Abstract
Na,K‐ATPase is a ubiquitous molecule contributing to the asymmetrical distribution of Na+ and K+ ions across the plasma membrane and maintenance of the membrane potential, a prerequisite of neuronal activity. Na,K‐ATPase comprises three subunits (α, β, and FXYD). The α subunit has four isoforms in mice, with three of them (α1, α2, and α3) expressed in the brain. However, the functional and biological significances of the different brain isoforms remain to be fully elucidated. Recent studies have revealed the association of Atp1a3, a gene encoding α3 subunit, with neurological disorders. To map the cellular distributions of the α subunit isoforms and their coexpression patterns, we evaluated the mRNA expression of Atp1a1, Atp1a2, and Atp1a3 by in situ hybridization in the mouse brain. Atp1a1 and Atp1a3 were expressed in neurons, whereas Atp1a2 was almost exclusively expressed in glial cells. Most neurons coexpressed Atp1a1 and Atp1a3, with highly heterogeneous expression levels across the brain regions and neuronal subtypes. We identified parvalbumin (PV)‐expressing GABAergic neurons in the hippocampus, somatosensory cortex, and retrosplenial cortex as an example of a neuronal subtype expressing low Atp1a1 and high Atp1a3. The expression of Atp1b isoforms was also heterogeneous across brain regions and cellular subtypes. The PV‐expressing neurons expressed a high level of Atp1b1 and a low level of Atp1b2 and Atp1b3. These findings provide basic information on the region‐ and neuronal‐subtype‐dependent expression of Na,K‐ATPase α and β subunit isoforms, as well as a rationale for the selective involvement of neurons expressing high levels of Atp1a3 in neurological disorders.
Keywords: immunohistochemistry; in situ hybridization; Na,K‐ATPase alpha subunit isoform; Na,K‐ATPase beta subunit isoform; neuron
Na,K‐ATPase comprises α (Atp1a), β (Atp1b), and FXYD subunits, and each subunit has isoforms. In situ hybridization for α and β subunit isoforms revealed regional‐ and neuronal‐subtype‐specific expression of the isoforms in the mouse brain. The photograph shows Atp1a1 (red) and Atp1a3 (green) expression in the hippocampal dentate gyrus (stained with DAPI, blue).

1. INTRODUCTION
Na,K‐ATPase is an essential membrane protein ubiquitously expressed in mammalian cells (Skou & Esmann, 1992). It uses one ATP molecule to transport three Na+ ions out of the cell in exchange for two K+ ions taken in across the plasma membrane. This pump function is a key contributor to the asymmetrical distribution of Na+ and K+ ions and the resting membrane potential; moreover, it accounts for typically 30% and up to 70% in nerve cells of the cellular ATP expenditure (Howarth, Gleeson, & Attwell, 2012). Na,K‐ATPase comprises three subunits: a catalytic α subunit and regulatory β and FXYD subunits (Forbush, Kaplan, & Hoffman, 1978; Skou & Esmann, 1992; Sweadner & Rael, 2000). The primary role of the α subunit is to bind and transport Na+ and K+. The α subunit has four isoforms (α1, α2, α3, and α4), encoded by Atp1a1, Atp1a2, Atp1a3, and Atp1a4, respectively (Shamraj & Lingrel, 1994; Shull, Greeb, & Lingrel, 1986; Sverdlov et al., 1987). The α subunit genes are expressed in a tissue‐ and cell type‐specific manner (Blanco, 2005). Specifically, isoforms α1, α2, and α3 are expressed in the brain (Shull et al., 1986). The α2 subunit is exclusively expressed in glial cells, whereas the α1 and α3 subunits are expressed in neurons (Hieber, Siegel, Fink, Beaty, & Mata, 1991; Mata, Hieber, Beaty, Clevenger, & Fink, 1992). The α3 subunit is functionally different from the α1 subunit in that it has a lower affinity for Na+ and K+ and is thought to enable the rapid restoration of ion gradients after intense neuronal firing (Azarias et al., 2013; Munzer, Daly, Jewell‐Motz, Lingrel, & Blostein, 1994; Zahler, Zhang, Manor, & Boron, 1997).
Neurons in the brain can be classified into distinct subtypes according to their morphology and firing properties. This heterogeneity raises the possibility that distinct neuronal subtypes may use specific isoforms of the α subunit to achieve appropriate membrane potentials, which is essential for neuronal membrane excitability and firing activity (Blanco & Mercer, 1998; Dobretsov & Stimers, 2005). Comprehensive profiling of the regional and cellular distributions of the different α subunits in the brain will aid in addressing this possibility. However, previous reports regarding the distribution of the α subunits in the brain are limited to specific regions (Hieber et al., 1991). Moreover, there have been scarce reports that demonstrated multiple histochemical labeling of mRNAs of the α subunit isoforms, which have limited our knowledge of whether and how every single neuron uses specific isoforms of the α subunits.
Although Na,K‐ATPase was previously considered not to be associated with specific neurological disorders, various recent studies have reported that Atp1a3 mutations may cause neurological disorders such as rapid‐onset dystonia‐parkinsonism (RDP), alternating hemiplegia of childhood (AHC), and cerebellar ataxia, areflexia, pes cavus, optic nerve atrophy, and sensorineural hearing loss (CAPOS) (de Carvalho Aguiar et al., 2004; Demos et al., 2014; Heinzen et al., 2012; Rosewich et al., 2012). Furthermore, amyloid β oligomers induce cell death in cultured neurons by directly inhibiting NAKα3‐derived pump activity (Ohnishi et al., 2015). Therefore, identifying the neurons that are critically dependent on Atp1a3 expression is important for elucidating the vulnerable neuronal populations in neurodegenerative disorders as well as understanding the physiological significance of selective Na,K‐ATPase isoform usage by neurons.
In the present study, we performed in situ hybridization for Atp1a1, Atp1a2, and Atp1a3, targeting the mRNA coding sequence (CDS) regions in the mouse brain to reveal the regional and neuronal‐subtype‐specific expression of the isoforms. Our results confirmed the heterogeneous expression of the previously reported isoforms (Hieber et al., 1991) and expanded our understanding of the cellular populations expressing specific Na,K‐ATPase α subunit isoforms. We identified parvalbumin (PV)‐expressing neurons in the hippocampus and somatosensory cortex as an example of a neuronal subtype which predominantly expresses Atp1a3 and scarcely expresses Atp1a1. We then evaluated the expression of Na,K‐ATPase β subunit isoforms (Atp1b1, Atp1b2, and Atp1b3) in the whole brain and found that PV neurons in the hippocampus and somatosensory cortex expressed Atp1b1 at a high level, and Atp1b2 and Atp1b3 at an undetectable level.
2. METHODS
2.1. Animals
All experiments were conducted in accordance with the Guidelines for Animal Experimentation in Neuroscience proposed by the Japan Neuroscience Society and were approved by the Experimental Animal Research Committee of the University of Fukui. C57BL/6J male mice were purchased from Japan SLC and fixed at the age of 8–12 weeks to evaluate the Atp1a and Atp1b isoform expression in the mouse brain after completing development and before starting aging.
2.2. Sample preparation for histochemistry
Mice were deeply anesthetized by intraperitoneal injection of sodium pentobarbital and transcardially perfused with phosphate buffer saline (PBS) followed by 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB, not containing NaCl). The brain was removed from the skull, immersed in 4% PFA in 0.1 M PB overnight, and transferred to 30% sucrose in 0.1 M PB. The testis, lung, and kidney were also removed after 4% PFA perfusion, immersed in 4% PFA in 0.1 M PB overnight, and transferred to 30% sucrose in 0.1 M PB. The brain and other organs were then embedded in optimal cutting temperature compound (Sakura Finetek), frozen at −80°C, and sliced into sections with a thickness of 20 μm using a cryotome (CM 3050S, Leica). The brain sections were rinsed in PBS and 0.1 M PB, mounted on glass slides (CREST coat, Matsunami) using a paintbrush. The sections of other organs were directly mounted on glass slides after cryosectioning. The sections were then dried overnight in a vacuum desiccator and stored at 4°C until histochemical staining.
2.3. RNA probe preparation for in situ hybridization
Mouse Atp1a1, Atp1a2, Atp1a3, Atp1b1, Atp1b2, and Atp1b3 cDNAs were subcloned by conventional PCR with the following primers: Atp1a1 (full‐length CDS), 5′‐ATGGGGAAGGGGGTTGG‐3′ – 5′‐CTAGTAGTAGGTTTCCTTCTCC‐3′; Atp1a2 (full‐length CDS), 5′‐ATGGGTCGTGGGGCAG‐3′ – 5′‐TCAGTAGTACGTCTCCTTCTCC‐3′; and Atp1a3 (243–3146), 5′‐GGATGACCTCAAGAAGGAAGTG‐3′ – 5′‐CGAAGATGAGGAAACTGTAGGG‐3′; Atp1b1 (513–1336), 5′‐AGCCAAGGAGGAAGGCAG‐3′ – 5′‐ACGCCTTACACTCGACGC; Atp1b2 (589–1461), 5′‐ATGGTCATCCAGAAAGAGAA‐3′ – 5′‐TCAGGTTTTGTTGATCCGGAG3′; Atp1b3 (576–1439), 5′‐GTCAGTTTCCAGTCTCCTTGCT‐3′ – 5′‐CCTTTTCCCTCCTCACATACAG‐3′. We used plasmids provided by Dr Nobuyuki Shiina (Shiina, Yamaguchi, & Tokunaga, 2010) as amplification templates for Atp1a1 and Atp1a3, a commercial mouse brain cDNA library (MD‐01, Genostaff) for Atp1a2, and an in‐house mouse brain cDNA library produced with RNeasy (Qiagen) and ReverTra Ace (Toyobo) for Atp1b isoforms. The PCR products were subcloned into pGEM‐T Easy (Promega) for Atp1a isoforms or pBluescript KS(−) vector for Atp1b isoforms. Plasmids for in vitro transcription of RNA probes for Vglut1, Gad65, and Gad67 mRNAs were kindly provided by Drs Katsuhiko Ono and Yuchio Yanagawa (Asada et al., 1997; Makinae et al., 2000; Ono et al., 2008). We prepared digoxigenin (DIG)‐ and fluorescein (FLU)‐labeled RNA probes using the in vitro transcription kit (Roche) according to the manufacturer's protocol. We prepared both antisense and sense probes for Atp1a and Atp1b isoforms and confirmed that in situ hybridization using sense probes yielded no detectable signals (data not shown). All data were obtained from in situ hybridization using antisense probes.
2.4. Antibody characterization
The antibodies used in the present study are listed in Table 1.
TABLE 1.
Antibodies used in the present study
| Name | Catalog number | Manufacturer | RRID | Species | Clonality | Concentration |
|---|---|---|---|---|---|---|
| Calbindin antibody | ab11426 | Abcam | AB_298031 | Rabbit | Polyclonal | ×400 |
| Calretinin antibody | AB5054 | Millipore | AB_2068506 | Rabbit | Polyclonal | ×400 |
| PV antibody | ab11427 | Abcam | AB_298032 | Rabbit | Polyclonal | ×400 |
| DIG fab fragments antibody, AP‐conjugated | 11093274910 | Roche | AB_514497 | Sheep | Polyclonal | ×500 or ×1,000 |
| FLU HRP‐conjugated antibody | NEF710 | PerkinElmer | AB_2314403 | Sheep | Polyclonal | ×500 |
| DNP HRP‐conjugated antibody | FP1129 | PerkinElmer | AB_2629439 | Rabbit | Polyclonal | ×500 |
| Dinitrophenyl‐KLH polyclonal antibody, Alexa Fluor 488 | A‐11097 | Thermo Fisher Scientific | AB_2534131 | Rabbit | Polyclonal | ×500 |
| Alexa Fluor 488 AffiniPure donkey anti‐rabbit IgG (H + L) | 711‐545‐152 | Jackson Immuno Research Labs | AB_2313584 | Donkey | Polyclonal | ×400 |
Abbreviations: DIG, digoxigenin; FLU, fluorescein.
