Abstract
The large‐scale and label‐free molecular characterization of single cells in their natural tissue habitat remains a major challenge in molecular biology. We present a method that integrates morphometric image analysis to delineate and classify individual cells with their single‐cell‐specific molecular profiles. This approach provides a new means to study spatial biological processes such as cancer field effects and the relationship between morphometric and molecular features.
Keywords: imaging, lipids, mass spectrometry, morphometry, single-cell analysis
Understanding how cells regulate their bioenergetic demands, adjust to the microenvironment, or respond to stimuli is the aim of molecular biology. Yet, methods able to describe the molecular content, histological identity, and spatial context of a single cell do not exist. The presented method integrates morphometric image analysis to delineate and classify individual cells with their cell‐specific molecular profiles obtained by MALDI‐MSI.
Matrix‐assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) combines the molecular sensitivity and specificity of mass spectrometry with the capability to spatially resolve the molecular information within tissues.1 It has therefore become an invaluable tool in biological research for the label‐free imaging of otherwise inaccessible molecular classes, such as metabolites, lipids, or drugs, in tissues.2, 3 Despite advances in sample preparation and instrumentation that enabled MALDI‐MSI to retrieve chemical information at single‐cell resolution a very limited number of high‐spatial‐resolution MALDI‐MSI biomedical studies has been reported.4, 5, 6, 7
Detailed histological tissue annotations to contextualize the obtained molecular signals from individual cells are considered a major limitation for comprehensive biomedical research using MALDI‐MSI. Typically, the MSI‐analyzed tissue section is histologically stained, co‐registered to the MALDI‐MSI data, and annotated by a pathologist.8 Annotation of every single cell is a tedious and time‐consuming process, even for a trained pathologist. A manual annotation that matches the spatial resolution of MALDI‐MSI at the single cell level is therefore infeasible and hampers single‐cell MALDI‐MSI applications in translational research.
Recently, the field of digital image analysis has emerged and aims to support the evaluation and annotation of microscopic images using machine learning algorithms.9, 10 In particular, the morphometric analysis of cells in histological images11 has gained interest as it can be used for the automatic detection and classification of cells.12 Morphometry uses quantitative statistical models to learn about size and shape (e.g. nucleus size, cell eccentricity, cell circumference, etc.) of each cell. These models are used to instantaneously detect and classify whole tissues according to the learned cell‐types.
The continuous increase in performance in resolution, sensitivity and specificity of MS‐based molecular imaging has always offered the potential of complete integration in digital pathology. Yet, comprehensive methods that allow this at the single‐cell level, in an automated fashion where both morphometric information and MSI information of a single cell are brought together, are lacking. Here, we present a method that uses machine learning for semi‐automated image analyses to obtain single‐cell morphometric annotations, which are then accurately correlated with the MALDI‐MSI data from the same tissue section. This workflow enables us to determine a detailed molecular profile for each individual cell in its complex tissue context. This results in innovative applications in basic biology, translational research and clinical diagnostics.
We demonstrate this approach with high‐spatial‐resolution MALDI‐MSI data acquired from a porcine colon and a human gastric cancer specimen using a timsTOF flex and a RapifleX tissue typer (both Bruker Daltonik, Bremen, Germany), respectively. Because cell sizes in mammals usually range between 5–20 μm,13 we performed all experiments at 10‐μm pixel size to ensure a well‐balanced chemical sensitivity and sufficient spatial resolution to obtain single‐cell molecular information. As sample preparation steps such as washing, enzyme application, or derivatization can delocalize molecules and hamper the achievable spatial resolution,14 we opted for MALDI‐MSI of lipids, which does not require any additional sample pre‐treatment steps after tissue sectioning and mounting. But, our method is applicable to all other MSI sample preparation protocols, within the limits of potential delocalization beyond the pixel resolution that may result from them. The tissue sections were stained by conventional hematoxylin & eosin (H&E) and scanned with a digital slide scanner (Aperio CS2, Leica Biosystems) after MALDI‐MSI acquisition (Supplementary Figures 1 A and 8A).
The H&E image was subsequently co‐registered to the MSI dataset via an optical image of the slide, which was taken prior to the MSI experiment. This optical image guides the MS acquisition to the defined measurement regions based on a manual control point co‐registration of the optical image with fiducial markers visible in the camera within the mass spectrometer (Supplementary Figures 1 B, 2, 8B and 9). This process links the spatial coordinates of the mass spectrometer with the pixel positions in the optical image. As the same fiducial markers can be used to co‐register the H&E image to the optical image, the H&E image can be superimposed with the MSI data.
