Abstract
CTX‐M‐type extended‐spectrum β‐lactamase (ESBL)‐producing Escherichia coli clones have been increasingly reported worldwide. In this regard, although discussions of transmission routes of these bacteria are in evidence, molecular data are lacking to elucidate the epidemiological impacts of ESBL producers in wild animals. In this study, we have screened 90 wild animals living in a surrounding area of São Paulo, the largest metropolitan city in South America, to monitor the presence of multidrug‐resistant (MDR) Gram‐negative bacteria. Using a genomic approach, we have analysed eight ceftriaxone‐resistant E. coli. Resistome analyses revealed that all E. coli strains carried bla CTX‐M‐type genes, prevalent in human infections, besides other clinically relevant resistance genes to aminoglycosides, β‐lactams, phenicols, tetracyclines, sulphonamides, trimethoprim, fosfomycin and quinolones. Additionally, E. coli strains belonged to international sequence types (STs) ST38, ST58, ST212, ST744, ST1158 and ST1251, and carried several virulence‐associated genes. Our findings suggest spread and adaptation of international clones of CTX‐M‐producing E. coli beyond urban settings, including wildlife from shared environments.
Keywords: Enterobacterales, ESBL, MDR bacteria, resistome, wildlife
1. INTRODUCTION
The spread of extended‐spectrum β‐lactamase (ESBL)‐producing Enterobacterales has been broadly reported worldwide (Brolund, 2014; Fernandes et al., 2018; Pardon et al., 2015). In this respect, a number of interlinked factors, such as food animals, environmental sources, human migration and access to basic sanitation in highly populated cities, are contributing for the accelerated dissemination of these bacteria in urban and wild environments (Radhouani et al., 2014; Sacramento et al., 2018; Sellera, Fernandes, Moura, Carvalho, & Lincopan, 2018).
While the exposure to polluted environments constitutes a risk factor for humans to acquire multidrug‐resistant (MDR) bacteria, recent studies have pointed out that it could also have implications for wildlife (Cerdà‐Cuéllar et al., 2019; Sellera, 2019; Wang et al., 2017). In fact, although this matter remains poorly addressed under ecological perspectives, the scientific community and nature conservation authorities have begun to see wild animals as reservoirs and potential disseminators of ESBL‐producing bacteria (Ardiles‐Villegas, González‐Acuña, Waldenström, Olsen, & Hernández, 2011; Cerdà‐Cuéllar et al., 2019; Sellera, 2019; Wang et al., 2017). Nowadays, most ESBL‐producing Escherichia coli circulating at the human–animal–environment interface belong to international sequence types (STs) such as ST10, ST38, ST58, ST131, ST212, ST648, ST744, ST1158 and ST1251 (Borges, Tarlton, & Riley, 2019; Cao et al., 2014; Castellanos et al., 2017; Haenni et al., 2018; Nüesch‐Inderbinen et al., 2019; Pitout, 2012; Tacão et al., 2017; Tafoukt, Touati, Leangapichart, Bakour, & Rolain, 2017; Vignoli et al., 2016; Zurfluh et al., 2017), suggesting a broad host adaptation of these pathogens. In this study, we report the occurrence of pandemic clones of CTX‐M‐producing E. coli recovered from a diversity of peri‐urban wild animals in Brazil, highlighting the transmission of this sort of bacteria in anthropogenic‐shared environments.
2. MATERIALS AND METHODS
Between June 2017 and July 2018, a local surveillance study was conducted to monitor the presence of MDR Gram‐negative bacteria in urbanized wild animals, in São Paulo, Brazil, the largest metropolitan city in South America. For this purpose, we sampled rectal or cloacal swabs from 90 wild animals, including reptiles, birds and mammals’ species rescued by authorities (firefighters and environmental police) and delivered to wildlife rehabilitation centres. The sampled species included Alouatta guariba (n = 4), Asio clamator (n = 7), Asio stygius (n = 1), Caracara plancus (n = 2), Coragyps atratus (n = 27), Didelphis aurita (n = 11), Egretta thula (n = 1), Hydrochoerus hydrochaeris (n = 14), Hydromedusa tectifera (n = 2), Megascops choliba (n = 2), Nasua nasua (n = 13), Nycticorax nycticorax (n = 1), Sapajus apellla (n = 1), Tapirus terrestris (n = 1), Tupinambis merianae (n = 2) and Tyto furcata (n = 1). Biological sample collections were authorized by the Authorization System and Information on Biodiversity (SISBIO licence number 55804–2).
Swab samples were streaked onto MacConkey agar plates supplemented with ceftriaxone (2 µg/ml), colistin (2 µg/ml) or meropenem (2 µg/ml), and the grown bacteria were identified by matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI‐TOF). Antimicrobial susceptibility was determined by disc diffusion and/or E‐test methods, using breakpoints approved by the Clinical and Laboratory Standards Institute (CLSI, 2015, 2017). Twenty‐two antibiotics were tested including amikacin, amoxicillin/clavulanic acid, ampicillin, aztreonam, ceftazidime, cephalothin, ciprofloxacin, chloramphenicol, ceftriaxone, ceftiofur, cefotaxime, doxycycline, enrofloxacin, cefepime, gentamicin, nalidixic acid, sulphonamide, trimethoprim/sulphamethoxazole, tetracycline, kanamycin, tobramycin and streptomycin. Additionally, the presence of CTX‐M‐type (bla CTX‐M‐1, bla CTX‐M‐2, bla CTX‐M‐8 and bla CTX‐M‐9) groups, carbapenemase (bla KPC‐2) and mobilized colistin resistance (mcr‐1) genes was evaluated by PCR analysis (Dropa et al., 2016; Liu et al., 2016; Minarini, Poirel, Trevisani, Darini, & Nordmann, 2009; Muzaheed et al., 2008; Poirel, Walsh, Cuvillier, & Nordmann, 2011; Saladin et al., 2002).
The isolates confirmed positive by PCR were whole‐genome sequenced. Genomic DNA was extracted from overnight cultures using the PureLink® Genomic DNA Mini Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Whole‐genome sequencing (WGS) was performed using Illumina NextSeq500 platform (Illumina, San Diego, CA) (150 bp paired‐end), and the reads were de novo assembled using Velvet 1.2.10 (Zerbino & Birney, 2008) or SPAdes 3.9 (Bankevich et al., 2012). Sequence types, serotypes, plasmid replicon types, antimicrobial resistance genes and virulence genes were identified using MLST 2.0, SerotypeFinder 2.0 (identity ≥ 85%; coverage ≥ 60%), PlasmidFinder 2.1 (identity ≥ 95%; coverage ≥ 60%), ResFinder 3.2 (identity ≥ 90%; coverage ≥ 60%) and VirulenceFinder 2.0 (identity ≥ 90%; coverage ≥ 60%) tools, respectively, available from the Center for Genomic Epidemiology (http://genomicepidemiology.org/). Analysis of the genetic context of bla CTX‐M genes was performed with BLASTn and ISFinder analyses (Siguier, Perochon, Lestrade, Mahillon, & Chandler, 2006) followed by manual curation using Geneious 10.2.6.
Plasmid transfer was performed by conjugation using streptomycin‐resistant E. coli C600 or azide‐resistant E. coli J53 recipient strains in LB broth assays, ratio 3:1 (recipient:donor). Transconjugants were selected using MacConkey agar supplemented with ceftriaxone (2 µg/ml) and streptomycin (2000 µg/ml), or ceftriaxone (2 µg/ml) and sodium azide (200 µg/ml). In transformation assays, plasmids were extracted by the alkaline lysis method (Sambrook & Russel, 2001), and ultra‐competent E. coli TOP10 was heat shock transformed as previously described (Inoue, Nojima, & Okayama, 1990), increasing the thermal shock time at 42°C to 1.5 min. Transformants were selected using MacConkey agar supplemented with ceftriaxone (2 µg/ml). Positive transconjugants and transformants strains were confirmed by bla CTX‐M genes using PCR.
3. RESULTS AND DISCUSSION
In this study, eight ceftriaxone‐resistant E. coli isolates (8/90; 8.88%) were recovered from five birds (one owl and four vultures) and three mammals (coatis). MDR profiles, defined as resistant to three or more classes of antibiotics (Magiorakos et al., 2012), were evidenced in six isolates (ECPET11, ECPET31, ECPET36, ICBUR6, ICBUR15 and ICBUR20). ECPET3 displayed resistance only to cephalosporins and aztreonam, whereas ECPET13 was resistant to cephalosporins, aztreonam and nalidixic acid. Additionally, ESBL production was confirmed by double‐disc synergy test (DDST), and PCR analysis revealed the presence of bla CTX‐M‐type genes in all eight bacterial isolates (Table 1). No MCR‐1‐positive or carbapenemase‐producing bacteria were identified.
