To the Editor,
Recent genome‐wide association studies (GWAS) have identified multiple candidate variants for food allergy (FA). 1 , 2 , 3 In February 2019, we convened to develop an International FA Consortium (InFAC) for genetic research (Vancouver, Canada). The main goal of InFAC was to enable large‐scale international collaborations on FA genetics, since polygenic diseases require large samples to identify associated genetic variants. One of the barriers addressed was the need for consensus phenotype definitions, as direct comparisons of previous findings are complicated by heterogeneity in the FA definitions applied. In particular, asymptomatic sensitized individuals and self‐reported FA without laboratory testing pose significant challenges. Another key issue is the definition of appropriate controls which range from biobanked general population controls to non‐atopic controls with negative sensitization testing or absence of clinical history.
The double‐blind, placebo‐controlled food challenge (DBPCFC) is the gold standard for FA diagnosis, but some large‐scale genetic resources may not have these data and it may be difficult to find an adequate number of individuals meeting this stringent FA definition. Some investigators have performed placebo‐controlled food challenges without double‐blinding or utilized open oral food challenges (OFC), both of which still provide strong clinical support for a diagnosis of FA. Use of standard cutoffs for in vivo and in vitro diagnostic tests as a surrogate for DBPCFC is not well established, because positive predictive value (PPV) cutoffs vary greatly across populations. We perceive the definition of FA phenotype is a major barrier toward making progress in FA genetic research. A key outcome from this meeting was to develop consensus around FA inclusion criteria for large‐scale genetic research.
Two key consensus statements emerged regarding large‐scale genetic FA research and are presented in Table 1a.
Table 1a.
InFAC 2019 statements regarding food allergy phenotypes for genetic research collaborations
| 1 |
Initial collaborations should concentrate on peanut, hen's egg, and cow's milk allergy due to difficulties with other types of FA in regard to low prevalence and lack of consensus regarding diagnostic criteria (ie, 95% PPV cutoffs for allergen‐specific IgE (sIgE) and skin prick test (SPT)) outside of DBPCFC Statement 1 is made with the assumption that the underlying genetic model involves specific risk loci for specific foods. If one assumes that all FA are influenced by the same genetic risk loci, a study design grouping all FA may be more powerful, but would require DBPCFC for all cases that do not have well‐established diagnostic criteria outside of DBPCFC. |
| 2 |
Consider 4 tiers of FA diagnostic criteria, from most stringent to more moderate. These include the following, in order of decreasing stringency (Figure 1A): Tier 1: Convincing reaction confirmed by oral food challenge Tier 2: Convincing clinical history of a reaction with objective symptoms and confirmed exposure (Table 1b and below) with evidence of sensitization at or above 95% PPV cutoffs for SPT OR sIgE (Table 1c, Table S1, online supplement) Tier 3: Convincing clinical history of a reaction with objective symptoms and confirmed exposure (Table 1b), with any sensitization ≥ 3mm SPT OR sIgE ≥ 0.35kUA/L Tier 4: No convincing history with sensitization at or above 95% PPV cutoffs for SPT or sIgE |
What determines a convincing history of an IgE‐mediated allergic reaction to a specific food is controversial. In practice, allergists use experienced clinical judgment regarding history, symptoms, and testing to make a diagnosis of FA. Several definitions and scores describing an allergic response were considered, including the World Allergy Organization definition of anaphylaxis (online supplement) 4 and the Astier score for reaction severity 5 (Table S2), but neither included all criteria deemed to be essential for a convincing history of FA. We therefore agreed that for the purposes of phenotyping for genetic studies, a convincing history should involve the following: a) exposure by ingestion of the food allergen b) onset of symptoms within 2 hours of exposure with resolution within 24 hours, and c) only objective symptoms (Table 1b). Two‐organ system involvement is not required and treatment is not considered. Local symptoms through contact of the food allergen with the skin or oral mucosa are not sufficient. Reactions solely identified by subjective symptoms (Table 1b) or linked to unclear exposure (ie, after consuming multiple food allergens and cross‐contamination) (Table 1b) are not considered convincing. Some subjective symptoms were identified by allergists in the group as having weight in a clinical history of FA, but most felt that these symptoms and anaphylaxis would be accompanied by objective symptoms.
