Abstract
Microbial biofilms have become increasingly recognized as a cause of wound chronicity. There are several topical antimicrobial wound care products available for use; however, their effectiveness has routinely been demonstrated with planktonic microorganisms. There is no target reference value for antimicrobial effectiveness of wound care products in biofilm models. In addition, data on antimicrobial activity of products in biofilm models are scattered across many test methods in a variety of studies. The aim of this work is to directly compare commercial products containing the commonly used topical antimicrobial agents iodine, silver, polyhexamethylene biguanide, octenidine, hypochlorous acid, benzalkonium chloride, and a surfactant‐based topical containing poloxamer 188. Five different in vitro biofilm models of varied complexity were used, incorporating several bacterial pathogens such as Staphylococcus, Enterococcus, Streptococcus, Pseudomonas, Acinetobacter, Klebsiella, and Enterobacter. The fungal pathogens Candida albicans and Candida auris were also evaluated. A multispecies bacterial biofilm model was also used to evaluate the products. Additionally, C. albicans was used in combination with S. aureus and P. aeruginosa in a multikingdom version of the polymicrobial biofilm model. Statistically significant differences in antimicrobial performance were observed between treatments in each model and changing microbial growth conditions or combinations of organisms resulted in significant performance differences for some treatments. The iodine and benzalkonium chloride‐containing products were overall the most effective in vitro and were then selected for in vivo evaluation in an infected immunocompromised murine model. Unexpectedly, the iodine product was statistically (P > .05) no different than the untreated control, while the benzalkonium chloride containing product significantly (P < .05) reduced the biofilm compared to untreated control. This body of work demonstrates the importance of not only evaluating antimicrobial wound care products in biofilm models but also the importance of using several different models to gain a comprehensive understanding of products' effectiveness.
1. INTRODUCTION
The evidence implicating microbial biofilms as a cause for wound chronicity is substantial and growing.1, 2, 3, 4 Studies have reported that the costs of managing chronic wounds to Medicare alone is approaching $100 billion, 5 and recent consensus documents call for early and aggressive treatment of microbial biofilms in chronic wounds.6, 7 It is known that microorganisms in biofilms tolerate several orders of magnitude higher antimicrobial concentrations than when in the planktonic state, yet treatment decisions for wound care have been historically based on planktonic paradigms.8, 9, 10 There are numerous biofilm models available for evaluating antibiofilm effectiveness but correlating in vitro performance data to clinical effectiveness is difficult due to the complexity of how biofilms exist and interact with their hosts.9, 11 There are also several antimicrobial wound care products touting effectiveness in biofilms, but these claims are made in the absence of standardized methodology and a specification defining adequate performance. 9 Differences in model design and test conditions have led to conflicting laboratory data about effectiveness of antimicrobial products in biofilms, 12 so it is challenging for clinicians to predict which technologies may provide a clinical benefit. This challenge also affects the development and evaluation of new antimicrobial technologies. Several laboratory biofilm models from in vitro to in vivo have been reported in the literature, and each has benefits and limitations. Less complex models are valuable for rapidly screening large numbers of treatments and can give a decisive rank order of effectiveness. They can be more amenable to changes such as media concentration, flow rates, and the organism to be tested. However, these models use biofilms grown on artificial substrates, which do not represent wound‐like conditions. More complex models can give better representation of wound conditions; however, they tend to not be high throughput, developed with defined media and growth conditions suited to specific organisms. Some models are closed and static systems, wherein treatments can remain concentrated as compared to open and dynamic model systems where treatments may be diluted or inactivated by fluid replenishment over time. 9 Within any model, the variables are numerous, and small modifications can have profound effects on effectiveness of antimicrobial products. With the lack of a single reference method, we employed a regime of in vitro microbial biofilm models, both newly developed and adapted from others' publications, to test antimicrobial performance of commercially available wound care products.
The research described here evaluated seven commercially available topical wound gels representing some of the most commonly used antimicrobial agents. The models selected each lend themselves to addressing specific questions and areas of need during the development or evaluation process. For example, early discovery work requires a model that is high throughput where hundreds of candidates can be evaluated efficiently. However, this model may require biofilms to be grown as a single species on an artificial substrate, which is not reflective of the polymicrobial biofilms associated with the tissue of a chronic wound. The value of a simple, high‐throughput model lies in its ability to filter out viable candidates or gain understanding of how a product ranks against its peers in antimicrobial performance in biofilms. Each subsequent model either evaluated the product in a different condition or added complexity in growth conditions to allow for elimination of nonviable solutions or gain resolution between products' antimicrobial performance. In addition to evaluating performance of each topical wound gel in various model systems, manipulations of several biofilm models were examined.
2. MATERIALS AND METHODS
2.1. Wound care products and evaluation methodology
Leading wound gel products representing the most commonly used antimicrobials or presenting claims affecting biofilm were evaluated for effectiveness in microbial biofilms: BlastX Antimicrobial Wound Gel (Next Science LLC, Jacksonville, Florida) containing benzalkonium chloride (BAC), Iodosorb Cadexomer Iodine Gel (Smith & Nephew Medical Ltd, Hull, England) containing cadexomer iodine (CI), Prontosan Wound Gel X (BBraun Medical, Inc., Melsunge, Germany) containing polyhexamethylene biguanide (PHMB), Octenilin Wound Gel (Schülke & Mayr GmbH, Norderstedt, Germany) containing octenidine (OCT), SilvaSorb Silver Antimicrobial Wound Gel (Medline Industries, Inc., Mundelein, IL) containing silver (AG), Anasept Antimicrobial Skin & Wound Gel (Anacapa Technologies, Inc., San Dimas, CA) containing hypochlorous acid (HCA), and PluroGel Burn and Wound Dressing (Medline Industries, Inc., Northfield, Illinois) containing poloxamer 188, a nonionic surfactant (POL). Equivalent volumes of each product were evaluated side by side for antimicrobial effectiveness in a variety of biofilm models using different microbial species. The microbial species and strains used are described in Table 1. Log reduction values (LRV) were calculated as follows: LRV = Average log(surviving CFU/treated biofilm) − average log(CFU/nontreated biofilm). At least three replicates were tested for each condition in each experiment. Statistically significant differences between treatment groups for the in vitro models were determined by one‐way ANOVA analysis of log survival values using Tukey's pairwise comparisons with P values of less than .05 considered statistically significant.