TABLE 1.
Phenotypic and genotypic features of ESBL‐producing E. coli strains isolated from peri‐urban wild animals, Brazil
| ID strains | Animal sources | ST/CC | Serotype | Virulence genes | Resistance phenotype | Resistance genotype | Plasmid type | Accession number |
|---|---|---|---|---|---|---|---|---|
| ECPET3 | Black Vulture (Coragyps atratus) | 212/− | O18/O18ac:H49 | gad, iss, lpfA | CRO, CTX, CAZ, CPM, ATM | bla CTX‐M‐55, bla TEM‐1B, mdf(A) | FII, N | PQET00000000 |
| ECPET11 | South American Coati (Nasua nasua) | 744/10 | O89/O162:H10 | gad, iss, cma, iroN | CRO, CTX, CAZ, CPM, ATM, CIP, NAL, GEN, KAN, TOB, STR, CLO, SUT, TET | bla CTX‐M‐55, bla TEM‐1B, aadA1, aadA2, aadA5, aac(3)‐IId, aph(3′)‐Ia, aph(3″)‐Ib, aph(6)‐Id, fosA3, catA1, cmlA1, sul1, sul2, dfrA17, tet(B), mdf(A) | Q1, FIB, FII, N, X1 | PQEU00000000 |
| ECPET13 | Striped Owl (Asio clamator) | 212/‐ | O18/O18ac:H49 | iss, lpfA | AMC, CRO, CTX, CAZ, CPM, ATM, NAL | bla CTX‐M‐55, bla TEM‐1B, mdf(A) | FII, N | PQEV00000000 |
| ECPET31 | South American Coati (Nasua nasua) | 58/155 | O78:H21 | gad, iss, lpfA | CRO, CTX, CAZ, CPM, ATM, CIP, GEN, KAN, TOB, STR, CLO, SUT, TET | bla CTX‐M‐2, aadA1, aac(3)‐IV, aph(3″)‐Ib, aph(3′)‐Ia, aph(4)‐Ia, aph(6)‐Id, sul1, sul2, dfrA7, tet(A), mdf(A) | FIA, HI2, HI2A, Q1, FII | PQEW00000000 |
| ECPET36 | South American Coati (Nasua nasua) | 1251/‐ | O130:H26 | gad | AMC, CRO, CTX, CAZ, CPM, ATM, CIP, NAL, GEN, KAN, TOB, STR, CLO, SUT | bla CTX‐M‐15, bla TEM‐1B, qnrB1, aac(3)‐IIa, aac(6′)Ib‐cr, aadA1, aph(3″)‐Ib, aph(6)‐Id, catB3, catA1, sul2, dfrA1, dfrA14, mdf(A) | HI2, HI2A, p0111 | PQEX00000000 |
| ICBUR6 | Black Vulture (Coragyps atratus) | 1158/31 | O17/O44/O77:H34 | iss, astA, eilA, celb, iha, air, ireA | CRO, CTX, CPM, ATM, KAN, TOB, CLO, SUL, TET | bla CTX‐M‐2, qnrB19, aadA1, aph(3′)‐Ia, aph(6)‐Id, aph(3″`)‐Ib, catA1, sul1, sul2, tet(B), mdf(A) | FIB, FII, Col156 | PPCS00000000 |
| ICBUR15 | Black Vulture (Coragyps atratus) | 38/38 | O86:H18 | gad, iss, astA, eilA | CRO, CTX, CPM, ATM, CIP, NAL, TOB, STR, SUL | bla CTX‐M‐14, aph(6)‐Id, aph(3″)‐Ib, sul2, mdf(A) | l2 | PPCU00000000 |
| ICBUR20 | Black Vulture (Coragyps atratus) | 38/38 | O86:H18 | iss, astA, eilA, air | CRO, CTX, CPM, ATM, CIP, NAL, AMK, STR, SUL | bla CTX‐M‐14, aph(6)‐Id, aph(3″)‐Ib, sul2, mdf(A) | l2 | PPCT00000000 |
Abbreviations: CC, clonal complex; ST, sequence type. Resistance phenotype: AMC, amoxicillin/clavulanic acid; AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CIP, ciprofloxacin; CLO, chloramphenicol; CPM, cefepime; CRO, ceftriaxone; CTX, cefotaxime; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SUL, sulfonamide; SUT, trimethoprim‐sulfamethoxazole; TET, tetracycline; TOB, Tobramycin.
WGS analysis revealed six different serotypes (i.e. O18/O18ac:H49, O89/O162:H10, O78:H21, O130:H26, O17/O44/O77:H34, O86:H18). In this regard, the O86:H18 has been previously identified in diarrhoeagenic E. coli isolated from humans, in Brazil (Ghilardi, Gomes, & Trabulsi, 2001; Piva et al., 2003), and in Asian and African countries (Sonda et al., 2018; Suzuki et al., 2009). On the other hand, while E. coli O89/O162:H10 has been associated with hospital‐acquired infections, in Asian countries (Lin, Kuroda, Suzuki, & Mu, 2019; Nguyen et al., 2019), E. coli O18:H49 and O78:H21 have been reported in wild animals from Europe and Asia (Bai et al., 2013; Eggert et al., 2013). Escherichia coli O130:H26 and O17/O44/O77:H34 have been identified in human and animal samples from Asia, Europe, Australia, Antarctica and South America (Bettelheim et al., 2003; Delgado‐Blas, Ovejero, Abadia‐Patino, & Gonzalez‐Zorn, 2016; Ho, Tan, Ooi, Yeo, & Thong, 2013; Mora et al., 2018; Müller et al., 2007).
Virulome analysis revealed a diversity of virulence determinants, including celb (endonuclease colicin E2), iha (irgA homolog adhesin), air (enteroaggregative immunoglobulin repeat protein), ireA (siderophore receptor), astA (EAST1 toxin), cma (colicin M), gad (glutamate decarboxylase), eilA (Salmonella HilA homolog), lpfA (long polar fimbriae), iroN (enterobactin siderophore receptor protein) and iss (increased serum survival) (Table 1). Interestingly, air, astA and eilA genes have been found in enteroaggregative E. coli (EAEC) causing acute and chronic diarrhoea (Konno, Yatsuyanagi, & Saito, 2012; Nüesch‐Inderbinen, Hofer, Hachler, Beutin, & Stephan, 2013; Sheikh et al., 2006). The lpfA gene has been identified in enteropathogenic E. coli (EPEC), the most important diarrhoeal pathogen in paediatric patients (Afset et al., 2006).
In addition to bla CTX‐M‐type genes, resistome analysis confirmed that the E. coli strains carried other clinically relevant resistance genes to β‐lactams [bla TEM‐1B], aminoglycosides [aadA1, aadA2, aadA5, aac(3)‐IId, aac(3)‐IIa, aac(3)‐IV, aac(6’)Ib‐cr, aph(3’)‐Ia, aph(3'')‐Ib, aph(3)‐Id, aph(4)‐Ia and aph(6)‐Id], phenicols [catA1, catB3 and cmlA1], tetracyclines [tet(A) and tet(B)], sulphonamides [sul1 and sul2], trimethoprim [dfrA1, dfrA7, dfrA14 and dfrA17], fosfomycin [fosA3], quinolones [qnrB1, qnrB19 and aac(6’)Ib‐cr] and macrolides [mdf(A)]. Interestingly, in Brazil, there are only three studies regarding the plasmid‐mediated quinolone resistance qnrB1 gene, which has been harboured by K. pneumoniae and Enterobacter cloacae (Scavuzzi et al., 2017; Viana et al., 2013), and Enterobacter hormaechei (Pereira et al., 2015). On the other hand, qnrB19, bla CTX‐M‐2, bla CTX‐M‐15 and bla CTX‐M‐55 genes have been carried by members of Enterobacterales genus isolated from human and non‐human hosts (Goldberg et al., 2019; Monte et al., 2019; Rocha, Pinto, & Barbosa, 2016; Sartori et al., 2017; Silva et al., 2018).
