Abstract
Molecular testing to select the appropriate targeted and standard of care therapies is essential for managing patients with colorectal cancer (CRC). The Japanese Society of Medical Oncology previously published clinical guidelines for molecular testing in CRC. In the third edition published in 2018, RAS and BRAF V600E mutations should be tested prior to first‐line chemotherapy to assess the benefit of anti–epidermal growth factor receptor (EGFR) antibody therapy in patients with unresectable CRC. Microsatellite instability (MSI) testing was recommended in patients with curatively resected stage II CRC because deficient mismatch repair is associated with low risk of recurrence. MSI testing was also recommended in patients with CRC suspected to be Lynch syndrome. The main aim of this fourth edition is to reflect recent advances in comprehensive genomic profiling (CGP) tests and liquid biopsy. Here, CGP tests performed on tumor tissues are strongly recommended to assess the benefit of molecular targeted drugs in patients with CRC. Circulating tumor DNA (ctDNA)‐based CGP tests are also proposed. ctDNA testing is recommended to determine the optimal treatment based on the risk of recurrence for curatively resected CRC and evaluate the suitability and monitor the therapeutic effects of anti–EGFR antibodies in patients with unresectable CRC. While both MSI testing and immunohistochemistry are strongly recommended to determine the indication of immune checkpoint inhibitors in patients with unresectable CRC, next‐generation sequencing‐based tests are weakly recommended because these tests have not been validated in clinical trials.
Keywords: circulating tumor DNA, colorectal cancer, comprehensive genomic profiling, guideline, microsatellite instability
Update to the molecular testing guideline for colorectal cancer defined by Japanese Society of Medical Oncology. The appropriate timing of each test and degree of recommendation are summarized.
1. INTRODUCTION
Colorectal cancer (CRC) is the most common cancer in Japan. Improved molecular understanding and development of molecular targeted therapies have led to significant improvements in care for CRC patients. The Japanese Society of Medical Oncology (JSMO) is dedicated to providing guidance regarding the proper use of genomic testing for the management of CRC through the publication and dissemination of clinical practice guidelines. The first JSMO guidelines, the “Japanese Guidelines for Testing of the KRAS Gene Mutation in Colorectal Cancer, First Edition,” was published in 2008, and them updated in 2014, with the publication of the “Japanese Society of Medical Oncology Clinical Guidelines: RAS (KRAS/NRAS) Mutation Testing in Colorectal Cancer Patients, Second Edition. 1 ” Both contributed to the proper use of KRAS and RAS mutation testing in clinical practice. The third edition, published in 2018, recommended BRAF V600E mutation testing prior to the initiation of first‐line therapy for patients with unresectable advanced or recurrent CRC, and MMR deficiency testing for cases suspected of having Lynch syndrome. 2
Since then, not only have tests for the BRAF V600E mutation and MMR deficiency received insurance coverage but there has also been approval of comprehensive genomic profiling (CGP) testing based on the rapid implementation of precision medicine. Furthermore, somatic gene testing by analyzing circulating tumor DNA (ctDNA) and ctDNA‐based profiling tests has also progressed rapidly. Therefore, the JSMO established a working group in November 2018 to revise the guidelines and published the revised Japanese version of guidelines (fourth edition) in October 2019 following the peer‐review process with an external review committee and public comments from JSMO members. 3 The updated guidelines set 10 basic requirements for molecular testing for CRC treatment. The degree of recommendation for each requirement was determined through votes by the working group members based on the evidence for each test and the expected balance between advantages and disadvantages for patients when the testing was performed (Table 1).
Table 1.
Degrees of recommendation and decision criteria
| Degree of recommendation | Decision criteria |
|---|---|
| Strong recommendation | Sufficient evidence and the benefits of testing outweigh the losses |
| Recommendation | Evidence considering the balance between benefits and losses |
| Expert consensus opinion | Consensus obtained although not enough evidence and information |
| No recommendation | Not recommended owing to the lack of evidence |
Sufficient evidence, consistent evidence from randomized control trials (RCT) without important limitations or exceptionally strong evidence from observational studies; evidence, evidence from RCT with important limitations or strong evidence from observational studies; consensus, evidence for at least 1 critical outcome from observational studies, case series, or RCT with serious flaws or indirect evidence.