2.5. In situ hybridization using a single probe
As shown in Figures 1, 2, 3, 4 and 12–15, in situ hybridization for Atp1a or Atp1b isoform mRNA was performed using a modification of previously described methods (Bepari, Watanabe, Yamaguchi, Tamamaki, & Takebayashi, 2012). In short, sections were fixed in 4% PFA for 20 min, digested with Proteinase K (10 μg/ml) for 30 min, and postfixed in 4% PFA for 15 min. After prehybridization, the sections were incubated overnight at 65°C with DIG‐labeled RNA probes. After stringent washing, the sections were blocked with 10% normal sheep serum, 1% bovine serum albumin, and 0.1% Triton X‐100 in PBS. Subsequently, the sections were incubated overnight at 4°C with alkaline phosphatase‐conjugated anti‐DIG antibody (1:1,000; Roche). The sections were washed in TNT (0.1 M Tris–HCl; pH, 7.5; 0.15 M NaCl; 0.1% Tween 20), followed by alkaline phosphatase buffer (100 mM NaCl; 100 mM Tris–HCl; pH, 9.5; 50 mM MgCl2; 0.1% Tween 20; 5 mM levamisole). The sections were treated overnight with NBT/BCIP (Roche) mixture at room temperature in a dark room for color development, then were rinsed in PBS and mounted in PermaFluor (Thermo Fisher Scientific).
FIGURE 1.
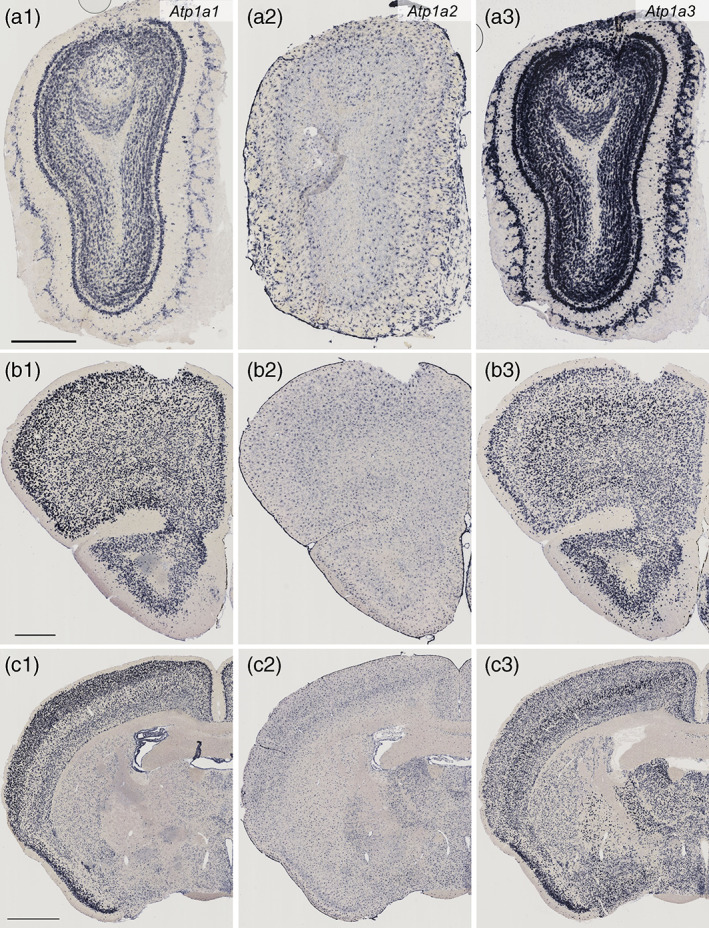
Distribution of Atp1a1, Atp1a2, and Atp1a3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 1, low magnification). Coronal mouse brain sections are shown. All sections through Figures 1, 2, 3, 4 were obtained from one mouse and stained in the same procedure to minimize the difference of signal intensity due to variations in experimental conditions. Atp1a1 mRNA (a1, b1, and c1), Atp1a2 mRNA (a2, b2, and c2), and Atp1a3 mRNA (a3, b3, and c3). The olfactory bulb (a1–a), the frontal cortex (b1–b3), and the striatum, globus pallidus and piriform cortex (c1–c3). Scale bars: 500 μm in a1 and b1, 1 mm in (c1)
FIGURE 2.

Distribution of Atp1a1, Atp1a2, and Atp1a3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 2, low magnification). Coronal mouse brain sections are shown. Atp1a1 mRNA (a1 and b1), Atp1a2 mRNA (a2 and b2), Atp1a3 mRNA (a3 and b3). The somatosensory cortex, hippocampus, and thalamus (a1–a3) and the cerebellum and brain stem (b1–b3). Scale bars: 1 mm
FIGURE 3.
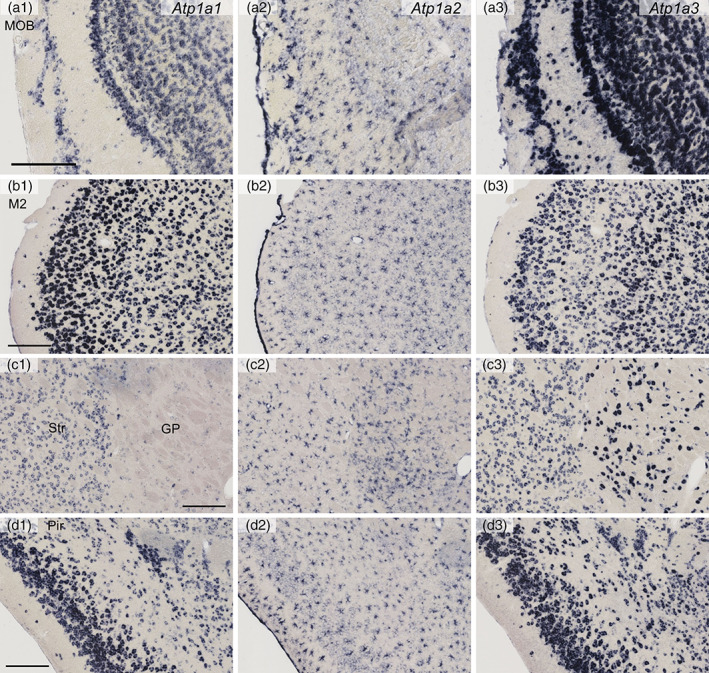
Distribution of Atp1a1, Atp1a2, and Atp1a3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 1, high magnification). Coronal mouse brain sections are shown. Atp1a1 mRNA (a1, b1, c1, and d1), Atp1a2 mRNA (a2, b2, c2, and d2), and Atp1a3 mRNA (a3, b3, c3, and d3). The main olfactory bulb (MOB; a1–a3), secondary motor cortex (M2; b1–b3), the striatum (Str) and globus pallidus (GP) (c1–c3), and the piriform cortex (Pir; d1–d3). Scale bars: 200 μm
FIGURE 4.

Distribution of Atp1a1, Atp1a2, and Atp1a3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 2, high magnification). Coronal mouse brain sections are shown. Atp1a1 mRNA (a1, b1, c1, and d1), Atp1a2 mRNA (a2, b2, c2, and d2) and Atp1a3 mRNA, (a3, b3, c3, and d3). The somatosensory cortex (S1; a1–a3), the hippocampus (CA1 and CA3), dentate gyrus (DG) and thalamic lateral geniculate nucleus (LGN) (b1–b3), the cerebellum (Cb; c1–c3), and the brain stem (d1–d3). Scale bars: 200 μm
2.6. Double fluorescent in situ hybridization using DIG‐ and FLU‐labeled probes
As shown in Figures 5, 6, 8, and 9, double in situ hybridization using DIG‐ and FLU‐labeled probes was performed using a modification of previously described methods (Watakabe, Komatsu, Ohsawa, & Yamamori, 2010). Hybridization and washing were performed as described above, except that both DIG and FLU probes were used for hybridization. After blocking in 1% blocking buffer (11096176001, Roche) for 1 hr, the DIG and FLU‐labeled probes were detected. For detection of the FLU‐labeled probes, the sections were incubated with an anti‐FLU antibody conjugated with horseradish peroxidase (1:500; PerkinElmer) for 1 hr at room temperature. After three 10‐min washes in TNT, the sections were treated with diluted (1:100) tyramide signal amplification (TSA)‐Plus dinitrophenol (DNP) reagents for 5 min according to the manufacturer's instructions (PerkinElmer), and the FLU signals were converted to DNP signals. To amplify the DNP signals, the sections were washed in TNT three times for 10 min each, incubated with an anti‐DNP antibody conjugated with horseradish peroxidase (1:500; PerkinElmer) for 1 hr at room temperature, and treated again with diluted TSA‐Plus DNP reagents (1:100) for 5 min. Subsequently, the sections were incubated overnight with an anti‐DNP antibody conjugated with Alexa 488 (1:500; Molecular Probes) in 1% blocking buffer at 4°C for fluorescence detection of DNP signals. At this point, an anti‐DIG antibody conjugated with alkaline phosphatase (1:500; Roche) was added to the incubation mixture for detection of the DIG probes. The sections were washed three times in TNT and once in Tris‐saline buffer (TS) 8.0 (0.1 M Tris–HCl; pH, 8.0; 0.1 M NaCl; 50 mM MgCl2), and alkaline phosphatase activity was detected using the 2‐hydroxy‐3‐naphtoic acid‐2′‐phenylanilide phosphate (HNPP) fluorescence detection set (11758888001, Roche) according to the manufacturer's instructions. The sections were incubated with the substrate three times for 30 min, and the reaction was stopped by washing with PBS. The sections were then counterstained with DAPI diluted in PBS (2 μg/ml) for 5 min. After washing with PBS, the sections were mounted in PermaFluor (Thermo Fisher Scientific).
FIGURE 5.

Differential expression of Atp1a1 and Atp1a3 mRNAs across neuronal subtypes in the mouse hippocampus. (a,b) Double fluorescent in situ hybridization for Atp1a1 and Atp1a3 mRNAs in the hippocampus with opposite combinations of detection probes (a, Atp1a1‐DIG and Atp1a3‐FLU; b, Atp1a1‐FLU and Atp1a3‐DIG). The panels show Atp1a1‐expressing cells (a1, a4, b1, and b4), Atp1a3‐expressing cells (a2, a5, b2, and,5), and a merged view (a3, a6, b3, and b6) in coronal sections of CA1 (a1–a3 and b1–b3) and the dentate gyrus (DG; a4–a6 and b4–b6). Scale bar: 100 μm. (c,d) Scatterplot of signal intensities by Atp1a1‐DIG and Atp1a3‐FLU (c1 and 2) or Atp1a1‐FLU and Atp1a3‐DIG (d). Data of c1 and c2 were obtained from different animals. Signal intensities in the pyramidal cell layer (PCL) of CA1 (blue dots), granule cell layer of the DG (orange dots), and the hilus (gray dots). Each dot represents a single cell (n = 20 cells in each region). Large plots and bars show means ± SD. Statistical significance was calculated using one‐way analysis of variance (ANOVA) with post hoc Tukey's test (n = 20 cells). **, p < .01; ***, p < .001
FIGURE 6.
