Abstract
The histone demethylase Jumonji domain‐containing 1A (JMJD1A) is overexpressed in multiple cancers and promotes cancer progression. However, the role and mechanism of JMJD1A in gastric cancer (GC) remains poorly understood. Here, we found that JMJD1A could suppress GC cell proliferation and xenograft tumor growth. Using RNA sequencing, we identified runt‐related transcription factor 3 (RUNX3) as a novel target gene of JMJD1A. Mechanistically, we identified that JMJD1A upregulated RUNX3 through co–activating Ets‐1 and reducing the H3K9me1/2 levels at the RUNX3 promoter in GC cells. Functionally, JMJD1A inhibits the growth of GC cells in vivo, which is partially dependent on RUNX3. Moreover, JMJD1A expression was decreased in GC and low expression of JMJD1A was correlated with an aggressive phenotype and a poor prognosis in patients with GC. Importantly, JMJD1A expression was positively associated with RUNX3 expression in GC samples. These studies indicated that JMJD1A upregulates RUNX3 expression via co–activation of transcription factor Ets‐1 to inhibit proliferation of GC cells. Our findings provide new insight into the mechanism by which JMJD1A regulates RUNX3 transcription and suggest that JMJD1A and/or RUNX3 may be used as a therapeutic intervention for GC.
Keywords: Ets‐1, histone demethylase, JMJD1A, RUNX3, transcriptional regulation
Using RNA sequencing, we identified RUNX3 as a novel target gene of JMJD1A. Mechanistically, we identified that JMJD1A upregulated RUNX3 through co–activating Ets‐1 and reducing the H3K9me1/2 levels at the RUNX3 promoter in gastric cancer cells. We found that low expression of JMJD1A was notably correlated to an aggressive phenotype and a poor prognosis in patients with GC.
1. INTRODUCTION
Gastric cancer (GC) is the most common digestive system cancer and has the second highest mortality rate worldwide. 1 Despite recent advances in the treatment of GC, including preoperative neoadjuvant chemotherapy and postoperative chemo‐radiotherapy, 2 , 3 the prognosis of GC remains poor because of the low rate of early diagnosis and recurrence after resection. 1 , 4 Therefore, a better understanding of the molecular mechanisms underlying GC initiation and development is necessary.
Histone methylation is a crucial epigenetic modification that determines whether a gene is transcriptionally active or silent. 5 H3K9 methylation is a well‐known repressive histone marker associated with silenced transcription. Jumonji domain‐containing 1A (JMJD1A), also known as KDM3A or JHDM2A, is a histone demethylase that promotes gene expression by removing histone H3K9 mono‐methylation and di‐methylation. 6 JMJD1A plays crucial roles in various cellular processes, including spermatogenesis, 7 energy metabolism, 8 , 9 sex determination, 10 stem cell self‐renewal, 11 , 12 and differentiation. 13 JMJD1A is shown to play a tumor‐promoting role in several types of cancers, such as colorectal cancer, prostate cancer, hepatocellular carcinoma, ovarian cancer, and bladder cancer. 14 , 15 , 16 , 17 , 18 , 19 , 20 Although JMJD1A has been shown to be a predictor for prognosis and a potential therapeutic target for GC, 21 the role and the underlying mechanism of JMJD1A in GC progression remain to be further elucidated.
Runt‐related transcription factor 3 (RUNX3) is a member of the RUNX family, which are developmental regulators that have an essential role in human cancers. 22 RUNX3 is a well‐characterized tumor suppressor that is frequently inactivated in GC due to hemizygous deletion or promoter hypermethylation. 23 , 24 RUNX3 functions as a gatekeeper linking oncogenic Wnt and anti–oncogenic TGF‐β/BMP signaling pathways in gastrointestinal cancers. 25 Besides its role as a tumor suppressor, RUNX3, as recently demonstrated, is a regulator in hypoxia and the tumor immune microenvironment, which highlights the critical role of RUNX3 in the initiation and progression of cancer. 26
Here we studied the effects of JMJD1A on GC cell proliferation and xenograft tumor growth. Using RNA‐seq, we identified RUNX3 as a novel target gene of JMJD1A. We investigated the mechanisms by which JMJD1A regulated RUNX3 transcription. In addition, we verified that JMJD1A inhibits the growth of GC cells in vivo, which is partially dependent on RUNX3. Importantly, we showed the positive correlation between JMJD1A and RUNX3 in GC samples. Moreover, our work indicated that low expression of JMJD1A was notably correlated with an aggressive phenotype and a poor prognosis in patients with GC. Our findings provide new insight into the mechanism by which JMJD1A regulates RUNX3 transcription.
2. MATERIALS AND METHODS
2.1. Cell lines and cell culture
SGC‐7901, MGC‐803, and HEK‐293 cells were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco). MKN‐45 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FBS (Gibco). Cells were cultured in a humidified incubator at 37°C with 5% CO2. Cell lines SGC‐7901, MGC‐803, and HEK‐293 were purchased from the Cell Bank of the Chinese Academy of Sciences.
2.2. Plasmids, transient transfection, and luciferase assays
Details can be found in the supporting information materials and methods Data S1.
2.3. Lentiviral transduction and generation of stable cell lines
Lentiviruses harboring JMJD1A, shJMJD1A, RUNX3, and shRUNX3 were purchased from GeneChem Company. To obtain stable cell lines, cells were infected with lentiviral supernatants for 24 hours and then selected with 1 μg/mL puromycin (Sigma) for 48 hours. The infected cells were passaged before use after identification by western blotting.
2.4. Cell cycle analysis
For this analysis, SGC‐7901 cells (4 × 105 per well) in which JMJD1A was stably knocked down or overexpressed were seeded into six‐well plates and incubated for 24 hours. The cells were digested and washed with PBS and then fixed in 70% cold ethanol at 4°C overnight. Cells were centrifuged at 1000 g for 5 minutes and resuspended in PBS. Then, cells were stained with propidium iodide at 37°C for 30 minutes in the dark. Cell cycle status was measured by flow cytometry (BD Biosciences).
2.5. MTT assays
Details can be found in the supporting information materials and methods Data S1.
2.6. Colony formation
For colony formation assays, 1 × 103 cells in which JMJD1A was stably knocked down or overexpressed were seeded into six‐well plates and cultured for two weeks. Two weeks later, the cells were fixed with ethanol and stained with trypan blue. The visible colonies were photographed and counted. All experiments were performed at least three times.
2.7. RNA extraction and quantitative RT‐PCR assays
TRIzol Reagent (Invitrogen) was used to extract total RNA in cells. Then RNA was reverse transcribed into cDNA by using a Reverse Transcription Kit (Takara). SYBR Premix Ex Taq (Takara) was used for quantitative RT‐PCR assays, which were conducted on a Stratagene Mx3000P real‐time PCR system. The primers for RUNX3 were 5ʹ‐ATACCTACCTCCCGCCAC‐3ʹ (sense) and 5ʹ‐CTCCACGCCATCACTCTG‐3' (antisense).
2.8. Immunoprecipitation and western blot
Details can be found in the supporting information materials and methods Data S1.
2.9. ChIP and ChIP re–ChIP
Details can be found in the supporting information materials and methods Data S1.
