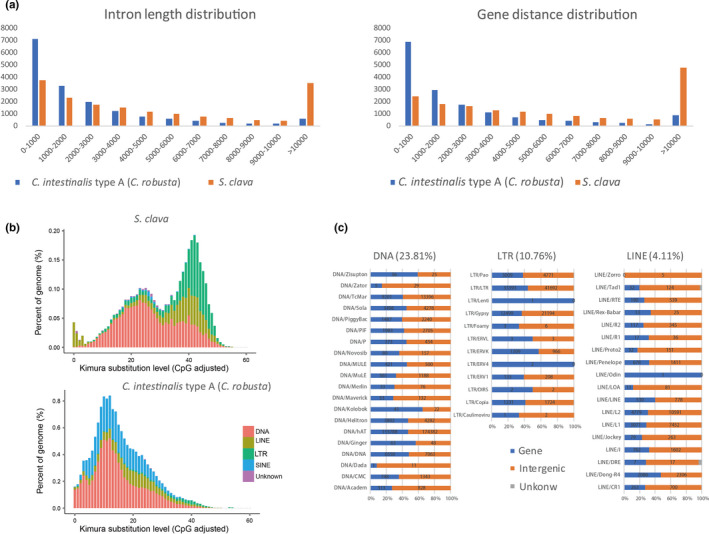
Gene structure and transposable element (TE) expansion in the S. clava genome. a, Intron length and gene distance distribution in the S. clava and C. intestinalis type A (C. robusta) genomes. The x‐axis indicates the length intervals, while the y‐axis indicates the number of genes. The blue and orange bars represent the intron length and gene distance distribution in C. intestinalis type A (C. robusta) and S. clava genomes, respectively. b, Classification and substitution level of TEs in the S. clava and C. intestinalis type A (C. robusta) genomes. The x‐axis represents the kimura substitution level (CpG adjusted) of TEs. The y‐axis represents the percentages of TEs in the genome. The different colors represent different types of transposons. The DNA elements are the main transposon types in both S. clava and C. intestinalis type A (C. robusta) genomes. However, the number of SINEs was decreased, while that of LTRs and LINEs was increased significantly in S. clava genomes. c, Distribution of the different types of transposons in the S. clava genome. The proportions of DNA, LTR, and LINE transposons in the S. clava genome were 23.81%, 10.76%, and 4.11%, respectively. The blue bars represent the numbers of transposons that are distributed in gene regions. The orange bars represent the number of transposons that are distributed in intergenic regions
Considering that 46.69% of the S. clava genome consisted of repeat sequences (Table S12), and that 45.92% of the sequences were annotated as transposable elements (TEs), we first compared the TEs between S. clava and C. intestinalis type A (C. robusta) to probe the mechanisms underlying the larger average gene length and gene distance observed in this species. The classification of TEs revealed that DNA elements, LTRs, and LINEs were the main TEs in S. clava, while DNA elements and SINEs were the major ones in C. intestinalis type A (C. robusta) (Chalopin, Naville, Plard, Galiana, & Volff, 2015; Figure 2b). The amount of TEs, such as SINEs, is strongly correlated with tunicate genome size in larvaceans (Naville et al., 2019). Here, we found that the composition of TEs also affected the genome size of a different species of ascidian. The appearance of LTRs and the absence of SINEs in the S. clava genome contributed to the genome size variation. A distribution analysis of TEs in the genome of S. clava showed that the major TEs were distributed in both intragenic and intergenic regions (Figure 2c). We inferred that insertions of TEs had a great effect on the doubling of the Styela gene size compared with in Ciona, and that insertions of TEs into intergenic regions resulted the more dispersive distribution of genes observed in S. clava. TEs are believed to evolve in the fashion of a molecular clock, as they are identical at the time of integration and then gradually diverge because of the accumulation of mutations (Bowen & McDonald, 2001; Carr & Suga, 2014). The LINEs dominated the lowest substitution level in S. clava (Figure 2b), indicating that they were active recently in the genome. Consistent with this expectation, LINEs were also identified from the transcriptome data and expressed at RNA‐Seq levels (Table S13; Figure S3).
A rapid genome evolution and, in particular, active TEs potentially facilitate the adaptation of invasive species to new environments (Stapley, Santure, & Dennis, 2015). It is possible that such plasticity in genome characteristics, particularly through transposon diversity, facilitates the observed invasive success of S. clava and other tunicate species.
3.2. Gene gain and gene loss shed light on the molecular basis of the environmental adaptation of S. clava
3.2.1. Hsp70 gene expansion
Among the known mechanisms underlying the rapid and adaptable responses to stress, heat‐shock proteins form the crucial molecular pathways. Heat‐shock proteins are a group of evolutionarily conserved protein families, which facilitate the coping with environmental fluctuations through the correction of protein folding or the maintenance of protein homeostasis (Feder & Hofmann, 1999). For example, the heat‐shock protein 70 (Hsp70) is central to the adaptation of oysters to sessile life in the highly stressful intertidal zone (Zhang et al., 2012).
The Hsp70 family of genes exhibited a remarkable genome‐wide gene duplication in S. clava (Figure 3a). Forty‐two Hsp70 genes were identified by searching the whole genome. Thirty‐two of them were located in the same chromosome (chromosome ID: Hic_asm_6) indicating that these genes were likely tandem duplicated. Thirteen of the genes were located on the same contig. In C. intestinalis type A (C. robusta), only eight genes are classified into the Hsp70 gene family (Fujikawa, Munakata, Kondo, Satoh, & Wada, 2010) and are dispersed in different scaffolds. The search for Hsp70 genes in other tunicate genomes also revealed that S. clava has expanded Hsp70 genes (Figure 3a). To investigate the temporal expression of Hsp70 genes, we analyzed the transcriptomic data during embryogenesis and larval development of S. clava. The results showed that most of the Hsp70 genes in S. clava were expressed during embryogenesis and larval development (Figure S4). Twelve Hsp70 genes were upregulated from the neurula to the tailbud stages, which indicated their participation for successful development (Figure S4). We inferred that the expression of Hsp70 at the tailbud stage was correlated with the adaptation to environmental stress, because the hatched embryos lost the protection from the envelope. The expansion and tandem duplication of Hsp70 genes may affect the adaptation to environmental stress in S. clava.
FIGURE 3.