2. BASIC REQUIREMENTS OF MOLECULAR TESTING FOR COLORECTAL CANCER TREATMENT (Table 2 and Figure 1)
2.1. RAS mutation testing
RAS mutation testing is recommended prior to first‐line chemotherapy to assess the benefit of anti–EGFR antibody therapy in patients with unresectable CRC. [Strong recommendation]
Anti–EGFR antibody therapy is not effective in patients with KRAS exons 2, 3 and 4 and NRAS exons 2, 3 and 4 mutations, regardless of the type of anti–EGFR antibody (cetuximab or panitumumab), treatment lines, use of combined chemotherapy or type of chemotherapy. While the addition of anti–EGFR antibody significantly improved the overall survival (OS) and progression‐free survival (PFS) in patients with RAS wild‐type left‐sided colon cancer, anti–EGFR antibody therapy was not beneficial in those with right‐sided colon cancer. The “Japanese Society for Cancer of the Colon and Rectum Guidelines 2019 for the Treatment of Colorectal Cancer” and the “Pan‐Asian Adapted ESMO Consensus Guidelines for the Management of Patients with Metastatic Colorectal Cancer” stratified the treatment strategies based on primary tumor location and RAS mutation status. 4
RAS mutation testing can determine the optimal perioperative chemotherapy based on the presumed recurrence risk in patients with resectable CRC. [Expert Consensus Opinion]
Table 2.
Summary of basic requirements
| Recommendation | |
|---|---|
| RAS mutation testing | |
| RAS mutation testing is recommended prior to first‐line chemotherapy to assess the benefit of anti–EGFR antibody therapy in patients with unresectable CRC. | Strong recommendation |
| RAS mutation testing can determine the optimal perioperative chemotherapy based on the presumed recurrence risk in patients with resectable CRC. | Expert Consensus Opinion |
| BRAF mutation testing | |
| BRAF V600E mutation testing is recommended prior to first‐line chemotherapy to determine the optimal treatment based on the prognosis of patients with unresectable CRC. | Strong recommendation |
| BRAF V600E mutation testing is recommended to determine the optimal perioperative chemotherapy based on the presumed recurrence risk in patients with resectable CRC. | Recommendation |
| BRAF V600E mutation testing is recommended to help diagnose Lynch syndrome. | Recommendation |
| Testing for mismatch repair deficiency | |
| MMR deficiency testing is recommended to evaluate the benefit of immune checkpoint inhibitors in patients with unresectable CRC. | Strong recommendation |
| MMR deficiency testing is recommended to assess the risk of recurrence and to stratify optimal perioperative chemotherapy in patients with resectable CRC. | Recommendation |
| MMR deficiency testing is recommended to screen for Lynch syndrome. | Strong recommendation |
| The following methods are recommended when assessing for MMR deficiency: | |
| MSI testing | Strong recommendation |
| IHC testing | Strong recommendation |
| NGS‐based testing | Recommendation |
| Next‐generation sequencing‐based comprehensive genomic profiling tests | |
| Comprehensive genomic profiling tests are recommended to assess the benefits of molecular targeted drugs in patients with unresectable CRC. | Strong recommendation |
| Liquid biopsy | |
| ctDNA testing is recommended to determine the optimal perioperative chemotherapy based on the presumed recurrence risk of patients with resectable CRC. | Recommendation |
| ctDNA testing is recommended to evaluate the suitability of and to monitor the therapeutic effects of anti–EGFR antibody therapy in patients with unresectable CRC. | Recommendation |
| ctDNA‐based comprehensive genomic profiling tests are recommended to assess the benefits of molecular targeted drugs for patients with unresectable CRC. | Recommendation |
| Angiogenic factors | |
| Measurement of VEGF‐D level is performed to identify the appropriate angiogenesis inhibitors for patients with unresectable CRC. | Expert Consensus Opinion |
| Samples for molecular testing | |
| FFPE tissue is suitable for genomictesting of somatic mutations. It is recommended to confirm that the samples have an adequate amount of tumor cells and expect sufficient quality of nucleic acids by assessing the matched reference hematoxylin and eosin stained slides. Selection of FFPE samples, decision on the need for macrodissection, and assessment of tumor cell content should be performed by a pathologist. | Strong recommendation |
| In ctDNA testing, use of collection tubes and the preservation and adjustment of plasma after blood collection should be performed in accordance with the manufacturer’s instructions. | Strong recommendation |
| Quality assurance requirements for testing | |
| Genomic testing for CRC treatment should be carried out under a quality assurance system. | Strong recommendation |
Abbreviations: CRC, colorectal cancer; ctDNA, circulating tumor DNA; EGFR, epidermal growth factor receptor; FFPE, formalin‐fixed paraffin‐embedded; IHC, immunohistochemistry; MMR, mismatch repair; NGS, next‐generation sequencing; VEGF, vascular endothelial growth factor.
