Abstract
The well‐known gene‐environment interaction between alcohol consumption and aldehyde dehydrogenase 2 (ALDH2) genotype in upper aerodigestive tract cancer risk may improve our ability to identify high‐risk subjects. Here, we developed and validated risk prediction models for this cancer in Japanese men and evaluated whether adding the gene‐environment interaction to the model improved the predictive performance. We developed two case‐cohort datasets in the Japan Public Health Center‐based Prospective Study: one from subjects in the baseline survey for model development (108 cases and 4049 subcohort subjects) and the second from subjects in the 5‐year follow‐up survey for model validation (31 cases and 1527 subcohort subjects). We developed an environmental model including age, smoking status, and alcohol consumption, and a gene‐environment interaction model including age, smoking status, and the combination of alcohol consumption and the ALDH2 genotype. We found a statistically significant gene‐environment interaction for alcohol consumption and the ALDH2 genotype. The c‐index for the gene‐environment interaction model (0.71) was slightly higher than that for the environmental model (0.67). The values of integrated discrimination improvement and net reclassification improvement for the gene‐environment interaction model were also slightly higher than those for the environmental model. Goodness‐of‐fit tests suggested that the models were well calibrated. Results from external model validation by the 5‐year follow‐up survey were consistent with those from the model development by the baseline survey. The addition of a gene‐environment interaction to a lifestyle‐based model might improve the performance to estimate the probability of developing upper aerodigestive tract cancer for Japanese men.
Keywords: absolute risk, gene‐environment interaction, risk prediction model, single‐nucleotide polymorphisms, upper aerodigestive tract cancer
We developed and validated risk prediction models for upper aerodigestive tract cancer in Japanese men based on a prospective cohort study. We demonstrated that the addition of the gene‐environment interaction between alcohol consumption and aldehyde dehydrogenase 2 genotype on upper aerodigestive tract cancer risk to a lifestyle‐based model improved the performance to estimate the probability of developing this risk.

Abbreviations
- ALDH2
aldehyde dehydrogenase 2
- CI
confidence interval
- HR
hazard ratio
- IDI
integrated discrimination improvement
- JPHC Study
Japan Public Health Center‐based Prospective Study
- NRI
net reclassification improvement
1. INTRODUCTION
A risk prediction model for cancer is a statistical tool for estimating the absolute risk that a currently healthy individual with specific risk factors will develop a cancer in the future. A model can be translated into more efficient risk‐based preventive measures and is necessary for the implementation of precision public health. While many risk prediction models have been developed, some constructed using nongenetic risk factors have shown only modest discriminatory accuracy. 1 , 2 , 3 , 4 To improve predictive performance, later intensive studies added genetic markers such as variants identified in genome‐wide association studies, but improvements have again been limited. 5 , 6 Further improvement therefore requires the incorporation of strong risk factors into the model. One candidate is the gene‐environment interaction, but so far very few models have included it. 7 , 8
A good example is the gene‐environment interaction between alcohol consumption and aldehyde dehydrogenase 2 (ALDH2) genotype on the risk of upper aerodigestive tract cancer. The ALDH2 genotype determines the individual's blood level of acetaldehyde, 9 which has been established as a carcinogen in experimental animals. 10 Previous studies, mainly case‐control studies, have shown a gene‐environment interaction between alcohol consumption and the ALDH2 genotype on the risk of upper aerodigestive tract cancer. 11 , 12 , 13 , 14 , 15 Considering this evidence, the IARC concluded that acetaldehyde derived from the metabolism of ethanol in alcohol beverages contributes to esophageal cancer. 10
Among the several prediction models for upper aerodigestive tract cancer reported to date, 7 , 8 , 16 , 17 only two incorporated the gene‐environment interaction between alcohol consumption and the ALDH2 genotype. 7 , 8 The first of these was a model for esophageal cancer developed from a case‐control study among Japanese men, which showed excellent discriminatory accuracy (area under the curve = 0.86). 7 The second involved validated models for upper aerodigestive tract cancer based on two hospital‐based case‐control studies among Japanese, and showed an improvement in discriminatory accuracy with the addition of the gene‐environment interaction. 8
The improvements arising from the inclusion of this gene‐environment interaction would be strengthened by a confirmation study using data from a prospective cohort. Further, determining the 10‐year absolute risk for developing upper aerodigestive tract cancer would aid the identification of individuals at high risk who might benefit from preventive measures. Accordingly, the aim of the present study was to develop and validate risk prediction models for upper aerodigestive tract cancer in Japanese men based on a population‐based prospective cohort study and to evaluate whether the addition of a gene‐environment interaction to the model improves the performance to estimate the 10‐year absolute risk of developing upper aerodigestive tract cancer.