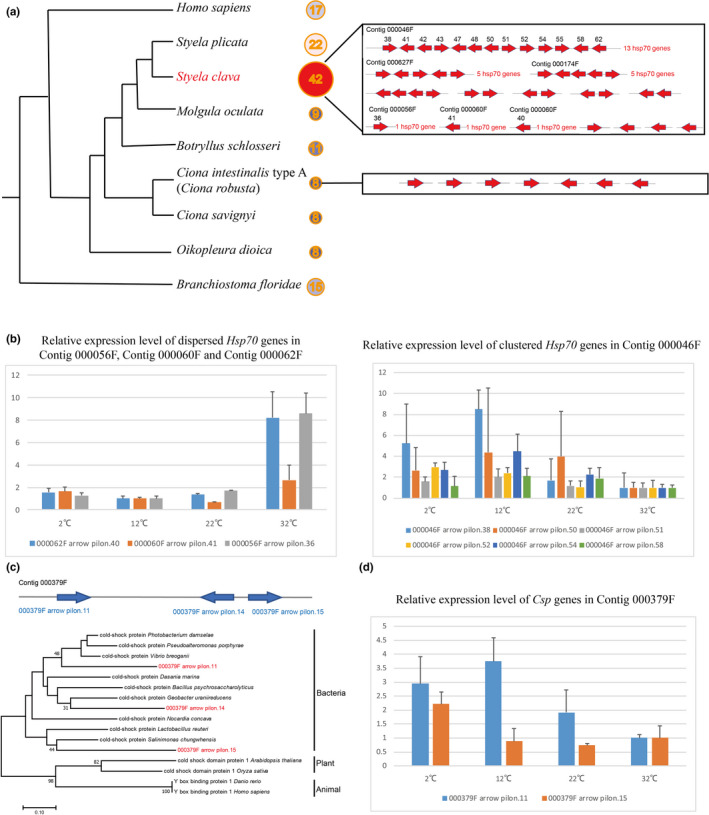
Expansion of heat‐shock protein 70 (Hsp70) genes and horizontally transferred cold‐shock protein (Csp) genes in the S. clava genome. a, Number of Hsp70 genes in different chordate animals. S. clava had the greatest number of Hsp70 genes among these species. The Hsp70 genes in the S. clava genome were both clustered and interspersed, while the Hsp70 genes in the C. intestinalis type A (C. robusta) genome were all interspersed. b, Expression levels of Hsp70 genes at different temperatures. The three dispersed Hsp70 genes were upregulated in high‐temperature stress, while the six‐tandem repeat Hsp70 genes were upregulated under low‐temperature stress and downregulated under high‐temperature stress. c, Positioning in the genome and phylogenetic analysis of Csp genes in S. clava. The three identified Csp genes were aligned in the same contig. The phylogenetic tree was constructed using the neighbor‐joining method. The three Csp genes in S. clava are labelled in red color and were clustered with bacteria Csp genes. d, The Csp genes were upregulated under lower‐temperature stress
To investigate whether the expansion of Hsp70 genes contributed to the plasticity of thermal stress adaptation, we examined their expression at different temperatures using qPCR. The relative expression levels revealed that the dispersive genes were upregulated at 32°C, while the clustered genes were upregulated at 12°C and 2°C (Figure 3b). Previous studies have focused on the expression dynamics of Hsp70 genes under acute temperature stress at both the transcription and translation levels (Fujikawa et al., 2010; Serafini, Hann, Kueltz, & Tomanek, 2011). The oyster genome contains 88 Hsp70 genes, which was regarded as crucial roles in protecting cells against heat and other stresses (Zhang et al., 2012). Although studies have shown that Hsp70 genes could be transcriptionally induced by heat stress in two ascidians (Fujikawa et al., 2010; Huang, Li, Gao, & Zhan, 2018; Huang, Li, & Zhan, 2019), our results showed that numerous, but not all, Hsp70 genes responded to either heat or cold temperature related‐environmental stresses.
3.2.2. Horizontally transferred genes
The living environment of S. clava is complex, with various parasitic or symbiotic microorganisms, and their interaction may offer a path for the horizontal transfer of functional genes in the course of their evolutionary history. Horizontal gene transfer (HGT) may spread rapidly evolutionarily success across lineages and facilitate the exploitation of recipient organisms of new niches or other resources. Although numerous cases of HGT have been documented in prokaryotes and unicellular eukaryotes (Soucy, Huang, & Gogarten, 2015), it is common belief that HGT is rare in multicellular eukaryotes, such as animals and plants (Kurland, Canback, & Berg, 2003). However, HGT occurs frequently in ascidians. Among all the protein‐coding genes in C. intestinalis type A (C. robusta), 169 were likely derived from algae, 92 of them with high probability (Ni et al., 2012). The best‐known example is the cellulose synthase genes in ascidians, which were transferred from bacteria responsible for tunic formation (Bhattachan & Dong, 2017; Matthysse et al., 2004; Sagane et al., 2010).
The methods used for detecting HGT generally rely on the analysis of phylogenetic conflict (Soucy et al., 2015). Here, we detected HGT events in S. clava at the whole‐genome level. Three cold‐shock protein‐like (Csp) genes in the same contig were screened. Their gene structures are shown in Table S14. Two of the three genes had introns. The phylogenic tree also revealed that they were horizontally transferred from bacteria (Figure 3c). In addition, we used blastx to search for their homologous genes in the NR database. The results showed that the top 10 annotations of the three genes all came from bacteria. The Illumina sequencing coverage analysis of Csp genes also showed that the genes had a coverage that was similar to that of the surrounding sequences.
Csps are small, abundant proteins that are essentially composed of one cold‐shock domain (CSD). Some members of the family are strongly induced after cold shock and are involved in adaptation to low temperatures (Horn, Hofweber, Kremer, & Kalbitzer, 2007; Mihailovich, Militti, Gabaldon, & Gebauer, 2010). CSDs have been found in many other bacterial and eukaryotic proteins, such as the well characterized eukaryotic Y‐box proteins (Ladomery & Sommerville, 1994; Mihailovich et al., 2010). However, reports of Csp genes in the animal kingdom are scarce. To validate the functions of the horizontally transferred Csp genes in the Styela genome, their expression levels under low‐temperature stress were also examined using qPCR. The results showed that the expression of Csp genes was upregulated at both 12°C and 2°C (Figure 3d). The HGT events of Csp genes provide one of the possible molecular mechanisms for S. clava adaptation to environmental stress, particularly low‐temperature stress.
3.2.3. The tunic structure of ascidians
The integument is an important defense system for sessile organisms, who cannot move from stressful environmental conditions, such as irradiation, drying, predation, infection, bio‐fouling (Euichi Hirose, 2009). The name Tunicata is derived from the integumentary tissue called the tunic, which covers the entire epidermis in ascidians (Zhao & Li, 2014). As the tunic has multiple functional roles (E. Hirose, Ohtake, & Azumi, 2009), it is hypothesized to be one of the features underlying the high adaptability and tolerance observed in ascidians. Cellulose constitutes the main structure of the tunic that surrounds the adult body. Tunicates are the only animal population that can produce cellulose by themselves (Hirose, Kimura, Itoh, & Nishikawa, 1999; Inoue, Nakashima, & Satoh, 2019; Kimura, Ohshima, Hirose, Nishikawa, & Itoh, 2001; Ranby, 1952).
The hardness of the tunic was associated with protection from the environmental stresses. However, there is great variation in the hardness and transparency of the tunic among the different species of ascidians. We inferred that the varying hardness of the tunic was caused by its composition. According to a previous study, the tunic is mainly composed of complex carbohydrates (Zhao & Li, 2014). The tunic structures could be described as cellulose‐protein fibrils cemented by sulfated mucopolysaccharides or sulfated glycans and lipids. In tunics, cellulose is the dominant carbohydrate (Hirose et al., 1999; Zhao & Li, 2014). Therefore, we assessed the content of different saccharides in the tunic of different species of ascidians. The results showed that the sulfated polysaccharide contents, particularly galactose, is more abundant in the order Phlebobranchia (e.g., C. intestinalis type A (C. robusta) and C. savignyi) than in Stolidobranchia (e.g., S. clava and H. roretzi) (Figure 4a). Reasonably, the higher contents of other sugars detected in the soft tunics imply higher content of sulfated glycans, which renders the structures more flexible compared with hard tunics.