Table 1b.
Objective versus subjective symptoms occurring within 2 h of ingestion of allergen
| System | Objective Symptoms | Subjective Symptoms |
|---|---|---|
| Airway | Angioedema of eyes, lips, tongue, oral cavity, and pharynx | Isolated pruritus (skin, eyes, lips, tongue, throat, and nose) with no visible symptoms |
| Dysphonia, hoarseness, and laryngeal edema | Throat congestion | |
| Shortness of breath | ||
| Wheezing | Chest tightness/pain | |
| Coughing | ||
| Sneezing episodes | ||
| Rhinorrhea | Nasal congestion | |
| Cutaneous | Conjunctivitis, tearing | Isolated perioral redness |
| Erythema/flushing | Skin discomfort | |
| Urticaria | Contact urticaria from food allergen contact, without ingestion | |
| Neurologic | Loss of consciousness/fainting | Dizziness, light‐headedness |
| Seizures | Feeling of impending doom | |
| Cardiac | Tachycardia | |
| Hypotension | ||
| Gastrointestinal | Emesis/Diarrhea | Abdominal pain |
| Nausea | ||
| Genitourinary | Uterine cramping/contractions |
Adapted from Pajno et al 2017, Allergy. 8
Tier 4 of statement 2 is the most controversial as the challenge is to define “minimal criteria” for FA. Many large‐scale genetic studies lack a detailed clinical history but have laboratory testing, allowing the allergic individuals in these studies to be captured by Tier 4. While we realize that this may lead to the inclusion of some individuals with asymptomatic sensitization, there was consensus that Tier 4, using the currently accepted 95% PPV cutoffs for SPT or sIgE (Table 1c, Table S1 online supplement), will generate a reasonable level of certainty to allow international collaboration at the most inclusive level. However, there is a trade‐off as categories become less stringent—available samples will increase, but at the cost of potential misclassification.
Table 1c.
Proposed 95% PPV cutoffs for skin prick test (SPT) and specific IgE (sIgE)
| Food type and age | SPT size (mm) | sIgE levels (kUA/mL) |
|---|---|---|
| Peanut | ≥8 | ≥14 |
| Peanut in ≤2 years | ≥8 | ≥14 |
| Cow's milk | ≥8 | ≥15 |
| Cow's milk in ≤2 years | ≥8 | ≥5 |
| Hen's egg white | ≥8 | ≥7 |
| Hen's egg white in ≤2 years | ≥8 | ≥2 |
The mode of inheritance, penetrance of risk genes, and their frequency in the study population determine sample size requirements. For GWAS, sample size requirements depend greatly upon the research question being asked, the effect size, and the comparator group. Risk variants with large effect sizes do not require as large of a sample size, but many polygenic diseases do not have risk variants with large effect sizes. The genetic model assumed in the experiment is key and could include the following: a) genetic risk variants exist for development of FA to a specific food, b) genetic risk variants exist that are common to developing all food allergies, but the specific allergy is determined by non‐genetic factors c) genetic risk variants exist that are common to the development of all atopic diseases (atopic dermatitis, allergic asthma, allergic rhinitis, and eosinophilic esophagitis) but which one develops is determined by interaction effects either genetic (gene‐gene) or non‐genetic (gene‐environment) factors, and d) a combination of all of these assumptions.