FIGURE 1.
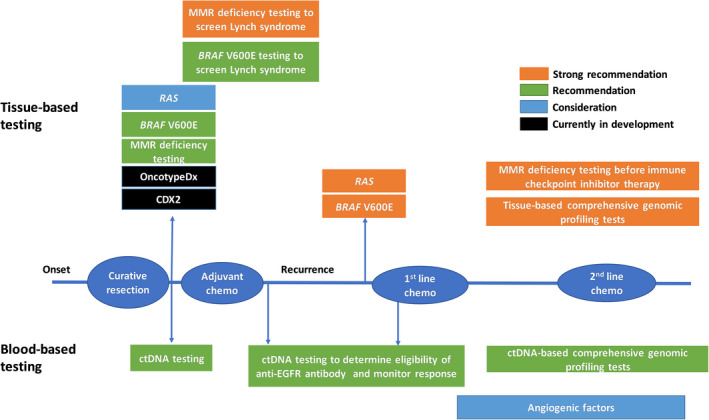
Timing for each genomic test
The benefit of cetuximab has not been shown in resectable CRC. In two phase III trials comparing FOLFOX with or without cetuximab as adjuvant therapy for curatively resected stage III colon cancer, the addition of cetuximab did not improve the recurrence‐free survival (RFS) or OS, even in patients with wild‐type KRAS exon 2. 5 , 6 In contrast, patients with KRAS mutations have significantly worse outcomes. Among patients with resected metastatic lesions such as liver metastases, patients with RAS mutations had shorter RFS and OS than those with RAS wild‐type. 7 Association was also reported between KRAS mutation and lung metastasis after curative resection in patients with stage II/III colon cancer. 8
2.2. BRAF mutation testing
BRAF V600E mutation testing is recommended prior to first‐line chemotherapy to determine the optimal treatment based on the prognosis of patients with unresectable CRC. [Strong recommendation]
The BRAF V600E mutation is a poor prognostic factor of advanced CRC. Recent studies demonstrated favorable therapeutic effects of FOLFOXIRI with bevacizumab as the first‐line treatment for patients with advanced CRC harboring the BRAF V600E mutation. 9 In addition, the BEACON CRC phase III trial showed that the combination of BRAF inhibitor with MEK inhibitor and anti–EGFR antibody significantly improved OS and objective response compared with standard care in patients with BRAF V600E mutant advanced CRC. 10 Therefore, the BRAF V600E mutation status is important in determining the optimal first‐line regimen in patients with metastatic CRC (mCRC).
BRAF V600E mutation testing is recommended to determine the optimal perioperative chemotherapy based on the presumed recurrence risk in patients with resectable CRC. [Recommendation]
The presence of the BRAF V600E mutation is associated with poor prognosis, especially in patients with microsatellite stable (MSS) resectable CRC. In a meta‐analysis of phase III trials of adjuvant chemotherapy in patients with stage II/III CRC, the presence of the BRAF V600E mutation was a risk factor for recurrence. 11 Moreover, the influence of the BRAF V600E mutation was examined between patients with microsatellite instability‐high (MSI‐H) and those with MSS tumors. MSS/BRAF mutant was a poor prognostic factor, while MSS/BRAF wild‐type and MSI‐H/BRAF wild‐type were comparable and favorable, respectively, and MSI‐H/BRAF mutant was moderate. 12
BRAF V600E mutation testing is recommended to help diagnose Lynch syndrome. [Recommendation]
BRAF V600E mutations are dominantly observed in patients with sporadic deficient mismatch repair (dMMR) CRC. Lynch syndrome harbors germline mutations in MMR genes, while most of the sporadic dMMR CRC are caused by promoter methylation, such as of the MLH1 gene. Among dMMR tumors, Lynch syndrome can be excluded with high probability if the BRAF V600E mutation is present, especially concomitant loss of MLH1 expression.