2.10. Tissue microarrays and immunohistochemistry
Gastric cancer tissue microarrays were purchased from Shanghai Outdo Biotechnology. These arrays contained 90 cancerous and 90 adjacent noncancerous specimens. Clinical parameters and follow‐up information were available for all 90 cases. Immunohistochemistry (IHC) for the target molecules was performed on tissue microarray chips and single serial sections made from xenograft tumor samples. IHC staining was performed using primary antibodies against JMJD1A (Abcam, ab107234), RUNX3 (Abcam, ab224641) and Ki67 (Cell Signaling Technology, 2586), appropriate secondary antibodies, and the ABC Elite immunoperoxidase kit according to the manufacturer’s instructions. Immunoreactivity was quantified using a combined “H score,” which assesses both the staining intensity (0, negative; 1, weak; 2, moderate; 3, strong) and the percentage of positively stained cells (0, <5%; 1, 5%‐25%; 2, 26%‐50%; 3, 51%‐75%; 4, 76%‐100%).
2.11. RNA‐seq and data analysis
Details can be found in the supporting information materials and methods Data S1.
2.12. Data availability statement
RNA‐seq data that support the findings of this study have been deposited in GEO with the accession code GSE145105 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145105).
2.13. Tumor xenograft analysis
All animal experiments were performed following the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Ethics Committee of China Medical University. The stably transfected cells were harvested, washed with PBS, and resuspended at a concentration of 1 × 107 cells/mL, then 200 μL of suspended cells were injected subcutaneously into the flanks of 5‐week‐old female BALB/c nude mice. Tumor growth was examined every 2 days; 22 days after injection, mice were killed, and the tumors were obtained, imaged, dissected, and analyzed. The volume of tumors was calculated by 1/2 length × width2.
2.14. Statistical analysis
The statistical analysis was carried out using SPSS (17.0) software (SPSS). Data of JMJD1A and RUNX3 expression in GC were analyzed by Spearman’s rank correlation coefficient test. The Mann‐Whitney U test was used to analyze the association between JMJD1A or RUNX3 expression and clinical features. The survival curve was estimated using the Kaplan‐Meier method. The statistical significance of difference was analyzed by ANOVA. Statistical significance was defined as P < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001). All experiments were repeated three times, and data were expressed as the mean ± SD from a representative experiment.
3. RESULTS
3.1. Jumonji domain‐containing 1A inhibits gastric cancer cell proliferation in vitro
To investigate the effects of JMJD1A on GC cell proliferation, we performed MTT, flow cytometry, and colony formation assays. First, we knocked down JMJD1A expression via lentiviral transduction in SGC‐7901 and MGC‐803 cells (Figure 1A). In contrast, JMJD1A was overexpressed in SGC‐7901 and MGC‐803 cells using lentiviral vector (Figure 1B). MTT assays showed that depletion of JMJD1A significantly promoted cell growth compared to control (Figure 1C). CCK8 assays showed that knockdown of JMJD1A promoted the cell proliferation of MGC‐803 cells (Figure 1D) and overexpression of JMJD1A blocked the cell proliferation (Figure 1E, F). Similarly, knockdown of JMJD1A reduced the G1‐phase ratio and increased the S‐phase and G2/M‐phase ratios of SGC‐7901 cells (Figure 1G). In contrast, overexpression of JMJD1A increased the G1‐phase ratio and decreased the S‐phase ratio of SGC‐7901 cells (Figure 1H). In addition, depletion of JMJD1A enhanced and overexpression of JMJD1A decreased the colony formation ability of SGC‐7901 and MGC‐803 cells (Figure 1I,J). These data indicate that JMJD1A inhibits GC cell growth in vitro.
Figure 1.
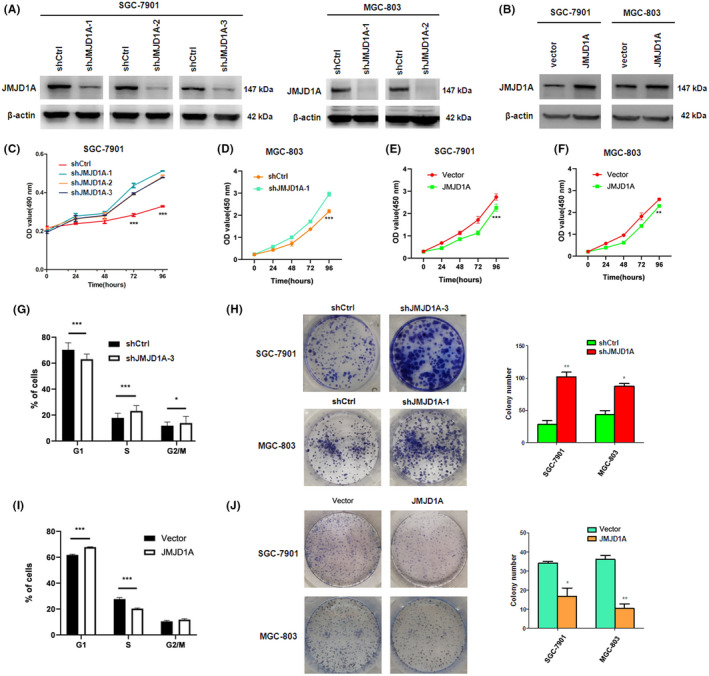
Jumonji domain‐containing 1A (JMJD1A) inhibits gastric cancer (GC) cell proliferation in vitro. Knockdown (A) or overexpression (B) of JMJD1A in SGC‐7901 cells or MGC‐803 cells as determined by western blot. (C) Cell viability was monitored by MTT assays. CCK8 was used to detect cell proliferation at different times using JMJD1A knockdown MGC‐803 cells (D), JMJD1A overexpression SGC‐7901 cells (E), or JMJD1A overexpression MGC‐803 cells (F). Cell cycle was analyzed by flow cytometry assays using JMJD1A knockdown (G) or overexpression (H) SGC‐7901 cells. Knockdown of JMJD1A increased (I) and overexpression of JMJD1A decreased (J) the colony formation of SGC‐7901 and MGC‐803 cells
3.2. Jumonji domain‐containing 1A inhibits gastric cancer cell proliferation in vivo
To further determine whether JMJD1A could inhibit GC cell tumorigenesis in vivo, SGC‐7901 cells stably transfected with shJMJD1A or shControl were subcutaneously implanted into nude mice. After 22 days, tumors were completely stripped. Photographs and measured weights of the tumors indicated that knockdown of JMJD1A in SGC‐7901 cells markedly increased xenograft tumor growth (Figure 2A,B). Moreover, JMJD1A depletion significantly increased the volumes of the tumors (Figure 2C). Consistently, histopathologic analyses revealed weak Ki‐67 staining in shCtrl xenografts, while shJMJD1A xenografts displayed strong Ki‐67 staining (Figure 2D). The expression of JMJD1A in tumor tissues was also analyzed by immunohistochemical staining (Figure 2D). These data indicate that JMJD1A inhibits GC cell proliferation in vivo.
Figure 2.