Schematic representations of the genetic contexts surrounding bla CTX‐M‐type genes in E. coli strains are presented in Figure 1. International genetic contexts of bla CTX‐M‐14 (ISEcp1‐bla CTX‐M‐14 ‐IS903D) (Lartigue, Poirel, & Nordmann, 2004) and bla CTX‐M‐15 (ISEcp1‐bla CTX‐M‐15 ‐orf477) (Dhanji et al., 2011) were identified in E. coli strains ST38 (ICBUR15 and ICBUR20) and ST1251 (ECPET36) isolated from C. atratus and N. nasua, respectively. In addition, two different contexts were found surrounding the bla CTX‐M‐55. The typical structure ISEcp1‐bla CTX‐M‐55 ‐orf477 (2,956 bp) was present in E. coli belonging to ST744 (ECPET11), whereas a similar array exhibiting a 243 bp with ISEcp1 truncated by an IS26 upstream of the bla CTX‐M‐55 gene was found in E. coli strains ECPET3 and ECPET13 (ST212). Similar genetic contexts of bla CTX‐M‐55 have been reported in Enterobacteriaceae isolated from humans, animals and food animals (Hu et al., 2018; Lv et al., 2013). Furthermore, bla CTX‐M‐2 gene from E. coli strains ST58 (ECPET31) and ST1158 (ICBUR6) was present into complex class 1 integrons (9,456 and 8,879 bp, respectively), sharing 99.7% and 99.9% nucleotide identity with partial integrons of E. coli (GenBank: AM040710) (8,133 bp) and K. pneumoniae (GenBank: KY286109) (7,824 bp) isolated from French and Chilean hospitals, respectively.
FIGURE 1.
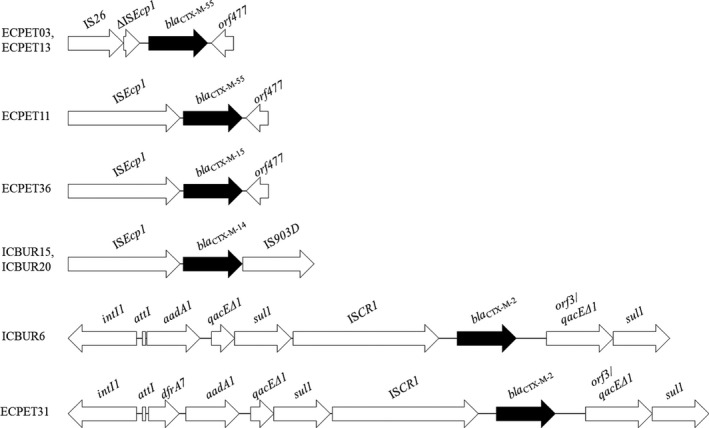
Schematic representation of the genetic context surrounding bla CTX‐M genes in Escherichia coli strains from peri‐urban wild animals in Brazil. For ECPET03 and ECPET13 E. coli strains, the genetic array IS26‐ΔISEcp1‐bla CTX‐M‐55‐orf477 was identified. In this regard, IS26 belongs to IS6 family, whereas orf477 encodes a cupin fold metalloprotein of WbuC family. For ECPET11 and ECPET36 E. coli strains, carrying bla CTX‐M‐55 or bla CTX‐M‐15, respectively, ISEcp1 was upstream, whereas orf477 was downstream of bla CTX‐M genes. E. coli strains ICBUR15 and ICBUR20, and ECPET36 displayed international genetic arrays ISEcp1‐bla CTX‐M‐14‐IS903D and ISEcp1‐bla CTX‐M‐15‐orf477, respectively
Although different plasmid replicon types were found among CTX‐M‐producing E. coli strains, bla CTX‐M genes were carried on IncF (FIA, FIB, and FII) plasmids, except bla CTX‐M‐14, which were carried on IncI2 plasmids. Most plasmids harbouring bla CTX‐M genes were successfully transferred by conjugation (from E. coli donors ECPET3, ECPET13, ECPET36 and ICBUR6), or by transformation assays using plasmids from ICBUR15 and ICBUR20 strains. As previously reported, IncF plasmids have been widely associated with the spread of bla CTX‐M‐15, whereas IncF, IncK and IncI are commonly associated with bla CTX‐M‐14 and other bla CTX‐M‐type genes (Zhao & Hu, 2013). Regarding other plasmids identified in this study, IncN and IncHI2 have been related to the spread of bla CTX‐M‐1 and bla CTX‐M‐9, respectively, and IncQ1 or IncX plasmid has been responsible by dissemination of carbapenemase encoding genes (Cerdeira et al., 2019; Mollenkopf et al., 2017; Paul et al., 2017; Zhao & Hu, 2013).
In this study, genomic analysis identified E. coli strains belonging to international ST38, ST58, ST212, ST744, ST1158 and ST1251 (Table 1). The global distribution of these E. coli clones is presented in Figures 2 and 3. The broadly distributed E. coli ST38 and ST744 have been reported in wildlife, farm animals and human samples from Europe, Africa, Asia, Australia and America, in general associated with the production of clinically significant beta‐lactamases (i.e. carbapenemases or ESBL) (Abraham et al., 2015; Belmahdi, Bakour, Al Bayssari, Touati, & Rolain, 2016; Guenther et al., 2017; Hasan et al., 2012; Ho et al., 2016; Mshana et al., 2011; Pitout, 2012; Poirel, Bernabeu, et al., 2011; Sellera et al., 2018; Stoesser et al., 2012; Yamamoto, Takano, Iwao, & Hishinuma, 2011). Escherichia coli ST38 has been frequently reported causing extraintestinal diseases, mainly bloodstream and urinary tract infections (Cao et al., 2014; Mendes, Jones, Woosley, Cattoir, & Castanheira, 2019; Pitout, 2012). In some cases, E. coli ST744 has been associated with plasmid‐mediated colistin resistance genes (mcr‐1 and mcr‐3) (Haenni et al., 2018; Tacão et al., 2017). Furthermore, ESBL or CMY‐2‐producing E. coli ST212 and ST1158 were previously isolated from farm animals, animal production chain and humans (Cadona, Bustamante, Gonzalez, & Sanso, 2016; Castellanos et al., 2017; Maamar et al., 2016; Mo, Slettemeas, Berg, Norstrom, & Sunde, 2016; Steinsland, Lacher, Sommerfelt, & Whittam, 2010; Vignoli et al., 2016; Zurfluh et al., 2014). Carbapenemase or CMY‐2‐producing E. coli ST212 was also recovered from diseased companion animal and water environments (Tafoukt et al., 2017; Vingopoulou et al., 2014), whereas E. coli ST1158 carrying bla CTX‐M was recovered from food animals (Vogt et al., 2014). Regarding E. coli ST1251, fluoroquinolone‐resistant strains have been reported in animal faeces and wastewater (Jamborova et al., 2015; Varela, Macedo, Nunes, & Manaia, 2015), as well as mcr‐1‐harbouring strains from food animals (Zurfluh et al., 2017). Escherichia coli belonging to ST58 has been globally reported from a variety of sources including food (Ben Said et al., 2015), polluted mangrove (Sacramento et al., 2018), poultry, hospital‐ and community‐acquired infections (Borges et al., 2019; McKinnon, Chowdhury, & Djordjevic, 2018) and bovine mastitis (Nüesch‐Inderbinen et al., 2019). Interestingly, ST58/CC155 frequently shares identical antimicrobial resistance patterns in both animal and human populations. Such evidence may significantly explain the successful establishment of this international lineage (Borges et al., 2019).
FIGURE 2.
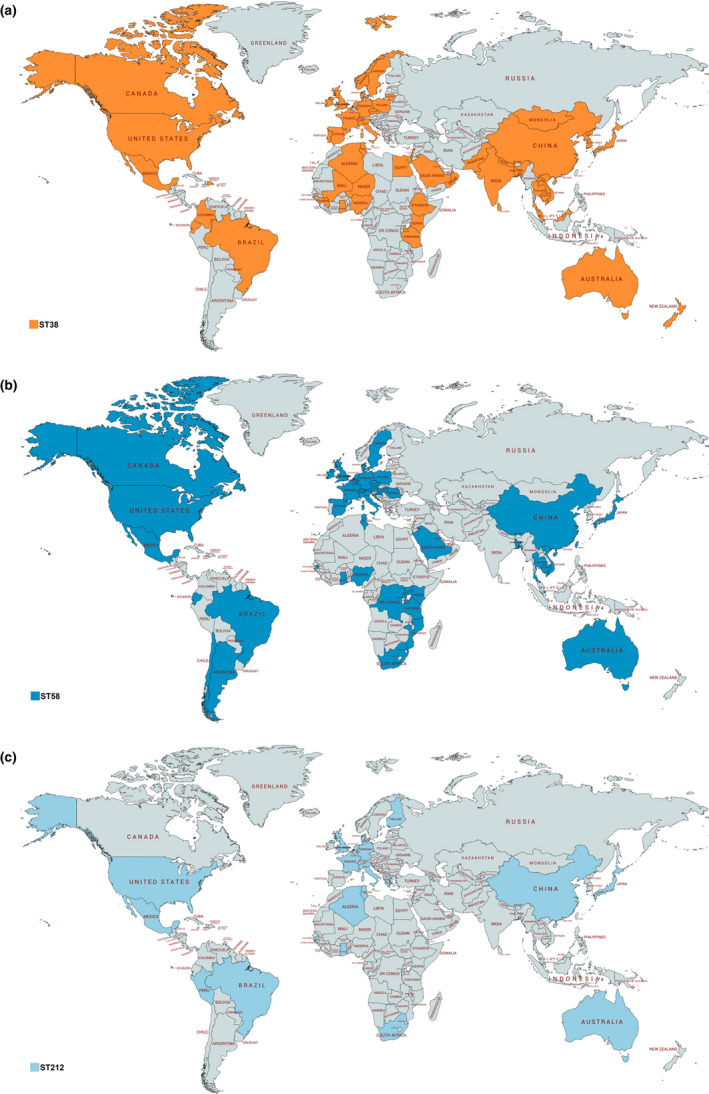
Global distribution of Escherichia coli belonging to sequence types (a) ST38, (b) ST58 and (c) ST212. This map was created using an online service (https://mapchart.net/) [Colour figure can be viewed at wileyonlinelibrary.com]
FIGURE 3.