2.3. Testing for mismatch repair deficiency
Mismatch repair deficiency testing is recommended to evaluate the benefit of immune checkpoint inhibitors in patients with unresectable CRC. [Strong recommendation]
Pembrolizumab was approved in Japan in December 2018 for the treatment of MSI‐H advanced solid tumors, including CRC, with an MSI testing kit (FALCO) as its companion diagnostic tool. For CRC, pembrolizumab is approved as a second‐line or subsequent treatment. Nivolumab also demonstrated favorable therapeutic effects in pre–treated patients with MSI‐H recurrent or metastatic CRC. 13 However, patients with MSI‐H CRC have poor prognoses, regardless of the presence of the BRAF V600E mutation. To avoid missing opportunities to use immune checkpoint inhibitors, it is recommended that MMR deficiency testing be assessed at an early stage of mCRC treatment. Notably, MMR deficiency and RAS/BRAF mutations are not mutually exclusive.
Mismatch repair deficiency testing is recommended to assess the risk of recurrence and to stratify optimal perioperative chemotherapy in patients with resectable CRC. [Recommendation]
Survival of dMMR was better than that of MSS among patients with curatively resected stage II and III CRC. However, fluoropyrimidine‐based adjuvant chemotherapy could increase the risk of recurrence for dMMR stage II colon cancer. 14 BRAF V600E mutations are more frequently observed in patients with dMMR than in those with mismatch repair‐proficient (pMMR) cancer. When BRAF V600E or KRAS exon 2 mutations are present with pMMR, the recurrence risk is significantly higher with poor prognosis in patients with stage III colon cancer. 15 Therefore, MMR deficiency testing is recommended to determine the regimen and duration of adjuvant chemotherapy stratified by the risk of recurrence in patients with resected colon cancer.
Mismatch repair deficiency testing is recommended to screen for Lynch syndrome. [Strong recommendation]
Lynch syndrome is an autosomal dominant inherited disorder caused by germline mutations in one of the MMR genes: MLH1, MSH2, MSH6 and PMS2. Lynch syndrome is a rare disease occurring in 2%–4% of Caucasian and 0.7% of Japanese patients with CRC. However, patients and their families have an increased risk of many types of malignancies. Therefore, MMR deficiency testing is strongly recommended in patients with CRC with suspected Lynch syndrome. Notably, MMR deficiency is also observed in a subset of sporadic CRC, such as tumors with hypermethylation of the MLH1 promoter.
The following methods are recommended when assessing for MMR deficiency:
| MSI testing | (Strong recommendation) |
| Immunohistochemistry (IHC) testing | (Strong recommendation) |
| Next‐generation sequencing (NGS)‐based testing | (Recommendation) |
The typical methods for evaluating MMR deficiency include MSI testing, assessing the MSI by the shift of microsatellite markers, IHC testing, assessing the expression of MMR proteins (MLH1, MSH2, MSH6 and PMS2) in cancer tissues, and NGS‐based evaluation of mismatch repair function.
Although dinucleotide markers, including the Bethesda panel, are valid for determining MSI‐low, it sometimes fails to detect MSI‐H in Lynch syndrome, caused by germline mutations in MSH6 or PMS2. In contrast, mononucleotide markers are more sensitive and specific than dinucleotide markers for the detection of MSI and are also less influenced by polymorphisms. Moreover, a mononucleotide marker panel identified patients with MSH6 deficiency at a relatively high rate (62.5%). 16 The MSI test kit (FALCO) determines the MSI status based on five mononucleotide markers.