Table 1.
Microbial strains used in these studies
| Species and strain | Source | Assays performed in this study |
|---|---|---|
| P. aeruginosa ATCC 15442 | ATCC | Colony biofilm, modified drip flow |
| A. baumanii ATCC BAA‐747 | ATCC | Colony biofilm |
| K. pneumoniae ATCC BAA‐1705 | ATCC | Colony biofilm |
| E. cloacae ATCC 35549 | ATCC | Colony biofilm |
| S. aureus 15981 | Clinical isolate (Valle et al., 2003. Mol Micro (48(4) 1075‐1087) | Colony biofilm, modified drip flow |
| E. faecalis ATCC 51299 | ATCC | Colony biofilm |
| S. pyogenes ATCC 49 399 | ATCC | Colony biofilm |
| C. albicans ATCC 10231 | ATCC | Colony biofilm |
| C. auris ATCC B11903 | ATCC | Colony biofilm |
| C. albicans SC3514 | Disseminated candidiasis isolate (Gillum et al., 1984. Mol Gen Genetics, 198(2):179‐82) | Multikingdom Lubbock |
| S. aureus Xen29 | Perkin Elmer | Mouse infection |
| P. aeruginosa PAO1 | Laboratory strain derived from wound isolate (Holloway 1955. J Gen Micro, 13(572‐581), (generous gift from Greg Schulz, University of Florida) | Multispecies Lubbock, multikingdom Lubbock |
| S. aureus HCMC 6‐1 | Wound isolate, 3M | Multispecies Lubbock, multikingdom Lubbock |
2.2. Colony biofilm model
The colony biofilms were grown as described in the study by Anderl et al 13 with modifications. Overnight cultures were prepared in trypticase soy broth (TSB) (Becton Dickinson, Sparks, Maryland) for bacteria and on Sabaraoud's dextrose agar (SDA) (Becton Dickinson) for yeasts. The bacterial cultures were diluted 1:100 in phosphate buffered water (PBW) (3M, St. Paul, Minnesota) and the yeast cultures were prepared to a MacFarland Turbidity Standard of 4.0 (MTS) (Remel, Lenexa, Kansas) in PBW and not diluted further. Sterile 0.2 μm polycarbonate membranes (Whatman, Florham Park, New Jersey) were placed onto TSA plates for bacteria and SDA plates for yeast. These plates were prepared as per the manufacturer's instructions as the 100% nutrient condition. About 10 μL of the diluted bacteria or yeast suspension was spotted to the center of the membrane. Plates were placed in a 35 ± 2°C incubator for bacteria and 30°C for yeast for 24 hours to grow biofilms. The membranes containing biofilms were then transferred to fresh TSA or SDA plates. A sterile silicone ring was placed around each biofilm and then 200 μL of treatment gel was placed onto biofilms within the ring to hold the treatment in place. Plates were then returned to the same incubation conditions for another 24 hours. At the end of treatment, each membrane, with biofilm and treatment, was transferred to 10 mL of Dey Engley (DE) neutralizer broth (Hardy Diagnostics, Santa Maria, California) in a sterile 50 mL centrifuge tube. The tubes were then sonicated in a water bath for 1 minute and vortexed at maximum speed for 3 minutes. Samples were then serially diluted 1:10 and plated onto Petrifilm Aerobic Count Plates (3M, St. Paul) for bacteria and SDA for yeasts. Plates were incubated at 35 ± 2°C for 48 hours and then enumerated. For modified nutrient conditions (2% TSA), TSB was diluted to 2% of the manufacturer's recommended concentration and combined with 1.5% agar (Becton Dickinson). Biofilm growth and treatment was then performed on 2% TSA plates.
2.3. Modified drip flow reactor model
Attributes of the modified drip flow biofilm reactor models published by Lipp et al 14 and Woods et al 15 were adapted for these studies. Cellulose pads were adhered to glass slides in the drip flow reactor (Biosurface Technologies, Bozeman, Montana) and the assembly was autoclaved. Overnight cultures were prepared in TSB and then diluted 1:100 in PBW. Sterile 0.2 μm polycarbonate membranes were placed onto TSA plates and then 10 μL of diluted bacteria suspension was spotted to the center of the membrane. The plates were then incubated for 4 hours at 35 ± 2°C before the membranes were transferred to the cellulose pads in the reactor and the entire assembly was placed into a 35 ± 2°C incubator. Each channel was then fed with a separate syringe of dilute brain heart infusion broth prepared at 20% or 40% of manufacturer's recommended concentration (BHI; Hardy Diagnostics), and delivered with a syringe pump at 0.25 mL/hr. Flow was initiated, and biofilms were grown for 24 hours. A sterile silicone ring was placed around each biofilm and 200 μL of a treatment gel was placed onto each biofilm as flow was continued for another 24 hours. At the end of treatment, biofilms were recovered and enumerated as in the colony biofilm model. BHI was prepared at 20% and 40% of the manufacturer's recommended concentration. BHI was chosen over TSB because it is a more complex medium with the addition of beef heart and calf brains.