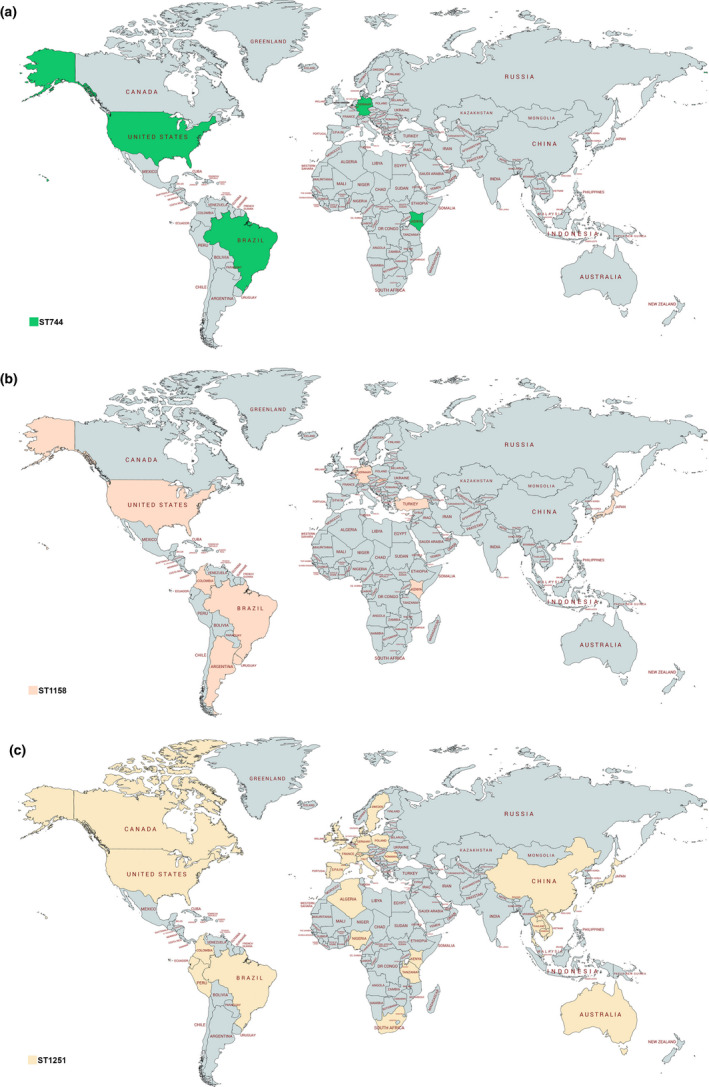
Global distribution of Escherichia coli belonging to sequence types (a) ST744, (b) ST1158 and (c) ST1251. This map was created using an online service (https://mapchart.net/) [Colour figure can be viewed at wileyonlinelibrary.com]
ESBL‐producing E. coli in wild animals begun to be documented in 2006, in Portugal (Costa et al., 2006), and then were rapidly observed in other countries from Europe, Africa, Asia, South America, North America and Australia (Allen et al., 2010; Wang et al., 2017). Predominantly, E. coli‐ and K. pneumoniae‐producing CTX‐M seem to be the most adapted to these hosts; however, the identification of ESBL genes in other different species of Enterobacterales has already been reported (Wang et al., 2017). In most of cases, animals became colonized in gastrointestinal tract without any evidences of infection, contributing for the silent dissemination of these critically important pathogens in natural environments.
A widely debated example is the occurrence of ESBL‐producing bacteria in migratory birds, which are probably involved in the spread of these pathogens through long distances, including natural reserves and pelagic areas with low anthropogenic impact (Ardiles‐Villegas et al., 2011; Cerdà‐Cuéllar et al., 2019). Otherwise, the role of peri‐urban wild animals as disseminators of bacterial pathogens has been so far neglected. In this study, all animals sampled lived in the transboundary area of São Paulo city, the most populated metropolitan region of Brazil, with about 21.5 million inhabitants, and one of the ten most populous metropolitan regions in the world. Even though the source of these bacterial isolates remains uncertain, wildlife is not directly exposed to antibiotics in most cases and other anthropogenic pathways of transmission, such as contact to contaminated water and predation of infected animals, should be considered (Wang et al., 2017). Yet, it is important to take in account that some highly polluted rivers cross this area, where KPC‐2‐ and ESBL‐producing K. pneumoniae isolates from water samples were previously reported (Cerdeira et al., 2017; Oliveira et al., 2014).
Since, in this study, ESBL‐positive isolates were recovered from predators with different ecological behaviours [i.e. vultures are scavengers and diurnal predators; owls are nocturnal hunters of small rodents; and coatis are remarkably well adapted predators, feeding on fruits, insects and small vertebrates], in order to investigate in more detail the genetic relatedness among these E. coli strains, core genome multilocus sequence typing (cgMLST) was performed by uploading the sequencing reads of the eight strains into cgMLSTFinder 1.1 (https://cge.cbs.dtu.dk/services/cgMLSTFinder/). Interestingly, two E. coli ST212 strains (ECPET3 and ECPET13) isolated from different hosts (black vulture and striped owl) were nested together (Figure S1). Remarkably, these strains also shared identical serotype, resistome and plasmidome. These findings suggest an adaptation of CTX‐M‐producing E. coli into the wildlife food chain and the versatility of these bacteria to colonize different hosts. Indeed, interspecific interactions among wild animals colonized by ESBL producers represent an incommensurable threat to ecosystem maintenance, since Enterobacteriaceae constitutes the gut microbiota of most endothermic animals (Madoshi et al., 2016). Thus, antimicrobial resistance must also be viewed as an ecological problem (Fuentes‐Castillo et al., 2019).
In conclusion, anthropogenic activities have been contributing for the dissemination of ESBL‐producing bacteria in wildlife. The occurrence of ESBL‐producing bacteria in peri‐urban wild animals from highly populated cities is a critical issue and deserves special attention. Therefore, continuous epidemiological and genomic surveillance studies are urgently required to determine routes of transmission of these bacteria in wildlife. Finally, while humans can negatively affect nature environments for contributing to the spread of MDR bacteria, animals could also disseminate these pathogens to humans in a continuous cycle.
ETHICAL STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Supporting information
Fig S1
ACKNOWLEDGEMENTS
This work was funded by research grants from Bill & Melinda Gates Foundation ‐ Grand Challenges Explorations Brazil – New approaches to characterize the global burden of antimicrobial resistance [grant OPP1193112], Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP 2016/08593‐9, 2015/13527‐2 and 2016/03044‐7], Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq 462042/2014‐6, 443819/2018‐1, 433128/2018‐6, 312249/2017‐9] and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [F.P.S. Finance Code 001]. N.L. is a research fellow of CNPq [312249/2017‐9]. We thank Cefar Diagnóstica Ltda. (Brazil), which kindly supplied antibiotic discs for antimicrobial susceptibility testing, and the CEFAP‐Genial by whole‐genome sequencing support.
de Carvalho MPN, Fernandes MR, Sellera FP, et al. International clones of extended‐spectrum β‐lactamase (CTX‐M)‐producing Escherichia coli in peri‐urban wild animals, Brazil. Transbound Emerg Dis. 2020;67:1804–1815. 10.1111/tbed.13558
Marcelo P. N. de Carvalho, Miriam R. Fernandes and Fábio P. Sellera are equally contributed to this article.