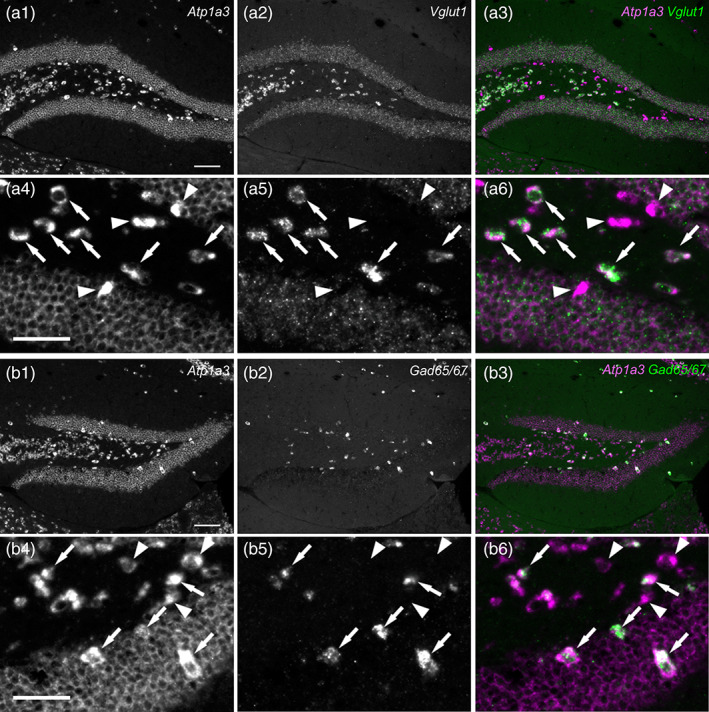
Glutamatergic and GABAergic cells expressing high levels of Atp1a3 mRNA in the mouse DG and hilus. (a) Double fluorescent in situ hybridization for Atp1a3 (a1 and a4) and Vglut1 (a2 and a5) mRNAs in the DG at low magnification (a1–a3), high magnification (a4–a6), and merged views (a3 and a6: magenta, Atp1a3; green, Vglut1). The arrows show Atp1a3(++) and Vglut1(++) cells. The arrowheads show Atp1a3(+) cells. (b) Double fluorescent in situ hybridization for Atp1a3 (b1 and b4) and Gad65/67 (b2 and b5) in the DG at low magnification (b1–b3), high magnification (b4–b6) and merged views (b3 and b6: magenta, Atp1a3; green, Gad65/67). The arrows show Atp1a3(++) and Gad65/67(++) cells. The arrowheads show Atp1a3(+) cells. Scale bars: 100 μm in a1 and b1, 50 μm in a4 and b4
2.7. mRNA/protein double fluorescent labeling
As shown in Figures 7, 10, 11, 17, and 18, we performed fluorescent mRNA labeling and protein immunostaining of the same sections using a modification of previously described methods (Watakabe et al., 2010). Hybridization with DIG‐labeled probes and washing were performed as described above. Subsequently, the sections were incubated in 1% blocking buffer (11096176001, Roche) for 1 hr. Primary antibodies for PV (1:400; Abcam), calbindin (1:400; Abcam), or calretinin (1:400; Millipore) and an anti‐DIG antibody conjugated with alkaline phosphatase (1:500; Roche) were included in the incubation mixture. The sections were washed three times with TNT and incubated with an Alexa Fluor 488‐conjugated secondary antibody (1:400; Jackson ImmunoResearch Labs) for 2 hr. After three washes with TNT and one wash with TS 8.0, alkaline phosphatase activity was detected using the HNPP fluorescence detection set (11758888001, Roche) according to the manufacturer's instructions. The sections were incubated with the substrate three times for 30 min, and the reaction was stopped by washing with PBS. The sections were then counterstained with DAPI diluted in PBS (2 μg/ml) for 5 min. After washing with PBS, the sections were mounted in PermaFluor (Thermo Fisher Scientific).
FIGURE 7.
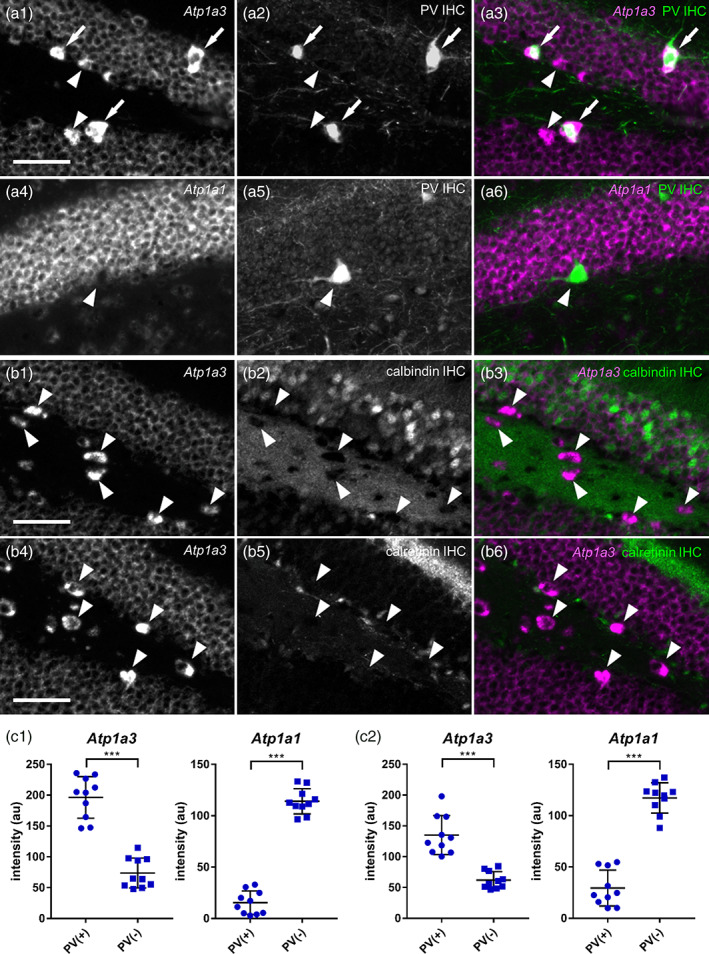
High levels of Atp1a3 expression and low levels of Atp1a1 expression by parvalbumin (PV) neurons in the mouse DG and hilus. (a) Double fluorescent labeling of Atp1a mRNAs (isoform 1 or 3; a4 and a1, respectively) and PV protein (a2 and a5) and a merged view (a3 and a6: magenta, Atp1a; green, PV) in the DG. Atp1a3 (a1–a3) and Atp1a1 (a4–a6). The arrows show PV(+) and Atp1a3(++) cells. The arrowheads show PV(+) cells. (b) Double fluorescent labeling of Atp1a3 (b1 and b4) and calbindin (b2) or calretinin (b5) and a merged view (b3 and b6: magenta, Atp1a3; green, calbindin or calretinin) in the DG. The arrowheads show Atp1a3(+) cells. Scale bars: 50 μm. (c) Signal intensities of Atp1a3 and Atp1a1 in a single PV(+) or PV(−) cell. Data of c1 and c2 were obtained from different animals. Data represent means ± SD and each dot represents data from a single cell (n = 10 cells). Statistical significance was calculated using unpaired t tests (n = 10 cells). ***, p < .001
2.8. Image acquisition and analysis
The sections were examined using a bright‐field virtual slide system (Hamamatsu Photonics, NanoZoomer) and are shown in Figures 1, 2, 3, 4 and 12–15, while a fluorescent microscope (Olympus, BX51WI) was used to obtain the images seen in Figures 5, 6, 7, 8, 9, 10, 11, 17, and 18. Fluorescent signal intensity (Figures 5cd, 7c, 8cd, 10b, 11b, 17b, and 18b) was quantified with ImageJ software. Fluorescent images of Atp1a1, Atp1a3, PV, Atp1b1, Atp1b2, and DAPI staining in the same field were acquired. We were cautious not to bleach fluorescent signals by restricting the section exposure to light as much as possible. To compare signal intensity between different image files (Figures 5cd and 8cd), the images were taken at the same excitation light intensity, exposure time, and gain setting. The soma of a single neuron was randomly selected and delineated according to in situ signals using the Polygon selection tool. This was followed by the confirmation that the region of interest contained a cell nucleus by DAPI images. In Figures 5cd and 8cd, the same region of interest was applied to the Atp1a1 and Atp1a3 images, and the Atp1a1 and Atp1a3 signal intensity were measured. In Figures 7c, 10b, 11b, 17b, and 18b, signal intensity of Atp1a1, Atp1a3, Atp1b1, and Atp1b2 was measured as mentioned above and PV image was used to identify PV(+) and PV(−) neurons. Multiple neurons were analyzed, and the mean value of the signal intensities from the individual cells was used for statistical tests and scatterplots. Fluorescence intensity of tissue background was also measured by delineating a neuropil region in the same image examined that contained no cellular signals. We used the intensity value of Atp1a and Atp1b isoforms subtracted by the intensity value of tissue background as representative of one cell.
FIGURE 8.

Differential expression of Atp1a1 and Atp1a3 mRNAs across neuronal subtypes in the mouse somatosensory cortex. (a) Double fluorescent in situ hybridization for Atp1a1 and Atp1a3 mRNAs in the somatosensory cortex (a1–a3, Atp1a1‐DIG and Atp1a3‐FLU; a4–a6, Atp1a1‐FLU and Atp1a3‐DIG). The panels show Atp1a1‐expressing cells (a1 and a4), Atp1a3‐expressing cells (a2 and a5), and a merged view (a3 and a6) in a coronal section of the somatosensory cortex. (b) Magnified view of Layer 2/3 (b1–b3) and Layer 5 (b4–b6) in a1–a3. The arrows show Atp1a1(++)/Atp1a3(++) cells. The arrowheads show Atp1a1(−) Atp1a3(++) cells. Scale bar: 100 μm in a1, 50 μm in b1. (c,d) Scatterplot of Atp1a1‐DIG and Atp1a3‐FLU (c) or Atp1a1‐FLU and Atp1a3‐DIG (d) signal intensities in Layer 2/3 (blue dots) and Layer 5 (orange dots) of the somatosensory cortex. Each dot represents a single cell (n = 50 cells in each layer). Large plots and bars show median with interquartile. Data of c1 and c2 were obtained from different animals. Statistical significance was calculated using Mann–Whitney U test (n = 50 cells). ***, p < .001
FIGURE 9.

Atp1a3‐expressing cells in the mouse superficial somatosensory cortex and deep neocortex. Double fluorescent in situ hybridization for Atp1a3 and Vlgut1 or Gad65/67 mRNAs in the somatosensory cortex. The Atp1a3 (+ and ++) cells in the superficial somatosensory cortex were primarily inhibitory, whereas those in the deep neocortex were both excitatory and inhibitory. (a) Low‐magnification views of Atp1a3 (a1 and a4) and Vglut1 (a2) or Gad65/67 (a5) labeling and a merged view (a3 and a6: magenta, Atp1a3; green, Vglut1 or Gad65/67). (b) High‐magnification views of Layer 2/3 (b1–b6) and Layer 5 (b7–b12) in (a). The arrows show Atp1a3(+) cells positive for the corresponding marker. The arrowheads show Atp1a3(+) cells negative for the corresponding marker. Scale bar: 100 μm in a1 and a4, 50 μm in b1
FIGURE 10.