2. MATERIALS AND METHODS
2.1. Study population
The Japan Public Health Center‐based Prospective Study (JPHC Study), initiated in 1990 for cohort I and in 1993 for cohort II, includes 140 420 subjects (68 722 men and 71 698 women) aged 40‐69 years living in municipalities supervised by 11 public health centers throughout Japan. In the baseline survey from 1990 to 1994, approximately 113 000 subjects returned the questionnaire, giving a response rate of 81%, and 48 000 provided blood samples or health checkup data, with most providing both. The 5‐year follow‐up survey was conducted to update information on lifestyle habits and health conditions between 1995 and 2000. Approximately 104 000 subjects returned the questionnaire, giving a response rate of 74%, and 34 000 provided blood samples or health checkup data, with most providing both. Details of the JPHC Study have been provided elsewhere. 18 To obtain genotyping data in a cost‐efficient manner, given the fixed maximum cost for our study, a case‐cohort design was applied to select an appropriate subsample of subjects from the JPHC Study.
2.2. Follow‐up and case‐cohort selection from the baseline survey for model development
In this study, we defined a base cohort of 33 736 subjects who responded to the baseline questionnaire and provided a blood sample during a health checkup after excluding subjects in two public health centers, ineligible subjects, and subjects declining to participate as described in Figure 1. From this base cohort, we randomly chose a subcohort of 13 024 subjects for model development.
FIGURE 1.
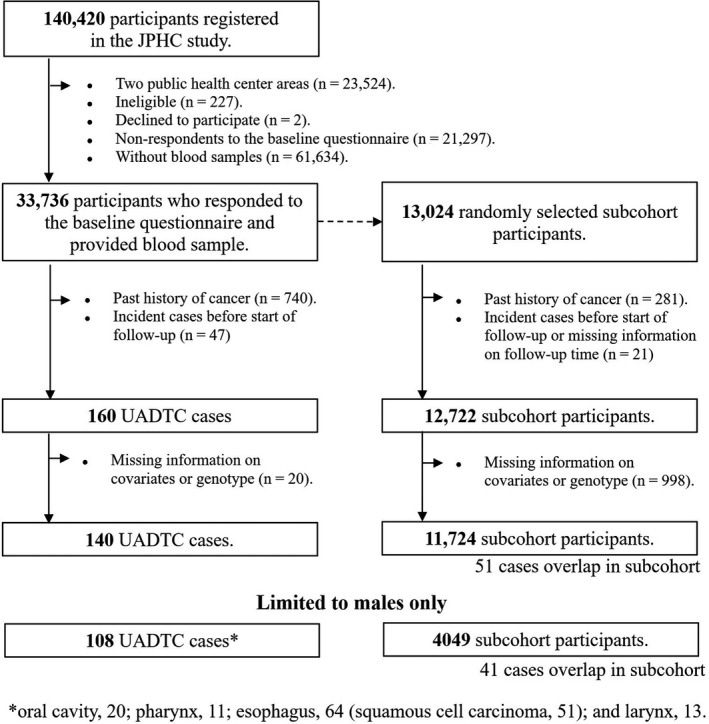
Selection of upper aerodigestive tract cancer (UADTC) cases and subcohort subjects from the baseline survey in the Japan Public Health Center‐based Prospective Study (JPHC Study)
All subjects were followed until 31 December, 2009. Changes in residence status and survival status were ascertained annually through the residential registry from each public health center area. Newly diagnosed cancer cases were identified by active patient notification from local major hospitals in the study area and data linkage with population‐based cancer registries. Death certificates were used as a supplementary information source. Upper aerodigestive tract cancer was defined according to the following codes of the International Classification of Diseases for Oncology, Third Edition: oral cavity (C00‐C08), pharynx (C10‐C13), esophagus (C15), and larynx (C32).
Among the 33 736 baseline cohort subjects, 140 cases (108 men and 32 women) of upper aerodigestive tract cancer were newly identified during the follow‐up after the exclusion of subjects with a past history of cancer in the baseline questionnaire, subjects diagnosed with cancer before the start of the follow‐up, and subjects with missing information on covariates and genotyping data (Figure 1). Similarly to the case subjects, 11 724 subcohort subjects (4049 men and 7675 women) among the 13 024 randomly selected subjects remained after excluding subjects using the same criteria as for the case selection described above.
2.3. Case‐cohort selection from the 5‐year follow‐up survey for external validation
To evaluate the validity of the developed prediction model based on case‐cohort subjects from the baseline survey, we constructed data that were independent of the subjects in the 5‐year follow‐up survey (Figure 2). To ensure that we did not duplicate subjects, we focused on subjects who responded to the self‐administered questionnaire and provided blood samples at the 5‐year follow‐up survey but did not respond to the self‐administered questionnaire and did not provide blood samples at the baseline survey. Thanks to this, the case‐cohort subjects from the 5‐year follow‐up survey who were used to assess the external validity were completely independent from the case‐cohort subjects from the baseline survey who were used to develop the prediction models. After excluding subjects using the same criteria as in the baseline survey, we randomly selected a subcohort sample of 4000 subjects from the 10 950 5‐year follow‐up cohort subjects for the validation study.
FIGURE 2.