Funding information
Bill & Melinda Gates Foundation‐ Grand Challenges Explorations Brazil – New approaches to characterize the global burden of antimicrobial resistance [grant OPP1193112], Fundação de Amparo à Pesquisa do Estado de São Paulo [grant 2016/08593‐9], Conselho Nacional de Desenvolvimento Científico e Tecnológico [grant 443819/2018‐1, 433128/2018‐6, 312249/2017‐9].
Contributor Information
Fábio P. Sellera, Email: fsellera@usp.br.
Nilton Lincopan, Email: lincopan@usp.br.
REFERENCES
- Abraham, S. , Jordan, D. , Wong, H. S. , Johnson, J. R. , Toleman, M. A. , Wakeham, D. L. , … Trott, D. J. (2015). First detection of extended‐spectrum cephalosporin‐ and fluoroquinolone‐resistant Escherichia coli in Australian food‐producing animals. Journal of Global Antimicrobial Resistance, 3(4), 273–277. 10.1016/j.jgar.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Afset, J. E. , Bruant, G. , Brousseau, R. , Harel, J. , Anderssen, E. , Bevanger, L. , & Bergh, K. (2006). Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. Journal of Clinical Microbiology, 44(10), 3703–3711. 10.1128/JCM.00429-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, K. H. , Donato, J. , Wang, H. H. , Cloud‐Hansen, K. A. , Davies, J. , & Handelsman, J. (2010). Call of the wild: Antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8(4), 251–259. 10.1038/nrmicro2312 [DOI] [PubMed] [Google Scholar]
- Ardiles‐Villegas, K. , González‐Acuña, D. , Waldenström, J. , Olsen, B. , & Hernández, J. (2011). Antibiotic resistance patterns in fecal bacteria isolated from Christmas shearwater (Puffinus nativitatis) and masked booby (Sula dactylatra) at remote Easter Island. Avian Diseases, 55(3), 486–489. 10.1637/9619-122010-ResNote.1 [DOI] [PubMed] [Google Scholar]
- Bai, X. , Zhao, A. , Lan, R. , Xin, Y. , Xie, H. , Meng, Q. , … Xiong, Y. (2013). Shiga toxin‐producing Escherichia coli in yaks (Bos grunniens) from the Qinghai‐Tibetan Plateau, China. PLoS ONE, 8(6), e65537 10.1371/journal.pone.0065537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A. A. , Dvorkin, M. , Kulikov, A. S. , … Pevzner, P. A. (2012). SPAdes: A new genome assembly algorithm and its applications to single‐cell sequencing. Journal of Computational Biology, 19(5), 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmahdi, M. , Bakour, S. , Al Bayssari, C. , Touati, A. , & Rolain, J. M. (2016). Molecular characterisation of extended‐spectrum β‐lactamase‐ and plasmid AmpC‐producing Escherichia coli strains isolated from broilers in Béjaïa, Algeria. Journal of Global Antimicrobial Resistance, 6, 108–112. 10.1016/j.jgar.2016.04.006 [DOI] [PubMed] [Google Scholar]
- Ben Said, L. , Jouini, A. , Klibi, N. , Dziri, R. , Alonso, C. A. , Boudabous, A. , … Torres, C. (2015). Detection of extended‐spectrum beta‐lactamase (ESBL)‐producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. International Journal of Food Microbiology, 16(203), 86–92. 10.1016/j.ijfoodmicro.2015.02.023 [DOI] [PubMed] [Google Scholar]
- Bettelheim, K. A. , Beutin, L. , Gleier, K. , Pearce, J. L. , Luke, R. K. , & Zimmermann, S. (2003). Serotypes of Escherichia coli isolated from healthy infants in Berlin, Germany and Melbourne, Australia. Comparative Immunology, Microbiology & Infectious Diseases, 26(1), 55–63. 10.1016/s0147-9571(02)00015-2 [DOI] [PubMed] [Google Scholar]
- Borges, C. A. , Tarlton, N. J. , & Riley, L. W. (2019). Escherichia coli from commercial broiler and backyard chickens share sequence types, antimicrobial resistance profiles, and resistance genes with human extraintestinal pathogenic Escherichia coli . Foodborne Pathogens and Disease, 16(12), 813–822. 10.1089/fpd.2019.2680 [DOI] [PubMed] [Google Scholar]
- Brolund, A. (2014). Overview of ESBL‐producing enterobacteriaceae from nordic perspective. Infection Ecology and Epidemology, 4(1), 24555 10.3402/iee.v4.24555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadona, J. S. , Bustamante, A. V. , Gonzalez, J. , & Sanso, A. M. (2016). Genetic relatedness and novel sequence types of non‐O157 shiga toxin‐producing Escherichia coli strains isolated in Argentina. Frontiers in Cellular and Infection Microbiology, 6, 93 10.3389/fcimb.2016.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X. , Zhang, Z. , Shen, H. , Ning, M. , Chen, J. , Wei, H. , & Zhang, K. (2014). Genotypic characteristics of multidrug‐resistant Escherichia coli isolates associated with urinary tract infections. Acta Pathologica, Microbiologica, Et Immunologica Scandinavica, 122(11), 1088–1095. 10.1111/apm.12260 [DOI] [PubMed] [Google Scholar]
- Castellanos, L. R. , Donado‐Godoy, P. , León, M. , Clavijo, V. , Arevalo, A. , Bernal, J. F. , … Hordijk, J. (2017). High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC Genes on IncI1 plasmids in the colombian poultry chain. PLoS ONE, 12(1), e0170777 10.1371/journal.pone.0170777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdà‐Cuéllar, M. , Moré, E. , Ayats, T. , Aguilera, M. , Muñoz‐González, S. , Antilles, N. , … González‐Solís, J. (2019). Do humans spread zoonotic enteric bacteria in Antarctica? Science of the Total Environment, 654, 190–196. 10.1016/j.scitotenv.2018.10.272 [DOI] [PubMed] [Google Scholar]
- Cerdeira, L. , Fernandes, M. R. , Ienne, S. , Souza, T. A. , de O. Garcia, D. , & Lincopan, N. (2017). Draft genome sequence of an environmental multidrug‐resistant Klebsiella pneumoniae ST340/CC258 harbouring blaCTX‐M‐15 and blaKPC‐2 genes. Journal of Global Antimicrobial Resistance, 8, 108–109. 10.1016/j.jgar.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Cerdeira, L. T. , Lam, M. M. C. , Wyres, K. L. , Wick, R. R. , Judd, L. M. , Lopes, R. , … Lincopan, N. (2019). Small IncQ1 and Col‐Like plasmids harboring blaKPC‐2 and non‐Tn4401 elements (NTEKPC‐IId) in high‐risk lineages of Klebsiella pneumoniae CG258. Antimicrobial Agents and Chemotherapy, 63(3), e02140–e2218. 10.1128/AAC.02140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . (2017). Performance standards for antimicrobial susceptibility testing; twenty‐seven informational supplement. CLSI document M100–S27. Wayne, PA: CLSI. [Google Scholar]
- Clinical Laboratory Standards Institute . (2015). Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals (3rd edition). CLSI Supplement VET01S. Wayne, PA: CLSI. [Google Scholar]
- Costa, D. , Poeta, P. , Sáenz, Y. , Vinué, L. , Rojo‐Bezares, B. , Jouini, A. , … Torres, C. (2006). Detection of Escherichia coli harbouring extended‐spectrum beta‐lactamases of the CTX‐M, TEM and SHV classes in faecal samples of wild animals in Portugal. Journal of Antimicrobial Chemotherapy, 58(6), 1311–1312. 10.1093/jac/dkl415 [DOI] [PubMed] [Google Scholar]
- Delgado‐Blas, J. F. , Ovejero, C. M. , Abadia‐Patino, L. , & Gonzalez‐Zorn, B. (2016). Coexistence of mcr‐1 and blaNDM‐1 in Escherichia coli from Venezuela. Antimicrobial Agents and Chemotherapy, 60(10), 6356–6358. 10.1128/AAC.01319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanji, H. , Patel, R. , Wall, R. , Doumith, M. , Patel, B. , Hope, R. , … Woodford, N. (2011). Variation in the genetic environments of blaCTX‐M‐15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. Journal of Antimicrobial Chemotherapy, 66(5), 1005–1012. 10.1093/jac/dkr041 [DOI] [PubMed] [Google Scholar]
- Dropa, M. , Lincopan, N. , Balsalobre, L. C. , Oliveira, D. E. , Moura, R. A. , Fernandes, M. R. , … Matté, M. H. (2016). Genetic background of novel sequence types of CTX‐M‐8‐ and CTX‐M‐15‐producing Escherichia coli and Klebsiella pneumoniae from public wastewater treatment plants in São Paulo, Brazil. Environmental Science and Pollution Research, 23(5), 4953–4958. 10.1007/s11356-016-6079-5 [DOI] [PubMed] [Google Scholar]