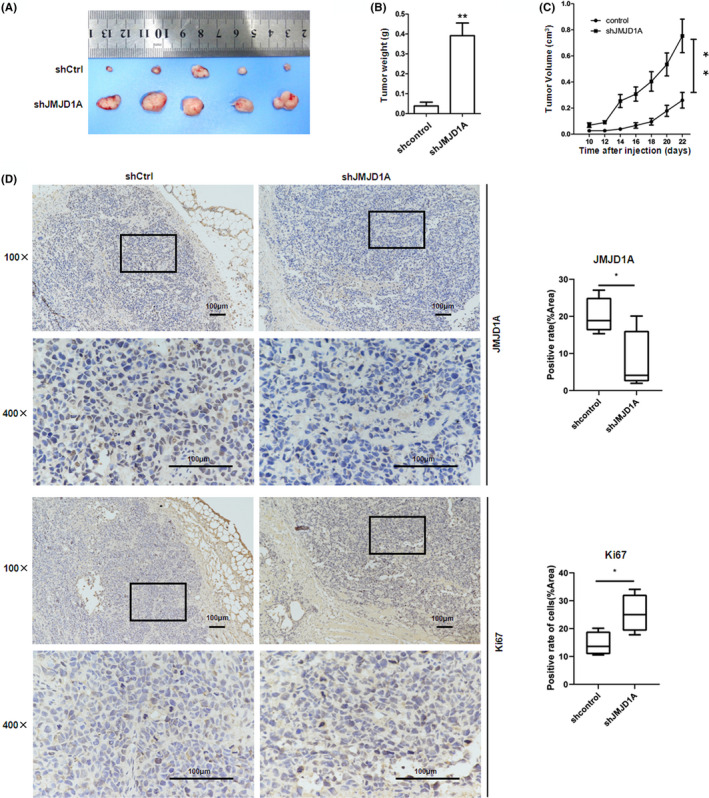
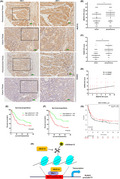
Jumonji domain‐containing 1A (JMJD1A) inhibits gastric cancer (GC) cell proliferation in vivo. Tumor images (A) and weights (B) at experimental endpoints in shControl (shCtrl) and shJMJD1A SGC‐7901 xenografts. (C) Tumor volumes, measured every 2 days after injection. (D) Immunohistochemistry (IHC) staining showed that the expression of JMJD1A differed. Scale bar, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001
3.3. Jumonji domain‐containing 1A upregulates RUNX3 expression in gastric cancer cells
To identify possible downstream targets of JMJD1A, we performed RNA sequencing to investigate the transcriptomic changes in JMJD1A stable knockdown GC cells. Analysis of the RNA sequencing results demonstrated that 1550 genes showed differential expression (fold change >2, P < 0.05); 577 of them showed increased expression in JMJD1A‐depleted SGC‐7901 cells, while 973 of them were downregulated (Figure 3A and Table S1 and S2). Gene ontology enrichment analysis indicated that these genes were enriched for broad categories of biological processes, among which we found the canonical Wnt signaling pathway (Figure 3B). Next, we found the Wnt and TGF‐β signaling pathways in the KEGG enrichment top 20 (Figure 3C). The genes shown in the heatmap (Figure 3D) were selected from the affected genes in the Wnt and TGF‐β pathways after JMJD1A knockdown, as determined by KEGG enrichment analysis. Emerging evidence has shown that dysregulations of Wnt and TGF‐β signaling pathways are important in gastric carcinogenesis. 27 Moreover, RUNX3 functions as a gatekeeper linking Wnt and TGF‐β/BMP signaling pathways in gastrointestinal cancers. 25 As a tumor suppressor, the inactivation or decreased expression of RUNX3 is closely associated with the tumorigenesis and progression of GC. 25 , 28 Therefore, we focused on RUNX3 as one of the potential targets of JMJD1A in GC. JMJD1A knockdown decreased the RUNX3 mRNA level (Figure 3E). Moreover, overexpression of JMJD1A increased the RUNX3 mRNA level (Figure 3F). In contrast, knockdown of JMJD1A reduced the RUNX3 protein level and overexpression of JMJD1A increased the RUNX3 protein level (Figure 3G,H). These data suggest that JMJD1A upregulates RUNX3 expression in GC cells.
Figure 3.
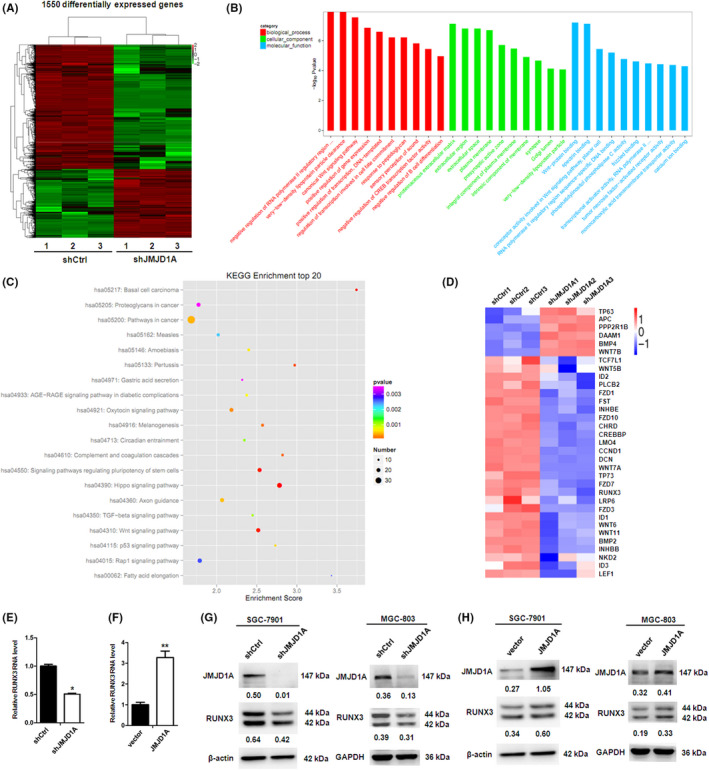
Jumonji domain‐containing 1A (JMJD1A) upregulates RUNX3 expression in gastric cancer (GC) cells. (A) Heatmap showing differentially expressed genes in SGC‐7901/shControl and SGC‐7901/shJMJD1A cells, analyzed by RNA‐seq (three biological replicates). (B) Gene ontology (GO) terms (biological processes, cellular components, and molecular functions) significantly enriched among differentially expressed genes, as determined by DAVID functional annotation analysis. (C) KEGG enrichment analysis of differentially expressed genes. (D) Heatmap of selected differentially expressed genes (the affected genes in Wnt and TGF‐β signaling pathways after JMJD1A knockdown, as determined by KEGG enrichment analysis) in JMJD1A knockdown (shJMJD1A) or control (shCtrl) SGC‐7901 cells. RUNX3 mRNA level of SGC‐7901 cells was measured by quantitative RT‐PCR after JMJD1A knockdown (E) or overexpression (F). RUNX3 protein level of SGC‐7901 and MGC‐803 cells was measured by western blot after JMJD1A knockdown (G) or overexpression (H)
3.4. Jumonji domain‐containing 1A stimulates runt‐related transcription factor 3 promoter through co–activation of transcription factor Ets‐1
The two promoter regions of the RUNX3 gene have been identified and designated P1 and P2. 29 To identify the mechanism by which JMJD1A upregulates RUNX3 transcription, we constructed RUNX3 P1 and P2‐driven luciferase reporter plasmids and performed luciferase assays. The results showed that JMJD1A was able to increase the P1 activity in a dose‐dependent manner (Figure 4A) but not P2 (Figure 4B). The reported potential transcription factor binding sites in the P1 region are shown in Figure 4C. 29 We mutated the Ets‐1 or CREB1 binding site and performed luciferase assays. Mutation or deletion of the Ets‐1 binding site impaired the increase of P1 activity by Ets‐1 (Figure 4D). Importantly, the promotion of P1 activity by JMJD1A was also impaired when the Ets‐1 binding site was mutated or deleted (Figure 4E), suggesting that Ets‐1 plays an important role in the activation of RUNX3 mediated by JMJD1A. When the CREB1 binding site was mutated, the increase of P1 activity by CREB1 was impaired (Figure S1). Moreover, luciferase