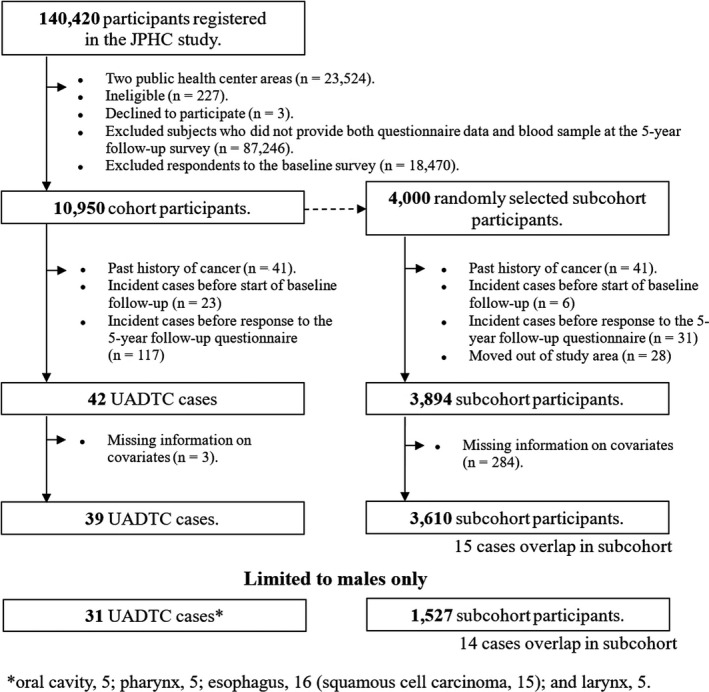
Selection of upper aerodigestive tract cancer (UADTC) cases and subcohort subjects from the 5‐y follow‐up survey in the Japan Public Health Center‐based Prospective Study (JPHC Study)
Based on follow‐up data obtained up to 31 December, 2009, we identified 39 cases (31 men and 8 women) of upper aerodigestive tract cancer and 3610 subcohort subjects (1527 men and 2083 women) after excluding subjects using the same criteria as in the baseline survey.
2.4. Definition of risk factors
We selected well‐established risk factors for upper aerodigestive tract cancer used in a previous risk prediction model. 8 Subjects were categorized by age into three groups of <50 years, 50‐59 years, and ≥60 years old; by smoking status into three groups of never, past, and current smokers; and by alcohol consumption into two groups of nondrinkers or drinkers who consumed <5 days per week or <23 g of ethanol per day, and drinkers who consumed ≥5 days per week and ≥23 g of ethanol per day.
2.5. Laboratory analysis
DNA samples of the subjects were analyzed using the HumanOmniExpressExome‐8 v1.2 BeadChip, HumanOmniExpress‐12 BeadChip, or HumanOmni2.5‐8 BeadChip arrays (Illumina Inc). Genotyping was conducted at the Genetics Division or Department of Clinical Genomics, Fundamental Innovative Oncology Core (FIOC), National Cancer Center Research Institute or at the RIKEN Center for Integrative Medical Sciences. In the present study, we extracted genotyping information on rs671 (ALDH2 Glu504Lys) from the above data.
2.6. Statistical analysis
Considering the lower incidence rate of upper aerodigestive tract cancer and lower prevalence of heavy smoking and drinking in women than in men in Japan, 19 , 20 the present analysis was limited to data from men only, and finally included 108 cases and 4049 subcohort subjects for model development and 31 cases and 1527 subcohort subjects for external validation (Figures 1 and 2).
We developed two models based on the case‐cohort subjects from the baseline survey: an environmental model which included age, smoking status, and alcohol consumption and a gene‐environment interaction model which included age, smoking status, and the combination of alcohol consumption and the ALDH2 genotype (Table 1). Hazard ratios (HRs) and 95% confidence intervals (CIs) for upper aerodigestive tract cancer according to variables included in the two models were estimated by inverse‐probability‐weighted Cox proportional hazard models stratified by study area, with the weights defined as 1 for cases and the inverse of the sampling probability for subcohort controls. 21 The interaction between alcohol consumption and the ALDH2 genotype was assessed by the Wald test of the product term. The 10‐year absolute risk for upper aerodigestive tract cancer in men was calculated by means of the Breslow estimator of cumulative hazards using the estimates of the weighted Cox regression analysis. 22
TABLE 1.