- Eggert, M. , Stuber, E. , Heurich, M. , Fredriksson‐Ahomaa, M. , Burgos, Y. , Beutin, L. , & Martlbauer, E. (2013). Detection and characterization of Shiga toxin‐producing Escherichia coli in faeces and lymphatic tissue of free‐ranging deer. Epidemiology & Infection, 141(2), 251–259. 10.1017/S0950268812000246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, M. R. , Sellera, F. P. , Moura, Q. , Gaspar, V. C. , Cerdeira, L. , & Lincopan, N. (2018). International high‐risk clonal lineages of CTX‐M‐producing Escherichia coli F‐ST648 in free‐roaming cats, South America. Infection, Genetics and Evolution, 66, 48–51. 10.1016/j.meegid.2018.09.009 [DOI] [PubMed] [Google Scholar]
- Fuentes‐Castillo, D. , Farfán‐López, M. , Esposito, F. , Moura, Q. , Fernandes, M. R. , Lopes, R. , … Lincopan, N. (2019). Wild owls colonized by international clones of extended‐spectrum β‐lactamase (CTX‐M)‐producing Escherichia coli and Salmonella Infantis in the Southern Cone of America. Science of the Total Environment, 674, 554–562. 10.1016/j.scitotenv.2019.04.149 [DOI] [PubMed] [Google Scholar]
- Ghilardi, A. C. , Gomes, T. A. , & Trabulsi, L. R. (2001). Production of cytolethal distending toxin and other virulence characteristics of Escherichia coli strains of serogroup O86. Memórias do Instituto Oswaldo Cruz, 96(5), 703–708. 10.1590/S0074-02762001000500022 [DOI] [PubMed] [Google Scholar]
- Goldberg, D. W. , Fernandes, M. R. , Sellera, F. P. , Costa, D. G. C. , Loureiro Bracarense, A. P. , & Lincopan, N. (2019). Genetic background of CTX‐M‐15‐producing Enterobacter hormaechei ST114 and Citrobacter freundii ST265 co‐infecting a free‐living green turtle (Chelonia mydas). Zoonoses Public Health, 66(5), 540–545. 10.1111/zph.12572 [DOI] [PubMed] [Google Scholar]
- Guenther, S. , Semmler, T. , Stubbe, A. , Stubbe, M. , Wieler, L. H. , & Schaufler, K. (2017). Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. Journal of Antimicrobial Chemotherapy, 72(5), 1310–1313. 10.1093/jac/dkx006 [DOI] [PubMed] [Google Scholar]
- Haenni, M. , Beyrouthy, R. , Lupo, A. , Chatre, P. , Madec, J. Y. , & Bonnet, R. (2018). Epidemic spread of Escherichia coli ST744 isolates carrying mcr‐3 and blaCTX‐M‐55 in cattle in France. Journal of Antimicrobial Chemotherapy., 73(2), 533–536. 10.1093/jac/dkx418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, B. , Sandegren, L. , Melhus, A. , Drobni, M. , Hernandez, J. , Waldenstrom, J. , … Olsen, B. (2012). Antimicrobial drug‐resistant Escherichia coli in wild birds and free‐range poultry, Bangladesh. Emerging Infectious Diseases, 18(12), 2055–2058. 10.3201/eid1812.120513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, P. L. , Cheung, Y. Y. , Wang, Y. , Lo, W. U. , Lai, E. L. , Chow, K. H. , & Cheng, V. C. (2016). Characterization of carbapenem‐resistant Escherichia coli and Klebsiella pneumoniae from a healthcare region in Hong Kong. European Journal of Clinical Microbiology & Infectious Diseases, 35(3), 379–385. 10.1007/s10096-015-2550-3 [DOI] [PubMed] [Google Scholar]
- Ho, W. S. , Tan, L. K. , Ooi, P. T. , Yeo, C. C. , & Thong, K. L. (2013). Prevalence and characterization of verotoxigenic‐Escherichia coli isolates from pigs in Malaysia. BMC Veterinary Research, 9, 109 10.1186/1746-6148-9-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Gou, J. , Guo, X. , Cao, Z. , Li, Y. , Jiao, H. , … Tian, F. (2018). Genetic contexts related to the diffusion of plasmid‐mediated CTX‐M‐55 extended‐spectrum beta‐lactamase isolated from Enterobacteriaceae in China. Annals of Clinical Microbiology and Antimicrobials, 17(1), 12 10.1186/s12941-018-0265-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H. , Nojima, H. , & Okayama, H. (1990). High efficiency transformation of Escherichia coli with plasmids. Gene, 96(1), 23–28. 10.1016/0378-1119(90)90336-p [DOI] [PubMed] [Google Scholar]
- Jamborova, I. , Dolejska, M. , Vojtech, J. , Guenther, S. , Uricariu, R. , Drozdowska, J. , … Literak, I. (2015). Plasmid‐mediated resistance to cephalosporins and fluoroquinolones in various Escherichia coli sequence types isolated from rooks wintering in Europe. Applied Environmental Microbiology, 81(2), 648–657. 10.1128/AEM.02459-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno, T. , Yatsuyanagi, J. , & Saito, S. (2012). Virulence gene profiling of enteroaggregative Escherichia coli heat‐stable enterotoxin 1‐harboring E. coli (EAST1EC) derived from sporadic diarrheal patients. FEMS Immunology and Medical Microbiology, 64(3), 314–320. 10.1111/j.1574-695X.2011.00913.x [DOI] [PubMed] [Google Scholar]
- Lartigue, M. F. , Poirel, L. , & Nordmann, P. (2004). Diversity of genetic environment of blaCTX‐M genes. FEMS Microbiology Letters, 234(2), 201–207. 10.1016/j.femsle.2004.01.051 [DOI] [PubMed] [Google Scholar]
- Lin, Y. C. , Kuroda, M. , Suzuki, S. , & Mu, J. J. (2019). Emergence of an Escherichia coli strain co‐harbouring mcr‐1 and blaNDM‐9 from a urinary tract infection in Taiwan. Journal of Global Antimicrobial Resistance, 16, 286–290. 10.1016/j.jgar.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Liu, Y.‐Y. , Wang, Y. , Walsh, T. R. , Yi, L.‐X. , Zhang, R. , Spencer, J. , … Shen, J. (2016). Emergence of plasmid‐mediated colistin resistance mechanism MCR‐1 in animals and human beings in China: A microbiological and molecular biological study. The Lancet Infectious Diseases, 16(2), 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Lv, L. , Partridge, S. R. , He, L. , Zeng, Z. , He, D. , Ye, J. , & Liu, J. H. (2013). Genetic characterization of IncI2 plasmids carrying blaCTX‐M‐55 spreading in both pets and food animals in China. Antimicrobial Agents and Chemotherapy, 57(6), 2824–2827. 10.1128/AAC.02155-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar, E. , Hammami, S. , Alonso, C. A. , Dakhli, N. , Abbassi, M. S. , Ferjani, S. , … Boutiba‐Ben Boubaker, I. (2016). High prevalence of extended‐spectrum and plasmidic ampc beta‐lactamase‐producing Escherichia coli from poultry in Tunisia. International Journal of Food Microbiology, 231, 69–75. 10.1016/j.ijfoodmicro [DOI] [PubMed] [Google Scholar]
- Madoshi, B. P. , Kudirkiene, E. , Mtambo, M. M. , Muhairwa, A. P. , Lupindu, A. M. , & Olsen, J. E. (2016). Characterisation of commensal Escherichia coli isolated from apparently healthy cattle and their attendants in Tanzania. PLoS ONE, 11(12), e0168160 10.1371/journal.pone.0168160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos, A.‐P. , Srinivasan, A. , Carey, R. B. , Carmeli, Y. , Falagas, M. E. , Giske, C. G. , … Monnet, D. L. (2012). Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection, 18(3), 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- McKinnon, J. , Roy Chowdhury, P. , & Djordjevic, S. P. (2018). Genomic analysis of multidrug‐resistant Escherichia coli ST58 causing urosepsis. International Journal of Antimicrobial Agents, 52(3), 430–435. 10.1016/j.ijantimicag.2018.06.017 [DOI] [PubMed] [Google Scholar]