FIGURE 4.
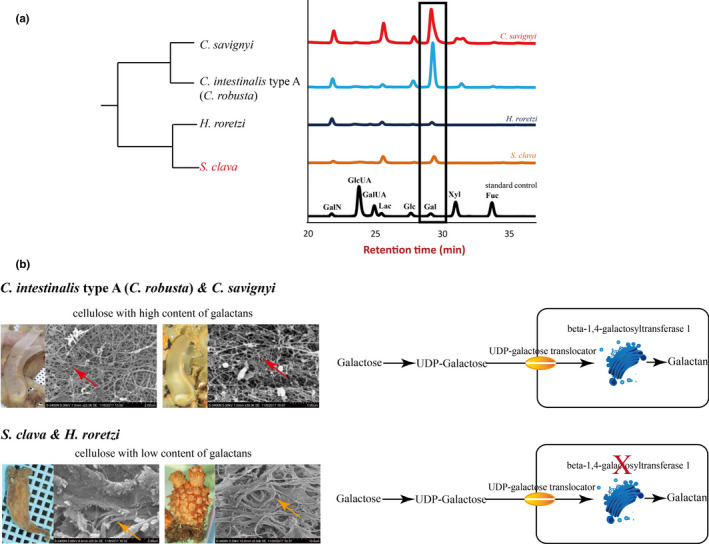
Tunic structure, monosaccharide composition, and galactan synthesis among different ascidians. a, Monosaccharide composition of the tunic in different ascidians. Both C. savignyi and C. intestinalis type A (C. robusta) had high levels of galactose, while H. roretzi and S. clava had relatively low levels of galactose. The rectangle represents the levels of galactose in different ascidians, as detected using mass spectrometry. The black line indicates the standard control. b, Tunic structures and galactan synthesis pathway in different ascidians. The main constituent of the tunic was cellulose. However, the cellulose structures with high galactans content were more scattered (red arrows) in C. intestinalis type A (C. robusta) and C. savignyi, while the cellulose structures with low galactan content were more bundled in H. roretzi and S. clava (orange arrows). In both C. intestinalis type A (C. robusta) and S. clava, galactose was first transferred into UDP‐galactose and then transported inside the cells through a UDP‐galactose translocator located in the cell membrane. The enzymes in the Golgi apparatus were responsible for the synthesis of polysaccharides. The key gene in this process, beta‐1,4‐galactosyltransferase 1, could only be identified in the genome of C. intestinalis type A (C. robusta) and C. savignyi, and not in the genome of S. clava
We used a scanning electron microscope to observe the detailed structures of tunics from C. intestinalis type A (C. robusta), C. savignyi, S. clava, and H. roretzi. The fibers in C. intestinalis type A (C. robusta) and C. savignyi were much thinner, while the fibers in S. clava and H. roretzi were bundled together (Figure 4b). Our results are consistent with results of previous studies of the morphology of a H. roretzi tunic, which reported that the tunic surface was entirely covered with a cuticular layer that had a very dense structure of bundled fibers, and that the tunic matrix was distributed between the fibers (Zhao & Li, 2016). To compare the molecular basis of the tunic, a whole‐genome level of GlycosylTransferase (GT) domain analysis was conducted (Figure S5). The cellulose synthase genes in different ascidians were similar among the various ascidians (Figures S5 and S6). Considering the difference in galactose contents between Phlebobranchia and Stolidobranchia, we compared the galactan synthesis‐related genes in the species and found conserved UDP‐galactose translocator genes in the four species, implying that the galactose transport ability is conserved (Figure 4b). After transportation into cells, many glycan structures of glycoproteins and glycolipids are assembled in the Golgi apparatus by families of glycosyltransferases (Inoue et al., 2019). The beta‐1,4‐galactosyltransferase (beta4GalT) is responsible for the synthesis of sulfated galactans. Each subfamily member is expressed in a tissue‐specific manner and exhibits differences in oligosaccharide acceptor specificity and tissue‐specific expression (Rautengarten et al., 2014). The comparison of the family members of beta4GalT genes showed that beta4GalT1 was only observed in C. intestinalis type A (C. robusta) and C. savignyi, and not in S. clava and H. roretzi (Figure 4b). The gene is unique among the beta4GalT genes because it encodes an enzyme that participates in the biosynthesis of both glycoconjugates and lactose. The loss of beta4GalT1 in Phlebobranchia may be related to the low level of sulfated galactans in the tunic structure. These findings suggest a molecular basis for the formation of distinct tunic structures among different tunicate species.
3.3. Thyroid hormone pathway and complement system genes participated in the regulation of the metamorphosis of S. clava
3.3.1. Transcriptome sequencing of different developmental stages of S. clava
Ascidians have a distinctive pattern of embryogenesis and larval development: zygotes first develop into larvae with a notochord, then, the notochord regresses after the attachment of larvae, thus providing a suitable model for developmental studies of metamorphosis. The larval developmental process is also referred to as retrogressive metamorphosis. To explore the molecular basis of the process, we performed a transcriptome analysis of 2–8–cell embryos (2–8 cells), gastrula embryos (gast), neurula embryos (neu), tailbud‐stage embryos (tb), hatched swimming larvae (hsl), tail‐regressed larvae (trl), and metamorphic juveniles (mj) (Figure 5a). There were three replicates at each stage (Table S15). The correlations between samples are shown in Figure S7. According to the results of a weighted correlation network analysis (WGCNA), (Figure 5b), genes were grouped using a clustering approach. Gene expression heat maps for each module were shown in Figure 5b and Figure S8. The GO enrichment analysis of genes in each module also was shown in Figure S9.
FIGURE 5.