Control group phenotypes also play a key role in sample size requirements: hypercontrols—individuals who have no history of FA, or atopic diseases, and laboratory testing that indicates that they are not sensitized to foods—are helpful to increase power to detect genetic variants. However, it would be difficult to separate genetic risk factors for atopic tendency from those that cause FA, and there may be an age discrepancy and generational differences between cases. Population‐based controls may also be used, with consideration of potential misclassification; prevalence of the FA being studied may be low in the population, but the combined effect of multiple atopic conditions, some of which may be self‐resolving, may not be negligible. Use of controls with atopic diseases for investigation of FA may be difficult given the waxing and waning conditions of the atopic march.
Case and control recruitment can be costly (both monetarily and in time) due to the diagnostic history, testing, and follow‐up required to achieve well‐defined allergic phenotypes. Smaller sample sizes may have the power to detect genetic variants of interest in circumstances where the genetic risk factor has a large effect size and there are well‐phenotyped cases and hypercontrols, but sample size requirements will vary depending upon the genetic model, the population studied, and the question asked.
For genetic studies, samples can be taken from a variety of tissue types. However, for epigenetic studies, sample type is key as each tissue will exhibit a specific epigenetic signature. Some sample types, for example blood samples, may be more difficult to use in epigenetic studies due to the variety of cell populations within the sample. For FA, which presents with a diverse variety of effects in various systems and thus has no single tissue affected, most studies have focused on blood samples. For epigenetic studies, the sample size is dependent on the purity of the tissue and how enriched that tissue is for the disease‐associated signal, in addition to the variables mentioned previously for GWA studies.
Recent publications on FA genetics have identified many potential loci for atopic diseases. However, the underlying genetic model for these diseases is unknown. Atopic conditions may share common genetic susceptibility, and environmental factors may determine which specific atopic disease develops; alternatively, each atopic disease may have its own risk variants, with transient and persistent forms differing in genetic susceptibility (Figure 1B).
Figure 1.
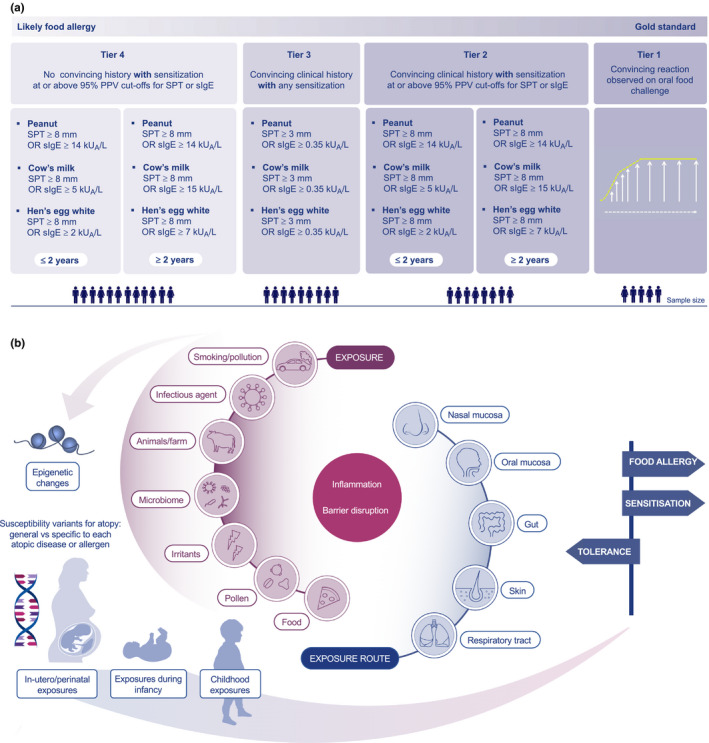
A, Proposed groups of food allergy phenotypes for genetic research. B, Summary of factors involved in development of food allergy
These complex questions cannot be answered by studies with small samples, and genetic and molecular studies are inherently costly endeavors. Pooling data across studies is a cost‐effective method to increase power. This will require a harmonization of phenotyping to ensure that efforts are not diluted by varying phenotypes. A balance between the gold standard and well‐accepted global clinical and laboratory surrogate markers is required, as well as data on the transferability of IgE values from different platforms. While the gold standard of phenotyping of FA is DBPCFC, for research purposes we propose various levels of phenotyping to maximize potential collaborations. Findings can be subsequently verified in FA diagnosed by the gold standard.