2.4. Porcine explant model
The biofilm model published by Phillips et al 16 was adapted for use in these studies. 3M's animal research program complies with the Animal Welfare Act and follows recommendations in the Guide for the Care and Use of Laboratory Animals. All animal studies were reviewed and approved by 3 M's Institutional Animal Care and Use Committee (IACUC), including acquisition of skin explants from Yucatan mini‐pigs. Explants were prepared and sterilized, and gentamicin (Sigma‐Aldrich Corp, Milwaukee, Wisconsin) soft agar (GSA) was prepared as previously described. 15 A culture of P. aeruginosa PAO1 was prepared by inoculating TSB and incubating overnight at 35 ± 2°C. The culture was diluted 1:100 in fresh TSB and returned to the incubator for 4 hours. About 10 μL of this culture was spotted onto the explants, which had been placed onto GSA plates before incubation for 72 hours at 35 ± 2°C. After the biofilm formation, the explants were submerged into TSB + 200 μg/mL gentamicin and incubated at 35 ± 2°C for 24 hours. The medium was then aspirated, and each explant was washed three times in in phosphate buffered saline (PBS) + 5 ppm Tween 20 (Thermo Scientific, Rockford, Illinois). The explants were then transferred to a 24‐well plate (Thermo Scientific ) containing GSA in each well, and then 500 μL of each treatment gel was applied. The plate was incubated at 35 ± 2°C for 24 hours after which the explant and treatment were transferred to 20 mL of DE broth. Samples were sonicated in a water bath for 1 minute, then vortexed at maximum speed for 3 minutes, serially diluted 1:10, plated onto 3M Petrifilm Aerobic Count Plates, incubated at 35 ± 2°C for 48 hours, and then enumerated.
2.5. Lubbock multispecies biofilm model
The Lubbock biofilm model is a biofilm growth method published by Sun et al 17 and was adapted as follows. Cultures of S. aureus HCMC 6, E. faecium ATCC 700221, and P. aeruginosa PA01 were prepared in TSB and incubated overnight at 35 ± 2°C and cell density was estimated by comparison with MTS 0.5 to 4.0. For approximately equal cell densities, an inoculum ratio was then determined and prepared by combining 300 μL of S. aureus and P. aeruginosa with 1 mL of E. faecium overnight cultures. The growth medium was prepared by combining fresh porcine plasma, lysed porcine red blood cells, and Bolton broth (Oxoid, Basingstoke, Hants, UK) at a ratio of 50:5:45. Porcine blood was collected under a protocol approved by 3 M' IACUC. About 3 mL of the growth medium was transferred into a sterile, round bottom, culture tube. The 20 μL of the inoculum was added to the broth, and the pipette tip was ejected into the tube for biofilm attachment and growth. The tubes were then incubated at 35 ± 2°C for 18‐20 hours shaking at 200 RPM. The biofilm was removed from the pipette tip with a sterile forceps and transferred to a graduated microcentrifuge tube where it was centrifuged 10 seconds at 10 000 RPM to separate the solid biofilm from excess liquid. The 500 μL of the gel treatment was spread evenly across the bottom of triplicate wells in a sterile 24‐well plate. Biofilms were submerged into the gel in the wells and incubated for 24 hours at 35 ± 2°C. Untreated controls were kept moist by adding 100 μL of PBS. The entire well contents were recovered into 20 mL of DE neutralizer, sonicated for 1 minute, and vortexed at maximum speed for 3 minutes. Samples were serially diluted 1:10 in PBW and plated on Columbia nalidixic acid agar (CNA) (Hardy Diagnostics) to select for gram positive organisms, and cetrimide agar (Hardy Diagnostics) to select for Pseudomonas. On CNA, Staphylococcus and Enterococci were differentiated based on colony morphology.
Multikingdom Lubbock (MKL) biofilms were prepared and treated similarly except that the cultures were seeded with 20 μL of a mixture of equal parts overnight cultures of P. aeruginosa strain PAO1 (about 9.7 log[CFU/ml]), S. aureus strain HCMC 6 (about 9.5 log[CFU/ml]), and C. albicans SC3514 (about 8.2 log[CFU/ml]) grown in TSB at 35 ± 2°C. Heparinized bovine plasma (Innovative Research Inc., Novi, Michigan) and laked horse blood (Remel) were used to make the growth medium. Biofilms were homogenized after treatment with an IKA T25 Ultra Turrax homogenizer at 20 000 RPM and potato dextrose agar was used to select for C. albicans.
2.6. Infected murine model
Murine studies were reviewed and approved by 3M's IACUC. A culture of S. aureus strain Xen29 (Perkin Elmer, Waltham, Massachusetts) was incubated overnight in TSB at 35 ± 2°C. The culture was then diluted 1:10 in fresh TSB and incubated at 35 ± 2°C for 60 minutes. Bacteria were washed three times with PBS and resuspended in PBS to a final concentration of about 108 CFU/mL. Athymic, nude mice were obtained from Charles River Laboratories. After administration of anesthesia, full thickness, 1 cm diameter wounds were created on the dorsum of the animals, seeded with 10 μL of the bacterial suspension, and then covered with 3M Tegaderm Transparent Dressing. Biofilms were allowed to form for 24 hours. Animals were then anesthetized, the Tegaderm Transparent Dressing was removed, and 150 μL of treatment gel was added directly to the wound of each animal. A 12 mm diameter Kendall Telfa Non‐Adherent Dressing (Covidien, Dublin, Ireland) dressing was placed on top of the gel, and then secured with 3M Tegaderm Transparent Dressing. Animals were then revived and returned to cages. Half of the animals in treatment groups with antimicrobial wound care products were anesthetized and their dressings changed after 24 hours of treatment so that they received two treatments in 48 hours. The other group received one treatment for 48 hours. At the end of the treatment phase, dressings were removed and the tissue at the base of the wound was excised and placed into a separate tube containing 10 mL of DE. Samples were vortexed at maximum speed, serially diluted 10‐fold and plated for enumeration on 3M Petrifilm Aerobic Count Plates. The mean log(CFU/tissue sample) in the treated groups was compared to the mean log(CFU/tissue sample) in the untreated, control group with Dunnett's test and P values of less than .05 were considered statistically significant.
3. RESULTS
3.1. Colony biofilm model
Nine different microorganisms were evaluated in this model (Table 1) to assess broad spectrum effectiveness. All organisms were evaluated at 100% of the media manufacturer's recommended concentration of nutrients, while S. aureus and P. aeruginosa were also challenged after growth at the 2% of manufacturer's recommended nutrient content.