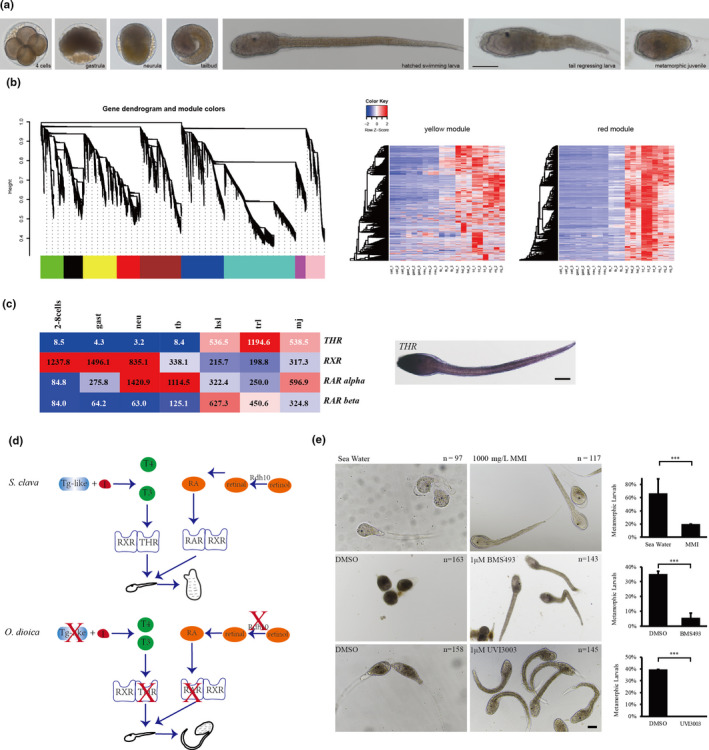
Thyroid hormone and retinoic acid signaling pathways. a, The seven developmental stages of S. clava embryos and larvae; i. e., 4‐cell stage, gastrula, neurula, tailbud, hatched swimming larvae, tail‐regressing larvae, and metamorphic juvenile. b, Weighted correlation network analysis (WGCNA) of the genes based on the transcriptomics data at different developmental stages. Different colors indicated different gene modules. The gene expression heat maps for yellow and red modules were also shown. c, The gene expression heatmap of thyroid hormone receptor (THR), retinoic acid receptor (RAR alpha), retinoic acid receptor beta (RAR beta) and retinoid X receptor (RXR) in S. clava. The numbers inside the grids indicate the FPKM values of each gene at different stages. The red color indicates genes that are highly expressed, while the blue color indicates genes that are relatively weakly expressed. THR was expressed in the epithelia of the head and terminal parts of the tail in the hatched larvae, according to in situ hybridization. d, Comparison of the thyroid hormone (TH) and retinoic acid (RA) signaling pathways between the tail‐regressing S. clava and the tail‐preserved O. dioica. The key genes for TH and RA synthesis and their receptors could be identified and were expressed in S. clava, while the retinol dehydrogenase 10 (rdh10) gene, the TH, and RA receptor genes were absent in the O. dioica genome. e, Inhibition of the THR, RAR and RXR signaling pathways led to the failture of tail regression in S. clava larvae. The groups treated with MMI (an inhibitor of TH synthesis), BMS493 (an inhibitor of RAR), and UVI3003 (an inhibitor of RXR) exhibited significantly reduced the metamorphic levels compared with the controls
3.3.2. Thyroid hormone receptor (THR) was enriched in tail regression transcriptomic data
One of these modules, here depicted in red, yielded some interesting results: 1,725 genes were clustered into this module (Figure 5b), which contained genes specifically expressed in hatched swimming larvae, tail‐regressing larvae, and metamorphic juveniles. Among these genes, the thyroid hormone receptor (THR) gene was highly upregulated during tail regression (Figure 5c). In addition, RAR was also highly expressed during tail regression (Figure 5c). The connectivity value of THR ranked the second among the transcription factors and in the top 10% among all genes in this module, representing a potential hub gene for network regulation. To understand this potentially central role of THR during metamorphosis, we decided to investigate the expression and function of THR.
3.3.3. TH signaling plays essential roles in the metamorphosis of S. clava
The THR gene was upregulated by 64‐fold in hatched swimming larvae and by 142‐fold in tail‐regressing larvae compared with tailbud‐stage embryos. In situ results showed that THR was expressed in the epithelia of the head and tail part of the larvae (Figure 5c). It has been demonstrated that TH signals are obligatory for vertebrate metamorphosis and function through the activation of a complex hierarchical cascade of target genes (Qasba, Ramakrishnan, & Boeggeman, 2008). Moreover, comparative genomics and transcriptomics analyses of flatfish identified the cross‐talk role between TH and retinoic acid signaling during metamorphosis (Laudet, 2011). These data suggest that TH and RA are required for tail regression in tunicates too. To test this hypothesis at the genomic level, we investigated the genome of O. dioica, which belongs to the class Appendicularian, keeps its notochord throughout its lifespan. This provides a suitable comparative study model. After searching its genome, we found no hits for the THR and retinoic acid receptor (RAR) genes in the genome of O. dioica (Figure 5d). Moreover, the key genes that are related to the synthesis of TH and retinoic acid, such as rdh10 genes, were also absent in the genome of O. dioica (Figure 5d). These results support our hypothesis that the THR and RAR genes are associated with the processes of tail regression in tunicates.
We substantiated further our hypothesis using inhibitor experiments. MMI (an inhibitor of TH synthesis, 1 g/L) (Shao et al., 2017) and BMS493 (an inhibitor of RAR signaling, 1 μm/L) (Brown, 1997) treatment resulted in significantly lower larval metamorphic rates compared with the rates observed in the control, while UVI3003 (an inhibitor of the retinoid X receptor (RXR), 1 μm/L) (Germain, Iyer, Zechel, & Gronemeyer, 2002) signaling treatment inhibited metamorphosis in S. clava (Figure 5e). The results presented above suggest that TH and RA signaling play essential roles in the metamorphosis of Styela larvae.
3.3.4. Complement system genes
Among the genes that were highly expressed during metamorphosis (e.g. in hatched swimming larvae, tail regressed larvae, and metamorphic juveniles), many of them were immune‐related genes. Although there is no evidence of adaptive immunity in tunicates, a search of the S. clava genome revealed a variety of genes that could mediate innate immunity, particularly complement system genes. There is also a high number of potential complement genes in C. intestinalis type A (C. robusta), including the C1q and C6 genes (Nahoum et al., 2007). However, a comparison between S. clava and other four tunicate species revealed a varying composition in complement system genes (Figure 6a). Pattern‐recognition genes, including the Mannose‐binding lectin (MBL) and ficolin genes, were expanded in S. clava. Twenty‐five MBL genes and 54 ficolin genes were identified. For proteases and receptors, 17 factor B genes and 16 complement receptor 1 genes were identified. Moreover, a great proportion of the expanded genes in S. clava exhibited high levels of expression during metamorphosis (Figure 6b). The results of whole‐mount in situ hybridization showed that component 3 (C3) and complement receptor 1 (CR1) genes were both highly expressed in epithelia cells of the trunk and the posterior part of the tail in the hatched larvae. C3 was also expressed in the neck region while CR1 expression was not seen in the neck region of swimming Styela larvae (Figure 6b).
FIGURE 6.