Hazard ratios (HR) and 95% confidence intervals (CIs) for upper aerodigestive tract cancer risk according to variables included in the models
| Model | Variable and categories | Number of cases | Number of subcohort subjects | HR | 95% CIs | P value |
|---|---|---|---|---|---|---|
| Environmental model | ||||||
| Age, y | ||||||
| <50 | 20 | 1300 | 1.00 | Reference | ||
| 50‐<60 | 52 | 1699 | 2.28 | 1.36‐3.81 | 1.8 × 10−3 | |
| ≥60 | 36 | 1050 | 3.62 | 2.08 ‐ 6.32 | 5.9 × 10−6 | |
| Smoking | ||||||
| Never smoker | 13 | 1184 | 1.00 | Reference | ||
| Past smoker | 29 | 1053 | 2.20 | 1.12‐4.30 | 2.1 × 10−2 | |
| Current smoker | 66 | 1812 | 3.14 | 1.71‐5.77 | 2.3 × 10−4 | |
| Alcohol consumption | ||||||
| Nondrinkers/<5 d per week or < 23 g of ethanol/day | 43 | 2490 | 1.00 | Reference | ||
| ≥5 d per week and ≥23 g of ethanol/day | 65 | 1559 | 2.15 | 1.45‐3.20 | 1.5 × 10−4 | |
| Gene‐environment interaction model | ||||||
| Age, y | ||||||
| <50 | 20 | 1300 | 1.00 | Reference | ||
| 50‐<60 | 52 | 1699 | 2.24 | 1.33‐3.77 | 2.3 × 10−3 | |
| ≥60 | 36 | 1050 | 3.71 | 2.13‐6.48 | 3.9 × 10−6 | |
| Smoking | ||||||
| Never smoker | 13 | 1184 | 1.00 | Reference | ||
| Past smoker | 29 | 1053 | 2.07 | 1.05‐4.08 | 3.5 × 10−2 | |
| Current smoker | 66 | 1812 | 2.94 | 1.58‐5.45 | 6.4 × 10−4 | |
| Combination of alcohol consumption and ALDH2 genotype | ||||||
|
Nondrinkers/<5 d per week or <23 g of ethanol/day + Glu/Glu Nondrinkers/<5 d per week or <23 g of ethanol/day + Glu/Lys + Lys/Lys |
43 | 2490 | 1.00 | Reference | ||
| ≥5 d per week and ≥ 23 g ethanol/day + Glu/Glu | 41 | 1351 | 1.58 | 1.02‐2.46 | 4.1 × 10−2 | |
| ≥5 d per week and ≥ 23 g ethanol/day + Glu/Lys + Lys/Lys | 24 | 208 | 6.00 | 3.53‐10.19 | 3.4 × 10−11 | |
Abbreviation: ALDH2, aldehyde dehydrogenase 2.
The prediction models were evaluated with regard to their ability to improve model discrimination. To compare model discrimination, we calculated the c‐index, the integrated discrimination improvement (IDI), and the continuous net reclassification improvement (NRI) based on the case‐cohort subjects from the baseline survey. 23 We also evaluated the classification accuracy using quantified differences in classification by NRI. The risk category of NRI was arbitrarily defined as <0.25%, 0.25%≤ & <1%, and ≥1% 10‐year absolute risk of developing upper aerodigestive tract cancer. Because of the case‐cohort study design, we calculated all of these measures weighted, using the weight of inversed sampling probability in a similar way to the method used for the weighted Cox regression model. 24
Calibration was assessed based on the case‐cohort subjects from the baseline survey by comparing the predicted and observed number of events with the Grønnesby and Borgan goodness‐of‐fit test (GB test), 25 , 26 which is similar to the Hosmer‐Lemeshow test for logistic regression models. To implement the GB test, all subjects were divided into five groups according to the weighted quintiles of the predicted absolute risks in the subcohort subjects. We then added the group indicator variables into the Cox proportional hazards model with the risk factors and performed an overall test for the group indicator variables using the Wald test with robust variance. The GB test for the case‐cohort study design is based on weighted martingale residuals. The 10‐year observed and predicted absolute risks in each group were also estimated by the weighted Kaplan‐Meier method and the weighted average of the predicted absolute risks, respectively.
To further evaluate the performance of the developed models, we conducted external validation based on the case‐cohort subjects from the 5‐year follow‐up survey, who were an independent population from the model development, by calculating the c‐index, IDI, and NRI for discrimination and performing the GB test for calibration using the predicted values from the developed models. For the GB test, all subjects were divided into three groups according to the weighted tertiles of the predicted absolute risks in the subcohort subjects, in consideration of the small number of cases (n = 31).
All reported P values are two‐sided, and the significance level was set at P < .05. All statistical analyses were performed using SAS 9.4 (SAS Institute Inc).
3. RESULTS
Table 1 shows the HRs and CIs for the variables included in the environmental and gene‐environment interaction models. Increasing age, smoking, and alcohol consumption were significantly associated with increased risk of upper aerodigestive tract cancer in the environmental model. Similarly, statistically significant associations were found for age, smoking status, and combination of alcohol consumption and the ALDH2 genotype in the gene‐environment interaction model. Compared with subjects who were nondrinkers or drank less than 5 days per week or 23 g of ethanol per day, regardless of the ALDH2 genotype, the HR (95% CI) was 6.00 (3.53‐10.19) among subjects who drank 5 days or more per week and more than 23 g of ethanol per day and had the Glu/Lys or Lys/Lys genotype. Of note, the test of interaction was statistically significant (P = 4.0 × 10−4). Similarly, we observed a statistically significant interaction between alcohol consumption and the ALDH2 genotype on the risk of esophageal cancer (data not shown).
In the development of prediction models based on case‐cohort subjects from the baseline survey, the c‐index for the gene‐environment interaction model (0.71) was slightly higher than that for the environmental model (0.67) (Table 2). The gene‐environment interaction model was also preferable to the environmental model with regard to the IDI (0.0036) and continuous NRI (0.033). The categorical NRI value (0.13) showed an improvement when the environmental and gene‐environment interaction models were compared (Table 2). The differences between the observed and predicted absolute risk by quintile category are shown in Figure 3. The P values for the GB test were 0.34 for the environmental model and 0.19 for the gene‐environment interaction model. Similarly, the gene‐environment interaction model for esophageal cancer also improved the model discriminations assessed by the NRI and IDI compared to the environmental model, and the two models were well calibrated (data not shown).