- Mendes, R. E. , Jones, R. N. , Woosley, L. N. , Cattoir, V. , & Castanheira, M. (2019). Application of next‐generation sequencing for characterization of surveillance and clinical trial isolates: Analysis of the distribution of β‐lactamase resistance genes and lineage background in the United States. Open Forum Infectious Diseases, 6(1), S69–S78. 10.1093/ofid/ofz004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minarini, L. A. , Poirel, L. , Trevisani, N. A. , Darini, A. L. , & Nordmann, P. (2009). Predominance of CTX‐M‐type extended‐spectrum beta‐lactamase genes among enterobacterial isolates from outpatients in Brazil. Diagnostic Microbiology and Infectious Disease, 65(2), 202–206. 10.1016/j.diagmicrobio.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Mo, S. S. , Slettemeas, J. S. , Berg, E. S. , Norstrom, M. , & Sunde, M. (2016). Plasmid and host strain characteristics of Escherichia coli resistant to extended‐spectrum cephalosporins in the Norwegian broiler production. PLoS ONE, 11(4), e015401911 10.1371/journal.pone.0154019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenkopf, D. F. , Stull, J. W. , Mathys, D. A. , Bowman, A. S. , Feicht, S. M. , Grooters, S. V. , … Wittum, T. E. (2017). Carbapenemase‐producing Enterobacteriaceae recovered from the environment of a swine farrow‐to‐finish operation in the United States. Antimicrobial Agents and Chemotherapy, 61(2), e01298–e1316. 10.1128/AAC.01298-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, D. F. , Lincopan, N. , Berman, H. , Cerdeira, L. , Keelara, S. , Thakur, S. , … Landgraf, M. (2019). Genomic features of high‐priority Salmonella enterica serovars circulating in the food production chain, Brazil, 2000–2016. Scientific Reports, 9(1), 11058 10.1038/s41598-019-45838-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora, A. , García‐Peña, F. J. , Alonso, M. P. , Pedraza‐Diaz, S. , Ortega‐Mora, L. M. , Garcia‐Parraga, D. , … Blanco, J. (2018). Impact of human‐associated Escherichia coli clonal groups in Antarctic pinnipeds: Presence of ST73, ST95, ST141 and ST131. Scientific Reports, 8(1), 4678 10.1038/s41598-018-22943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mshana, S. E. , Imirzalioglu, C. , Hain, T. , Domann, E. , Lyamuya, E. F. , & Chakraborty, T. (2011). Multiple st clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX‐M‐15 in a tertiary hospital in Tanzania. Clinical Microbiology and Infection, 17(8), 1279–1282. 10.1111/j.1469-0691.2011.03518.x [DOI] [PubMed] [Google Scholar]
- Müller, D. , Greune, L. , Heusipp, G. , Karch, H. , Fruth, A. , Tschape, H. , & Schmidt, M. A. (2007). Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single‐step multiplex PCR. Applied and Environmental Microbiology, 73(10), 3380–3390. 10.1128/AEM.02855-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzaheed, , Doi, Y. , Adams‐Haduch, J. M. , Endimiani, A. , Sidjabat, H. E. , Gaddad, S. M. , & Paterson, D. L. (2008). High prevalence of CTX‐M‐15‐producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in Southern India. Journal of Antimicrobial Chemotherapy, 61(6), 1393–1394. 10.1093/jac/dkn109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, L. P. , Pinto, N. A. , Vu, T. N. , Mai, H. , Pham, A. H. T. , Lee, H. , … Yong, D. (2019). Resistome profiles, plasmid typing, and whole‐genome phylogenetic tree analyses of blaNDM‐9 and mcr‐1 co‐harboring Escherichia coli ST617 from a patient without a history of farm exposure in Korea. Pathogens, 8(4), E212 10.3390/pathogens8040212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuesch‐Inderbinen, M. T. , Hofer, E. , Hachler, H. , Beutin, L. , & Stephan, R. (2013). Characteristics of enteroaggregative Escherichia coli isolated from healthy carriers and from patients with diarrhoea. Journal of Medical Microbiology, 62(12), 1828–1834. 10.1099/jmm.0.065177-0 [DOI] [PubMed] [Google Scholar]
- Nüesch‐Inderbinen, M. , Käppeli, N. , Morach, M. , Eicher, C. , Corti, S. , & Stephan, R. (2019). Molecular types, virulence profiles and antimicrobial resistance of Escherichia coli causing bovine mastitis. Veterinary Record Open, 6(1), e000369 10.1136/vetreco-2019-000369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, S. , Moura, R. A. , Silva, K. C. , Pavez, M. , McCulloch, J. A. , Dropa, M. , … Lincopan, N. (2014). Isolation of KPC‐2‐producing Klebsiella pneumoniae strains belonging to the high‐risk multiresistant clonal complex 11 (ST437 and ST340) in urban rivers. Journal of Antimicrobial Chemotherapy, 69(3), 849–852. 10.1093/jac/dkt431 [DOI] [PubMed] [Google Scholar]
- Pardon, B. , Smet, A. , Butaye, P. , Argudín, M. A. , Valgaeren, B. , Catry, B. , … Deprez, P. (2015). Nosocomial intravascular catheter infections with extended‐spectrum beta‐lactamase‐producing Escherichia coli in calves after strain introduction from a commercial herd. Transboundary and Emerging Diseases, 64(1), 130–136. 10.1111/tbed.12352.64:130-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, D. , Bhattacharjee, A. , Ingti, B. , Choudhury, N. A. , Maurya, A. P. , Dhar, D. , & Chakravarty, A. (2017). Occurrence of blaNDM‐7 within IncX3‐type plasmid of Escherichia coli from India. Journal of Infection and Chemotherapy, 23(4), 206–210. 10.1016/j.jiac.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Pereira, P. S. , Borghi, M. , Albano, R. M. , Lopes, J. C. , Silveira, M. C. , Marques, E. A. , … Carvalho‐Assef, A. P. (2015). Coproduction of NDM‐1 and KPC‐2 in Enterobacter hormaechei from Brazil. Microbiology Drug Resistance, 21(2), 234–236. 10.1089/mdr.2014.0171 [DOI] [PubMed] [Google Scholar]
- Pitout, J. D. (2012). Extraintestinal pathogenic Escherichia coli: A combination of virulence with antibiotic resistance. Frontiers in Microbiology, 3, 9 10.3389/fmicb.2012.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva, I. C. , Pereira, A. L. , Ferraz, L. R. , Silva, R. S. N. , Vieira, A. C. , Blanco, J. E. , … Giugliano, L. G. (2003). Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasilia, Brazil. Journal of Clinical Microbiology, 41(5), 1827–1832. 10.1128/jcm.41.5.1827-1832.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel, L. , Bernabeu, S. , Fortineau, N. , Podglajen, I. , Lawrence, C. , & Nordmann, P. (2011). Emergence of OXA‐48‐producing Escherichia coli clone ST38 in France. Antimicrobial Agents and Chemotherapy, 55(10), 4937–4938. 10.1128/AAC.00413-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel, L. , Walsh, T. R. , Cuvillier, V. , & Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagnostic Microbiology and Infectious Disease, 70(1), 119–123. 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Radhouani, H. , Silva, N. , Poeta, P. , Torres, C. , Correia, S. , & Igrejas, G. (2014). Potential impact of antimicrobial resistance in wildlife, environment, and human health. Frontiers in Microbiology, 23, 1–12. 10.3389/fmicb.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, F. R. , Pinto, V. P. , & Barbosa, F. C. (2016). The Spread of CTX‐M‐type extended‐spectrum β‐lactamases in Brazil: A systematic review. Microbial Drug Resistance, 22(4), 301–311. 10.1089/mdr.2015.0180 [DOI] [PubMed] [Google Scholar]
- Sacramento, A. G. , Fernandes, M. R. , Sellera, F. P. , Muñoz, M. E. , Vivas, R. , Dolabella, S. S. , & Lincopan, N. (2018). Genomic analysis of MCR‐1 and CTX‐M‐8 co‐producing Escherichia coli ST58 isolated from a polluted mangrove ecosystem in Brazil. Journal of Global Antimicrobial Resistance, 15, 288–289. 10.1016/j.jgar.2018.10.024 [DOI] [PubMed] [Google Scholar]