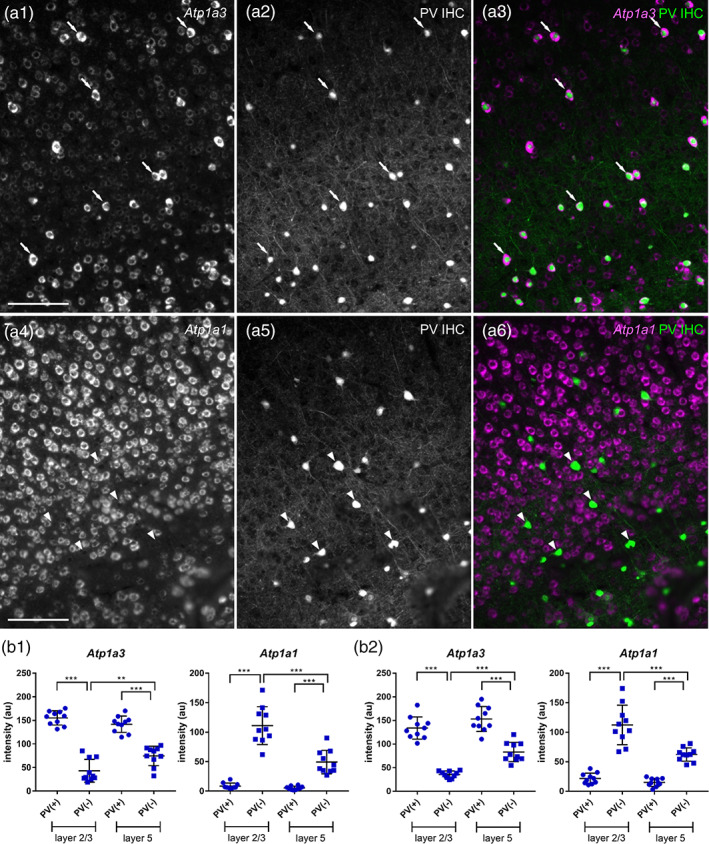
High levels of Atp1a3 expression and low levels of Atp1a1 expression by PV neurons in the mouse somatosensory cortex. (a) Double fluorescent labeling of Atp1a isoform mRNAs (a1 and a4) and PV protein (a2 and a5) and merged views (a3 and a6: magenta, Atp1a mRNAs; green, PV protein) in the somatosensory cortex. Atp1a3 (a1–a3) and Atp1a1 (a4–a6). The arrows show PV(+) Atp1a(+) cells. The arrowheads show PV(+) Atp1a(−) cells. Scale bar: 100 μm. (b) Signal intensities of Atp1a1 and Atp1a3 in a single PV(+) and PV(−) cell in Layers 2/3 and 5. Data represent means ± SD and each dot represents data from a single cell (n = 10 cells). Data of b1 and b2 were obtained from different animals. Statistical significance was calculated using one‐way analysis of variance (ANOVA) with post hoc Tukey's test (n = 10 cells). **, p < .01; ***, p < .001
FIGURE 11.

High levels of Atp1a3 expression and low levels of Atp1a1 expression by PV neurons in the retrosplenial cortex. (a) Double fluorescent labeling of Atp1a isoform mRNAs (a1 and a2) and PV protein (a3 and a4) and merged views (a5 and a6) in the retrosplenial cortex. Atp1a3 (a1, a3, and a5) and Atp1a1 (a2, a4, and a6). The arrows show PV(+) Atp1a(+) cells. The arrowheads show PV(+) Atp1a(−) cells. Scale bar: 100 μm. (b) Signal intensities of Atp1a1 and Atp1a3 in PV(+) and PV(−) cells. Data represent means ± SD and each dot represents data from a single cell (n = 10 cells). Data of b1 and b2 were obtained from different animals. Statistical significance was calculated using unpaired t test (n = 10 cells). ***, p < .001
2.9. Statistical analysis
The normality of data was at first tested by the Kolmogorov–Smirnov test (MATLAB R2018b). The data were presented as means ± SD (other than Figure 8cd) or median with interquartile (Figure 8cd), and the values were analyzed for statistical differences using one‐way analysis of variance with post hoc Tukey's test (Figures 5c, 10b, and 18b), Student's t test (Figures 7c, 11b, and 17b), or Mann–Whitney U test (Figure 8cd). All statistical analyses were performed using GraphPad Prism 7.
FIGURE 18.
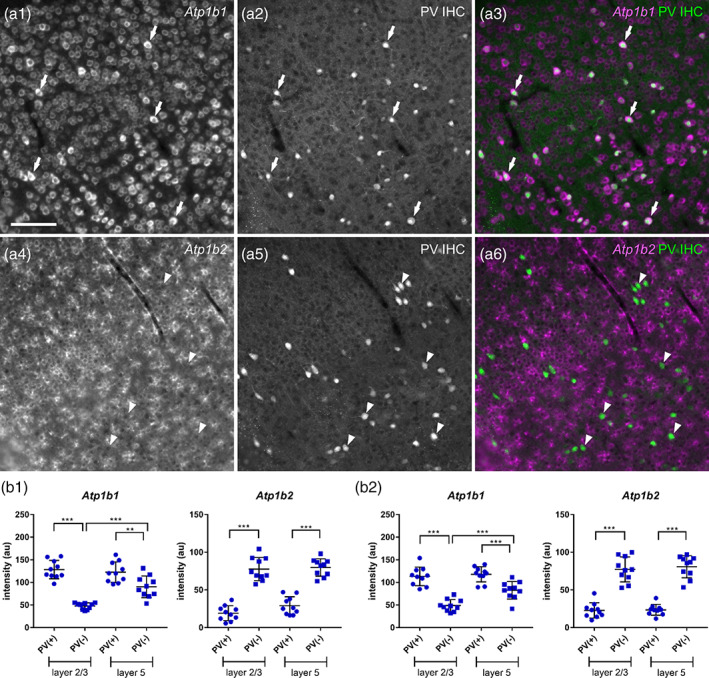
High levels of Atp1b1 expression and low levels of Atp1b2 expression by PV neurons in the somatosensory cortex. (a) Double fluorescent labeling of Atp1b isoform 1 or 2 mRNA (a1 and a4, respectively) and PV protein (a2 and a5), and merged views (a3 and a6: magenta, Atp1b; green, PV) in the somatosensory cortex. Atp1b1 (a1–a3) and Atp1b2 (a4–a6). The arrows show PV(+) Atp1b(+) cells. The arrowheads show PV(+) Atp1b(−) cells. Scale bar: 100 μm. (b) Signal intensities of Atp1b1 and Atp1b2 in a single PV(+) and PV(−) cell in Layers 2/3 and 5. Data represent means ± SD and each dot represents data from a single cell (n = 10 cells). Data of b1 and b2 were obtained from different animals. Statistical significance was calculated using one‐way analysis of variance (ANOVA) with post hoc Tukey's test (n = 10 cells). **, p < .01; ***, p < .001
FIGURE 17.

High levels of Atp1b1 expression and low levels of Atp1b2 expression by PV neurons in the DG and hilus in the mouse brain. (a) Double fluorescent labeling of Atp1b isoform 1 or 2 mRNA (a1 and a4, respectively) and PV protein (a2 and a5), and merged views (a3 and a6: magenta, Atp1a; green, PV) in the DG. Atp1b1 (a1–a3) and Atp1b2 (a4–a6). The arrows show PV(+) and Atp1b1(++) cells. The arrowheads show PV(+) cells. Scale bars: 100 μm. (b) Signal intensities of Atp1b1 and Atp1b2 in a single PV(+) or PV(−) cell. Data represent means ± SD and each dot represents data from a single cell (n = 10 cells). Data of b1 and b2 were obtained from different animals. Statistical significance was calculated using unpaired t test (n = 10 cells). ***, p < .001
3. RESULTS
3.1. Heterogeneous expression levels of Atp1a isoforms in the mouse brain
To identify the cellular populations expressing the different Na,K‐ATPase α subunit isoforms, we performed in situ hybridization for Atp1a1, Atp1a2, and Atp1a3 mRNAs in mouse brain sections. We observed heterogeneous isoform expression across multiple cellular subtypes (Figures 1, 2, 3, 4). In general, neurons expressed Atp1a1 and Atp1a3, whereas the glia, including the Bergmann glia in the cerebellum, expressed Atp1a2. The choroid plexus expressed Atp1a1 and Atp1a2. The signal intensities of Atp1a1 and Atp1a3 were highly heterogeneous across the brain regions and neuronal subtypes. We identified the neuronal subtypes expressing Atp1a1 and Atp1a3 and qualitatively classified them according to the Atp1a1 and Atp1a3 expression levels (Figures 1, 2, 3, 4 and Table 2).
TABLE 2.
Atp1a1 and Atp1a3 signal intensities across neuronal subtypes
| Brain region | Subregions/cell types | Atp1a1 | Atp1a3 |
|---|---|---|---|
| Olfactory bulb | Periglomerular cells and granule cells | + | ++ |
| Mitral cells and tufted cells | − | ++ | |
| Frontal cortex | Superficial layer (2/3) | ++ | + |
| Deep layer (5) | + | ++ | |
| Striatum | + | + | |
| Globus pallidus | − | ++ | |
| Piriform cortex | Layer 2 | ++ | ++ |
| Somatosensory cortex | Glutamatergic cells in superficial layer (2/3) | ++ | +/− |
| Glutamatergic cells in deep layer (5) | + | + | |
| PV neurons in superficial layer (2/3) | − | ++ | |
| PV neurons in deep layer (5) | − | ++ | |
| Hippocampus | Granule cells in the dentate gyrus | ++ | +/− |
| Pyramidal cells in CA1/CA3 | + | + | |
| Mossy cells in the hilus | − | ++ | |
| PV neurons in dentate gyrus/hilus | − | ++ | |
| Thalamus | LGN | − | ++ |
| Cerebellum | Purkinje cells | +/− | ++ |
| Granule cells | ++ | +/− | |
| Stellate cells and basket cells | +/− | ++ | |
| Golgi cells | +/− | ++ | |
| Cerebellar nuclei neurons | +/− | ++ | |
| Brainstem | Reticular nucleus neurons | + | ++ |
| Facial nucleus neurons | ++ | + |
Abbreviations: Atp1a, Na,K‐ATPase α subunit; LGN, lateral geniculate nucleus; PV, parvalbumin.
3.1.1. Olfactory bulb
Atp1a1 expression was moderate in the glomerular layer (GL), mitral cell layer (MCL), and granule cell layer (GCL; Figures 1a and 3a). The somata of the Atp1a1‐expressing cells were small (10–16 μm in soma size), suggesting that they were interneurons (periglomerular and granule cells) (Shepherd, 2004). Atp1a3 expression was high in GL, MCL, GCL, and the external cell layer (ECL). In addition to the interneuron‐like small cells, large cells (20–36 μm) in GL, EPL, and MCL also expressed Atp1a3; this indicated that both interneurons and projection neurons (mitral cells and tufted cells) expressed Atp1a3. Atp1a2 expression was observed in glia at a high level.
3.1.2. Frontal cortex
Both Atp1a1 and Atp1a3 were expressed in all six layers of the frontal cortex (Figure 1b). The Atp1a1 signal was stronger in the superficial layer than in the deep layer of the secondary motor cortex (Figure 3b). In contrast, the Atp1a3 signal was stronger in the deep layer than in the superficial layer. Atp1a2 expression was at a high level in glia.