This registration, however, is not sufficiently accurate because the fiducial markers are usually off‐tissue and thereby too far away for precise co‐registration within tissue. We performed a series of manual and automated registrations in order to fine‐tune the alignment and improve the co‐registration precision of the H&E image, the optical image, and the MALDI‐MSI dataset. To do so, all image datasets were imported into MATLAB R2018b. First, we manually co‐registered the H&E image to the optical image (Figure 1 A). At least five prominent morphological features were selected as control points that were clearly visible in both images (Supplementary Figures 2 and 9). An automated, intensity‐based co‐registration between the MALDI‐MSI data and the H&E image was then performed to fine‐tune the previous manual alignments (Figure 1 B,C).
Figure 1.
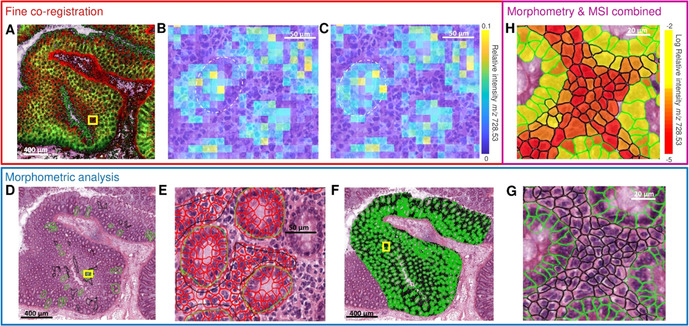
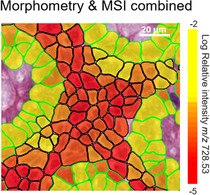
Step‐by‐step illustration of the integral classification workflow using a lipid MALDI‐MSI dataset of a porcine colon. A) High‐spatial‐resolution MALDI‐MSI dataset at 10‐μm resolution is manually co‐registered to the hematoxylin and eosin (H&E) image. B) Zoom of the area indicated by yellow square in (A), showing that manual co‐registration lacks accuracy as displayed by an overlay of an intensity image of MSI (m/z 728.53±0.15 Da) specific for glandular cells and H&E. The dashed white line indicates the glandular cells in the H&E. C) The result of fine‐tuned co‐registration using thresholded images of MSI (m/z 728.53±0.15 Da) and H&E. D) In parallel, regions of interest containing the cell types of interest are manually defined in the H&E image (green=glandular cells, black=lamina propria cells). E) Enlargement of the area indicted by yellow square in (D), which illustrates the automated cell detection and morphometric feature extraction (delineated in red) for the training of a multivariate classifier. F) Result of the application of the trained classifier to the entire remaining specimen. G) Illustration of the single‐cell morphometric classification magnified from the yellow square highlighted in (F). H) Final integration of single‐cell automatic morphometric annotation and features with MALDI‐MSI lipid classification. Average intensity of m/z 728.53±0.15 Da per cell is calculated.
All image registrations were performed using affine geometric transformations15 using MATLAB built‐in functions, and the overall co‐registration error was estimated based on the difference between the calculated (using our co‐registration approach) and the visible positions of the laser shot burns in the H&E image (Supplementary Figure 6). An average co‐registration error of 4.5±1.45 μm standard deviation (Supplementary Table 1) was considered acceptable as it constitutes less than half a pixel size.
In parallel, the scanned H&E image was analyzed in QuPath (v.0.2.0.m8), an open‐source software platform for digital pathology image annotation and analysis.16 There, representative regions for each cell type of interest were manually defined (Figure 1 D). Automated cell detection and morphometric feature extraction of the detected cells were then used to train a Random Forest classifier to distinguish the annotated cell types (Figure 1 E). This classifier was then applied to the area of the MSI measurement (Figure 1 F) and validated by a trained pathologist.
These cell annotations were then geometrically transformed using the previously calculated co‐registration between H&E image and MALDI‐MSI data. As a result, the ≈130 000 individual cell annotations could be overlaid with the MSI images and linked to their cell‐specific lipid profiles (Figure 1 H, Supplementary Figure 7).
