Abstract
Ovarian cancer is the most lethal gynecological cancer due to lack of early screening methods and acquired drug resistance. MicroRNAs (miRNAs) are effective post‐transcriptional regulators that are transferred by extracellular vesicles, such as exosomes. Numerous studies have revealed that miRNAs are differentially expressed in epithelial ovarian cancer and act either as oncogenes or tumor suppressor genes. Cancer cells secrete exosomes containing miRNAs, which exert various effects on the components of the tumor microenvironment, including cancer‐associated fibroblasts, macrophages, and adipocytes. Conversely, cancer cells also receive exosomes from these cells. As a result of cell‐to‐cell communication, epithelial ovarian cancer acquires a more aggressive phenotype and resistance to multiple drugs. In addition, some circulating miRNAs are protected from RNase degradation in the peripheral blood and can be potential non‐invasive biomarkers. In particular, the combination of several circulating miRNAs enhances the accuracy of cancer screening. Likewise, comprehensive analyses revealed specific miRNA signatures in non‐epithelial ovarian tumors and several miRNAs contributing to alterations of carcinogenic pathways. Overall, miRNAs play a crucial role in ovarian cancer progression. In this review, we discuss the emerging roles of intra‐ and extracellular miRNAs in ovarian cancers. In the near future, miRNAs will be practical biomarkers and computational deep learning will help in the clinical application of miRNAs. Moreover, miRNAs are potential therapeutic targets and agents, and there are ongoing clinical trials of miRNA replacement therapy. Therefore, accelerating research on miRNA might improve the prognosis of patients with ovarian cancer.
Keywords: epithelial ovarian cancer, miRNAs, non‐epithelial ovarian tumor, non‐invasive biomarkers, treatment resistance
In this review, we discuss the emerging roles of intra‐ and extracellular miRNAs in ovarian cancers. In the near future, miRNAs will be practical biomarkers and computational deep learning will help in the clinical application of miRNAs.
Abbreviations
- CAF
cancer‐associated fibroblast
- EMT
epithelial‐mesenchymal transition
- EOC
epithelial ovarian carcinoma
- MET
mesenchymal‐epithelial transition
- miR‐200f
miR‐200 family
- miRNA
microRNA
- PARPi
PARP inhibitors
- TAM
tumor‐associated macrophages
1. INTRODUCTION
Ovarian cancer is the most lethal gynecological cancer, and approximately 300 000 new cases and 18 000 deaths were reported worldwide in 2018. 1 Ovarian tumors show a variety of histological representations and it is now evident that EOC consists of different histological subtypes with distinct molecular biology, tissues of origin, and clinical presentation. 2 Treatment strategies for EOC have dramatically changed with the approval of drugs such as bevacizumab and olaparib. 2 , 3 , 4 However, clinical challenges such as accurate screening and treatment for drug‐resistant EOC remain persistent. Moreover, for non‐epithelial malignant tumors, elucidation of the pathology and development of effective therapeutic methods are highly demanded.
Research relating to microRNAs (miRNAs) has been growing exponentially since it was discovered in C elegans in 1993. 5 There are 2654 annotated mature miRNAs in the human genome; miRNAs modulate the expression of their target genes by interacting with partial complementary 3′‐untranslated regions of the genes. 6 , 7 , 8 Moreover, miRNAs can be encapsulated and delivered to targets by extracellular vesicles such as exosomes, which are small vesicles composed of a lipid bilayer, and mediate cell‐to‐cell communication in the local and distant microenvironment. 9 , 10 Through exosomal transfer, the delivered miRNAs and mRNAs may demonstrate oncogenic functions in recipient cells; thus miRNAs play pivotal roles in cancer progression. 11
In this review, we discuss the recent findings on miRNA in ovarian cancers, including non‐epithelial tumors, and the future prospects of miRNAs for clinical applications.