In IHC testing, tumors without MMR deficiency express all four proteins, while proteins that correspond to the inactivated MMR genes are not expressed in patients with dMMR tumors. The results of testing showed high concordance between IHC testing and MSI testing. Currently, IHC is used in screening Lynch syndrome; however, it is expected to become a companion diagnostic assay for immune checkpoint inhibitors (ICI). In a pooled analysis of the five KEYNOTE studies and the CheckMate 142 trial, the efficacy of anti–PD‐1 antibody was demonstrated in patients with dMMR determined by IHC. 17
FoundationOne CDx detects MSI status by evaluating 95 intronic microsatellite markers, showing a more than 95% concordance rate with MSI testing and IHC. There are other algorithms to analyze MSI status, such as the MSIsensor algorithm in MSK‐IMPACT and the MOSAIC and MANTIS algorithms with whole exome sequencing. Each method uses different microsatellite markers and algorithms. Although these NGS‐based tests are found to be useful, the tests were not validated in clinical trials of ICIs.
2.4. Next‐generation sequencing‐based comprehensive genomic profiling tests
Comprehensive genomic profiling (CGP) tests are recommended to assess the benefits of molecular targeted drugs in patients with unresectable CRC. [Strong recommendation]
Patients with driver mutations receiving a matched targeted agent showed better PFS and OS than those whose tumors did not harbor druggable driver mutations. 18 The CGP test can identify rare driver abnormalities in CRC, such as NTRK fusion, HER2 amplification, BRAF non–V600E mutations and ALK translocations. Currently, CGP tests are approved for use in CRC patients with disease progression after receiving standard chemotherapy. Although off‐label drug use based on CGP results is not permitted in the Japanese healthcare system, patients harboring these mutations may be eligible for clinical trials.
2.5. Liquid biopsy
Circulating tumor DNA (ctDNA) testing is recommended to determine the optimal perioperative chemotherapy based on the presumed recurrence risk of patients with resectable CRC. [Recommendation]
Circulating tumor DNA (genetically mutated allele) has an extremely short half‐life in plasma (ie, within 2 hours). NGS‐based ctDNA analysis is being developed to assess minimal residual disease (MRD) and to monitor recurrence in cancer. Postoperative ctDNA analysis was useful for the stratification of recurrence risk independent of conventional clinicopathological risk factors. 19 Moreover, longitudinal ctDNA analysis after surgery could identify CRC recurrence earlier than radiologic imaging. Therefore, ctDNA testing for the detection of MRD is recommended to identify patients at high risk of recurrence and to optimize the treatment strategies for patients with resectable CRC.
Circulating tumor DNA testing is recommended to evaluate the suitability of and to monitor the therapeutic effects of anti–EGFR antibody therapy in patients with unresectable CRC. [Recommendation]
The fraction of KRAS‐mutated alleles quantified by digital PCR was inversely correlated with response to anti–EGFR therapy. Furthermore, the rechallenge treatment of cetuximab and irinotecan after resistance to first‐line irinotecan and cetuximab‐based therapy was effective only in patients with RAS and BRAF wild‐type mCRC confirmed by ctDNA analysis. 20 The OncoBEAM RAS CRC kit, which uses BEAMing to detect RAS mutations in ctDNA, showed an 86% concordance rate with RAS testing by tissue analysis, and the kit was approved for use in July 2019.
Circulating tumor DNA‐based comprehensive genomic profiling tests are recommended to assess the benefits of molecular targeted drugs for patients with unresectable CRC. [Recommendation]
Circulating tumor DNA‐based CGP analysis detects multiple genomic abnormalities that are associated with resistance to anti–EGFR therapy, including EGFR, KRAS, NRAS and BRAF mutations, and HER2 and MET amplifications. In addition, ctDNA‐negative patients showed a better response to rechallenge treatment with anti–EGFR antibody therapy. 21 ctDNA‐based CGP analyses were used to screen eligible patients and/or assess the efficacy of molecular targeted agents in clinical trials. For example, the reduction of allele frequency after treatment was correlated with the degree of response in a phase 1b trial of vemurafenib with irinotecan and cetuximab in patients with BRAF V600E mutant mCRC. 22
2.6. Angiogenic factors
Measurement of vascular endothelial growth factor (VEGF)‐D levels is performed to identify the appropriate angiogenesis inhibitors for patients with unresectable CRC. [Expert Consensus Opinion]
The phase III RAISE trial demonstrated that ramucirumab, a VEGF receptor 2 binding monoclonal antibody, plus 5‐fluorouracil, leucovorin and irinotecan (FOLFIRI) significantly improved OS and PFS compared with placebo plus FOLFIRI as second‐line treatment. 23 Biomarker analysis showed that VEGF‐D is a potential predictive biomarker for ramucirumab efficacy.