assays showed that JMJD1A was able to enhance P1 activity independently and cumulatively with Ets‐1 (Figure 4F). ChIP assays showed that endogenous JMJD1A or Ets‐1 was associated with the RUNX3 promoter P1 (Figure 4G‐H). ChIP assays also indicated that exogenous Ets‐1 was associated with P1 (Figure 4I). Importantly, ChIP Re‐ChIP assays indicated that Ets‐1 and JMJD1A acted in a combined manner on the RUNX3 promoter P1 (Figure 4J). We further observed that JMJD1A decreased the H3K9me1 level at the P1 and P2 region of RUNX3 promoter and H3K9me2 level at the P1 region (Figure 4K). Furthermore, our ChIP results showed that Ets‐1 knockdown increased the H3K9me2 level at promoter P1 (Figure 4L), indicating that Ets‐1 is fundamental for the H3K9me2 level at promoter P1. Immunoprecipitation assay confirmed that there was an interaction between JMJD1A and Ets‐1 in SGC‐7901 cells (Figure 4M). These data suggest that JMJD1A activates RUNX3 transcription through co–activating Ets‐1 and decreasing the H3K9me1/2 levels at the RUNX3 promoter.
Figure 4.
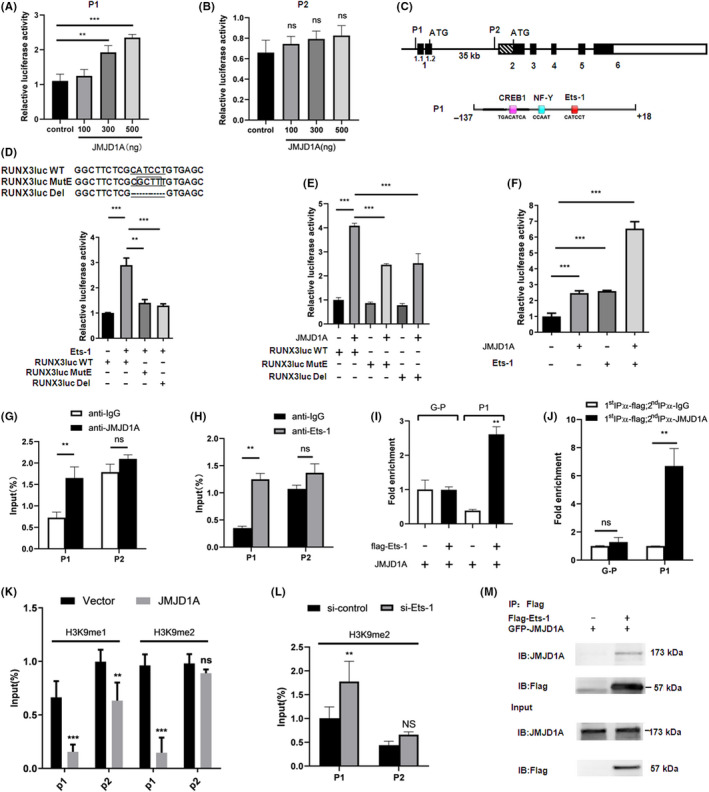
Jumonji domain‐containing 1A (JMJD1A) stimulates RUNX3 promoter through co–activation of transcription factor Ets‐1. The activities of pGL3‐P1 (A) or pGL3‐P2 (B) were measured by luciferase assays in cells transfected with increasing amounts of JMJD1A expression vector. (C) Schematic diagram depicting genomic organization of the human RUNX3 gene and the three potential transcription factor binding sites in the P1 promoter of RUNX3. Black boxes represent coding exons, whereas the other boxes represent UTR. The effect of Ets‐1 (D) or JMJD1A (E) on the activities of RUNX3 promoter P1 of wild type (WT) or the mutant (MutE) or the delete (Del) was examined by luciferase assays in HEK293 cells. (F) JMJD1A and Ets‐1 were transiently transfected into HEK293 cells as indicated, and the P1 promoter activity was estimated by luciferase assays. ***P < 0.001. (G,H) SGC‐7901 cells were subjected to ChIP assays with antibodies as indicated, followed by quantitative PCR with primers amplifying the RUNX3 promoter P1 and P2. (I) The interaction of Ets‐1 with the P1 region of RUNX3 promoter was examined by ChIP assays. SGC‐7901 cells were transfected with flag‐Ets‐1 and JMJD1A expression vector as indicated. ChIP was carried out using antibody against flag, followed by PCR with primers amplifying the RUNX3 promoter P1 (P1) or GAPDH promoter (G‐P). J, SGC‐7901 cells were transiently transfected with JMJD1A expression vector and flag‐Ets‐1 vector. Then ChIP re‐ChIP assays were carried out to examine whether JMJD1A and Ets‐1 were assembled on the same promoter (P1). Soluble chromatin was first immunoprecipitated with antibody against flag (1st IP). The complexes eluted from the 1st IP were divided into two aliquots, followed by reimmunoprecipitation with antibody against IgG or JMJD1A (2nd IP), respectively. The PCR primers were specific for amplifying the RUNX3 promoter P1 (P1) or GAPDH promoter (G‐P). (K) ChIP‐quantitative PCR (qPCR) was carried out using H3K9me1 or H3K9me2 Abs, and negative control Abs (IgG) in SGC‐7901 cells, followed by qPCR with primers amplifying the RUNX3 promoter P1 and P2 region. (L) ChIP assays were carried out with antibody against H3K9me2 in control and Ets‐1 knockdown MGC‐803 cells, followed by qPCR with primers amplifying the RUNX3 promoter P1 and P2. (M) The interaction of JMJD1A with Ets‐1 was examined by immunoprecipitation (IP) assays in SGC‐7901 cells
To determine whether JMJD1A had a role in the regulation of Ets‐1, we performed quantitative real‐time PCR and western blot analysis using JMJD1A‐overexpressed SGC‐7901 cells. The results showed that JMJD1A overexpression increased Ets‐1 mRNA and protein levels, indicating that JMJD1A upregulated Ets‐1 expression in SGC‐7901 cells (Figure S2). Moreover, our western blot results showed that transient overexpression of Ets‐1 increased the RUNX3 protein level in SGC‐7901 cells (Figure S3). Using the TCGA database, we analyzed the correlation between Ets‐1 relative expression and RUNX3 relative expression. The result showed that the Ets‐1 expression level was positively correlated with the RUNX3 expression level in GC patients (Figure S4A). Using the Kaplan‐Meier plotter database, we made survival curves of GC patients based on Ets‐1 expression. Unfortunately, although it has a tendency that low expression of Ets‐1 was correlated to a poor prognosis in patients with GC, it has no statistical significance (P > 0.05, Figure S4B).