2. RECONSIDERING THE IMPACT OF miRNAS IN EOC PROGRESSION
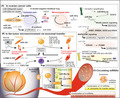
One of the most well known miRNAs is the miR‐200 family (miR‐200f), which is composed of the miR‐200ba/429 and miR‐200c/141 clusters. 6 The expression of miR‐200f is stimulated by oxidative stress and is upregulated in EOC tissues compared with normal tissues (Figure 1A and Table 1). 12 , 13 Importantly, miR‐200f is also associated with EMT, and it is demonstrated as a double‐negative feedback loop between miR‐200f and its target ZEB1/2. 14 This study reported that high levels of miR‐200f inhibit ZEB1 in the epithelial state but, conversely, high levels of ZEB1 inhibit miR‐200f in the mesenchymal state. In general, cancer cells gradually acquire more‐mesenchymal characteristics, which confer more aggressive invasion‐metastatic potential. 15 However, it cannot be concluded that low expression of miR‐200f is associated with more malignant behavior. According to a meta‐analysis, high expression of miR‐200f was associated with poor survival of patients with EOC. 16 Moreover, miR‐141 and miR‐200a promote proliferation, whereas miR‐200b inhibits it in EOC cell lines. 12 , 17 Therefore, the functions of miR‐200f can be different depending on the miRNA subtype and cell type.
FIGURE 1.
The role of miRNAs in epithelial ovarian cancer
TABLE 1.
Direct target genes and functions of miRNAs
| miRNA | Direct target gene | Function | Category | Ref |
|---|---|---|---|---|
| miR‐9 | BRCA1 | Sensitization to cisplatin | Anti‐ | 42 |
| miR‐21‐3p | SOCS4, SOCS5 | Remodeling of macrophages to TAMs | Pro‐ | 27 |
| miR‐21‐5p | STAT3 | Treg/Th17 imbalance | Pro‐ | 28 |
| miR‐21 | APAF1 | Enhancement of resistance to paclitaxel | Pro‐ | 49 |
| miR‐21 | unknown | Promotion of angiogenesis | Pro‐ | 55 |
| miR‐29a‐3p | STAT3 | Treg/Th17 imbalance | Pro‐ | 28 |
| miR‐92a | DKK1 | Maintenance of stemness | Pro‐ | 20 |
| miR‐99a‐5p | FN1, VTN | Promotion of cancer cell invasion by remodeling peritoneal mesothelial cells | Pro‐ | 30 |
| miR‐141 | p38α | Promotion of cell proliferation/sensitization to paclitaxel | Pro‐/anti‐ | 12 |
| miR‐141 | CXCL12β | Maintenance of immunocompetence | Anti‐ | 26 |
| miR‐141 | unknown | Sensitization to paclitaxel/enhancement of resistance to carboplatin | Anti‐/pro‐ | 39 |
| miR‐141 | KEAP1 | Enhancement of resistance to cisplatin | Pro‐ | 41 |
| miR‐141‐3p | unknown | Promotion of angiogenesis | Pro‐ | 54 |
| miR‐124 | SPHK1 | Dedifferentiation of CAFs to normal fibroblasts | Anti‐ | 25 |
| miR‐125b‐5p | SOCS4 | Remodeling of macrophages to TAMs | Pro‐ | 27 |
| miR‐125b | ERBB3 | Inhibition of angiogenesis | Anti‐ | 51 |
| miR‐126‐5p | DKK3, AXIN1, BACH1, NFAT5 | Enhancement of resistance to cisplatin | Pro‐ | 46 |
| miR‐181a | SMAD7 | Promotion of cellular survival, migration, invasion, and drug resistance | Pro‐ | 18 |
| miR‐181d‐5p | SOCS5 | Remodeling of macrophages to TAMs | Pro‐ | 27 |
| miR‐182 | BRCA1 | Sensitization to radiation and PARP inhibitor | Anti‐ | 60 |
| miR‐182 | unknown | Promotion of cell proliferation, invasion, and metastasis | Pro‐ | 61 |
| miR‐193a‐3p | GRB7 | Suppression of cell proliferation, migration, and invasion | Anti‐ | 22 |
| miR‐195‐5p | PSAT1 | Sensitization to cisplatin and inhibition of angiogenesis | Anti‐ | 47 |
| miR‐199a | ERBB2 | Inhibition of angiogenesis | Anti‐ | 51 |
| miR‐200a | p38α | Promotion of cell proliferation/sensitization to paclitaxel | Pro‐/anti‐ | 12 |
| miR‐200a | CXCL12β | Maintenance of immunocompetence | Anti‐ | 26 |
| miR‐200a | Unknown | Sensitization to paclitaxel/enhancement of resistance to carboplatin | Anti‐/pro‐ | 39 |
| miR‐200b | Unknown | Promotion of cell proliferation | Pro‐ | 17 |