TABLE 2.
Comparison of predictive performance between risk prediction models for upper aerodigestive tract cancer based on case‐cohort subjects from the baseline survey
| C‐index (95% confidence interval) | IDI | Continuous NRI | Categorical NRI a | |
|---|---|---|---|---|
| Model | ||||
| Environmental model | 0.67 (0.52‐0.80) | Reference | Reference | Reference |
| Gene‐environment interaction model | 0.71 (0.57‐0.84) | 0.0036 | 0.033 | 0.13 |
Abbreviations: IDI, integrated discrimination improvement; NRI, net reclassification improvement.
Risk category of NRI was defined as <0.25%, ≥0.25%, and <1% and ≥1% 10‐y absolute risk of developing upper aerodigestive tract cancer.
FIGURE 3.
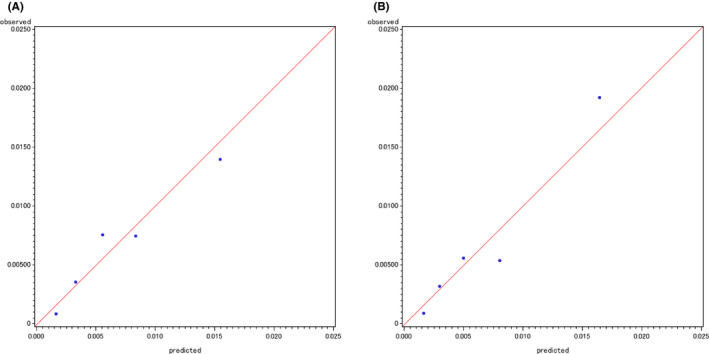
Observed versus predicted absolute risks for the development of upper aerodigestive tract cancer by quintile categories for an environmental model (A) and gene‐environment interaction model (B) based on case‐cohort subjects from the baseline survey
In the assessment of the external validity of the developed risk prediction models based on case‐cohort subjects from the 5‐year follow‐up survey, the c‐index for the gene‐environment interaction model (0.76) was slightly higher than that for the environmental model (0.72) (Table 3). The superiority of the gene‐environment interaction model was also shown by the IDI (0.0089), continuous NRI (0.37), and categorical NRI (0.16). In terms of calibration, we did not observe significant differences between the observed and predicted absolute risk by tertile category (Figure 4).
TABLE 3.
External validity of the developed risk prediction models based on the case‐cohort subjects from the 5‐y follow‐up survey
| C‐index (95% confidence interval) | IDI | Continuous NRI | Categorical NRI a | |
|---|---|---|---|---|
| Model | ||||
| Environmental model | 0.72 (0.44‐0.93) | Reference | Reference | Reference |
| Gene‐environment interaction model | 0.76 (0.50‐0.96) | 0.0089 | 0.37 | 0.16 |
Abbreviations: IDI, integrated discrimination improvement; NRI, net reclassification improvement.
Risk category of NRI was defined as <0.25%, ≥0.25%, and <1%, and ≥1% 10‐y absolute risk of developing upper aerodigestive tract cancer.
FIGURE 4.
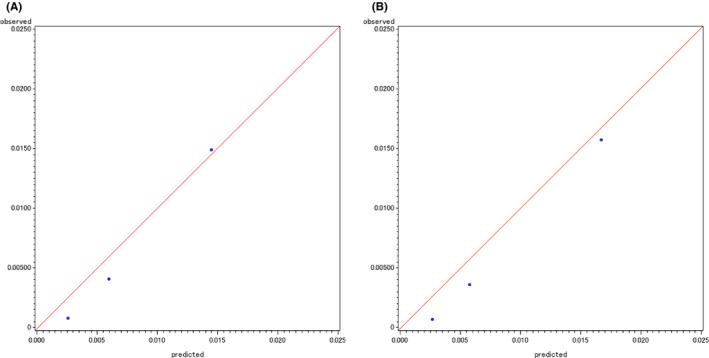
Observed versus predicted absolute risks for the development of upper aerodigestive tract cancer by tertile categories for an environmental model (A) and gene‐environment interaction model (B) based on case‐cohort subjects from the 5‐year follow‐up survey
We estimated the absolute risks for developing upper aerodigestive tract cancer for men according to risk factor profiles based on the case‐cohort subjects from the baseline survey (Table 4). The 10‐year absolute risks for developing upper aerodigestive tract cancer varied by risk factor profile from 0.09% to 6.05%. Of note, the 10‐year absolute risks substantially differed between subjects who had the Glu/Glu genotype and Glu/Lys or Lys/Lys genotype among subjects who drank 5 days or more per week and more than 23 g of ethanol per day (eg, 1.59% vs 6.05% for current smokers at age 60 years or older).
TABLE 4.