Application of our method to the porcine colon dataset enabled the extraction of molecular lipid profiles specific for glandular cells and lamina propria cells (Figure 1 H). Simultaneously, Figure 1 H revealed that MSI data further differentiates lamina propria cells based on their lipid content. Cells with the same overall morphological classification are molecularly differentiated by a variable expression of Hexosyl‐Ceramide 36:0 with possible contamination of PS 32:3 (m/z 728.53) among the lamina propria cells. Ceramides are regarded as important signaling molecules in apoptosis as well as precursors in biosynthesis of glycosphingolipids and gangliosides.17
Having established the accurate co‐registration workflow for porcine colon, we wished to validate our method on a gastric disease model. Diffuse‐type gastric carcinoma (DGC), unlike intestinal‐type gastric carcinoma, does not grow in tumor agglomerates or larger masses. This is a direct result of loss of cellular adhesion properties, where individual tumor cells or small tumor assemblies infiltrate nearby tissues (Figure 2 A).18 Molecular pathology studies of cases like DGC are challenging because of a) the time‐consuming manual annotation of each tumor cell in large specimens, and b) the need for high‐spatial‐resolution MALDI‐MSI experiments in order to differentiate the individual tumor cells from other cells. Thus, DGC is a prime example where the combination of morphometric tissue analysis combined with high‐resolution MALDI‐MSI may enable molecular profiling to differentiate the different individual cells involved.
Figure 2.
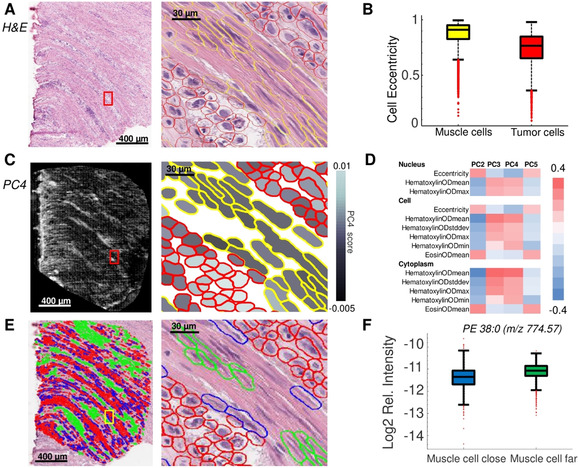
Spatial statistics enabled by our method for the investigation of diffuse‐type gastric carcinoma. A) Histological images (H&E): full tissue section (left) and magnification of the highlighted region (red square) after cell detection and classification (right). B) Box‐plot shows cell eccentricity as a differential morphometric feature to discern tumor from muscle cells of the muscularis propria. C) MALDI‐MSI at 10‐μm resolution was performed and the average scores of the principal component (PC) 4 for each cell are shown (left) and overlaid with the co‐registered cell classification shown in the magnification denoted by a red square (right). D) Correlations of morphometric features with PCs from the MALDI‐MSI lipid data. E) Cell detection provides the spatial coordinates of every cell, which allows distinguishing muscle cells far away from (green) and close to (blue) tumor cells (red). Full tissue section (left), magnification of the area indicated by a yellow square (right). F) The lipid PE 38:0 (m/z 774.57±0.3 Da) exhibits a differential molecular abundance in muscle cells located close to tumor cells compared to muscle cells far away from tumor cells.
Here, we apply our workflow to investigate the lipid molecular profiles of the different cell types in a DGC specimen by MALDI‐MSI at a spatial resolution of 10 μm. The H&E image was subjected to a semi‐automated cell classification to discern tumor cells from surrounding muscle cells of the muscularis propria (Figure 2 A) based on the cells’ morphometric features such as cell and nuclear eccentricity (Figure 2 B). This classification allowed the isolation of lipidomic information of DGC cells in MALDI‐MSI (Figure 2 C). The morphometric characteristics of individual cell types could be related with their molecular content as determined by principal component analysis of the MALDI‐MSI data (Figure 2 D). Moreover, the cancer field effect implies that tumor cells influence the molecular profiles of other cells in their vicinity.19 Having the coordinates of every cell and the corresponding lipidomic information, our method permits the molecular profiles of muscle cells to be compared and investigated based on proximity to tumor cells (Figure 2 E). For example, the lipid PE 38:0 differentiated muscle cells located far away from tumor cells from to those located close to tumor cells (Figure 2 F, Supplementary Figure 13). PE 38:0 has many cellular functions, for example, it is a lipid chaperone assisting in the folding of membrane proteins, it plays a role in lipid‐induced stress in the endoplasmic reticulum and in the initiation of autophagy.20 This shows that our method opens new means to relate molecular information to spatial functional information.
In order to correctly interpret the MALDI‐MSI information in the histological context of the tissue, precise co‐registration is crucial. Using a second, intensity‐based co‐registration, we corrected for most sources of alignment errors. These imprecise alignments could be due to manual co‐registration during setup of an MSI experiment or the potential mismatch between the recorded pixel position and actual laser impact due to laser misalignment or imprecise stage movement. Here, we used the visible laser burn marks in the tissue to estimate the alignment error between the MALDI‐MSI dataset and H&E image, which was determined to be well below half a pixel size. We did not use the laser burns as a part of the co‐registration workflow, since laser burns might not be visible during multimodal image acquisition at lower laser power. While we considered this error negligible with current instrumentation capabilities, precise co‐registration will be the next bottleneck in MSI when higher spatial resolutions below 5 μm will become routinely available. This anticipated problem further highlights the need for novel co‐registration approaches in the field of MSI based on new, fine‐structured fiducial markers, which are visible in both MSI and brightfield microscopy, as well as the introduction of better optical visualization systems in commercial MSI instrumentation (e.g. IMScope, Shimadzu).