3.5. Jumonji domain‐containing 1A inhibits the growth of gastric cancer cells in vivo, partially dependent on RUNX3
As RUNX3 plays important roles in GC 30 and JMJD1A upregulates RUNX3 expression in GC cells we wondered whether the inhibition of GC cell growth by JMJD1A is RUNX3‐dependent. We used nude mouse models to examine the effect of RUNX3 on JMJD1A‐mediated GC cell proliferation in vivo. As expected, silencing of JMJD1A by shRNA significantly promoted the growth of GC cells in mice and RUNX3 overexpression impaired the growth promotion induced by shJMJD1A (Figure 5A‐C), suggesting that RUNX3 plays an important role in the inhibition of GC cell proliferation induced by JMJD1A. Meanwhile, the expression levels of JMJD1A and RUNX3 in tumor samples from mice were validated by western blot analysis (Figure 5D). Conversely, JMJD1A overexpression remarkably inhibited the growth of GC cells in mice and silencing of RUNX3 by shRNA partially reversed the inhibitory effects of JMJD1A overexpression on tumor volume and weight (Figure S5), indicating that the inhibition of tumorigenicity by JMJD1A is partially RUNX3‐dependent. Thus, we conclude that RUNX3 plays an important role in JMJD1A‐mediated GC cell growth inhibition in vivo.
Figure 5.
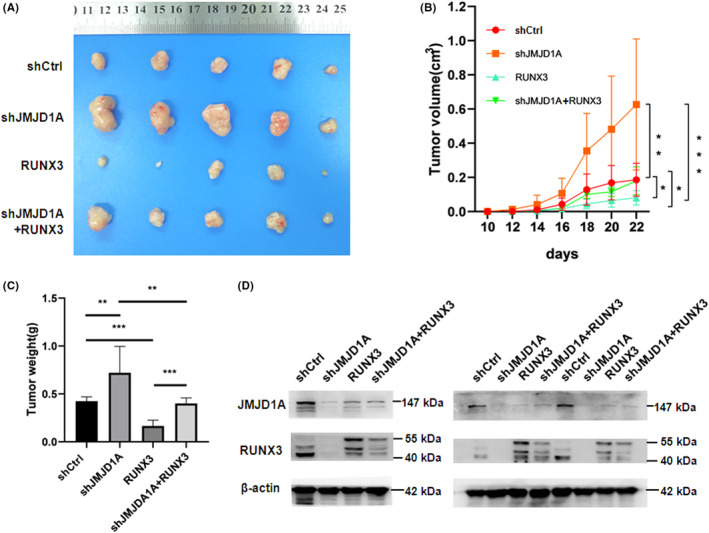
Jumonji domain‐containing 1A (JMJD1A) inhibits the growth of gastric cancer (GC) cells in vivo, which is partially dependent on RUNX3. (A) Xenograft tumors were obtained from different groups of nude mice transfected with shCtrl, shJMJD1A, RUNX3, and shJMJD1A + RUNX3, respectively. The growth curves (B) and the average weights (C) of tumors from different groups of nude mice were shown. (D) The expression levels of JMJD1A and RUNX3 were examined by western blot analysis in tumor tissues from mice. The results of three independent samples are shown. RUNX3 was overexpressed by lentiviral transduction. We used lentiviral vector Ubi‐MCS‐3FLAG‐SV40‐Cherry to construct recombinant vector. The approximately 55 kD band in the RUNX3 blotting (which was upper) was due to the Cherry tag
3.6. Jumonji domain‐containing 1A expression is positively associated with runt‐related transcription factor 3 expression in gastric cancer samples
To further explore the role of JMJD1A in GC as well as to substantiate the functional link between JMJD1A and RUNX3, expression levels of JMJD1A and RUNX3 were examined by immunohistochemical staining in GC tissue microarrays. Representative images are shown in Figure 6A. Unlike adjacent noncancerous tissue, low expression levels of JMJD1A and RUNX3 were observed in GC tissues (Figure 6A‐C). Importantly, the JMJD1A expression level was positively correlated with the RUNX3 expression level (Figure 6D). To further investigate the important role of JMJD1A or RUNX3 clinically, we explored the correlation between JMJD1A or RUNX3 expression and clinicopathological characteristics of GC patients. These samples were classified into two groups based on JMJD1A or RUNX3 levels (histological score). The data showed that low expression of JMJD1A was significantly correlated with clinical stages (P = .017), lymph node metastasis (P = 0.029) and pathological stages (P = 0.033; Table 1). The data also showed that decreased expression of RUNX3 was notably correlated with clinical stages (P = 0.003), depth of invasion (P = 0.039), and lymph node metastasis (P = 0.009; Table 2). Furthermore, the Kaplan‐Meier analysis showed that the low JMJD1A expression or low RUNX3 expression significantly reduced overall survival (Figure 6E‐F). Survival curves using the Kaplan‐Meier plotter database showed that GC patients with low JMJD1A expression had a shorter overall survival (Figure 6G). Taken together, these data indicate that the expression of JMJD1A is decreased in GC tissues, leading to the decreased expression of RUNX3. The low expression of JMJD1A correlates with poor prognosis in GC patients.
Figure 6.