Ten‐year absolute risk (95% confidence interval) (%) of developing upper aerodigestive tract cancer for men according to risk factor profile based on case‐cohort subjects from the baseline survey
| Age group | Smoking status | Combination of alcohol consumption and ALDH2 genotype | ||
|---|---|---|---|---|
|
Nondrinkers/<5 d per week or <23 g of ethanol/day + Glu/Glu Nondrinkers/<5 d per week or <23 g of ethanol/day + Glu/Lys + Lys/Lys |
≥5 d per week and ≥23 g ethanol/day + Glu/Glu | ≥5 d per week and ≥23 g ethanol/day + Glu/Lys + Lys/Lys | ||
| <50 y old | Never smoker | 0.09 (0.05, 0.18) | 0.15 (0.07, 0.31) | 0.55 (0.23, 1.31) |
| Past smoker | 0.19 (0.10, 0.37) | 0.30 (0.17, 0.55) | 1.15 (0.57, 2.32) | |
| Current smoker | 0.27 (0.16, 0.47) | 0.43 (0.25, 0.74) | 1.63 (0.86, 3.10) | |
| 50‐<60 y old | Never smoker | 0.21 (0.11, 0.40) | 0.33 (0.16, 0.66) | 1.24 (0.55, 2.79) |
| Past smoker | 0.43 (0.23, 0.80) | 0.68 (0.40, 1.16) | 2.58 (1.38, 4.82) | |
| Current smoker | 0.61 (0.40, 0.93) | 0.96 (0.65, 1.42) | 3.65 (2.22, 6.01) | |
| ≥60 y old | Never smoker | 0.34 (0.19, 0.62) | 0.54 (0.27, 1.11) | 2.06 (0.94, 4.50) |
| Past smoker | 0.71 (0.41, 1.23) | 1.13 (0.65, 1.94) | 4.27 (2.38, 7.65) | |
| Current smoker | 1.01 (0.64, 1.58) | 1.59 (0.95, 2.66) | 6.05 (3.49, 10.47) | |
Abbreviation: ALDH2: aldehyde dehydrogenase 2.
4. DISCUSSION
This study confirmed a statistically significant gene‐environment interaction for alcohol consumption and the ALDH2 genotype on the risk of upper aerodigestive tract cancer in a population‐based prospective cohort of Japanese men. We developed two risk prediction models of upper aerodigestive tract cancer for Japanese men and found that a further improvement of the performance to estimate the absolute risk was achieved by the addition of the gene‐environment interaction to the environmental model. This finding was supported by the external model validation. Our findings provide support for the inclusion of information on gene‐environment interaction into risk assessment models of upper aerodigestive tract cancer to more accurately stratify men into low‐ to high‐risk groups.
This is the first study to show a statistically significant gene‐environment interaction for alcohol consumption and the ALDH2 genotype on the risk of upper aerodigestive tract cancer in a population‐based prospective cohort study among Japanese men. In contrast, a recent cohort study in China failed to detect a statistically significant interaction. 15 Findings from cohort studies are generally less likely to be affected by recall bias and/or reverse causality caused by a change in alcohol drinking habits due to the symptoms of prediagnosed cancer. Considered together with evidence for significant interaction from case‐control studies, 11 , 12 , 13 , 14 therefore, the consistent findings of our present cohort study warrant recognition as convincing evidence.
The discriminatory accuracy of our developed models was assessed by the c‐index, IDI, and NRI. The results of the IDI and NRI based on the external model validation by the 5‐year follow‐up survey showed an improvement when the environmental and gene‐environment interaction models were compared although a remarkable difference in the c‐index between the two models was not observed. This finding was consistent with the results from the model development by the baseline survey. The results of the c‐index indicated that the performance to discriminate the occurrence of cases and noncases was limited in our models. Meanwhile, the results of the IDI and NRI suggested that subjects with higher or lower probability of upper aerodigestive tract cancer occurrence were reclassified more correctly to higher or lower risk groups in our models. Considering not only the above results from the c‐index, IDI, and NRI but also the other results including calibration comprehensively, our findings suggested that the addition of the gene‐environment interaction to the environmental model improved the performance to estimate the 10‐year absolute risk of developing upper aerodigestive tract cancer according to the status of risk factors.
Overall, this finding is consistent with a previous study based on case‐control studies by Koyanagi et al. 8 Despite using almost the same variables in the models, however, the c‐index values in our present study were lower than those of Koyanagi et al. 8 For example, the c‐index values in the model development phase of the present study were 0.67 for the environmental model and 0.71 for the gene‐environment interaction model, versus respective values of 0.76 and 0.82 in Koyanagi's study. 8 This might be explained by the difference in the strength of observed associations in the high‐exposure group. In our study, the highest HR (6.00) was found for subjects who drank 5 days or more per week and more than 23 g of ethanol per day and had the Glu/Lys or Lys/Lys genotype; the P value for interaction was 4.0 × 10−4. Meanwhile, the highest odds ratio (12.17) was observed in heavy drinkers (consumption on 5 days or more per week of more than 46 g ethanol on each occasion) with the Glu/Lys genotype; the P value for interaction was markedly small (1.6 × 10−9). 8 This indicates that a strong risk factor substantially contributes to improving discriminatory accuracy.