In the presented examples, the morphometric differences between the individual cells were clearly visible to the human eye, and thus the morphometric classification performed was accordingly accurate. Because the primary aim was to ease and expedite the annotation workload of a researcher or pathologist for single‐cell MALDI‐MSI purposes, semi‐automated classifications were used. The user trains the system first, validates the results, and is thus relieved from extensive, manual single‐cell annotations. While single‐cell MALDI‐MSI has been available for a decade, its integration with histological images for large‐scale single cell profiling has not been exploited. Here, we present a new method that accelerates histological annotation for rapid correlation with the chemical information provided by high‐spatial‐resolution MALDI‐MSI. It results in 105 single‐cell morphometric and molecular profiles. This workflow allows for true molecular histology at the single‐cell level and as such this methodology lays out the future of high throughput, automated digital pathology‐based diagnosis. Our method accelerates research in cellular molecular science that aims at understanding how spatially structured communities of individual cells act and interact in the context of their environment.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
The authors would like to thank Prof. dr. N. D. Bouvy from MUMC+, Maastricht, The Netherlands, for providing the porcine colon tissue. This work was financially supported by the Dutch province of Limburg as part of the “LINK” program, as well as funding from the National Institutes of Health (NIH) of the United States of America (NIH R01 CA213492). BB and FD acknowledge the financial support of the European Union (ERA‐NET TRANSCAN 2; Grant No. 643638). FD received support from the Joint Imaging Valley program of the University of Liège and the Maastricht University.
K. Ščupáková, F. Dewez, A. K. Walch, R. M. A. Heeren, B. Balluff, Angew. Chem. Int. Ed. 2020, 59, 17447.
References
- 1. Chughtai K., Heeren R. M. A., Chem. Rev. 2010, 110, 3237–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swales J. G., Hamm G., Clench M. R., Goodwin R. J. A., Int. J. Mass Spectrom. 2019, 437, 99–112. [Google Scholar]
- 3. Vaysse P.-M., Heeren R. M. A., Porta T., Balluff B., Analyst 2017, 142, 2690–2712. [DOI] [PubMed] [Google Scholar]
- 4. Gilmore I. S., Heiles S., Pieterse C. L., Annu. Rev. Anal. Chem. 2019, 12, 201–224. [DOI] [PubMed] [Google Scholar]
- 5. Neumann E. K., Comi T. J., Rubakhin S. S., Sweedler J. V., Angew. Chem. Int. Ed. 2019, 58, 5910–5914; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 5971–5975. [Google Scholar]
- 6. Kompauer M., Heiles S., Spengler B., Nat. Methods 2017, 14, 1156–1158. [DOI] [PubMed] [Google Scholar]
- 7. Ščupáková K., et al., Clin. Chem. Lab. Med. 2020, 58, 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chaurand P., et al., Anal. Chem. 2004, 76, 1145–1155. [DOI] [PubMed] [Google Scholar]
- 9. Komura D., Ishikawa S., Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Madabhushi A., Lee G., Med. Image Anal. 2016, 33, 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collan Y., Torkkeli T., Pesonen E., Jantunen E., Kosma V. M., Anal. Quant. Cytol. Histol. 1987, 9, 79–88. [PubMed] [Google Scholar]
- 12. Kumar M., Chatterjee K., Purkait S. K., Samaddar D., J. Oral Maxillofac. Pathol. 2017, 21, 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ginzberg M. B., Kafri R., Kirschner M., Science 2015, 348, 1245075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huizing L. R. S., et al., Clin. Mass Spectrom. 2019, 12, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdelmoula W. M., et al., Anal. Chem. 2014, 86, 3947–3954. [DOI] [PubMed] [Google Scholar]
- 16. Bankhead P., et al., Sci. Rep. 2017, 7, 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tserng K.-Y., Griffin R. L., Biochem. J. 2004, 380, 715–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laurén P., Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [DOI] [PubMed] [Google Scholar]
- 19. Slaughter D. P., Southwick H. W., Smejkal W., Cancer 1953, 6, 963–968. [DOI] [PubMed] [Google Scholar]
- 20. Patel D., Witt S. N., Oxid. Med. Cell. Longevity 2017, 4829180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