3.1.3. Striatum, globus pallidus, and piriform cortex
In the striatum, moderate signals of both Atp1a1 and Atp1a3 were observed (Figures 1c and 3c). In the globus pallidus, the Atp1a1 signal was barely detectable, whereas a strong Atp1a3 signal was observed (Figures 1c and 3c). In the piriform cortex, strong signals of both Atp1a1 and Atp1a3 were observed especially in the Layer 2 (Figures 1c and 3d). Atp1a2 expression was observed in glia in both gray matter and white matter (internal capsule) at a high level.
3.1.4. Somatosensory cortex
Both Atp1a1 and Atp1a3 were expressed in all six layers (Figure 2a). Similar to the findings in the frontal cortex, the Atp1a1 signal was stronger in the superficial layer than in the deep layer (Figure 4a). In contrast, the Atp1a3 signal was stronger in the deep layer than in the superficial layer. In addition, the superficial layer contained sparsely distributed cells with high levels of Atp1a3 expression. Atp1a2 expression was at a high level in glia in gray matter and white matter (corpus callosum).
3.1.5. Hippocampus
The expression level of Atp1a1 was high in the dentate gyrus (DG) cell layer, moderate in the CA1 and CA3 cell layers, and scarce in the hilus (Figures 2a and 4b). The Atp1a3 signal was strong in the CA1 and CA3 cell layers and the hilus and moderate in the DG cell layer. Neurons in neuropils of the CA1, CA3, DG, and hilus, which were putative interneurons, showed moderate or strong signals for Atp1a3, but not for Atp1a1. Atp1a2 expression was at a high level in glia.
3.1.6. Thalamus
In the lateral geniculate nucleus, Atp1a1 expression was scarcely observed, whereas the Atp1a3 signal was strong (Figures 2a and 4b). Atp1a2 expression was at a high level in glia.
3.1.7. Cerebellum
The expression level of Atp1a1 was high in the GCL and weak and scarce in the molecular layer, Purkinje cell layer, and cerebellar nuclei (Figures 2b and 4c). The Atp1a3 signal was strong in the molecular layer, Purkinje cell layer, and cerebellar nuclei and relatively weak in GCL. However, medium‐sized neurons (15–21 μm in soma size) in GCL that appeared to be Golgi cells showed high Atp1a3 expression levels. Atp1a2 expression was at a high level in glia including putative Bergman glia in the Purkinje cell layer.
3.1.8. Brain stem
The reticular formation showed moderate Atp1a1 and high Atp1a3 expression levels (Figures 2b and 4d). The facial nucleus showed strong Atp1a1 and Atp1a3 signals. Atp1a2 expression was at a high level in glia.
3.2. Atp1a1 and Atp1a3 mRNA expression levels revealed three subpopulations of neurons in the hippocampus
We observed heterogeneous expression of Atp1a1 and Atp1a3 across the brain regions and neuronal subtypes (Table 2). Neurons appeared to coexpress Atp1a1 and Atp1a3 with different ratios of Atp1a1 and Atp1a3 across neuronal subtypes. To confirm the coexpression of Atp1a1 and Atp1a3 with varied ratios, we performed double fluorescent labeling of Atp1a1 and Atp1a3 mRNAs using both combinations of signal detection (Atp1a1‐DIG–Atp1a3‐FLU (Figure 5a,c) and Atp1a1‐FLU–Atp1a3‐DIG (Figure 5b,d)). We used the hippocampus as an example because of the reliable identification of neuronal subtypes due to homogeneous cell populations within a given subregion or layer. The Atp1a1 and Atp1a3 signals were colocalized in each cell in the CA1, DG, and hilus; this indicated that Atp1a1 and Atp1a3 were coexpressed in the same neurons (Figure 5a,b). Consistent with the results of single‐gene in situ hybridization (Figures 2 and 4), quantification of the signal intensities of Atp1a1 and Atp1a3 in a single cell revealed that the expression level of Atp1a1 was moderate in the pyramidal cell layer (PCL) of the CA1, high in GCL of the DG (DG‐GCL), and low in the hilus (Figure 5c,d). In contrast, the expression level of Atp1a3 was moderate in the PCL of the CA1, low in the DG‐GCL, and high in the hilus (Figure 5c,d). Putative nongranule cell neurons in the DG‐GCL showed high Atp1a3 expression levels (arrowhead in Figure 5a middle‐low panel, and Figure 6).
Next, we compared the relative signals of both Atp1a1 and Atp1a3 in a single cell among the PCL of the CA1, hilus, and DG (Figure 5c,d). The Atp1a1/Atp1a3 signal intensity scatterplot revealed three large clusters: cells expressing moderate levels of Atp1a1 and Atp1a3 [Atp1a1(+) Atp1a3(+)], cells expressing high levels of Atp1a1 and low levels of Atp1a3 [Atp1a1(++) Atp1a3(+/−)], and cells expressing undetectable levels of Atp1a1 and high levels of Atp1a3 [Atp1a1(−) Atp1a3(++)]. These clusters corresponded to the PCL of the CA1, the DG‐GCL, and the hilus, respectively. These results raised the possibility that the hippocampal neurons can be classified into three subpopulations with distinct dependencies on Atp1a1 and Atp1a3 for maintenance of the low intracellular sodium ion activity. We note that the distribution of the three large clusters was independent of the fluorescence labeling method (Figure 5c vs. d), supporting the idea that the difference of relative fluorescence signal between Atp1a1 and Atp1ab3 was not due to the difference in labeling methods (DIG vs. FLU) or fluorescence filter combinations.
Atp1a3 has been shown to be a target of amyloid β oligomers and suggested to be related to neurodegenerative diseases such as Alzheimer's disease (Ohnishi et al., 2015), and this suggests that Atp1a1(−) Atp1a3(++) cells are selectively affected in neurological disorders. To identify the neuronal subtypes of the Atp1a3 (++) cells in the DG and the hilus, we performed double in situ hybridization for Atp1a3 and Vglut1 or Gad65/67 mRNAs (Figure 6). More than half the Atp1a3(++) cells in the hilus were Vglut1‐positive cells, while the rest were Vglut1‐negative cells (Figure 6a). Since the majority of the glutamatergic neurons in the hilus are mossy cells (Scharfman, 2016), the Vglut1(+) Atp1a3(++) cells were likely to be mossy cells (Figure 6a, white arrows). We also observed Gad65/67(+) Atp1a3(+) cells in the hilus and the DG‐GCL (Figure 6b, white arrows).
GABAergic interneurons in the hippocampus and neocortex have numerous anatomical, physiological, and biochemical diversity (Pelkey et al., 2017). The histochemical analysis allowed us to classify the GABAergic neurons based on the expression of calcium‐binding proteins. To further identify the neuronal subtypes of the Atp1a3(++) GABAergic neurons, we performed double labeling of Atp1a isoform mRNAs and calcium‐binding proteins (PV, calbindin, and calretinin, Figure 7). We observed that Atp1a3(++) cells also expressed PV (Figure 7a top, white arrows), but not calretinin or calbindin (Figure 7b), suggesting that the Atp1a3(++) GABAergic neurons in the DG‐GCL and the hilus were PV neurons. Notably, these PV neurons did not show significant Atp1a1 signals (Figure 7a, bottom; Figure 7c, right). Collectively, these data suggest that the Atp1a1(−) Atp1a3(++) cells in the DG and the hilus are mossy cells and PV neurons.
3.3. The classification of neurons according to Atp1a1 and Atp1a3 mRNA expression levels was applicable to the somatosensory cortex
Double fluorescent labeling of Atp1a1 and Atp1a3 mRNAs in the hippocampus suggested that neurons could be classified into subpopulations according to their expression levels. Next, we tested whether the classification is applicable to the somatosensory cortex (Figures 8 and 9) and PV neurons in the somatosensory cortex were Atp1a1(−) Atp1a3(++) similar to those in the hippocampus (Figure 10). Consistent with the results of single‐gene in situ hybridization (Figures 2 and 4), Atp1a1 was strongly expressed in the superficial layer and weakly expressed in the deep layer (Figures 8a,b). In contrast, the expression levels of Atp1a3 were low in the superficial layer and high in the deep layer. In both the superficial and deep layers, some neurons show high and low Atp1a3 and Atp1a1 expression levels, respectively (Figure 8b, upper arrowheads). To confirm that neurons in the somatosensory cortex could be classified into three subpopulations, namely Atp1a1(++) Atp1a3(+/−), Atp1a1(+) Atp1a3(+), and Atp1a1(−) Atp1a3(++), we quantified the signal intensities of both Atp1a1 and Atp1a3 in the same cells in Layers 2/3 and 5 (Figure 8c). Neurons in Layers 2/3 and 5 roughly formed three subpopulations; Atp1a1(+) Atp1a3(+) cells were primarily observed in Layer 5, Atp1a1(++) Atp1a3(+/−) cells in Layer 2/3, and Atp1a1(−) atp1a3(++) cells in both Layers 2/3 and 5. We note that the distribution of the three subpopulations was observed by both fluorescent labeling combination (Atp1a1‐DIG–Atp1a3‐FLU (Figure 8a, upper row and c) and Atp1a1‐FLU–Atp1a3‐DIG (Figure 8a, lower row and d)).
To identify the neuronal subtypes with distinct expression levels of Atp1a3, we performed double in situ hybridization for Atp1a3 and Vglut1 or Gad65/67 mRNAs (Figure 9). In Layer 2/3, 4% of the sum of Atp1a3 (+) and Atp1a3 (++) cells were Vglut1(+), and 91% were Gad65/67(+) (n = 100 cells; Figure 9b, white arrows). In Layer 5, 63% of the sum of Atp1a3 (+) and Atp1a3 (++) cells were Vglut1(+), while 31% were Gad65/67(+) (n = 100 cells, Figure 9b, white arrows). These results suggest that Atp1a3 (+) and Atp1a3 (++) cells in Layer 2/3 of the somatosensory cortex are primarily GABAergic, whereas those in Layer 5 are both glutamatergic and GABAergic. We further confirmed that the Atp1a3 (++) GABAergic neurons were PV neurons via fluorescent labeling of Atp1a1 or Atp1a3 mRNAs in combination with immunostaining for PV (Figure 10a, white arrows; Figure 10b). None of the examined PV neurons expressed detectable Atp1a1 mRNA (Figure 10a, white arrowheads in lower panel; Figure 10b). These results suggest that pyramidal neurons in Layers 2/3 and 5 are the Atp1a1(++) Atp1a3(+/−) and Atp1a1(+) Atp1a3(+) cells in the somatosensory cortex, respectively. Moreover, PV neurons in the somatosensory cortex are Atp1a1(−) Atp1a3(++) cells, similar to those in the hippocampus.
3.4. PV neurons in the retrosplenial cortex showed high Atp1a3 expression and low Atp1a1 expression
Our finding regarding PV neurons in the somatosensory cortex expressing Atp1a3 at a high level and Atp1a1 at a low level raises the question of how applicable this feature of PV neurons is to other cortical regions. We extended the analysis to the retrosplenial cortex, which plays an essential role in spatial learning, memory, and navigation in human and rodents (Mitchell, Czajkowski, Zhang, Jeffery, & Nelson, 2018) by double labeling of Atp1a3 or Atp1a1 mRNAs and PV protein (Figure 11). Consistent with the somatosensory cortex (Figure 10), PV(+) neurons in the retrosplenial cortex expressed a higher level of Atp1a3 and a lower level of Atp1a1 than PV(−) neurons.