Our developed models were evaluated for discriminatory accuracy and calibration in an independent population. The c‐index values were somewhat higher in the external validation phase than in the model development phase. The reason for this difference is unclear, but one possibility may be the wider 95% CI due to the small number of cases in the external validation phase. This is indeed one of the limitations of the present study and requires a further external validation study with a large number of cases.
A second limitation is the generalizability of the models for possible use as an absolute risk estimate for Japanese men. In particular, the absolute risk provided by the models was based on a baseline risk of upper aerodigestive tract cancer estimated by participants in the JPHC Study, which would be applicable only to populations with comparable rates. In this regard, while incidence rates of upper aerodigestive tract cancer from a nationwide cancer registry would be representative of the Japanese population, and therefore desirable for estimating the absolute risk, the only incidence rates available in Japan were in fact estimated using high‐quality data from selected population‐based cancer registries. Indeed, the crude incidence rate of upper aerodigestive tract cancer estimated from cancer registries in 2000 was 53.5 per 100 000 population in Japanese men aged 40‐69 years, versus a corresponding rate in the JPHC Study of 64 per 100 000 person‐years in men aged 40‐69 years. 19 This might at least suggest the lack of any extreme discrepancy between rates.
Nevertheless, the absolute risk estimates varied greatly depending on risk profile. Our gene‐environment interaction model estimated the 10‐year absolute risk from 0.09% to 6.05% in the present study. In particular, the substantial difference in the absolute risk between subjects with the Glu/Glu genotype and Glu/Lys or Lys/Lys genotype among subjects who drank 5 days or more per week and more than 23 g of ethanol per day might prompt lifestyle modification. Moreover, these findings suggest that our model might help identify subgroups of Japanese men at elevated risk of upper aerodigestive tract cancer and who might benefit most from preventive measures. One potential approach is upper aerodigestive tract endoscopy combined with esophageal iodine staining to detect early‐stage cancer in a target population determined by the individual's absolute risk. 7
In summary, this study suggested that the addition of a gene‐environment interaction to a lifestyle‐based model for Japanese men improved the performance to estimate the 10‐year absolute risk of developing upper aerodigestive tract cancer. This model might allow the stratification of Japanese men into low‐ and high‐risk groups.
DISCLOSURE
The authors have declared no conflict of interest.
ETHICAL CONSIDERATIONS
This study was approved by the institutional review board of the National Cancer Center, Tokyo, Japan.
ACKNOWLEDGMENT
We thank the participants of the JPHC Study and all the staff members in each study area as well as the central offices for their cooperation and technical assistance. We are indebted to the Aomori, Iwate, Akita, Ibaraki, Niigata, Osaka, Kochi, Nagasaki, and Okinawa Cancer Registries for providing their incidence data. We thank Drs. Hiromi Sakamoto and Teruhiko Yoshida and the staff at the National Cancer Center Research Core Facility and the RIKEN Center for Integrative Medical Sciences for some of the genotyping analyses in our study. The Core Facility was supported in part by the National Cancer Center Research and Development Fund (29‐A‐2). This study was supported by the National Cancer Center Research and Development Fund (23‐A31[toku], 26‐A‐2, and 29‐A‐4 since 2011, and 25‐A‐14, 28‐A‐19, and 31‐A‐18), a Grant‐in‐Aid for Cancer Research from the Ministry of Health, Labor and Welfare of Japan (from 1989 to 2010), and the Practical Research for Innovative Cancer Control (JP16ck0106095h0003 and JP19ck0106266h0003) from the Japan Agency for Medical Research and Development (AMED).
Iwasaki M, Budhathoki S, Yamaji T, et al; for the Japan Public Health Center‐based Prospective Study (JPHC Study) Group . Inclusion of a gene‐environment interaction between alcohol consumption and the aldehyde dehydrogenase 2 genotype in a risk prediction model for upper aerodigestive tract cancer in Japanese men. Cancer Sci. 2020;111:3835–3844. 10.1111/cas.14573
REFERENCES
- 1. Ma E, Sasazuki S, Iwasaki M, Sawada N, Inoue M. 10‐Year risk of colorectal cancer: development and validation of a prediction model in middle‐aged Japanese men. Cancer Epidemiol. 2010;34(5):534‐541. [DOI] [PubMed] [Google Scholar]
- 2. Win AK, Macinnis RJ, Hopper JL, Jenkins MA. Risk prediction models for colorectal cancer: a review. Cancer Epidemiol Biomarkers Prev. 2012;21(3):398‐410. [DOI] [PubMed] [Google Scholar]
- 3. Meads C, Ahmed I, Riley RD. A systematic review of breast cancer incidence risk prediction models with meta‐analysis of their performance. Breast Cancer Res Treat. 2012;132(2):365‐377. [DOI] [PubMed] [Google Scholar]