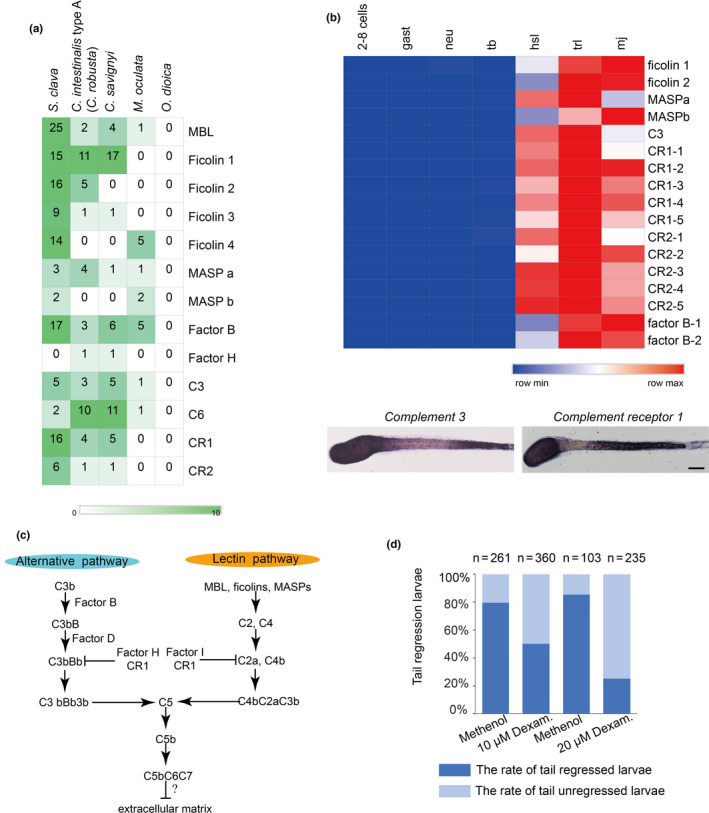
Expanded complement system genes and their upregulation in the hatched larvae of S. clava. a, The identified complement system‐related genes were expanded in S. clava, compared with other tunicate species. The rows represent the gene number in each genome. The numbers inside the grids indicate the number of genes in the genome. b, The gene expression heatmap of the selected complement pathway genes in S. clava transcriptomes. 2–8 cells, gast, neu, tb, hsl, trl, mj represent the gene expression of 2–8‐cell embryos, gastrula embryos, neurula embryos, tailbud‐stage embryos, hatched swimming larvae, tail‐regressing larvae, and metamorphic juveniles, respectively. The scale bar indicates the FPKM values. Most of the genes were highly expressed during tail regression. The complement 3 (C3) and complement receptor 1 (CR1) were expressed in epithelial cells of the trunk and terminal parts of the tail in the hatched larvae. c, Putative alternative and lectin complement pathways in S. clava. d, Inhibition of complement signaling pathways led to the failture of tail regression in S. clava larvae. Fertilized eggs were treated with 10 and 20 μm dexamethasone, and phenotypes were observed at 22 hpf. The larval tail‐regression rate was sigificantly decreased in drug‐treated groups under the two treatment concentrations
The expression patterns and upregulated expression of complement system genes in Styela support our speculation that complement system gene signaling may occur in response to high levels of stress linked to the death and reorganization of larval tissues. In another species of solitary ascidians, Boltenia villosa, innate immune signaling during metamorphosis has been reported to be involved in coordinating the resorption of larval tissues, the maturation of the tunic, or the adhesion and migration of mesenchymal cells (Nonaka & Kimura, 2006). The expanded MBL‐complement pathways in Styela could be important in the detection of the environmental cues that initiate settlement just like the situation in B. villosa (Davidson & Swalla, 2002) (Figure 6c).
The expression patterns of the complement 3 gene and its receptor resembled those of the fibroblast growth factor and Wnt Signaling genes (Roberts et al., 2007), indicating that complement pathways may participate in the regulation of larval tissue development. As complement molecules were highly expressed in the metamorphic stages, we speculated that they were involved in the regulation of larval metamorphosis in S. clava. Therefore, we used dexamethasone (Packard & Weiler, 1983; Pasini, Manenti, Rothbacher, & Lemaire, 2012) to inhibit the activity of the complement system in Styela larvae, and observed that inhibitor‐treated larvae failed to regress their tails (Figure 6d). These results suggest that the complement signaling pathway is essential for Styela larvae tail regression.
4. CONCLUSION
Here, we reported the chromosome‐level assembly of the genome of the leathery sea squirt, S. clava, and constructed the transcriptomic profiles in development of S. clava. Expansion of DNA, LTR, and LINE transposons was more closely related to the doubled genome size of S. clava than to that of C. intestinalis type A (C. robusta). Moreover, the expanded Hsp 70 family and horizontally transferred cold‐shock proteins responded to acute temperature stress, while different types of galactan synthesis genes were associated with the tunic structure and hardness. An integrated TH synthesis pathway was also found, and the associated genes were highly expressed during tail regression. A functional analysis demonstrated that the pathway was associated with ascidian metamorphosis. The complement system genes were expanded in the S. clava genome, and potentially participated in both immunology and the regulation of metamorphosis in this species. The present study provides insights into the mechanisms of adaptation to changes in the environment and the mechanisms of larval metamorphosis in invasive tunicates.
5. COMPETING INTERESTS
The authors declared no competing interests.
AUTHOR CONTRIBUTIONS
B.D. and J‐k. W. conceived and initiated the Styela genome project. B. D. designed major scientific objective and experiments. J‐k.W. directed final data analyses and interpretation. J. Z. participated the data analysis, sample collection, and performed inhibitor experiments of TH and RA signaling. Q. L. studied the development of Styela embryos and larvae, collected the samples, and performed inhibitor experiments of complement system genes. P. R. performed Q‐PCR experiments. X. G. and S. D. performed in situ experiments of THR and complement genes. J. W. and S. W. performed the WGCNA analysis on the transcriptome data. X. L. extracted genomic DNA for sequencing. Y. C. performed MS analysis of ascidian tunic. H. Y, X. Z., X. Y, and H. G. participated in the data analysis and discussion. J‐k. W. and B. D. made the figures, drafted, and revised the manuscript with input from other authors. All the authors read and proved the final manuscript.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Key Research and Development Program of China (2018YFD0900705, 2019YFE0190900), Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2018SDKJ0302‐1, 2018SDKJ0406‐5), the National Natural Science Foundation of China (Grant No. 31970487), the Fundamental Research Funds for the Central Universities (201822016), and the Taishan Scholar Program of Shandong Province, China (201502035).
Wei J, Zhang J, Lu Q, et al. Genomic basis of environmental adaptation in the leathery sea squirt (Styela clava). Mol Ecol Resour. 2020;20:1414–1431. 10.1111/1755-0998.13209
DATA AVAILABILITY STATEMENT
The genome sequence project was deposited in NCBI under the BioProject number PRJNA523448. The transcriptome data used for biological analysis were also deposited in the NCBI SRA database with the accession numbers SRR8599814 to SRR8599834. The genome resources of other ascidians were downloaded from ANISEED (https://www.aniseed.cnrs.fr/), and the genome resources of O. dioica were acquired from OikoBase (http://oikoarrays.biology.uiowa.edu/Oiko/).