2.7. Samples for molecular testing
Formalin‐fixed paraffin‐embedded (FFPE) tissue is suitable for genomic testing of somatic mutations. It is recommended to confirm that the samples have an adequate amount of tumor cells and expect sufficient quality of nucleic acids by assessing the matched reference hematoxylin and eosin stained slides. Selection of FFPE samples, decision on the need for macrodissection, and assessment of tumor cell content should be performed by a pathologist. [Strong recommendation]
In ctDNA testing, use of collection tubes and the preservation and adjustment of plasma after blood collection should be performed in accordance with the manufacturer’s instructions. [Strong recommendation]
2.8. Quality assurance requirements for testing
Genomic testing for CRC treatment should be carried out under a quality assurance system. [Strong recommendation]
CONFLICT OF INTEREST
Hideaki Bando received honoraria from Taiho and Eli Lilly, and research funding from AstraZeneca, Sysmex and Taiho. Hiroya Taniguchi received honoraria from Eli Lilly, Taiho, Takeda and Chugai, and research funding from Takeda. Yu Sunakawa received honoraria from Sanofi, Taiho, Takeda, Chugai, Merck Biopharma and Yakult. Yoshinaga Okugawa received scholarship funding from Kaken Phamaceutical, Shionogi, Taiho, Takeda, Tanabe Mitsubishi and Chugai. Yutaka Hatanaka received honoraria from MSD and Pfizer, and research funding from Eisai, MSD, Konica Minolta, Sysmex, Taiho, Nichirei Bioscience, RIKEN genesis and Roche Diagnostics, and scholarship funding from Qiagen, Konica Minolta, Sakura Fintek, Sawai, Genetic Lab, Taiho, Moroo and RIKEN genesys. Kentaro Yamazaki received honoraria from Takade and Chugai. The other authors have no conflicts of interests.
ACKNOWLEDGMENTS
This work was supported by JSMO. The author is grateful to Drs Hideyuki Ishida, Katsuya Tsuchihara, Kazuto Nishio and Eishi Baba for reviewing the Japanese version of this guideline, and to Dr Saori Misima for her insightful input.
Ebi H, Bando H, Taniguchi H, et al. Japanese Society of Medical Oncology Clinical Guidelines: Molecular Testing for Colorectal Cancer Treatment, 4th edition. Cancer Sci. 2020;111:3962–3969. 10.1111/cas.14567
This report is a digest of the “Japanese Society of Medical Oncology Clinical Guidelines: Molecular Testing for Colorectal Cancer Treatment, 4th Edition.” The English‐translated full text is available at https://www.jsmo.or.jp/about/doc/guideline_200225.pdf
REFERENCES
- 1. Taniguchi H, Yamazaki K, Yoshino T, et al. Japanese society of medical oncology clinical guidelines: RAS (KRAS/NRAS) mutation testing in colorectal cancer patients. Cancer Sci. 2015;106:324‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamazaki K, Taniguchi H, Yoshino T, et al. Japanese society of medical oncology clinical guidelines: molecular testing for colorectal cancer treatment, Third edition. Cancer Sci. 2018;109:2074‐2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Japanese Society of Medical Oncology Clinical Guidelines: Molecular Testing for Colorectal Cancer Treatment, Fourth Edition (Japanese).