- 4. Charvat H, Sasazuki S, Inoue M, et al. Prediction of the 10‐year probability of gastric cancer occurrence in the Japanese population: the JPHC study cohort II. Int J Cancer. 2016;138(2):320‐331. [DOI] [PubMed] [Google Scholar]
- 5. Iwasaki M, Tanaka‐Mizuno S, Kuchiba A, et al. Inclusion of a genetic risk score into a validated risk prediction model for colorectal cancer in Japanese men improves performance. Cancer Prevention Res. 2017;10:535‐541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fung SM, Wong XY, Lee SX, Miao H, Hartman M, Wee HL. Performance of single‐nucleotide polymorphisms in breast cancer risk prediction models: a systematic review and meta‐analysis. Cancer Epidemiol Biomarkers Prev. 2019;28(3):506‐521. [DOI] [PubMed] [Google Scholar]
- 7. Yokoyama T, Yokoyama A, Kumagai Y, et al. Health risk appraisal models for mass screening of esophageal cancer in Japanese men. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2846‐2854. [DOI] [PubMed] [Google Scholar]
- 8. Koyanagi YN, Ito H, Oze I, et al. Development of a prediction model and estimation of cumulative risk for upper aerodigestive tract cancer on the basis of the aldehyde dehydrogenase 2 genotype and alcohol consumption in a Japanese population. Eur J Cancer Prev. 2017;26(1):38‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mizoi Y, Yamamoto K, Ueno Y, Fukunaga T, Harada S. Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol. 1994;29(6):707‐710. [PubMed] [Google Scholar]
- 10. International Agency for Research on Cancer . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans A Review of Human Carcinogens, Part E: Personal Habits and Indoor Combustions. Vol. 100 Lyon: IARC Press; 2009. [PMC free article] [PubMed] [Google Scholar]
- 11. Matsuo K, Hamajima N, Shinoda M, et al. Gene‐environment interaction between an aldehyde dehydrogenase‐2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis. 2001;22(6):913‐916. [DOI] [PubMed] [Google Scholar]
- 12. Hiraki A, Matsuo K, Wakai K, Suzuki T, Hasegawa Y, Tajima K. Gene‐gene and gene‐environment interactions between alcohol drinking habit and polymorphisms in alcohol‐metabolizing enzyme genes and the risk of head and neck cancer in Japan. Cancer Sci. 2007;98(7):1087‐1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boccia S, Hashibe M, Galli P, et al. Aldehyde dehydrogenase 2 and head and neck cancer: a meta‐analysis implementing a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2009;18(1):248‐254. [DOI] [PubMed] [Google Scholar]
- 14. Zhao T, Wang C, Shen L, et al. Clinical significance of ALDH2 rs671 polymorphism in esophageal cancer: evidence from 31 case‐control studies. Onco Targets Ther. 2015;8:649‐659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu C, Guo YU, Bian Z, et al. Association of low‐activity ALDH2 and alcohol consumption with risk of esophageal cancer in Chinese adults: a population‐based cohort study. Int J Cancer. 2018;143:1652‐1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Collins GS, Altman DG. Identifying patients with undetected gastro‐oesophageal cancer in primary care: external validation of QCancer(R) (Gastro‐Oesophageal). Eur J Cancer. 2013;49(5):1040‐1048. [DOI] [PubMed] [Google Scholar]
- 17. Xie SH, Lagergren J. A model for predicting individuals' absolute risk of esophageal adenocarcinoma: moving toward tailored screening and prevention. Int J Cancer. 2016;138(12):2813‐2819. [DOI] [PubMed] [Google Scholar]
- 18. Tsugane S, Sawada N. The JPHC study: design and some findings on the typical Japanese diet. Jpn J Clin Oncol. 2014;44(9):777‐782. [DOI] [PubMed] [Google Scholar]
- 19. Hori M, Matsuda T, Shibata A, et al. Cancer incidence and incidence rates in Japan in 2009: a study of 32 population‐based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn J Clin Oncol. 2015;45(9):884‐891. [DOI] [PubMed] [Google Scholar]
- 20. Lu Y, Sobue T, Kitamura T, et al. Cigarette smoking, alcohol drinking, and oral cavity and pharyngeal cancer in the Japanese: a population‐based cohort study in Japan. Eur J Cancer Prev. 2018;27(2):171‐179. [DOI] [PubMed] [Google Scholar]
- 21. Barlow WE. Robust variance estimation for the case‐cohort design. Biometrics. 1994;50(4):1064‐1072. [PubMed] [Google Scholar]
- 22. Langholz B, Jiao J. Computational methods for case‐cohort studies. Comput Stat Data Anal. 2007;51(8):3737‐3748. [Google Scholar]
- 23. Pencina MJ, D' Agostino RB, D' Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27(2):157‐172; discussion 207–12. [DOI] [PubMed] [Google Scholar]
- 24. Ganna A, Reilly M, de Faire U, Pedersen N, Magnusson P, Ingelsson E. Risk prediction measures for case‐cohort and nested case‐control designs: an application to cardiovascular disease. Am J Epidemiol. 2012;175(7):715‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gronnesby JK, Borgan O. A method for checking regression models in survival analysis based on the risk score. Lifetime Data Anal. 1996;2(4):315‐328. [DOI] [PubMed] [Google Scholar]
- 26. May S, Hosmer DW. A simplified method of calculating an overall goodness‐of‐fit test for the Cox proportional hazards model. Lifetime Data Anal. 1998;4(2):109‐120. [DOI] [PubMed] [Google Scholar]