REFERENCE
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410. 10.1016/s0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P. T. , & Huber, W. (2015). HTSeq‐a python framework to work with high‐throughput sequencing data. Bioinformatics, 31(2), 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattachan, P. , & Dong, B. (2017). Origin and evolutionary implications of introns from analysis of cellulose synthase gene. Journal of Systematics and Evolution, 55(2), 142–148. 10.1111/jse.12235 [DOI] [Google Scholar]
- Blanchoud, S. , Rutherford, K. , Zondag, L. , Gemmell, N. J. , & Wilson, M. J. (2018). De novo draft assembly of the Botrylloides leachii genome provides further insight into tunicate evolution. Scientific Reports, 8, 18 10.1038/s41598-018-23749-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, N. J. , & McDonald, J. F. (2001). Drosophila euchromatic LTR retrotransposons are much younger than the host species in which they reside. Genome Research, 11(9), 1527–1540. 10.1101/gr.164201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D. D. (1997). The role of thyroid hormone in zebrafish and axolotl development. Proceedings of the National Academy of Sciences of the United States of America, 94(24), 13011–13016. 10.1073/pnas.94.24.13011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, M. , & Suga, H. (2014). The holozoan Capsaspora owczarzaki possesses a diverse complement of active transposable element families. Genome Biology and Evolution, 6(4), 949–963. 10.1093/gbe/evu068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalopin, D. , Naville, M. , Plard, F. , Galiana, D. , & Volff, J.‐N. (2015). Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biology and Evolution, 7(2), 567–580. 10.1093/gbe/evv005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon, J.‐P. , Nakayama, A. , Takamura, K. , McDougall, A. , & Satoh, N. (2007). ERK‐ and JNK‐signalling regulate gene networks that stimulate metamorphosis and apoptosis in tail tissues of ascidian tadpoles. Development, 134(6), 1203–1219. 10.1242/dev.002220 [DOI] [PubMed] [Google Scholar]
- Chen, J. Y. , Huang, D. Y. , Peng, Q. Q. , Chi, H. M. , Wang, X. Q. , & Feng, M. (2003). The first tunicate from the early Cambrian of South China. Proceedings of the National Academy of Sciences of the United States of America, 100(14), 8314–8318. 10.1073/pnas.1431177100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Yang, P. , Jiang, F. , Wei, Y. , Ma, Z. , & Kang, L. (2010). De Novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS One, 5(12), 10.1371/journal.pone.0015633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, C.‐S. , Peluso, P. , Sedlazeck, F. J. , Nattestad, M. , Concepcion, G. T. , Clum, A. , … Schatz, M. C. (2016). Phased diploid genome assembly with single‐molecule real‐time sequencing. Nature Methods, 13(12), 1050–1054. 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown, S. L. , Hodgins, K. A. , Griffin, P. C. , Oakeshott, J. G. , Byrne, M. , & Hoffmann, A. A. (2015). Biological invasions, climate change and genomics. Evolutionary Applications, 8(1), 23–46. 10.1111/eva.12234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloney, R. A. (1982). Ascidian larvae and the events of metamorphosis1. American Zoologist, 22(4), 817–826. 10.1093/icb/22.4.817 [DOI] [Google Scholar]
- Colombera, D. (1971). The chromosomes of ascidians. Bollettino Di Zoologia, 38(2), 187–191. 10.1080/11250007109436972 [DOI] [Google Scholar]
- Colombera, D. (1974). Chromosome number within the class Ascidiacea. Marine Biology, 26(1), 63–68. 10.1007/BF00389087 [DOI] [Google Scholar]
- Conklin, E. G. (1905). Mosaic development in ascidian eggs. Journal of Experimental Zoology, 2(2), 145–223. 10.1002/jez.1400020202 [DOI] [PubMed] [Google Scholar]
- Dardaillon, J. , Dauga, D. , Simion, P. , Faure, E. , Onuma, T. A. , DeBiasse, M. B. , … Lemaire, P. (2020). ANISEED 2019: 4D exploration of genetic data for an extended range of tunicates. Nucleic Acids Research, 48(D1), D668–d675. 10.1093/nar/gkz955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, B. , & Swalla, B. J. (2002). A molecular analysis of ascidian metamorphosis reveals activation of an innate immune response. Development, 129(20), 4739–4751. [DOI] [PubMed] [Google Scholar]
- De Bie, T. , Cristianini, N. , Demuth, J. P. , & Hahn, M. W. (2006). CAFE: A computational tool for the study of gene family evolution. Bioinformatics, 22(10), 1269–1271. 10.1093/bioinformatics/btl097 [DOI] [PubMed] [Google Scholar]
- Dehal, P. , Satou, Y. , Campbell, R. K. , Chapman, J. , Degnan, B. , De Tomaso, A. , … Rokhsar, D. S. (2002). The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science, 298(5601), 2157–2167. 10.1126/science.1080049 [DOI] [PubMed] [Google Scholar]
- Delsuc, F. , Philippe, H. , Tsagkogeorga, G. , Simion, P. , Tilak, M.‐K. , Turon, X. , … Douzery, E. J. P. (2018). A phylogenomic framework and timescale for comparative studies of tunicates. Bmc Biology, 16, 39 10.1186/s12915-018-0499-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoeud, F. , Henriet, S. , Mungpakdee, S. , Aury, J.‐M. , Da Silva, C. , Brinkmann, H. , … Chourrout, D. (2010). Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science, 330(6009), 1381–1385. 10.1126/science.1194167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, L. , Viard, F. , Dowell, M. J. , Wood, C. , & Bishop, J. D. D. (2009). Fine‐ and regional‐scale genetic structure of the exotic ascidian Styela clava (Tunicata) in southwest England, 50 years after its introduction. Molecular Ecology, 18(3), 442–453. 10.1111/j.1365-294X.2008.04045.x [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman, R. M. , Rousou, J. A. , Flack, J. E. , Deaton, D. W. , Kalfin, R. , & Das, D. (1995). Influence of steroids on complement and cytokine generation after cardiopulmonary bypass. Annals of Thoracic Surgery, 60(3), 801–804. 10.1016/0003-4975(95)00211-3 [DOI] [PubMed] [Google Scholar]
- Feder, M. E. , & Hofmann, G. E. (1999). Heat‐shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annual Review of Physiology, 61, 243–282. 10.1146/annurev.physiol.61.1.243 [DOI] [PubMed] [Google Scholar]
- Fujikawa, T. , Munakata, T. , Kondo, S.‐I. , Satoh, N. , & Wada, S. (2010). Stress response in the ascidian Ciona intestinalis: Transcriptional profiling of genes for the heat shock protein 70 chaperone system under heat stress and endoplasmic reticulum stress. Cell Stress & Chaperones, 15(2), 193–204. 10.1007/s12192-009-0133-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain, P. , Iyer, J. , Zechel, C. , & Gronemeyer, H. (2002). Co‐regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature, 415(6868), 187–192. 10.1038/415187a [DOI] [PubMed] [Google Scholar]
- Goldstien, S. J. , Dupont, L. , Viard, F. , Hallas, P. J. , Nishikawa, T. , Schiel, D. R. , … Bishop, J. D. D. (2011). Global phylogeography of the widely introduced North West Pacific Ascidian Styela clava . PLoS One, 6(2), e16755 10.1371/journal.pone.0016755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstien, S. J. , Schiel, D. R. , & Gemmell, N. J. (2010). Regional connectivity and coastal expansion: Differentiating pre‐border and post‐border vectors for the invasive tunicate Styela clava . Molecular Ecology, 19(5), 874–885. 10.1111/j.1365-294X.2010.04527.x [DOI] [PubMed] [Google Scholar]