- 4. Yoshino T, Arnold D, Taniguchi H, et al. Pan‐Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO–ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Annals of Oncology. 2018;29 (1):44–70. 10.1093/annonc/mdx738. [DOI] [PubMed] [Google Scholar]
- 5. Taieb J, Tabernero J, Mini E, et al. Oxaliplatin, fluorouracil, and leucovorin with or without cetuximab in patients with resected stage III colon cancer (PETACC‐8): an open‐label, randomised phase 3 trial. Lancet Oncol. 2014;15:862‐873. [DOI] [PubMed] [Google Scholar]
- 6. Sargent DJ. Effect of Oxaliplatin, Fluorouracil, and Leucovorin With or Without Cetuximab on Survival Among Patients With Resected Stage III Colon Cancer. JAMA. 2012;307:1383 10.1001/jama.2012.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schirripa M, Bergamo F, Cremolini C, et al. BRAF and RAS mutations as prognostic factors in metastatic colorectal cancer patients undergoing liver resection. Br J Cancer. 2015;112:1921‐1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tie J, Lipton L, Desai J, et al. KRAS mutation is associated with lung metastasis in patients with curatively resected colorectal cancer. Clin Cancer Res. 2011;17:1122‐1130. [DOI] [PubMed] [Google Scholar]
- 9. Cremolini C, Loupakis F, Antoniotti C, et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first‐line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open‐label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306‐1315. [DOI] [PubMed] [Google Scholar]
- 10. Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E‐Mutated Colorectal Cancer. N Engl J Med. 2019;381:1632‐1643. [DOI] [PubMed] [Google Scholar]
- 11. Zhu L, Dong C, Cao Y, et al. Prognostic Role of BRAF mutation in stage II/III colorectal cancer receiving curative resection and adjuvant chemotherapy: a meta‐analysis based on randomized clinical trials. PLoS One. 2016;11:e0154795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Y, Wang D, Jin L, et al. Prognostic value of the combination of microsatellite instability and BRAF mutation in colorectal cancer. Cancer Manag Res. 2018;10:3911‐3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair‐deficient or microsatellite instability‐high colorectal cancer (CheckMate 142): an open‐label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182‐1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite‐instability status as a predictor of benefit from fluorouracil‐based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247‐257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaanan A, Shi Q, Taieb J, et al. Role of deficient DNA mismatch repair status in patients with stage III colon cancer treated with FOLFOX adjuvant chemotherapy: a pooled analysis from 2 randomized clinical trials. JAMA Oncol. 2018;4:379‐383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goel A, Nagasaka T, Hamelin R, et al. An optimized pentaplex PCR for detecting DNA mismatch repair‐deficient colorectal cancers. PLoS One. 2010;5:e9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair‐deficient/microsatellite instability‐high metastatic colorectal cancer. J Clin Oncol. 2018;36:773‐779. [DOI] [PubMed] [Google Scholar]
- 18. Tsimberidou AM, Hong DS, Ye Y, et al. Initiative for molecular profiling and advanced cancer therapy (IMPACT): an MD anderson precision medicine study. JCO Precis Oncol. 2017:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell‐free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cremolini C, Rossini D, Dell'Aquila E, et al. Rechallenge for patients with RAS and BRAF wild‐type metastatic colorectal cancer with acquired resistance to first‐line cetuximab and irinotecan: a phase 2 single‐arm clinical trial. JAMA Oncol. 2019;5:343‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montagut C, Argiles G, Ciardiello F, et al. Efficacy of Sym004 in patients with metastatic colorectal cancer with acquired resistance to anti–EGFR therapy and molecularly selected by circulating tumor DNA analyses: a phase 2 randomized clinical trial. JAMA Oncol. 2018;4:e175245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hong DS, Morris VK, El Osta B, et al. Phase IB study of vemurafenib in combination with irinotecan and cetuximab in patients with metastatic colorectal cancer with BRAFV600E mutation. Cancer Discov. 2016;6:1352‐1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tabernero J, Yoshino T, Cohn AL, et al. Ramucirumab versus placebo in combination with second‐line FOLFIRI in patients with metastatic colorectal carcinoma that progressed during or after first‐line therapy with bevacizumab, oxaliplatin, and a fluoropyrimidine (RAISE): a randomised, double‐blind, multicentre, phase 3 study. Lancet Oncol. 2015;16:499‐508. [DOI] [PubMed] [Google Scholar]