In this model (Figure 1), treatment with BAC and CI resulted in greater than six‐log reduction for all organisms tested, showing the best broad spectrum antimicrobial activity. Treatment with OCT also resulted in greater than six log reduction for Gram positive bacteria but was not as effective in killing gram negative bacteria except for Enterobacter. PHMB and AG were the only other products which killed more than six logs of any organism, but for most organisms tested, these products killed less than two logs. HCA did not kill more than 1 log with exception of Acinetobacter where the LRV was 1.89. POL did not kill more than one log of any organism tested.
Figure 1.
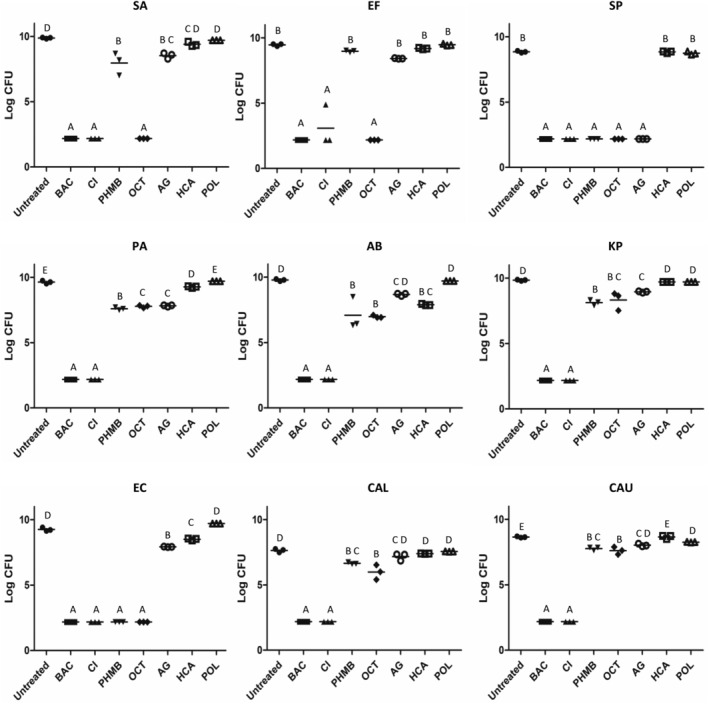
CBF Log CFU/membrane values tested at the 100% nutrient level. SA, S. aureus 15981; EF, E. faecalis 51 299; SP, S. pyogenes 49399; PA, P. aeruginosa 15442; AB, A. baumannii BAA‐747; KP, K. pneumoniae BAA‐1705; E. cloacae 35549; CAL, C. albicans 10231; CAU, C. auris B11903. Treatments that share the same letter are not significantly different (P < .05)
The cell density of the biofilms grown on 2% or 100% nutrient were all within 9.4‐9.9 log CFU/sample before treatment for P. aeruginosa and S. aureus. Nutrient concentration of biofilm growth media affected antimicrobial performance for some products. When the nutrient concentration was increased from 2% to 100%, there was a significant drop in antimicrobial effectiveness for AG in S. aureus (2% LRV = 3.39 ± 0.22; 100% LRV = 1.35 ± 0.13; P = .001) and for HCA in P. aeruginosa (2% LRV = 6.95 ± 0.49; 100% LRV = 0.68 ± 0.04; P < .001).
3.2. Modified drip flow reactor model
The modified biofilm drip flow method was used to grow biofilms for subsequent challenge with antimicrobial wound care gels. The treatments were used to challenge both S. aureus and P. aeruginosa biofilms grown at nutrient concentrations of 20% and 40% of the manufacturer's recommended concentration of BHI. For S. aureus biofilms grown at 40% nutrient concentration (Figure 2), a range of log reduction values were observed for the various products tested. POL and HCA were significantly less effective than all other treatments in this condition except for PHMB. BAC and CI killed P. aeruginosa grown in the 40% nutrient condition (Figure 2) to the limit of detection and LRVs for these products were significantly greater than for the other products, which were not significantly different from each other.
Figure 2.
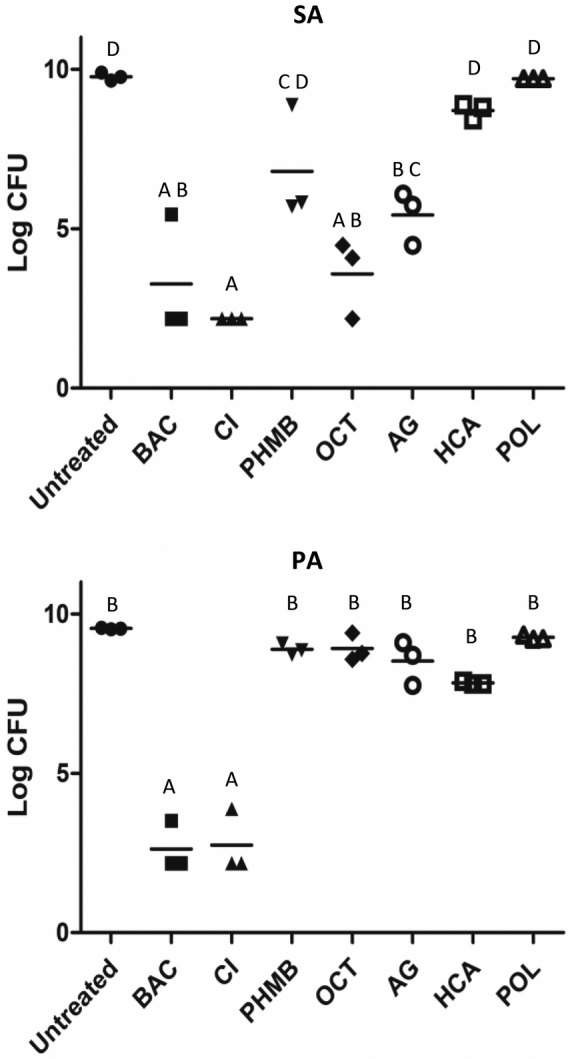
Log CFU/membrane values of SA, S. aureus 15981 and PA, P. aeruginosa 15442 tested in the modified drip flow reactor with 40% nutrient. Treatments that share the same letter are not significantly different (P < .05)
Only HCA was statistically, significantly less effective (P = .008) at killing P. aeruginosa biofilms grown at 40% nutrient concentration (LRV = 1.71 ± 0.03) compared to the 20% nutrient growth condition (LRV = 6.36 ± 0.95). Therefore, lowering the nutrient concentration in growth medium had no statistically significant effect on antimicrobial effectiveness with this exception for the modified drip flow model.