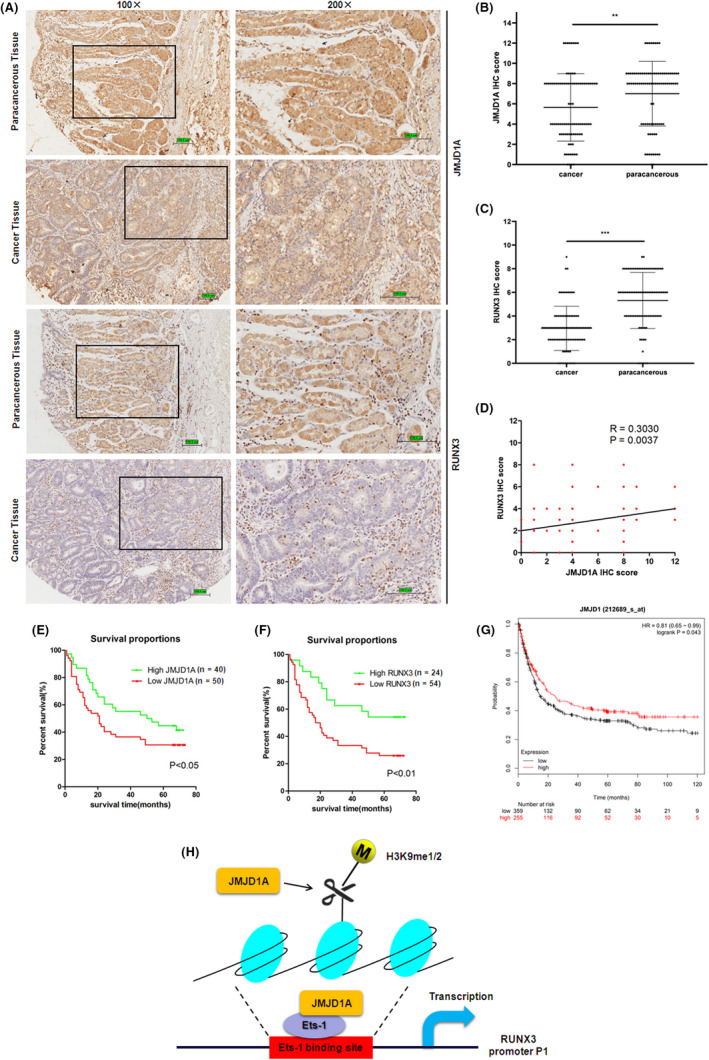
Jumonji domain‐containing 1A (JMJD1A) expression is positively associated with runt‐related transcription factor 3 (RUNX3) expression in gastric cancer (GC) samples. (A) The expression levels of JMJD1A and RUNX3 were examined by immunohistochemistry (IHC) staining analysis in paracancerous tissue and GC tissue of tissue microarray. Scale bar, 100.8 μm. The IHC scores of JMJD1A (B) and RUNX3 (C) were quantified and plotted as graphs. (D) Spearman’s rank test was used to analyze the correlation between JMJD1A and RUNX3 relative expression in GC samples. Kaplan‐Meier survival curves of gastric cancer patients based on JMJD1A expression (E) and RUNX3 expression (F). (G) Kaplan‐Meier survival curves of gastric cancer patients based on JMJD1A expression using Kaplan‐Meier plotter database. (H) Model shows the mechanism by which JMJD1A upregulates RUNX3 by Ets‐1 in gastric cancer cells
Table 1.
Correlation between JMJD1A expression and clinicopathological characteristics of gastric cancer patients
| Feature | n | JMJD1A expression | P‐value | |
|---|---|---|---|---|
| Weak | Strong | |||
| Age (y) | ||||
| <65 | 44 | 26 | 18 | 0.532 |
| ≥65 | 46 | 24 | 22 | |
| Gender | ||||
| Male | 53 | 28 | 25 | 0.667 |
| Female | 37 | 22 | 15 | |
| Tumor size (cm) | ||||
| <5 cm | 35 | 22 | 13 | 0.286 |
| ≥5 cm | 55 | 28 | 27 | |
| Clinical stages | ||||
| Stages Ⅰ, Ⅱ | 36 | 14 | 22 | 0.017 * |
| Stage Ⅲ | 54 | 36 | 18 | |
| Depth of invasion (pT) | ||||
| T1, T2 | 14 | 5 | 9 | 0.145 |
| T3, T4 | 76 | 45 | 31 | |
| Lymph node metastasis (pN) | ||||
| N0, N1 | 35 | 14 | 21 | 0.029 * |
| N2, N3 | 55 | 36 | 19 | |
| Distant metastasis (pM) | ||||
| M0 | 86 | 48 | 38 | 1.000 |
| M1 | 4 | 2 | 2 | |
| Pathological stage (pStage) | ||||
| Stages Ⅰ, Ⅱ | 40 | 17 | 23 | 0.033 * |
| Stages Ⅲ, Ⅳ | 50 | 33 | 17 | |
Bold values indicate statistical significance.
P < 0.05.
Table 2.
Correlation between RUNX3 expression and clinicopathological characteristics of gastric cancer patients
| Feature | n | RUNX3 expression | P‐value | |
|---|---|---|---|---|
| Weak | Strong | |||
| Age (y) | ||||
| <65 | 41 | 25 | 16 | 0.532 |
| ≥65 | 37 | 29 | 8 | |
| Gender | ||||
| Male | 45 | 31 | 14 | 1.000 |
| Female | 33 | 23 | 10 | |
| Tumor size(cm) | ||||
| <5 cm | 30 | 20 | 10 | 0.802 |
| ≥5 cm | 48 | 34 | 14 | |
| Clinical stages | ||||
| Stages Ⅰ, Ⅱ | 32 | 16 | 16 | 0.003 * |
| Stage Ⅲ | 46 | 38 | 8 | |
| Depth of invasion (pT) | ||||
| T1, T2 | 12 | 5 | 7 | 0.039 * |
| T3, T4 | 66 | 49 | 17 | |
| Lymph node metastasis (pN) | ||||
| N0, N1 | 30 | 16 | 14 | 0.009 * |
| N2, N3 | 50 | 40 | 10 | |
| Distant metastasis (pM) | ||||
| M0 | 75 | 51 | 24 | 0.549 |
| M1 | 3 | 3 | 0 | |
| Pathological stage (pStage) | ||||
| Stages Ⅰ, Ⅱ | 32 | 21 | 11 | 0.623 |
| Stages Ⅲ, Ⅳ | 46 | 33 | 13 | |
Bold values indicate statistical significance.
P < 0.05.
4. DISCUSSION
Increasing evidence has shown that JMJD1A promotes proliferation, survival, and metastasis in several types of cancers, including prostate cancer, colorectal cancer, hepatocellular carcinoma, ovarian cancer, bladder cancer, and Ewing sarcoma. 14 , 15 , 16 , 17 , 18 , 19 , 20 , 31 , 32 , 33 , 34 Moreover, it has been reported that JMJD1A is downregulated in nasopharyngeal carcinoma (NPC) and downregulation of JMJD1A is associated with poor prognosis of NPC patients. 35 JMJD1A is also downregulated in human germ cell‐derived tumors, such as seminomas, yolk sac tumors, and embryonal carcinomas, and acts as a tumor suppressor. 36 JMJD1A functions as either a tumor promoter 37 , 38 or tumor suppressor 39 in breast cancer, as reported previously. However, little is known about the role and the underlying mechanism of JMJD1A in tumorigenesis and progression of GC. Here we verified a critical role of JMJD1A in suppressing proliferation of GC cells. Consistently, JMJD1A is downregulated in GC tissues. Furthermore, the decreased expression of JMJD1A was demonstrated to be associated with an aggressive phenotype and a poor prognosis in patients with GC. Our results support JMJD1A being a potential tumor suppressor in GC.
Jumonji domain‐containing 1A functions in tumorigenesis and progression by controlling gene expression. Given that JMJD1A acts as a transcription co–activator to control cancer cell growth and survival, 16 , 40 we speculated that JMJD1A might be involved in the regulation of RUNX3 by co–activating some transcription factors. Although our data suggest that Ets‐1 is essential for the upregulation of RUNX3 induced by JMJD1A, we cannot rule out that other transcription factors play a role in JMJD1A‐mediated transcriptional control of RUNX3, such as CREB1. Further study is needed.