- Haas, B. J. , Salzberg, S. L. , Zhu, W. , Pertea, M. , Allen, J. E. , Orvis, J. , … Wortman, J. R. (2008). Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biology, 9(1), R7 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, E. (2009). Ascidian tunic cells: Morphology and functional diversity of free cells outside the epidermis. Invertebrate Biology, 128(1), 83–96. 10.1111/j.1744-7410.2008.00153.x [DOI] [Google Scholar]
- Hirose, E. , Kimura, S. , Itoh, T. , & Nishikawa, J. (1999). Tunic morphology and cellulosic components of pyrosomas, doliolids, and salps (Thaliacea, Urochordata). Biological Bulletin, 196(1), 113–120. 10.2307/1543173 [DOI] [PubMed] [Google Scholar]
- Hirose, E. , Ohtake, S. I. , & Azumi, K. (2009). Morphological characterization of the tunic in the edible ascidian, Halocynthia roretzi (Drasche), with remarks on 'soft tunic syndrome' in aquaculture. Journal of Fish Diseases, 32(5), 433–445. 10.1111/j.1365-2761.2009.01034.x [DOI] [PubMed] [Google Scholar]
- Horn, G. , Hofweber, R. , Kremer, W. , & Kalbitzer, H. R. (2007). Structure and function of bacterial cold shock proteins. Cellular and Molecular Life Sciences, 64(12), 1457–1470. 10.1007/s00018-007-6388-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Li, S. , Gao, Y. , & Zhan, A. (2018). Genome‐wide identification, characterization and expression analyses of heat shock protein‐related genes in a highly invasive Ascidian Ciona savignyi . Frontiers in Physiology, 9, 1043 10.3389/fphys.2018.01043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Li, S. , & Zhan, A. (2019). Genome‐wide identification and evaluation of new reference genes for gene expression analysis under temperature and salinity stresses in Ciona savignyi . Frontiers in Genetics, 10(71), 10.3389/fgene.2019.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, J. , Nakashima, K. , & Satoh, N. (2019). ORTHOSCOPE analysis reveals the presence of the cellulose synthase gene in all tunicate genomes but not in other animal genomes. Genes, 10(4), 294 10.3390/genes10040294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jue, N. K. , Batta‐Lona, P. G. , Trusiak, S. , Obergfell, C. , Bucklin, A. , O'Neill, M. J. , & O'Neill, R. J. (2016). Rapid Evolutionary Rates and Unique Genomic Signatures Discovered in the First Reference Genome for the Southern Ocean Salp, Salpa thompsoni (Urochordata, Thaliacea). Genome Biology and Evolution, 8(10), 3171–3186. 10.1093/gbe/evw215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaiskou, A. , Swalla, B. J. , Sasakura, Y. , & Chambon, J. P. (2015). Metamorphosis in Solitary Ascidians. Genesis, 53(1), 34–47. 10.1002/dvg.22824 [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. , & Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12(4), 357–U121. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, S. , Ohshima, C. , Hirose, E. , Nishikawa, J. , & Itoh, T. (2001). Cellulose in the house of the appendicularian Oikopleura rufescens. Protoplasma, 216(1–2), 71–74. 10.1007/bf02680133 [DOI] [PubMed] [Google Scholar]
- Korf, I. (2004). Gene finding in novel genomes. BMC Bioinformatics, 5, 59 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland, C. G. , Canback, B. , & Berg, O. G. (2003). Horizontal gene transfer: A critical view. Proceedings of the National Academy of Sciences of the United States of America, 100(17), 9658–9662. 10.1073/pnas.1632870100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladomery, M. , & Sommerville, J. (1994). Binding of y‐box proteins to RNA: involvement of different protein domains. Nucleic Acids Research, 22(25), 5582–5589. 10.1093/nar/22.25.5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, G. (2007). Invasive sea squirts: A growing global problem. Journal of Experimental Marine Biology and Ecology, 342(1), 3–4. 10.1016/j.jembe.2006.10.009 [DOI] [Google Scholar]
- Langfelder, P. , & Horvath, S. (2008). WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudet, V. (2011). The origins and evolution of vertebrate metamorphosis. Current Biology, 21(18), R726–R737. 10.1016/j.cub.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Li, H. , & Durbin, R. (2009). Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25(14), 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods, 25(4), 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Locke, A. , Hanson, J. M. , Ellis, K. M. , Thompson, J. , & Rochette, R. (2007). Invasion of the southern Gulf of St. Lawrence by the clubbed tunicate (Styela clava Herdman): Potential mechanisms for invasions of Prince Edward Island estuaries. Journal of Experimental Marine Biology and Ecology, 342(1), 69–77. 10.1016/j.jembe.2006.10.016 [DOI] [Google Scholar]
- Lowe, T. M. , & Eddy, S. R. (1997). tRNAscan‐SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 25(5), 955–964. 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros, W. H. , Pertea, M. , & Salzberg, S. L. (2004). TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene‐finders. Bioinformatics, 20(16), 2878–2879. 10.1093/bioinformatics/bth315 [DOI] [PubMed] [Google Scholar]
- Marcais, G. , & Kingsford, C. (2011). A fast, lock‐free approach for efficient parallel counting of occurrences of k‐mers. Bioinformatics, 27(6), 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A. G. , Deschet, K. , Williams, M. , Marry, M. , White, A. R. , & Smith, W. C. (2004). A functional cellulose synthase from ascidian epidermis. Proceedings of the National Academy of Sciences of the United States of America, 101(4), 986–991. 10.1073/pnas.0303623101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailovich, M. , Militti, C. , Gabaldon, T. , & Gebauer, F. (2010). Eukaryotic cold shock domain proteins: Highly versatile regulators of gene expression. BioEssays, 32(2), 109–118. 10.1002/bies.200900122 [DOI] [PubMed] [Google Scholar]
- Nahoum, V. , Perez, E. , Germain, P. , Rodriguez‐Barrios, F. , Manzo, F. , Kammerer, S. , … Bourguet, W. (2007). Modulators of the structural dynamics of the retinoid X receptor to reveal receptor function. Proceedings of the National Academy of Sciences of the United States of America, 104(44), 17323–17328. 10.1073/pnas.0705356104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville, M. , Henriet, S. , Warren, I. , Sumic, S. , Reeve, M. , Volff, J. N. , & Chourrout, D. (2019). Massive Changes of Genome Size Driven by Expansions of Non‐autonomous Transposable Elements. Current Biology, 29(7), 1161–1168.e6. 10.1016/j.cub.2019.01.080 [DOI] [PubMed] [Google Scholar]
- Nawrocki, E. P. , & Eddy, S. R. (2013). Infernal 1.1: 100‐fold faster RNA homology searches. Bioinformatics, 29(22), 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, T. , Yue, J. , Sun, G. , Zou, Y. , Wen, J. , & Huang, J. (2012). Ancient gene transfer from algae to animals: Mechanisms and evolutionary significance. Bmc Evolutionary Biology, 12, 10.1186/1471-2148-12-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, M. , & Kimura, A. (2006). Genomic view of the evolution of the complement system. Immunogenetics, 58(9), 701–713. 10.1007/s00251-006-0142-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard, B. D. , & Weiler, J. M. (1983). Steroids inhibit activation of the alternative‐amplification pathway of complement. Infection and Immunity, 40(3), 1011–1014. 10.1128/iai.40.3.1011-1014.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini, A. , Manenti, R. , Rothbacher, U. , & Lemaire, P. (2012). Antagonizing retinoic acid and FGF/MAPK pathways control posterior body patterning in the invertebrate chordate Ciona intestinalis . PLoS One, 7(9), 11 10.1371/journal.pone.0046193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasba, P. K. , Ramakrishnan, B. , & Boeggeman, E. (2008). Structure and function of beta‐1,4‐Galactosyltransferase. Current Drug Targets, 9(4), 292–309. 10.2174/138945008783954943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A. , Drummond, A. J. , Xie, D. , Baele, G. , & Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology, 67(5), 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranby, B. G. (1952). Physico‐chemical investinations on animal cellulose (Tunicin). Arkiv for Kemi, 4, 241–248. [Google Scholar]