3.3. Porcine explant model
Biofilms were grown on porcine skin explants and then challenged with antimicrobial wound care gels. The average total bacteria recovered from the explants after day 3 was 8.6 log CFU/tissue explants (n = 3), and after the 24 hour antibiotic treatment to reduce planktonic bacteria, it was reduced to 7.8 log CFU/tissue explant (n = 3). BAC and CI were the only treatments that killed at least an average of four logs of bacteria in this model (Figure 3). CI, BAC, and AG were significantly more effective than the other treatments with CI being significantly more effective than AG.
Figure 3.
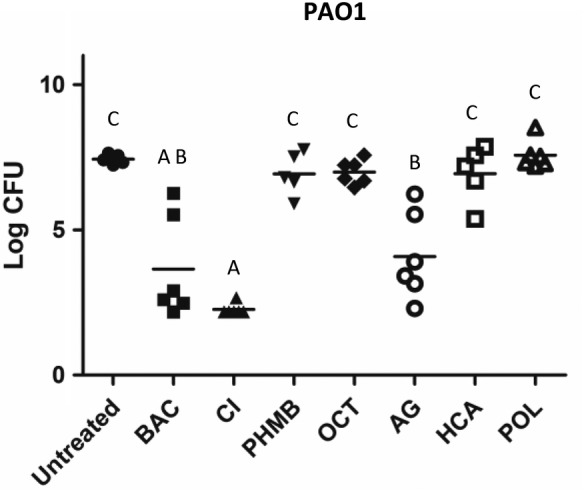
Log CFU/explant values of P. aeruginosa PAO1 tested in the porcine explant model. Treatments that share the same letter are not significantly different (P < .05)
3.4. Lubbock Multibacterial Biofilm Model
The Lubbock biofilm model was grown as described previously (Sun et al) 17 with modifications and treated with antimicrobial gel products. BAC and CI killed more than four logs of S. aureus in the biofilms, more than any other treatment (Figure 4). CI killed more E. faecium than any other treatment, whereas all other treatments killed a similar number of E. faecium with the exception of HCA, which supported some additional growth of E. faecium. P. aeruginosa was especially susceptible to antimicrobial treatment in this model, with all treatments except for OCT and HCA killing more than four logs of P. aeruginosa.
Figure 4.
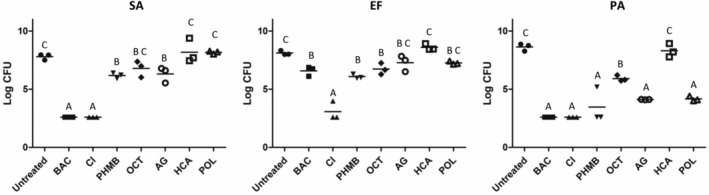
Log CFU/biofilm values of LMBM multispecies biofilm containing SA, S. aureus HCMC 6‐1, EF, E. faecium 700 221, and PA, P. aeruginosa PAO1. Treatments that share the same letter are not significantly different (P < .05)
3.5. Multikingdom Lubbock Model
The Lubbock biofilm model was modified to include P. aeruginosa, S. aureus, and C. albicans to mimic the mixed bacterial and fungal communities found in biofilms in some chronic wounds. Treatment with BAC and CI resulted in a statistically, significantly larger log reduction for all three microbial species compared to other treatments (Figure 5). However, S. aureus was not particularly susceptible to killing by any treatment, with BAC and CI killing less than two logs.
Figure 5.

Log CFU/biofilm values of MKL multispecies biofilm containing SA ‐ S. aureus HCMC 6‐1, PA, P. aeruginosa PAO1, and CA, Candida albicans SC3514. Treatments that share the same letter are not significantly different (P < .05)
3.6. Murine Model
Biofilms were grown in full thickness wounds on the dorsum of athymic, nude mice for 24 hours before treatment. Mice were treated with either CI or BAC for 48 hours and the mice received one or two treatments during that time period. A control group of infected mice was also left untreated except for covering the wound with a nonantimicrobial dressing. For both treatment schedules, BAC‐treated mice had statistically, significantly less bacterial counts in their wound bed tissue at the end of the study compared to the control group, while CI‐treated mice did not (Figure 6).
Figure 6.