Our results showed that Ets‐1 is essential for the upregulation of RUNX3 by JMJD1A. Having demonstrated that JMJD1A was able to interact with Ets‐1 (Figure 4M), we wondered whether JMJD1A had a role in the regulation of Ets‐1. JMJD1A bound and demethylated H3K9me2 at Ets‐1 promoter in Ewing Sarcoma cells, 41 indicating that JMJD1A might increase Ets‐1 expression. Our results showed that JMJD1A overexpression increased Ets‐1 mRNA and protein levels in SGC‐7901 cells (Figure S2), which was consistent with the previous report. 41 Further studies are needed to investigate the mechanism by which JMJD1A regulates Ets‐1.
Because Ets‐1 promotes invasion, metastasis and angiogenesis, it is known to be oncogenic. 42 Beside promoting cell invasion, Ets‐1 attenuates cell growth in breast tumors as well as metastases, 32 indicating that Ets‐1 plays a dual role in tumor biology. Moreover, Ets‐1 is a transcriptional activator of p16INK4a, a known tumor suppressor. 43 We showed the Ets‐1‐dependency in upregulation of RUNX3 in GC cells and the positive correlation between Ets‐1 and RUNX3 expression in GC patients (Figure S3 and Figure S4A), indicating that Ets‐1 is a positive regulator of RUNX3. As for the relationship between Ets‐1 expression and prognosis of GC patients, further study is needed.
Runt‐related transcription factor 3 functions as a gatekeeper linking the Wnt oncogenic and TGF‐β/BMP tumor‐suppressive pathways, and is involved in cell proliferation, apoptosis, angiogenesis, cell adhesion, and invasion. 25 , 44 , 45 A large amount of information indicates that RUNX3 may be a tumor suppressor in a multitude of epithelial cancers, including GC, colorectal cancer, breast cancer and esophageal cancer. 45 In this study, we showed the low expression of RUNX3 in GC tissues compared with adjacent noncancerous tissues, which is consistent with previous research. 23 Moreover, we demonstrated that the decreased expression of RUNX3 was correlated with an aggressive phenotype and a poor prognosis in patients with GC.
Runt‐related transcription factor 3 is regulated by two distantly located promoters, P1 and P2. 29 P2 is CpG island‐rich, but P1 is not. Because the expressed RUNX3 in gastric cancer cells is predominantly P2‐RUNX3, most studies of RUNX3 in gastric cancer focus on the transcript from P2. It has been shown that the transcripts generated by RUNX3 P2 have tumor suppressor activity. 44 RUNX3 P2 has been reported frequently to be hypermethylated in various cancers, 46 , 47 , 48 , 49 whereas the role for the alternate non‐CpG island RUNX3 P1 has been overlooked. In fact, genes with non–CpG island promoters share many epigenetic features that are associated with CpG island promoter genes, despite their low CpG density. For example, it has been reported that DNA methylation occurring at CpG poor RUNX3 P1 directly leads to transcriptional silencing in 623 melanoma cells. 50 Furthermore, using nucleosome occupancy methylome‐sequencing, NOMe‐Seq, and clonal analysis, it was shown that RUNX3 P1 is monoallelically methylated. 50 These results suggest that aberrant methylation patterns of RUNX3 P1 may also contribute to tumorigenesis and should, therefore, be included in analyses of cancer epigenetics. Moreover, Kurklu et al 51 uncovered a preneoplastic P1 hypomethylation signature reflecting altered cell‐type composition of the gastric epithelium/tumor microenvironment via immune cell infiltration. Here, we considered both P1 and P2, and found that JMJD1A mainly affected the activity of RUNX3 P1 (Figure 4A‐B) and endogenous JMJD1A or Ets‐1 was associated with RUNX3 P1 (Figure 4G‐H). Interestingly, we found that JMJD1A decreased the H3K9me1 level at both P1 and P2 regions of RUNX3 promoter (Figure 4K). We cannot rule out that P2 plays a role in the JMJD1A‐mediated transcriptional control of RUNX3. The role of P2 in JMJD1A‐mediated RUNX3 transcriptional regulation needs to be further studied.
Taken together, in this study we report a novel function of JMJD1A in the regulation of RUNX3 through transcription factor Ets‐1 in GC. We proposed a hypothesized model showing the mechanism by which JMJD1A upregulates RUNX3 through co–activating Ets‐1 in GC cells (Figure 6H). Our findings provide new insight into the mechanism of RUNX3 regulation mediated by JMJD1A in GC and suggest that JMJD1A and/or RUNX3 may be used as a therapeutic intervention for GC.
DISCLOSURE
Authors declare no conflicts of interest for this article.
Supporting information
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Data S1
ACKNOWLEDGMENTS
We would like to thank Dr Min Wu (Wuhan University, College of Life Sciences) for providing the JMJD1A expression vector, Dr Zhenguo Cheng (China Medical University, School of Life Sciences), PhD student Ruizhi Zhou (China Medical University, School of Life Sciences) and Dr Yang Li (China Medical University, School of Life Sciences) for technical support. This work was supported by grants from the National Natural Science Foundation of China (Nos. 31271389, 31571457, 31771553, and 31970741), the Natural Science Foundation of Liaoning Province of China (No. 20180550551), and the Scientific Research Fund of Liaoning Provincial Education Department (LK201630). All procedures for mouse experiments were approved by the Animal Ethics Committee of the China Medical University.
Ning K, Shao Y, He Y, et al. Histone demethylase Jumonji domain‐containing 1A inhibits proliferation and progression of gastric cancer by upregulating runt‐related transcription factor 3. Cancer Sci. 2020;111:3679–3692. 10.1111/cas.14594
Ning and Shao contributed equally to this work.
Contributor Information
Yangguang Shao, Email: ygshao@cmu.edu.cn.
Feng Li, Email: lifeng@cmu.edu.cn.