3.5. Heterogeneous expression levels of Atp1b isoforms in the mouse brain
The α subunit is not a sole determinant of Na,K‐ATPase function, but rather it depends on the combination of α and β subunit isoforms (Blanco, 2005). Our data of Atp1a1 isoforms raise the question of whether the expression of the β subunit isoforms in the mouse brain is also heterogeneous. To examine cellular profiles expressing the different β subunit isoforms, we performed in situ hybridization for Atp1b1, Atp1b2, and Atp1b3 mRNAs. Similar to Atp1a isoforms, Atp1b isoforms showed heterogeneous expression across brain regions and cellular subtypes (Figures 12, 13, 14, 15). Distribution of Atp1b1‐expressing cells resembled that of Atp1a3. Atp1b2 was preferential but not specific to the glia and expressed by some neuronal subtypes as follows. Signal of Atp1b3 was faint and limited to small population of neurons as described below (Figures 12, 13, 14, 15, 16).
FIGURE 12.

Distribution of Atp1b1, Atp1b2, and Atp1b3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 1, low magnification). Coronal mouse brain sections are shown. All sections through Figures 12, 13, 14, 15 were obtained from one mouse and stained in the same procedure to minimize the difference of signal intensity due to variations in experimental conditions. Atp1b1 mRNA (a1, b1, and c1), Atp1b2 mRNA (a2, b2, and c2), and Atp1b3 mRNA (a3, b3, and c3). The olfactory bulb (a1–a3), the frontal cortex (b1–b3) and the striatum, globus pallidus and piriform cortex (c1–c3). Scale bars: 500 μm in a1 and b1, 1 mm in c1
FIGURE 13.
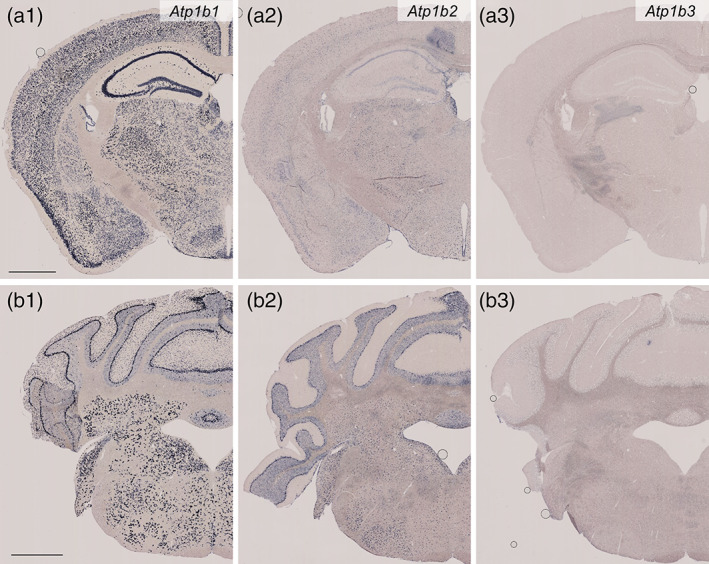
Distribution of Atp1b1, Atp1b2, and Atp1b3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 2, low magnification). Coronal mouse brain sections are shown. Atp1b1 mRNA (a1 and b1), Atp1b2 mRNA (a2 and b2), and Atp1b3 mRNA (a3 and b3). The somatosensory cortex, hippocampus, and thalamus (a1–a3) and the cerebellum and brain stem (b1–b3). Scale bars: 1 mm
FIGURE 14.
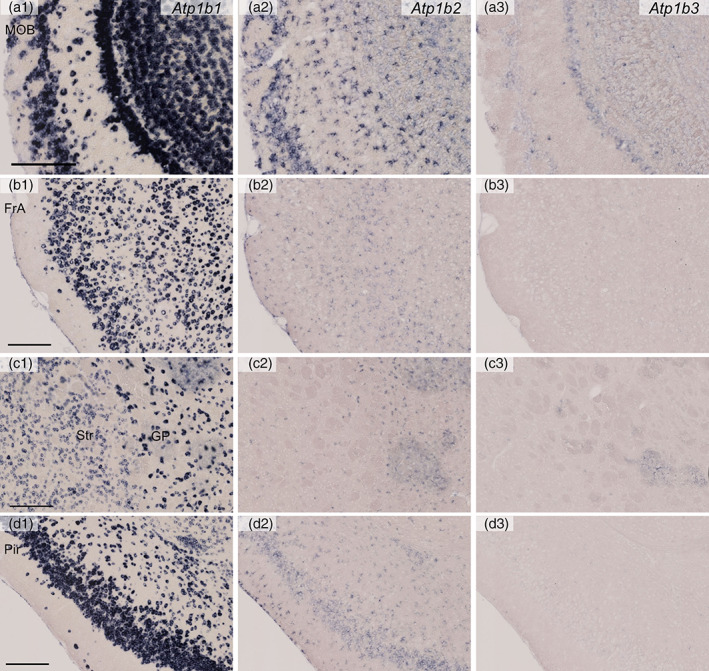
Distribution of Atp1b1, Atp1b2, and Atp1b3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 1, high magnification). Coronal mouse brain sections are shown. Atp1b1 mRNA (a1, b1, c1, and d1), Atp1b2 mRNA (a2, b2, c2, and d2), and Atp1b3 mRNA (a3, b3, c3, and d3). The main olfactory bulb (MOB; a1–a3), secondary frontal association cortex (FrA; b1–b3), the striatum (Str) and globus pallidus (GP) (c1–c3), and the piriform cortex (Pir; d1–d4). Scale bars: 200 μm
FIGURE 15.

Distribution of Atp1b1, Atp1b2, and Atp1b3 mRNAs in the mouse brain as revealed by in situ hybridization (Part 2, high magnification). Coronal mouse brain sections are shown. Atp1b1 mRNA (a1, b1, c1, and d1), Atp1b2 mRNA (a2, b2, c2, and d2), and Atp1b3 mRNA (a3, b3, c3, and d3). The somatosensory cortex (S1; a1–a3), the hippocampus (CA1 and CA3), dentate gyrus (DG) and thalamic lateral geniculate nucleus (LGN) (b1–b3), the cerebellum (Cb; c1–c3), and the brain stem (d1–d3). Scale bars: 200 μm
FIGURE 16.
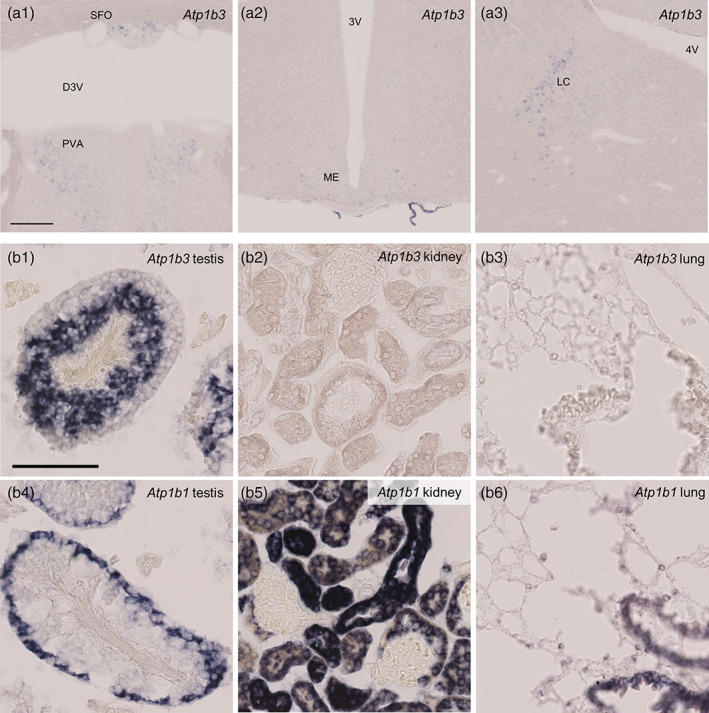
Atp1b3 expression was limited to specific neuronal population in the mouse brain as revealed by in situ hybridization. (a) Atp1b3 expression in anterior nucleus of paraventricular thalamus (PVA, a1), SFO (a1), ME (a2), and LC (a3) (b) Atp1b3 (b1–b3) or Atp1b1 (b4–b6) expression in testis (b1 and b4), kidney (b2 and b5), and lung (b3 and b6). 3V, third ventricle; 4V, fourth ventricle; D3V, dorsal third ventricle; LC, locus coeruleus; ME, median eminence; PVA, anterior nucleus of PVA; SFO, subfornical organ. Scale bars: 200 μm in a1, 100 μm in b1
3.5.1. Olfactory bulb
Atp1b1 expression was high in the GL, MCL, GCL, and ECL. Putative projection neurons (mitral and tufted cells, large‐sized somata) at a higher level of Atp1b1 than interneuron‐like small cells (Figures 12a and 14a). Atp1b2 was expressed by glia at a high level and by interneuron‐like small cells at a low level. Atp1b3 expression was at a low level in the GL, EPL, MCL, and GCL.
3.5.2. Frontal cortex
Atp1b1 was expressed in all six layers of the frontal cortex (Figure 12b). The Atp1b1 signal was stronger in the deep layer than in the superficial layer in general and some Atp1b1 highly expressing cells were distributed in the superficial layer (Figure 14b). Atp1b2 was expressed by glia at a moderate level and by neurons in the Layer 4 at a low level. Atp1b3 expression was faint or undetectable.
3.5.3. Striatum, globus pallidus, and piriform cortex
In the striatum, moderate signals of Atp1b1 were observed (Figures 12c and 14c). In the globus pallidus, a strong Atp1b1 signal was observed (Figures 12c and 14c). In the piriform cortex, strong signals of Atp1b1 were observed especially in the Layer 2 (Figures 12c and 14d). Atp1b2 was expressed by glia at a moderate level and by neurons in the piriform cortex at a low level. Atp1b3 expression was faint or undetectable.
3.5.4. Somatosensory cortex
Atp1b1 was expressed in all six layers (Figures 13a and 15a). Similar to the findings in the frontal cortex, the Atp1b1 signal was stronger in the deep layer than in the superficial layer. In addition, the superficial layer contained sparsely distributed cells with high levels of Atp1b1 expression. Atp1b2 was expressed by glia at a moderate level and by neurons in Layer 4 at a low level. Atp1b3 expression was faint or undetectable.
3.5.5. Hippocampus
The Atp1b1 signal was strong in the CA1 and CA3 cell layers and the hilus and moderate in the DG cell layer (Figures 13a and 15b). Neurons in neuropils of the CA1, CA3, DG, and hilus, which were putative interneurons, showed moderate or strong signals for Atp1b1. Atp1b2 was expressed by glia and CA1 pyramidal neurons at a moderate level and by neurons in the DG at a low level. Notably, Atp1b2 expression in CA3 pyramidal neurons was faint or undetectable. Atp1b3 expression was faint or undetectable.
3.5.6. Thalamus
In the lateral geniculate nucleus, the Atp1b1 signal was strong (Figures 13a and 15b). Atp1b2 was expressed by glia and neurons at a moderate level. Atp1b3 expression was faint or undetectable.