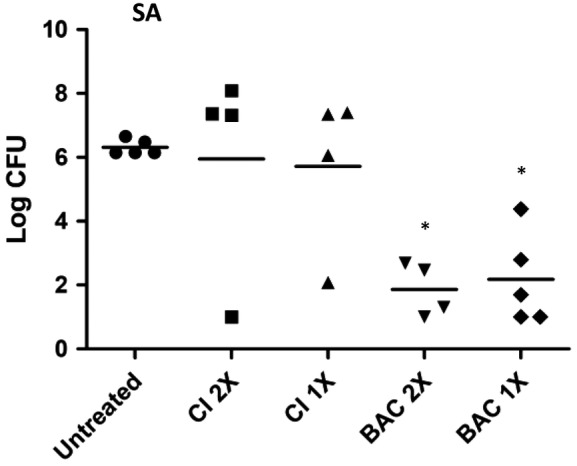
Log CFU/excision values of SA—S. aureus Xen29 biofilms in nude mice. For 2×, treatments were applied at T0 and T24 hours, then recovered at 48 hours. For 1×, treatments were applied at T0, then recovered at 48 hours. *Treatments are significantly different from untreated control (P < .05)
4. DISCUSSION
With the lack of standard biofilm methodologies for evaluating antimicrobial effectiveness of wound care products, it has been left up to the manufacturer or independent researchers to select appropriate models and the parameters within which to run them. 9 With numerous models described in the literature and each possessing multiple variables that are open to manipulations, it becomes nearly impossible to compare effectiveness data from different sources. The primary goal of this work was to evaluate representative products containing the most commonly used topical antimicrobials side‐by‐side in a suite of microbial biofilm models varied in complexity. In addition, several biofilm models were subjected to minor manipulations in biofilm growth conditions or combination of organisms in order to examine the effect on antimicrobial performance of the tested products. The products tested contain the antimicrobial agents iodine, silver, PHMB, octenidine, hypochlorous acid, benzalkonium chloride, or a surfactant‐based topical containing poloxamer 188. Iodine and silver kill microbes by interfering with cellular processes. PHMB, octenidine, and benzalkonium chloride are cationic molecules that disrupt cell membranes. Hypochlorous acid interferes with cellular process and has an effect on cell membranes. 18 Poloxamer 188 is a surfactant with no antimicrobial activity. 19
Multiple microbial species were evaluated among the models with strains of S. aureus and P. aeruginosa being tested prevalently because of their relevance in wound infections. Both organisms have some of the highest incidence rates in a variety of wound types.20, 21, 22 The complexity of each biofilm model was considered, and testing started at the most simple model (single species colony biofilm; CBF) and subsequently moved to increasingly complex models to address the limitations identified in the former models. The amount of treatment used in each model was dictated by the conditions of the model. For example, a 200 μL dose in the CBF was sufficient to cover the approximate 5 mm diameter biofilm. However, the 12 mm diameter explant in the PEM is entirely covered in biofilm and required a larger treatment volume. In contrast, a smaller volume was used in the MM to prevent excess treatment from migrating onto the periwound. This highlights the difficulty of trying to compare results between models and underscores the need to evaluate treatments in multiple models, side by side with identical conditions.
Every model evaluated used biofilms that were established for minimally 24 hours prior to applying treatment. This allowed for evaluation of disruption and killing of the biofilm microorganisms. Some of the existing data describe applying treatments immediately after or within a few hours of inoculation. Organisms in such studies are planktonic and have not yet formed a biofilm. Therefore, evaluations conducted as such examine prevention of biofilm formation.14, 23 It is likely that biofilm prevention is easier to achieve than kill of microorganisms in biofilms which have already been formed and differentiation of products tested in biofilm prevention models might be difficult to observe.
The CBF model is suitable for high throughput screening of numerous antimicrobial candidates in multiple species, which is important in early product development. A recent study of P. aeruginosa fitness in wound infections revealed that genetic determinants of fitness in a mouse infection model correlated with determinants of fitness in colony biofilm growth conditions. 24 This result suggests that the bacteria in the colony biofilm model may employ similar strategies for growth and survival as in the mouse wound model. However, the colony biofilm model is an assay that is closed and static.9, 11 All of the actives investigated are water soluble and can leach out of their carrier gels. They are likely to be diluted by wound fluid and potentially deactivated by nonspecific binding of macromolecules in the wound environment. Thus, the MDFR was employed which is an open and dynamic model.9, 11 It provides the additional challenge of constant medium flow, providing nutrients to the biofilm and providing molecules, which could potentially interact with and deactivate the antimicrobial agents. In addition, the constant flow of liquid through the system could dilute the antimicrobial products. It can be conceded that in any model such as CBF and MDFR, where the biofilm density is >9 log CFU/sample, is not representative of the microbial density for a wound biofilm observed in clinic. However, by making the test this stringent, one can easily discern which products have the best effectiveness when equal volumes of each product are used.
One limitation to CBF and MDFR models is that the biofilms are grown on an artificial substrate. Using the porcine explant model mitigates this limitation by evaluating antimicrobial product effectiveness in a biofilm grown on tissue. This model offers an additional challenge by employing an antibiotic treatment step to eliminate the planktonic bacteria, which are more susceptible to antimicrobial agents and create mature, antibiotic tolerant biofilms 16 such as those seen in clinical samples. This model requires several replicates per antimicrobial evaluated and takes 10 consecutive days to perform one assay. The complexity and length of time required to carry out this assay highlight the importance of employing more streamlined screening model systems such as CBF and MDFR for screening product candidates.
Wound biofilms are likely to be polymicrobial20, 21, 22 and testing antimicrobial products in a polymicrobial biofilm model may provide better indication of clinical effectiveness than single species models. There are several polymicrobial model options described in the literature. We chose the Lubbock multispecies biofilm because the fibrous biofilm matrix of the Lubbock biofilms provided differentiation from the biofilm models already employed in our studies. Recent examination of clinical samples has demonstrated that fungal species are also likely to be abundant in biofilms in chronic wounds. 25 The multikingdom modification of the Lubbock biofilm model allows for further assessment of broad‐spectrum effectiveness of antimicrobial wound care products. Interestingly, in the multispecies bacterial Lubbock biofilm model, several treatments including BAC, CI, PHMB, AG, and POL were able to kill over four logs of P. aeruginosa. In contrast, in the multikingdom variation of the Lubbock biofilm model, only BAC and CI were able to kill more than four logs of the same P. aeruginosa strain. It has been previously reported that multikingdom coinfections can produce altered phenotypes. 26 Mixed species biofilms containing C. albicans and bacterial species have been studied extensively, and a wide variety of interkingdom interactions have been described. 27 Many studies have reported increased resistance of microorganisms to antimicrobials in the presence of mixed species biofilms, including a study demonstrating increased resistance of both C. albicans and Staphylococcus epidermidis to antibiotics when the two organisms are living together in a biofilm. 28 Our observation of increased resistance of P. aeruginosa in the mixed kingdom biofilm model suggests that modification of the microbial community similarly affects resistance of P. aeruginosa to broad spectrum antimicrobial agents. BAC, like PHMB and OCT, is a cationic agent which disrupts microbial cell membranes. CI, like HCA and AG, interfere with the cell processes. The in vitro effectiveness of BAC and CI as compared to the other treatments suggests that the mechanism of action of the antimicrobials plays only a partial role and it is the overall composition of the gel system that drives performance. The BAC product is hyperosmotic and contains citric acid which chelate metal ions necessary for biofilm structure. The CI product has a sustained delivery mechanism, which may put continual antimicrobial pressure on the biofilm, especially in closed systems.