- Saladin, M. , Cao, V. T. , Lambert, T. , Donay, J. L. , Herrmann, J. L. , Ould‐Hocine, Z. , … Arlet, G. (2002). Diversity of CTX‐M beta‐lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiology Letters, 209(2), 161–168. 10.1111/j.1574-6968.2002.tb11126.x [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , & Russel, D. W. (2001). Molecular cloning: A laboratory manual (Vol. 3, 3rd ed., p. 2100). New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sartori, L. , Fernandes, M. R. , Ienne, S. , de Souza, T. A. , Gregory, L. , Cerdeira, L. , & Lincopan, N. (2017). Draft genome sequences of two fluoroquinolone‐resistant CTX‐M‐15‐producing Escherichia coli ST90 (ST23 complex) isolated from a calf and a dairy cow in South America. Journal of Global Antimicrobial Resistance, 11, 145–147. 10.1016/j.jgar.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Scavuzzi, A. M. L. , Maciel, M. A. V. , de Melo, H. R. L. , Alves, L. C. , Brayner, F. A. , & Lopes, A. C. S. (2017). Occurrence of qnrB1 and qnrB12 genes, mutation in gyrA and ramR, and expression of efflux pumps in isolates of Klebsiella pneumoniae carriers of blaKPC‐2. Journal of Medical Microbiology, 66(4), 477–484. 10.1099/jmm.0.000452 [DOI] [PubMed] [Google Scholar]
- Sellera, F. P. (2019). Epidemiological implications of drug‐resistant bacteria in wildlife rehabilitation centers. Journal of Infection and Public Health, 12(5), 748–749. 10.1016/j.jiph.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Sellera, F. P. , Fernandes, M. R. , Moura, Q. , Carvalho, M. P. N. , & Lincopan, N. (2018). Extended‐spectrum‐β‐lactamase (CTX‐M)‐producing Escherichia coli in wild fishes from a polluted area in the Atlantic Coast of South America. Marine Pollution Bulletin, 135, 183–186. 10.1016/j.marpolbul.2018.07.012 [DOI] [PubMed] [Google Scholar]
- Sheikh, J. , Dudley, E. G. , Sui, B. , Tamboura, B. , Suleman, A. , & Nataro, J. P. (2006). EilA, a HilA‐like regulator in enteroaggregative Escherichia coli . Molecular Microbiology, 61(2), 338–350. 10.1111/j.1365-2958.2006.05234.x [DOI] [PubMed] [Google Scholar]
- Siguier, P. , Perochon, J. , Lestrade, L. , Mahillon, J. , & Chandler, M. (2006). ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Research, 34, D32–36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, M. M. , Fernandes, M. R. , Sellera, F. P. , Cerdeira, L. , Medeiros, L. K. G. , Garino, F. , … Lincopan, N. (2018). Multidrug‐resistant CTX‐M‐15‐producing Klebsiella pneumoniae ST231 associated with infection and persistent colonization of dog. Diagnostic Microbiology and Infectious Disease, 92(3), 259–261. 10.1016/j.diagmicrobio.2018.06.012 [DOI] [PubMed] [Google Scholar]
- Sonda, T. , Kumburu, H. , van Zwetselaar, M. , Alifrangis, M. , Mmbaga, B. T. , Aarestrup, F. M. , … Lund, O. (2018). Whole genome sequencing reveals high clonal diversity of Escherichia coli isolated from patients in a tertiary care hospital in Moshi, Tanzania. Antimicrobial Resistance & Infection Control, 7, 72 10.1186/s13756-018-0361-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinsland, H. , Lacher, D. W. , Sommerfelt, H. , & Whittam, T. S. (2010). Ancestral lineages of human enterotoxigenic Escherichia coli . Journal of Clinical Microbiology, 48(8), 2916–2924. 10.1128/JCM.02432-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoesser, N. , Crook, D. W. , Moore, C. E. , Phetsouvanh, R. , Chansamouth, V. , Newton, P. N. , & Jones, N. (2012). Characteristics of CTX‐M ESBL‐producing Escherichia coli isolates from the Lao People's Democratic Republic, 2004–09. Journal of Antimicrobial Chemotherapy, 67(1), 240–242. 10.1093/jac/dkr434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, S. , Shibata, N. , Yamane, K. , Wachino, J. , Ito, K. , & Arakawa, Y. (2009). Change in the prevalence of extended‐spectrum‐beta‐lactamase‐producing Escherichia coli in Japan by clonal spread. Journal of Antimicrobial Chemotherapy, 63(1), 72–79. 10.1093/jac/dkn463 [DOI] [PubMed] [Google Scholar]
- Tacão, M. , Tavares, R. D. S. , Teixeira, P. , Roxo, I. , Ramalheira, E. , Ferreira, S. , & Henriques, I. (2017). mcr‐1 and blaKPC‐3 in Escherichia coli sequence type 744 after meropenem and colistin therapy. Portugal. Emerging Infectious Diseases, 23(8), 1419–1421. 10.3201/eid2308.170162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafoukt, R. , Touati, A. , Leangapichart, T. , Bakour, S. , & Rolain, J. M. (2017). Characterization of OXA‐48‐like‐producing Enterobacteriaceae isolated from river water in Algeria. Water Research, 120, 185–189. 10.1016/j.watres.2017.04.073 [DOI] [PubMed] [Google Scholar]
- Varela, A. R. , Macedo, G. N. , Nunes, O. C. , & Manaia, C. M. (2015). Genetic characterization of fluoroquinolone resistant Escherichia coli from urban streams and municipal and hospital effluents. FEMS Microbiology Ecology, 91(5), fiv015 10.1093/femsec/fiv015 [DOI] [PubMed] [Google Scholar]
- Viana, A. L. , Cayô, R. , Avelino, C. C. , Gales, A. C. , Franco, M. C. , & Minarini, L. A. (2013). Extended‐spectrum β‐lactamases in Enterobacteriaceae isolated in Brazil carry distinct types of plasmid‐mediated quinolone resistance genes. Journal of Medical Microbiology, 62(Pt 9), 1326–1331. 10.1099/jmm.0.055970-0 [DOI] [PubMed] [Google Scholar]
- Vignoli, R. , Garcia‐Fulgueiras, V. , Cordeiro, N. F. , Bado, I. , Seija, V. , Aguerrebere, P. , … Chabalgoity, J. (2016). Extended‐spectrum beta‐lactamases, transferable quinolone resistance, and virulotyping in extra‐intestinal E. coli in Uruguay. Journal of Infection in Developing Countries, 10(1), 43–52. 10.3855/jidc.6918 [DOI] [PubMed] [Google Scholar]
- Vingopoulou, E. I. , Siarkou, V. I. , Batzias, G. , Kaltsogianni, F. , Sianou, E. , Tzavaras, I. , … Miriagou, V. (2014). Emergence and maintenance of multidrug‐resistant Escherichia coli of canine origin harbouring a blaCMY‐2‐IncI1/ST65 plasmid and topoisomerase mutations. Journal of Antimicrobial Chemotherapy, 69(8), 2076–2080. 10.1093/jac/dku090 [DOI] [PubMed] [Google Scholar]
- Vogt, D. , Overesch, G. , Endimiani, A. , Collaud, A. , Thomann, A. , & Perreten, V. (2014). Occurrence and genetic characteristics of third‐generation cephalosporin‐resistant Escherichia coli in Swiss retail meat. Microbial Drug Resistance, 20(5), 485–494. 10.1089/mdr.2013.0210 [DOI] [PubMed] [Google Scholar]
- Wang, J. , Ma, Z. B. , Zeng, Z. L. , Yang, X. W. , Huang, Y. , & Liu, J. H. (2017). The role of wildlife (wildbirds) in the global transmission of antimicrobial resistance genes. Zoological Research, 38(2), 55–80. 10.24272/j.issn.2095-8137.2017.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T. , Takano, T. , Iwao, Y. , & Hishinuma, A. (2011). Emergence of NDM‐1‐positive capsulated Escherichia coli with high resistance to serum killing in Japan. Journal of Infection Chemotherapy, 17(3), 435–439. 10.1007/s10156-011-0232-3 [DOI] [PubMed] [Google Scholar]
- Zerbino, D. R. , & Birney, E. (2008). Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research, 18(5), 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. H. , & Hu, Z. Q. (2013). Epidemiology and genetics of CTX‐M extended‐spectrum beta‐lactamases in Gram‐negative bacteria. Critical Reviews in Microbiology, 39(1), 79–101. 10.3109/1040841X.2012.691460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh, K. , Nuesch‐Inderbinen, M. , Klumpp, J. , Poirel, L. , Nordmann, P. , & Stephan, R. (2017). Key features of mcr‐1‐bearing plasmids from Escherichia coli isolated from humans and food. Antimicrobial Resistance & Infection Control, 6(91), 1–6. 10.1186/s13756-017-0250-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh, K. , Wang, J. , Klumpp, J. , Nuesch‐Inderbinen, M. , Fanning, S. , & Stephan, R. (2014). Vertical transmission of highly similar blaCTX‐M‐1‐harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Frontiers in Microbiology, 5(519), 1–7. 10.3389/fmicb.2014.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1