3.5.7. Cerebellum
The Atp1b1 signal was strong in the molecular layer, Purkinje cell layer, and cerebellar nuclei and relatively weak in GCL (Figures 13b and 15c). Medium‐sized neurons (15–21 μm in soma size) in GCL that appeared to be Golgi cells showed high Atp1b1 expression levels. Atp1b2 was expressed by glia including putative Bergman glia at a high level and neurons in GCL at a moderate level. Atp1b3 expression was faint or undetectable.
3.5.8. Brain stem
The reticular formation showed high Atp1a3 expression levels (Figures 13b and 15d). The facial nucleus showed strong Atp1a3 signals. Atp1b2 was expressed by glia at a high level, whereas Atp1b2 expression was faint in neurons. Atp1b3 expression was faint or undetectable.
Atp1b3 expression was not ubiquitous through the mouse brain and observed in the olfactory bulb (Figures 12a3 and 14a3), anterior nucleus of paraventricular thalamus (Figure 16a1), subfornical organ (Figure 16a1), median eminence (Figure 16a2), and locus coeruleus (Figure 16a3) as far as we examined. Because Atp1b3 signals were weak compared to Atp1b1 and Atp1b2 (Figures 12, 13, 14, 15, 16), it remains possible that the detection sensitivity of the Atp1b3 probe was insufficient. To validate the sensitivity of the Atp1b3 probe, we examined Atp1b3 expression in the testis, kidney, and lung. In accordance with a previous report that the testis showed high Atp1b3 expression (Malik, Canfield, Beckers, Gros, & Levenson, 1996), we observed clear Atp1b3 signals in inner part of the seminiferous tubule (Figure 16b). The kidney and lung, which were suggested to weakly express Atp1b3 (Malik et al., 1996), showed very faint or undetectable Atp1b3 signals, although Atp1b1 expression in these organs showed clear signals. These observations support the idea that Atp1b3 expression in the mouse brain is limited to a small population of neurons at a low level.
3.6. PV neurons in the dentate gyrus and somatosensory cortex expressed Atp1b1 at a high level and Atp1b2 at a low level
The resemblance of cellular distribution of Atp1b1 expression to Atp1a3 and that of Atp1b2 expression to Atp1a2 suggests that PV neurons express high Atp1b1 and low Atp1b2. To confirm this point, we performed fluorescent labeling of Atp1b1 or Atp1b2 mRNAs in combination with immunostaining for PV in the DG and somatosensory cortex (Figures 17 and 18). Atp1b1 was highly expressed by PV(+) neurons than PV(−) neurons (Figures 17a and 18a white arrows in upper panel; Figures 17b and 18b) None of the examined PV neurons expressed detectable Atp1b2 mRNA (Figures 17a and 18a, white arrowheads in lower panel; Figures 17b and 18b). These results suggest that PV neurons in the dentate gyrus and somatosensory cortex highly express a combination of Atp1a3 and Atp1b1.
4. DISCUSSION
In the present study, we found distinct region‐ and neuronal subtype‐specific patterns of Na,K‐ATPase α subunit isoform expression in the mouse brain. The neuronal population showed highly variable coexpression of Atp1a1 and Atp1a3 (Table 2). The expression levels of Atp1a1 and Atp1a3 mRNAs revealed three large neuronal subpopulations in the hippocampus and somatosensory cortex (Figures 5b and 8c). As previously reported (Mata et al., 1992), Atp1a2 expression was almost exclusive to glial cells, including the Bergman glia in the cerebellum, and undetectable or faint if any in neurons (Figures 1, 2, 3, 4). The choroid plexus expressed high levels of Atp1a1, moderate levels of Atp1a2, and low levels of Atp1a3 (Figures 1, 2, 3, 4). Our results are mostly consistent with those of Hieber et al., who reported a differential distribution of the Atp1a isoform mRNAs using radioisotope‐based in situ hybridization (Hieber et al., 1991). A previous immunohistochemical study using anti‐NAKα3 antibody has shown that α3 subunit expression is restricted to neurons at a high level in projection neurons, GABAergic neurons in the basal ganglia including globus pallidus, and hippocampal GABAergic neurons, which is consistent with our observation of Atp1a3 mRNA expression (Bottger et al., 2011). In addition to the Atp1a isoforms, Atp1b isoforms showed heterogeneous expression levels across regional and cellular subtypes (Figures 12, 13, 14, 15, 16). Our data of cellular profiles in the cerebellum are primarily consistent with another previous immunohistochemical study using anti‐β isoform antibodies as well as anti‐α isoform antibodies in that Purkinje cells express α3 and β1 whereas granule cells express α1, β1, and β2, and lightly α3 (Peng, Martin‐Vasallo, & Sweadner, 1997). Our histochemical identification is also consistent with previous electrophysiological studies using ouabain that reported the differential contribution of Atp1a isoforms to interneurons and pyramidal neurons in the hippocampus and cortical Layer 5 neurons (Anderson, Huguenard, & Prince, 2010; Richards, Bommert, Szabo, & Miles, 2007). Our highly sensitive detection method enabled us to extend our understanding of the precise distribution of Atp1a‐ and Atp1b‐isoform‐expressing cells in the mouse brain with single‐cell resolution.
We observed no Atp1a3‐negative neurons, whereas Atp1a1 expression in some neurons was below the detection level (e.g., hippocampal mossy cells, PV neurons in the hippocampus and somatosensory cortex, pallidal, and thalamic neurons; Table 2). Due to the technical limitations of in situ hybridization analysis, we could not estimate the copy number of mRNAs or directly compare the expression levels between different RNA probes (namely, Atp1a1 vs. Atp1a3). Measurement of the density of NAKα1 and NAKα3 protein on the cell membrane and comparison of it among distinct neuronal subtypes would expand our understanding of the dependency of each neuronal subtype on NAKα1 and NAKα3. Nevertheless, our current results suggest that Atp1a3, rather than Atp1a1, is a ubiquitous neuronal isoform in the mouse brain.
The heterogeneous Atp1a1/Atp1a3 expression ratio across neuronal subtypes may suggest that neurons have different degrees of dependence on the Na,K‐ATPase α1 and α3 subunits for maintaining the asymmetrical distribution of Na+/K+ ions and membrane excitability. It is noteworthy that we identified neuronal subtypes that were highly dependent on Atp1a3 (i.e., neurons with no or low Atp1a1 signals and high levels of Atp1a3; Table 2). One of the Atp1a1(−) Atp1a3(++) cell types identified in the present study was PV neurons in the hippocampus and cerebral cortex, which are known as the fast‐spiking type of neurons (Hu, Gan, & Jonas, 2014; Kawaguchi & Kubota, 1993). In line with this idea, tufted cells in the olfactory bulb and neurons in the globus pallidus are also Atp1a1(−) Atp1a3(++) cell types, both of which show high‐frequency firing (Hikosaka, Takikawa, & Kawagoe, 2000; Nagayama, Takahashi, Yoshihara, & Mori, 2004). These findings raise a possibility that the fast‐spiking type of neurons may be dependent on NAKα3 rather than NAKα1 to maintain resting membrane potential (Anderson et al., 2010; Richards et al., 2007). We also found that cellular profiles of Atp1b1 expression were more similar to those of Atp1a3 expression than to those of Atp1a1 expression, which was prominent in PV neurons. Further studies are required to confirm whether the α3 subunit, but not the α1 subunit, is preferentially involved in the high‐frequency firing, as well as how each neuronal subtype uses distinct α and β subunit isoform combinations.
Recent studies have reported Atp1a3 involvement in neurological disorders such as RDP, AHC, and cerebellar ataxia, areflexia, pes cavus, optic nerve atrophy, and sensorineural hearing loss (CAPOS) (de Carvalho Aguiar et al., 2004; Demos et al., 2014; Heinzen et al., 2012; Rosewich et al., 2012). Atp1a3‐dependent neurons may be responsible for as well as selectively affected in such disorders. The α3 subunit is also a target of amyloid β oligomers which directly bind to the α3 subunit and inhibit the pump activity of Na,K‐ATPase (Ohnishi et al., 2015). Thus, Atp1a1(−) Atp1a3(++) cells such as PV neurons would be more vulnerable to Amyloid β oligomers than Atp1a1 positive cells. The expression level of Atp1a3 and Atp1a1 in the aged rat brain is different from that in the young rat brain (N. Chauhan & Siegel, 1997a, 1997b; N. B. Chauhan & Siegel, 1996). Since our current study used only young‐adult mice (8‐12‐week‐old), further analysis using elder animals will be necessary to identify the vulnerable neuronal subpopulation.
In conclusion, the present study determined the cellular profile of Na,K‐ATPase α and β subunit isoform expression in the mouse brain. Our findings further the understanding of the functional differences among the Atp1a and Atp1b isoforms and identify a vulnerable neural circuit potentially involved in Atp1a3‐related neurological disorders.
ACKNOWLEDGMENTS
The authors thank Drs Nobuyuki Shiina, Katsuhiko Ono, and Yuchio Yanagawa for the RNA probe plasmids, Drs Kotaro Konno and Masahiko Watanabe for technical advice of fluorescent in situ hybridization as well as Eri Murai, Noriko Funabashi, Hisayuki Miyagoshi, and members of the Life Science Research Laboratory at the University of Fukui for technical assistance. K. M. was supported by Japan Society for the Promotion of Science KAKENHI Grant Numbers 16K18377, 16H01671, 17KK0190, and 18H05005. Y. F. was supported by Japan Society for the Promotion of Science KAKENHI Grant Numbers 15H01281, 16H04662, 16H06280, 17K19446, 18H05120, and 19H03323.
Murata K, Kinoshita T, Ishikawa T, Kuroda K, Hoshi M, Fukazawa Y. Region‐ and neuronal‐subtype‐specific expression of Na,K‐ATPase alpha and beta subunit isoforms in the mouse brain. J Comp Neurol. 2020;528:2654–2678. 10.1002/cne.24924
Peer Review The peer review history for this article is available at https://publons.com/publon/10.1002/cne.24924.
Funding information Japan Society for the Promotion of Science, Grant/Award Numbers: 15H01281, 16H01671, 16H04662, 16H06280, 16K18377, 17K19446, 17KK0190, 18H05005, 18H05120, 19H03323
Contributor Information
Koshi Murata, Email: kmurata@u-fukui.ac.jp.