Each model discussed thus far covers a wide range of key attributes but is still limited by the fact that they are in vitro models and lack a living host. The in vitro model systems described here provide a strong regime of tests to narrow the number of products or candidate materials which need to be tested in animal models. An additional intent in developing this testing regime is to be able to differentiate and advance superior, broad spectrum antimicrobial treatments thus reducing the number of animals to be used for experimentation. When the in vitro data described here were reviewed, it was clear that CI and BAC were the most effective and worthy of evaluation in vivo.
The in vivo model is carried out in immunocompromised, athymic, nude mice which allows for quick establishment of a microbial biofilm with high log(CFU/wound) counts. The in vivo model was first developed using the S. aureus strain Xen29, which contains a bioluminescence‐encoding operon on its chromosome. We used this strain for infection because its behavior in the model was well understood, but we only counted total viable colony‐forming units recovered from the tissue at the base of the wound at the end of the study. The susceptibility of the Xen29 strain to the CI‐containing and the BAC‐containing products was examined in the colony biofilm model and results were similar to those observed when we used other S. aureus strains in a similar experiment. The infected, full thickness wounds produced large amounts of exudate. CI has a dark brown appearance at application from the iodine but had turned gray/white at 24 hours post‐application. This appearance suggests that the antimicrobial iodine had already been depleted from the product at that time point. The BAC‐containing product is a white cream at application but becomes transparent at body temperature and thus no observation could be made as to its state. Mice were treated either once at the beginning of a 48 hour treatment period, or twice in that period (one treatment every 24 hours). In both cases, BAC‐treated mice had significantly lower viable bacterial counts in the tissue at the base of the wound than the untreated mice. In contrast, the viable counts of bacteria recovered from wounds in both groups of the CI‐treated mice were not statistically different than the control group. This result is different from the results observed with S. aureus in several other model systems including the colony biofilm models, the MDFR models, the Lubbock multi species biofilm model, and the Lubbock multikingdom biofilm model. In these model systems, CI and BAC did not differ in their antimicrobial performance against S. aureus. These results suggest that the in vivo growth environment or the physical conditions of the wound in the mouse biofilm model renders the S. aureus biofilm less susceptible to the CI‐containing products than BAC‐containing product.
These studies demonstrate how model design and conditions play a role in the performance of an antimicrobial treatment. It was demonstrated in CBF and MDFR models that by changing the nutrient content, effectiveness of HCA and AG can be significantly altered. The HCA‐containing product killed over six logs more P. aeruginosa when grown in reduced nutrient conditions in the CBF model and killed over four logs more P. aeruginosa in the MDFR model with reduced nutrient growth conditions. The AG‐containing product killed over two logs more S. aureus when biofilms were grown in reduced nutrient conditions in the CBF model. These observations suggest that these products may be less effective in the presence of additional host protein which is present in wound exudate and slough.
Even though cell density did not vary widely in the CBF, the visual appearance of the biofilms was remarkably different between nutrient conditions. Biofilms grown in the nutrient‐rich condition appear thicker under magnification and had stronger pigmentation. At 2% nutrient concentration, even though the cell density was similar, biofilms lacked the pigmentation observed at 100% nutrient concentration and the biofilm center was more transparent. The difference in appearance of the biofilms in varied growth conditions also suggests that the biofilm extracellular matrices may be more or less robust. It is also possible that the production of additional extracellular proteins and/or polysaccharide in the presence of more nutrients results in nonspecific binding and deactivation of antimicrobial agents, rendering them less effective than in lower nutrient growth conditions. It is also important to consider static and closed models vs open and dynamic ones and how nutrients are delivered. In the static and closed CBF model the nutrient source is finite and leached antimicrobial will not leave the system. However, in the MDFR, the nutrient source is continuous and leached antimicrobial molecules may be driven away from the biofilm. The latter maybe more representative of highly exudating wounds.
When considering the entire data set, the in vitro superiority of BAC and CI was clearly demonstrated when compared to all other treatments. In any in vitro model, no other treatment demonstrated a significantly higher LRV compared to BAC or CI. BAC and CI performed statistically the same in every instance with exception of BAC killing significantly more Candida cells than CI in MKL and CI killing significantly more Enterococci than BAC in LMBM. In the in vivo model, a statistically significant difference was not demonstrated between CI and BAC, perhaps because of the large variability in CI performance. Only BAC treatment resulted in significantly fewer recoverable bacteria in the wound tissue than in the untreated control group. As shown in Figure 6, BAC was more consistent in performance always demonstrating decreased recovered CFU. Treatment with CI however, resulted in several instances where the recovered CFU was higher than the untreated control, suggesting that when the iodine is depleted, the product may support additional growth of the S. aureus.
While the organisms chosen for these studies are relevant wound pathogens, the list of organisms was not exhaustive. The same can be said for the products chosen for evaluation. Nutrient concentration was the adjusted variable studied in CBF and MDFR. However, several other important conditions may also have impact such as medium flow rate in the MDFR, additional organism combinations in the Lubbock model, or using an immunocompetent mouse strain in the murine model.
The antimicrobial gel products evaluated in these studies varied in their performance depending on the biofilm model used and the conditions used to grow the biofilms. The present studies demonstrate the need for using varied biofilm model systems for comparing product performance of antimicrobial wound care products, and for the development of such products. While animal models are likely the most relevant systems in which to test the effectiveness of antimicrobial wound care products, it is impractical and unethical to test large numbers of products or concepts in such models. Therefore, it is essential to use a variety of test methods to narrow the number of materials to test in an animal model.