Some may question the utility of genetic markers for FA due to ample evidence on the role of the environment in the development of allergies. 6 In an era of precision medicine, biologics, oral, epicutaneous, and sublingual immunotherapy, genetic markers may significantly enhance decision‐making. 7 Genetic susceptibility may have a role in the development of natural or treatment‐induced tolerance.
Understanding the genetic architecture of FA remains a research imperative as accurate polygenic models may inform differing natural histories (ie, resolution versus persistence of FA), biomarker discovery, and identifies patients most likely to benefit from targeted primary prevention and immunotherapy. This aligns with efforts to provide more specific recommendations among patient subgroups, as considerable heterogeneity exists even among similar FA phenotypes. Thus, it is of utmost importance to pursue joint global approaches to identify the genetic drivers of FA. To succeed, these efforts need to be based on a consensus of how to diagnose FA.
AUTHOR CONTRIBUTION
YA and DD organized the InFAC meeting. YA, DM, TE, KN, GK, AC, EC, ES, CL, BM, IM, DR, SE, CH, JG, AE, LS, JH, MA, AS, and DD participated in the InFAC meeting and contributed to the content and text of the paper. YA wrote the paper with valuable comments from DM, TE, KN, GK, AC, YL, EC, ES, CL, BM, IM, DR, SE, CH, JG, AE, LS, AS, and DD.
Supporting information
Table S1‐S3
REFERENCES
- 1. Asai Y, Eslami A, van Ginkel CD, et al. Genome‐wide association study and meta‐analysis in multiple populations identifies new loci for peanut allergy and establishes C11orf30/EMSY as a genetic risk factor for food allergy. J Allergy Clin Immunol. 2018;141(3):991‐1001. [DOI] [PubMed] [Google Scholar]
- 2. Marenholz I, Grosche S, Kalb B, et al. Genome‐wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat Commun. 2017;8(1):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martino DJ, Ashley S, Koplin J, et al. Genomewide association study of peanut allergy reproduces association with amino acid polymorphisms in HLA‐DRB1. Clin Exp Allergy. 2017;47(2):217‐223. [DOI] [PubMed] [Google Scholar]
- 4. Simons FE, Ardusso LR, Bilo MB, et al. World allergy organization guidelines for the assessment and management of anaphylaxis. World Allergy Organ J. 2011;4(2):13‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Astier C, Morisset M, Roitel O, et al. Predictive value of skin prick tests using recombinant allergens for diagnosis of peanut allergy. J Allergy Clin Immunol. 2006;118(1):250‐256. [DOI] [PubMed] [Google Scholar]
- 6. Sbihi H, Boutin RC, Cutler C, Suen M, Finlay BB, Turvey SE. Thinking bigger: How early‐life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy 2019;74(11):2103‐2115. [DOI] [PubMed] [Google Scholar]
- 7. Eiwegger T, Hung L, San Diego KE, O'Mahony L, Upton J. Recent developments and s in food allergy. Allergy. 2019;74 (12):2355–2367. 10.1111/all.14082. [DOI] [PubMed] [Google Scholar]
- 8. Pajno GB, Fernandez‐Rivas M, Arasi S, et al. EAACI Guidelines on allergen immunotherapy: IgE‐mediated food allergy. Allergy 2018;73(4):799‐815. [DOI] [PubMed] [Google Scholar]
- 9. Nowak‐Wegrzyn A, Assa'ad AH, Bahna SL, et al. Work Group report: oral food challenge testing. J Allergy Clin Immunol. 2009;123(6 Suppl):S365‐383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1‐S3