REFERENCES
- 1. Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet. 2016;388:2654‐2664. [DOI] [PubMed] [Google Scholar]
- 2. Macdonald JS, Smalley SR, Benedetti J, et al. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345:725‐730. [DOI] [PubMed] [Google Scholar]
- 3. Das M. Neoadjuvant chemotherapy: survival benefit in gastric cancer. Lancet Oncol. 2017;18:e307. [DOI] [PubMed] [Google Scholar]
- 4. Jacome AA, Coutinho AK, Lima EM, et al. Personalized medicine in gastric cancer: where are we and where are we going? World J Gastroenterol. 2016;22:1160‐1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dimitrova E, Turberfield AH, Klose RJ. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015;16:1620‐1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC‐containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483‐495. [DOI] [PubMed] [Google Scholar]
- 7. Okada Y, Scott G, Ray MK, et al. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119‐123. [DOI] [PubMed] [Google Scholar]
- 8. Tateishi K, Okada Y, Kallin EM, et al. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Inagaki T, Tachibana M, Magoori K, et al. Obesity and metabolic syndrome in histone demethylase JHDM2a‐deficient mice. Genes Cells. 2009;14:991‐1001. [DOI] [PubMed] [Google Scholar]
- 10. Kuroki S, Matoba S, Akiyoshi M, et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106‐1109. [DOI] [PubMed] [Google Scholar]
- 11. Loh YH, Zhang W, Chen X, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self‐renewal in embryonic stem cells. Genes Dev. 2007;21:2545‐2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma DK, Chiang CH, Ponnusamy K, et al. G9a and Jhdm2a regulate embryonic stem cell fusion‐induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131‐2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101:e115‐e123. [DOI] [PubMed] [Google Scholar]
- 14. Peng K, Su G, Ji J, et al. Histone demethylase JMJD1A promotes colorectal cancer growth and metastasis by enhancing Wnt/beta‐catenin signaling. J Biol Chem. 2018;293:10606‐10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang HY, Long QY, Tang SB, et al. Histone demethylase KDM3A is required for enhancer activation of hippo target genes in colorectal cancer. Nucleic Acids Res. 2019;47:2349‐2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan L, Peng G, Sahgal N, et al. Regulation of c‐Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene. 2016;35:2441‐2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park SJ, Kim JG, Son TG, et al. The histone demethylase JMJD1A regulates adrenomedullin‐mediated cell proliferation in hepatocellular carcinoma under hypoxia. Biochem Biophys Res Commun. 2013;434:722‐727. [DOI] [PubMed] [Google Scholar]
- 18. Ramadoss S, Sen S, Ramachandran I, et al. Lysine‐specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene. 2017;36:1537‐1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cho HS, Toyokawa G, Daigo Y, et al. The JmjC domain‐containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer. 2012;131:E179‐E189. [DOI] [PubMed] [Google Scholar]
- 20. Wan W, Peng K, Li M, et al. Histone demethylase JMJD1A promotes urinary bladder cancer progression by enhancing glycolysis through coactivation of hypoxia inducible factor 1alpha. Oncogene. 2017;36:3868‐3877. [DOI] [PubMed] [Google Scholar]
- 21. Yang H, Liu Z, Yuan C, et al. Elevated JMJD1A is a novel predictor for prognosis and a potential therapeutic target for gastric cancer. Int J Clin Exp Pathol. 2015;8:11092‐11099. [PMC free article] [PubMed] [Google Scholar]
- 22. Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15:81‐95. [DOI] [PubMed] [Google Scholar]
- 23. Li QL, Ito K, Sakakura C, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113‐124. [DOI] [PubMed] [Google Scholar]
- 24. Fan XY, Hu XL, Han TM, et al. Association between RUNX3 promoter methylation and gastric cancer: a meta‐analysis. BMC Gastroenterol. 2011;11:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito K. RUNX3 in oncogenic and anti–oncogenic signaling in gastrointestinal cancers. J Cell Biochem. 2011;112:1243‐1249. [DOI] [PubMed] [Google Scholar]
- 26. Manandhar S, Lee YM. Emerging role of RUNX3 in the regulation of tumor microenvironment. BMB Rep. 2018;51:174‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu WK, Cho CH, Lee CW, et al. Dysregulation of cellular signaling in gastric cancer. Cancer Lett. 2010;295:144‐153. [DOI] [PubMed] [Google Scholar]
- 28. Ju X, Ishikawa TO, Naka K, et al. Context‐dependent activation of Wnt signaling by tumor suppressor RUNX3 in gastric cancer cells. Cancer Sci. 2014;105:418‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bangsow C, Rubins N, Glusman G, et al. The RUNX3 gene–sequence, structure and regulated expression. Gene. 2001;279:221‐232. [DOI] [PubMed] [Google Scholar]
- 30. Nazir SU, Kumar R, Singh A, et al. Breast cancer invasion and progression by MMP‐9 through Ets‐1 transcription factor. Gene. 2019;711:143952. [DOI] [PubMed] [Google Scholar]
- 31. Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20‐38. [DOI] [PubMed] [Google Scholar]
- 32. Furlan A, Vercamer C, Bouali F, et al. Ets‐1 controls breast cancer cell balance between invasion and growth. Int J Cancer. 2014;135:2317‐2328. [DOI] [PubMed] [Google Scholar]
- 33. Furlan A, Vercamer C, Heliot L, et al. Ets‐1 drives breast cancer cell angiogenic potential and interactions between breast cancer and endothelial cells. Int J Oncol. 2019;54:29‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lotem J, Levanon D, Negreanu V, et al. Runx3 at the interface of immunity, inflammation and cancer. Biochim Biophys Acta. 2015;1855:131‐143. [DOI] [PubMed] [Google Scholar]
- 35. Du ZM, Hu LF, Wang HY, et al. Upregulation of MiR‐155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One. 2011;6:e19137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ueda J, Ho JC, Lee KL, et al. The hypoxia‐inducible epigenetic regulators Jmjd1a and G9a provide a mechanistic link between angiogenesis and tumor growth. Mol Cell Biol. 2014;34:3702‐3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao QY, Lei PJ, Zhang X, et al. Global histone modification profiling reveals the epigenomic dynamics during malignant transformation in a four‐stage breast cancer model. Clin Epigenetics. 2016;8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramadoss S, Guo G, Wang CY. Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non‐histone protein p53. Oncogene. 2017;36:47‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pedanou VE, Gobeil S, Tabaries S, et al. The histone H3K9 demethylase KDM3A promotes anoikis by transcriptionally activating pro‐apoptotic genes BNIP3 and BNIP3L. Elife. 2016;5:e16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wade MA, Jones D, Wilson L, et al. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res. 2015;43:196‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sechler M, Parrish JK, Birks DK, Jedlicka P. The histone demethylase KDM3A, and its downstream target MCAM, promote Ewing Sarcoma cell migration and metastasis. Oncogene. 2017;36:4150‐4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oikawa T. ETS transcription factors: possible targets for cancer therapy. Cancer Sci. 2004;95:626‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krishnamurthy J, Torrice C, Ramsey MR, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bae SC, Choi JK. Tumor suppressor activity of RUNX3. Oncogene. 2004;23:4336‐4340. [DOI] [PubMed] [Google Scholar]
- 45. Subramaniam MM, Chan JY, Yeoh KG, et al. Molecular pathology of RUNX3 in human carcinogenesis. Biochim Biophys Acta. 2009;1796:315‐331. [DOI] [PubMed] [Google Scholar]
- 46. Kang GH, Lee S, Lee HJ, Hwang KS. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202:233‐240. [DOI] [PubMed] [Google Scholar]
- 47. Kim TY, Lee HJ, Hwang KS, et al. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest. 2004;84:479‐484. [DOI] [PubMed] [Google Scholar]
- 48. Jiang Y, Tong D, Lou G, et al. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology. 2008;75:244‐251. [DOI] [PubMed] [Google Scholar]
- 49. Wolff EM, Liang G, Cortez CC, et al. RUNX3 methylation reveals that bladder tumors are older in patients with a history of smoking. Cancer Res. 2008;68:6208‐6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Han H, Cortez CC, Yang X, et al. DNA methylation directly silences genes with non‐CpG island promoters and establishes a nucleosome occupied promoter. Hum Mol Genet. 2011;20:4299‐4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurklu B, Whitehead RH, Ong EK, et al. Lineage‐specific RUNX3 hypomethylation marks the preneoplastic immune component of gastric cancer. Oncogene. 2015;34:2856‐2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Figure S4
Figure S5
Table S1
Table S2
Data S1
Data Availability Statement
RNA‐seq data that support the findings of this study have been deposited in GEO with the accession code GSE145105 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145105).