Yugo Fukazawa, Email: yugo@u-fukui.ac.jp.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Anderson, T. R. , Huguenard, J. R. , & Prince, D. A. (2010). Differential effects of Na+‐K+ ATPase blockade on cortical layer V neurons. The Journal of Physiology, 588(Pt 22), 4401–4414. 10.1113/jphysiol.2010.191858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada, H. , Kawamura, Y. , Maruyama, K. , Kume, H. , Ding, R. G. , Kanbara, N. , … Obata, K. (1997). Cleft palate and decreased brain gamma‐aminobutyric acid in mice lacking the 67‐kDa isoform of glutamic acid decarboxylase. Proceedings of the National Academy of Sciences of the United States of America, 94(12), 6496–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarias, G. , Kruusmagi, M. , Connor, S. , Akkuratov, E. E. , Liu, X. L. , Lyons, D. , … Aperia, A. (2013). A specific and essential role for Na,K‐ATPase alpha3 in neurons co‐expressing alpha1 and alpha3. The Journal of Biological Chemistry, 288(4), 2734–2743. 10.1074/jbc.M112.425785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bepari, A. K. , Watanabe, K. , Yamaguchi, M. , Tamamaki, N. , & Takebayashi, H. (2012). Visualization of odor‐induced neuronal activity by immediate early gene expression. BMC Neuroscience, 13, 140 10.1186/1471-2202-13-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco, G. (2005). Na,K‐ATPase subunit heterogeneity as a mechanism for tissue‐specific ion regulation. Seminars in Nephrology, 25(5), 292–303. 10.1016/j.semnephrol.2005.03.004 [DOI] [PubMed] [Google Scholar]
- Blanco, G. , & Mercer, R. W. (1998). Isozymes of the Na‐K‐ATPase: Heterogeneity in structure, diversity in function. The American Journal of Physiology, 275(5 Pt 2), F633–F650. [DOI] [PubMed] [Google Scholar]
- Bottger, P. , Tracz, Z. , Heuck, A. , Nissen, P. , Romero‐Ramos, M. , & Lykke‐Hartmann, K. (2011). Distribution of Na/K‐ATPase alpha 3 isoform, a sodium‐potassium P‐type pump associated with rapid‐onset of dystonia parkinsonism (RDP) in the adult mouse brain. The Journal of Comparative Neurology, 519(2), 376–404. 10.1002/cne.22524 [DOI] [PubMed] [Google Scholar]
- Chauhan, N. , & Siegel, G. (1997a). Differential expression of Na,K‐ATPase alpha‐isoform mRNAs in aging rat cerebellum. Journal of Neuroscience Research, 47(3), 287–299. [PubMed] [Google Scholar]
- Chauhan, N. , & Siegel, G. (1997b). Na,K‐ATPase: Increases in alpha1‐messenger RNA and decreases in alpha3‐messenger RNA levels in aging rat cerebral cortex. Neuroscience, 78(1), 7–11. [DOI] [PubMed] [Google Scholar]
- Chauhan, N. B. , & Siegel, G. J. (1996). In situ analysis of Na, K‐ATPase alpha1‐ and alpha3‐isoform mRNAs in aging rat hippocampus. Journal of Neurochemistry, 66(4), 1742–1751. [DOI] [PubMed] [Google Scholar]
- de Carvalho Aguiar, P. , Sweadner, K. J. , Penniston, J. T. , Zaremba, J. , Liu, L. , Caton, M. , … Ozelius, L. J. (2004). Mutations in the Na+/K+ ‐ATPase alpha3 gene ATP1A3 are associated with rapid‐onset dystonia parkinsonism. Neuron, 43(2), 169–175. 10.1016/j.neuron.2004.06.028 [DOI] [PubMed] [Google Scholar]
- Demos, M. K. , van Karnebeek, C. D. , Ross, C. J. , Adam, S. , Shen, Y. , Zhan, S. H. , … FORGE Canada Consortium . (2014). A novel recurrent mutation in ATP1A3 causes CAPOS syndrome. Orphanet Journal of Rare Diseases, 9, 15 10.1186/1750-1172-9-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov, M. , & Stimers, J. R. (2005). Neuronal function and alpha3 isoform of the Na/K‐ATPase. Frontiers in Bioscience, 10, 2373–2396. [DOI] [PubMed] [Google Scholar]
- Forbush, B., III , Kaplan, J. H. , & Hoffman, J. F. (1978). Characterization of a new photoaffinity derivative of ouabain: Labeling of the large polypeptide and of a proteolipid component of the Na, K‐ATPase. Biochemistry, 17(17), 3667–3676. [DOI] [PubMed] [Google Scholar]
- Heinzen, E. L. , Swoboda, K. J. , Hitomi, Y. , Gurrieri, F. , Nicole, S. , de Vries, B. , … Goldstein, D. B. (2012). De novo mutations in ATP1A3 cause alternating hemiplegia of childhood. Nature Genetics, 44(9), 1030–1034. 10.1038/ng.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieber, V. , Siegel, G. J. , Fink, D. J. , Beaty, M. W. , & Mata, M. (1991). Differential distribution of (Na, K)‐ATPase alpha isoforms in the central nervous system. Cellular and Molecular Neurobiology, 11(2), 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka, O. , Takikawa, Y. , & Kawagoe, R. (2000). Role of the basal ganglia in the control of purposive saccadic eye movements. Physiological Reviews, 80(3), 953–978. 10.1152/physrev.2000.80.3.953 [DOI] [PubMed] [Google Scholar]
- Howarth, C. , Gleeson, P. , & Attwell, D. (2012). Updated energy budgets for neural computation in the neocortex and cerebellum. Journal of Cerebral Blood Flow and Metabolism, 32(7), 1222–1232. 10.1038/jcbfm.2012.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. , Gan, J. , & Jonas, P. (2014). Interneurons. Fast‐spiking, parvalbumin(+) GABAergic interneurons: From cellular design to microcircuit function. Science, 345(6196), 1255263 10.1126/science.1255263 [DOI] [PubMed] [Google Scholar]
- Kawaguchi, Y. , & Kubota, Y. (1993). Correlation of physiological subgroupings of nonpyramidal cells with parvalbumin‐ and calbindinD28k‐immunoreactive neurons in layer V of rat frontal cortex. Journal of Neurophysiology, 70(1), 387–396. 10.1152/jn.1993.70.1.387 [DOI] [PubMed] [Google Scholar]
- Makinae, K. , Kobayashi, T. , Kobayashi, T. , Shinkawa, H. , Sakagami, H. , Kondo, H. , … Yanagawa, Y. (2000). Structure of the mouse glutamate decarboxylase 65 gene and its promoter: Preferential expression of its promoter in the GABAergic neurons of transgenic mice. Journal of Neurochemistry, 75(4), 1429–1437. [DOI] [PubMed] [Google Scholar]
- Malik, N. , Canfield, V. A. , Beckers, M. C. , Gros, P. , & Levenson, R. (1996). Identification of the mammalian Na,K‐ATPase 3 subunit. The Journal of Biological Chemistry, 271(37), 22754–22758. 10.1074/jbc.271.37.22754 [DOI] [PubMed] [Google Scholar]
- Mata, M. , Hieber, V. , Beaty, M. , Clevenger, M. , & Fink, D. J. (1992). Activity‐dependent regulation of Na+, K(+)‐ATPase alpha isoform mRNA expression in vivo. Journal of Neurochemistry, 59(2), 622–626. [DOI] [PubMed] [Google Scholar]
- Mitchell, A. S. , Czajkowski, R. , Zhang, N. , Jeffery, K. , & Nelson, A. J. D. (2018). Retrosplenial cortex and its role in spatial cognition. Brain and Neuroscience Advances, 2, 2398212818757098 10.1177/2398212818757098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzer, J. S. , Daly, S. E. , Jewell‐Motz, E. A. , Lingrel, J. B. , & Blostein, R. (1994). Tissue‐ and isoform‐specific kinetic behavior of the Na,K‐ATPase. The Journal of Biological Chemistry, 269(24), 16668–16676. [PubMed] [Google Scholar]
- Nagayama, S. , Takahashi, Y. K. , Yoshihara, Y. , & Mori, K. (2004). Mitral and tufted cells differ in the decoding manner of odor maps in the rat olfactory bulb. Journal of Neurophysiology, 91(6), 2532–2540. 10.1152/jn.01266.2003 [DOI] [PubMed] [Google Scholar]
- Ohnishi, T. , Yanazawa, M. , Sasahara, T. , Kitamura, Y. , Hiroaki, H. , Fukazawa, Y. , … Hoshi, M. (2015). Na, K‐ATPase alpha3 is a death target of Alzheimer patient amyloid‐beta assembly. Proceedings of the National Academy of Sciences of the United States of America, 112(32), E4465–E4474. 10.1073/pnas.1421182112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, K. , Takebayashi, H. , Ikeda, K. , Furusho, M. , Nishizawa, T. , Watanabe, K. , & Ikenaka, K. (2008). Regional‐ and temporal‐dependent changes in the differentiation of Olig2 progenitors in the forebrain, and the impact on astrocyte development in the dorsal pallium. Developmental Biology, 320(2), 456–468. 10.1016/j.ydbio.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Pelkey, K. A. , Chittajallu, R. , Craig, M. T. , Tricoire, L. , Wester, J. C. , & McBain, C. J. (2017). Hippocampal GABAergic inhibitory interneurons. Physiological Reviews, 97(4), 1619–1747. 10.1152/physrev.00007.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L. , Martin‐Vasallo, P. , & Sweadner, K. J. (1997). Isoforms of Na,K‐ATPase alpha and beta subunits in the rat cerebellum and in granule cell cultures. The Journal of Neuroscience, 17(10), 3488–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, K. S. , Bommert, K. , Szabo, G. , & Miles, R. (2007). Differential expression of Na+/K+‐ATPase alpha‐subunits in mouse hippocampal interneurones and pyramidal cells. The Journal of Physiology, 585(Pt 2), 491–505. 10.1113/jphysiol.2007.144733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewich, H. , Thiele, H. , Ohlenbusch, A. , Maschke, U. , Altmuller, J. , Frommolt, P. , … Gartner, J. (2012). Heterozygous de‐novo mutations in ATP1A3 in patients with alternating hemiplegia of childhood: A whole‐exome sequencing gene‐identification study. Lancet Neurology, 11(9), 764–773. 10.1016/S1474-4422(12)70182-5 [DOI] [PubMed] [Google Scholar]
- Scharfman, H. E. (2016). The enigmatic mossy cell of the dentate gyrus. Nature Reviews. Neuroscience, 17(9), 562–575. 10.1038/nrn.2016.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamraj, O. I. , & Lingrel, J. B. (1994). A putative fourth Na+,K(+)‐ATPase alpha‐subunit gene is expressed in testis. Proceedings of the National Academy of Sciences of the United States of America, 91(26), 12952–12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, G. M. (2004). The synaptic organization of the brain. New York: Oxford University Press. [Google Scholar]
- Shiina, N. , Yamaguchi, K. , & Tokunaga, M. (2010). RNG105 deficiency impairs the dendritic localization of mRNAs for Na+/K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. The Journal of Neuroscience, 30(38), 12816–12830. 10.1523/JNEUROSCI.6386-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull, G. E. , Greeb, J. , & Lingrel, J. B. (1986). Molecular cloning of three distinct forms of the Na+,K+‐ATPase alpha‐subunit from rat brain. Biochemistry, 25(25), 8125–8132. [DOI] [PubMed] [Google Scholar]
- Skou, J. C. , & Esmann, M. (1992). The Na,K‐ATPase. Journal of Bioenergetics and Biomembranes, 24(3), 249–261. [DOI] [PubMed] [Google Scholar]
- Sverdlov, E. D. , Monastyrskaya, G. S. , Broude, N. E. , Ushkaryov Yu, A. , Allikmets, R. L. , Melkov, … Ovchnikov Yu. A (1987). The family of human Na+,K+‐ATPase genes. No less than five genes and/or pseudogenes related to the alpha‐subunit. FEBS Letters, 217(2), 275–278. [DOI] [PubMed] [Google Scholar]
- Sweadner, K. J. , & Rael, E. (2000). The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics, 68(1), 41–56. 10.1006/geno.2000.6274 [DOI] [PubMed] [Google Scholar]
- Watakabe, A. , Komatsu, Y. , Ohsawa, S. , & Yamamori, T. (2010). Fluorescent in situ hybridization technique for cell type identification and characterization in the central nervous system. Methods, 52(4), 367–374. 10.1016/j.ymeth.2010.07.003 [DOI] [PubMed] [Google Scholar]
- Zahler, R. , Zhang, Z. T. , Manor, M. , & Boron, W. F. (1997). Sodium kinetics of Na,K‐ATPase alpha isoforms in intact transfected HeLa cells. The Journal of General Physiology, 110(2), 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