- Rautengarten, C. , Ebert, B. , Moreno, I. , Temple, H. , Herter, T. , Link, B. , … Orellana, A. (2014). The Golgi localized bifunctional UDP‐rhamnose/UDP‐galactose transporter family of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 111(31), 11563–11568. 10.1073/pnas.1406073111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rius, M. , Bourne, S. , Hornsby, H. G. , & Chapman, M. A. (2015). Applications of next‐generation sequencing to the study of biological invasions. Current . Zoology, 61(3), 488–504. 10.1093/czoolo/61.3.488 [DOI] [Google Scholar]
- Roberts, B. , Davidson, B. , MacMaster, G. , Lockhart, V. , Ma, E. , Smith Wallace, S. , & Swalla, B. J. (2007). A complement response may activate metamorphosis in the ascidian Boltenia villosa . Development Genes and Evolution, 217(6), 449–458. 10.1007/s00427-007-0157-0 [DOI] [PubMed] [Google Scholar]
- Sagane, Y. , Zech, K. , Bouquet, J.‐M. , Schmid, M. , Bal, U. , & Thompson, E. M. (2010). Functional specialization of cellulose synthase genes of prokaryotic origin in chordate larvaceans. Development, 137(9), 1483–1492. 10.1242/dev.044503 [DOI] [PubMed] [Google Scholar]
- Sambrook, J. (2001). Molecular cloning: A laboratory manual: Third edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Satou, Y. , Nakamura, R. , Yu, D. , Yoshida, R. , Hamada, M. , Fujie, M. , … Satoh, N. (2019). A nearly complete genome of Ciona intestinalis type A (C. robusta) reveals the contribution of inversion to chromosomal evolution in the genus Ciona. Genome Biology and Evolution, 11(11), 3144–3157. 10.1093/gbe/evz228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H. C. , Kube, M. , Edvardsen, R. B. , Jensen, M. F. , Beck, A. , Spriet, E. , … Chourrout, D. (2001). Miniature genome in the marine chordate Oikopleura dioica . Science, 294(5551), 2506 10.1126/science.294.5551.2506 [DOI] [PubMed] [Google Scholar]
- Serafini, L. , Hann, J. B. , Kueltz, D. , & Tomanek, L. (2011). The proteomic response of sea squirts (genus Ciona) to acute heat stress: A global perspective on the thermal stability of proteins. Comparative Biochemistry and Physiology D‐Genomics & Proteomics, 6(3), 322–334. 10.1016/j.cbd.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Shao, C. , Bao, B. , Xie, Z. , Chen, X. , Li, B. , Jia, X. , … Chen, S. (2017). The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nature Genetics, 49(1), 119–124. 10.1038/ng.3732 [DOI] [PubMed] [Google Scholar]
- Shu, D. G. , Chen, L. , Han, J. , & Zhang, X. L. (2001). An early Cambrian tunicate from China. Nature, 411(6836), 472–473. 10.1038/35078069 [DOI] [PubMed] [Google Scholar]
- Simakov, O. , Kawashima, T. , Marletaz, F. , Jenkins, J. , Koyanagi, R. , Mitros, T. , … Gerhart, J. (2015). Hemichordate genomes and deuterostome origins. Nature, 527(7579), 459–465. 10.1038/nature16150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao, F. A. , Waterhouse, R. M. , Ioannidis, P. , Kriventseva, E. V. , & Zdobnov, E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single‐copy orthologs. Bioinformatics, 31(19), 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Small, K. S. , Brudno, M. , Hill, M. M. , & Sidow, A. (2007). A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biology, 8(3), R41 10.1186/gb-2007-8-3-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy, S. M. , Huang, J. , & Gogarten, J. P. (2015). Horizontal gene transfer: Building the web of life. Nature Reviews Genetics, 16(8), 472–482. 10.1038/nrg3962 [DOI] [PubMed] [Google Scholar]
- Stanke, M. , Keller, O. , Gunduz, I. , Hayes, A. , Waack, S. , & Morgenstern, B. (2006). AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Research, 34, W435–W439. 10.1093/nar/gkl200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley, J. , Santure, A. W. , & Dennis, S. R. (2015). Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Molecular Ecology, 24(9), 2241–2252. 10.1111/mec.13089 [DOI] [PubMed] [Google Scholar]
- Stolfi, A. , Lowe, E. K. , Racioppi, C. , Ristoratore, F. , Brown, C. T. , Swalla, B. J. , & Christiaen, L. (2014). Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. Elife, 3, e03728 10.7554/eLife.03728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strydom, D. J. (1994). Chromatographic separation of 1‐phenyl‐3‐methyl‐5‐pyrazolone‐derivatized neutral, acidic and basic aldoses. Journal of Chromatography A, 678(1), 17–23. 10.1016/0021-9673(94)87069-1 [DOI] [Google Scholar]
- Taylor, K. M. (1967). The chromosomes of some lower chordates. Chromosoma, 21(1), 181–188. 10.1007/bf00343643 [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. , & Salzberg, S. L. (2009). TopHat: Discovering splice junctions with RNA‐Seq. Bioinformatics, 25(9), 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboynik, A. , Neff, N. F. , Sahoo, D. , Newman, A. M. , Pushkarev, D. , Koh, W. , … Quake, S. R. (2013). The genome sequence of the colonial chordate Botryllus schlosseri . Elife, 2, e00569 10.7554/eLife.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24(8), 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhan, A. , Briski, E. , Bock, D. G. , Ghabooli, S. , & MacIsaac, H. J. (2015). Ascidians as models for studying invasion success. Marine Biology, 162(12), 2449–2470. 10.1007/s00227-015-2734-5 [DOI] [Google Scholar]
- Zhang, G. , Fang, X. , Guo, X. , Li, L. , Luo, R. , Xu, F. , … Wang, J. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature, 490(7418), 49–54. 10.1038/nature11413 [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , & Li, J. (2014). Excellent chemical and material cellulose from tunicates: Diversity in cellulose production yield and chemical and morphological structures from different tunicate species. Cellulose, 21(5), 3427–3441. 10.1007/s10570-014-0348-6 [DOI] [Google Scholar]
- Zhao, Y. , & Li, J. (2016). Ascidian bioresources: Common and variant chemical compositions and exploitation strategy examples of Halocynthia roretzi, Styela plicata, Ascidia sp and Ciona intestinalis . Zeitschrift Fur Naturforschung Section C‐a Journal of Biosciences, 71(5–6), 165–180. 10.1515/znc-2016-0012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The genome sequence project was deposited in NCBI under the BioProject number PRJNA523448. The transcriptome data used for biological analysis were also deposited in the NCBI SRA database with the accession numbers SRR8599814 to SRR8599834. The genome resources of other ascidians were downloaded from ANISEED (https://www.aniseed.cnrs.fr/), and the genome resources of O. dioica were acquired from OikoBase (http://oikoarrays.biology.uiowa.edu/Oiko/).