CONFLICT OF INTEREST
3M is the exclusive distributor of BlastX Antimicrobial Wound Gel.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Greg Schulz and his laboratory at the University of Florida for their gift of the P. aeruginosa PAO1 strain and training on the porcine explant model. The authors also thank our institution's Preclinical Services Department personnel who provided their expertise in conducting the in vivo study. This study was funded by 3M.
Stoffel JJ, Kohler Riedi PL, Hadj Romdhane B. A multimodel regime for evaluating effectiveness of antimicrobial wound care products in microbial biofilms. Wound Rep Reg. 2020;28:438–447. 10.1111/wrr.12806
Funding information 3M
REFERENCES
- 1. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318‐1322. [DOI] [PubMed] [Google Scholar]
- 2. James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Rep Regen. 2008;16(1):37‐44. [DOI] [PubMed] [Google Scholar]
- 3. Percival SL, Bowler PG. Biofilms and their potential role in wound healing. Wounds. 2004;16(7):234‐240. [Google Scholar]
- 4. Bjarnsholt T, Kirketerp‐Moller K, Jensen PO, et al. Why chronic wounds will not heal: a novel hypothesis. Wound Rep Reg. 2008;16:2‐10. [DOI] [PubMed] [Google Scholar]
- 5. Landsman A, Masturzo A, Barbul A. Examining the real‐world healthcare costs of treating chronic wounds. Value Health. 2019;22:S213. [Google Scholar]
- 6. Schultz G, Bjarnsholt T, James GA, et al. Consensus guidelines for the identification and treatment of biofilms in chronic nonhealing wounds. Wound Rep Regen. 2017;25(5):744‐757. [DOI] [PubMed] [Google Scholar]
- 7. Leaper DJ, Schultz G, Carville K, Fletcher J, Swanson T, Drake R. Extending the TIME concept: what have we learned in the past 10 years? Int Wound J. 2014;9(Suppl. 2):1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167‐193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malone M, Goeres DM, Gosbell I, Vickery K, Jensen S, Stoodley P. Approaches to biofilm‐associated infections: the need for standardized and relevant biofilm methods for clinical applications. Expert Rev Anti Infect Ther. 2017;15(2):147‐156. [DOI] [PubMed] [Google Scholar]
- 10. Coenye T, Goeres D, Van Bambeke F, Bjarnsholt T. Should standardized susceptibility testing for microbial biofilms be introduced in clinical practice? Clin Microbiol Infect. 2018;24(6):570‐572. [DOI] [PubMed] [Google Scholar]
- 11. Coenye T, Nelis HJ. In vitro and in vivo model systems to study microbial biofilm formation. J Microbiol Meth. 2010;83(2):89‐105. [DOI] [PubMed] [Google Scholar]
- 12. Bourdillon K. Dressings and biofilms: interpreting evidence from in vitro biofilm models. Wounds Int. 2016;7(1):9‐15. [Google Scholar]
- 13. Anderl J, Franklin M, Stewart P. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44(7):1818‐1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lipp C, Kirker K, Agostinho A, James G, Stewart P. Testing wound dressings using an in vitro wound model. J Wound Care. 2010;19(6):220‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woods J, Boegli L, Kirker KR, et al. Development and application of a polymicrobial, in vitro, wound biofilm model. J Appl Microbiol. 2012;112(5):998‐1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phillips PL, Yang Q, Davis S, et al. Antimicrobial dressing efficacy against mature Pseudomonas aeruginosa biofilm on porcine skin explants. Int Wound J. 2015;12(4):469‐483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun Y, Dowd SE, Smith E, Rhoads DD, Wolcott RD. In vitro multispecies Lubbock chronic wound biofilm model. Wound RepReg. 2008;16:805‐813. [DOI] [PubMed] [Google Scholar]
- 18. Maris P. Modes of action of disinfectants. Rev Sci Tech off Int Epiz. 1995;14(1):47‐55. [DOI] [PubMed] [Google Scholar]
- 19. Patel HR, Patel RP, Patel MM. Poloxamers: a pharmaceutical excipients with therapeutic behaviors. Int J PharmTech Res. 2009;1(2):299‐303. [Google Scholar]
- 20. Bowler PG, Davies BJ. The microbiology of infected and non infected leg ulcers. Int J Dermatol. 1999;38:101‐106. [DOI] [PubMed] [Google Scholar]
- 21. Dowd SE, Sun Y, Secor PR, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8(43):1‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomsen TR, Aasholm MS, Rudkjobing VB, et al. The bacteriology of chronic venous leg ulcer examined by culture‐independent molecular methods. Wound Repair Regen. 2010;18(1):38‐49. [DOI] [PubMed] [Google Scholar]
- 23. Christiansen C, Huniche GB, Allesen‐Holm M. in vitro evaluation of a silver foam dressing with and without silicone adhesive against biofilms and a broad range of microorganisms. Proceedings of the EWMA; 2018; Krakow, Poland.
- 24. Morgan SJ, Lippman SI, Bautista GE, et al. Bacterial fitness in chronic wounds appears to be mediated by the capacity for high‐density growth, not virulence or biofilm functions. PLoS Pathog. 2019;15(3):e1007511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalan L, Loesche M, Hodkinson BP, et al. Redefining the chronic‐wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. MBio. 2016;7(5): e01058–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC Jr, Mylonakis E. Prokaryote‐eukaryote interactions identified by using Caenorhabditis elegans . Proc Natl Acad Sci U S A. 2008;105(38):14585‐14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lohse MB, Gulati M, Johnson AD, Nobile CJ. Development and regulation of single‐ and multi‐species Candida albicans biofilms. Nat Rev Microbiol. 2018;16(1):19‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adam B, Baillie GS, Douglas LJ. Mixed species biofilms of Candida albicans and Staphyloccocus epidermidis. J Med Microbiol. 2002;51(4):344‐349. [DOI] [PubMed] [Google Scholar]