| miR‐200c | NRP1 | Sensitization to PARP inhibitor | Anti‐ | 63 |
| miR‐200 family | ZEB1/2 | Maintenance of epithelial state | Anti‐ | 14 |
| miR‐200 family | IL8, CXCL1 | Inhibition of angiogenesis | Anti‐ | 56 |
| miR‐205 | Unknown | Promotion of angiogenesis | Pro‐ | 53 |
| miR‐216b | PARP1 | Sensitization to cisplatin | Anti‐ | 45 |
| miR‐223 | PTEN | Enhancement of resistance to cisplatin | Pro‐ | 50 |
| miR‐484 | VAGFB, VEGFR2 | Inhibition of angiogenesis | Anti‐ | 57 |
| miR‐506 | CDK4/6 | Suppression of cell proliferation | Anti‐ | 21 |
| miR‐506 | RAD51 | Sensitization to cisplatin and PARP inhibitor | Anti‐ | 43 |
| miR‐509‐3p | RAD51, HMGA2 | Sensitization to cisplatin and PARP inhibitor | Anti‐ | 44 |
| miR‐551b‐3p | STAT3 | Resistance to apoptosis and promotion of survival and proliferation | Pro‐ | 19 |
| miR‐591 | ZEB1 | Enhancement of resistance to paclitaxel | Pro‐ | 40 |
| miR‐622 | KU70, KU80 | Enhancement of resistance to cisplatin and PARP inhibitor | Pro‐ | 62 |
| miR‐1246 | CAV1 | Enhancement of resistance to paclitaxel | Pro‐ | 48 |
Abbreviations: anti‐, anti‐oncogenic; CAFs, cancer‐associated fibroblasts; pro‐, pro‐oncogenic; Ref, references; TAMs, tumor‐associated macrophages.
Other miRNAs also contribute to EOC progression, and miR‐181a is shown to promote EMT via suppression of SMAD7. 18 Chr3q26.2 amplification, a frequently observed DNA copy‐number alteration, leads to an increased expression of miR‐551b‐3p, and subsequently upregulates its target gene, STAT3. 19 STAT3 then activates Wnt/β‐catenin signaling through upregulation of miR‐92a, which confer stemness properties. 20 In contrast, miR‐193a‐3p acts as a tumor suppressor by modulating the MAPK/ERK signaling pathway, and miR‐506 inhibits proliferation by targeting CDK4 and CDK6. 21 , 22 Reflecting histological subtypes, several miRNAs in the chrXq27.3 cluster are highly expressed in clear cell carcinoma compared with high‐grade serous carcinoma. 23 Downregulation of the chrXq27.3 cluster is associated with an early relapse in patients with advanced EOC. 24
In addition, exosomal miRNAs modulate the tumor microenvironment (Figure 1B). Decrease in transfer of miR‐124 from EOC cells can result in the transition of quiescent fibroblasts into CAFs. 25 In CAFs, miR‐200f‐regulated CXCL12β is shown to promote immunosuppression. 26 Moreover, EOC‐derived miR‐21‐3p, ‐125b‐5p, and ‐181d‐5p remodel macrophages to TAM under hypoxic conditions. 27 TAM‐derived exosomes then confer a Treg/Th17 cell imbalance by transferring miR‐21‐5p and ‐29a‐3p. 28 Additionally, malignant ascites has also been reported to contain many cancer‐derived exosome. 29 Upon receiving exosomal miR‐99a‐5p, peritoneal mesothelial cells enhance the expression of FN1 and vitronectin (VTN) and make cancer invasion easy. 30
Therefore, miRNAs have been shown to play different important functions in physiology of cancer progression.
3. THE SIGNIFICANCE OF EXTRACELLULAR miRNAS AS NON‐INVASIVE BIOMARKERS IN EOC
Clinically, measurement of serum CA‐125 levels has been widely performed to diagnose EOC, although it is not enough to screen EOC precisely. 31 Despite the presence of RNases in the plasma, miRNAs exist stably in peripheral blood samples partly because they are encapsulated by exosomes. 32 Therefore, the application of circulating miRNAs as non‐invasive biomarkers has been investigated in various cancers. 33
Circulating cancer‐derived exosomes increase as EOC progresses, and the miRNA profile of these exosomes is similar to that of their corresponding cancers. 34 Thus, miR‐200f can be a diagnostic marker, as the level of miR‐200f is elevated in the peripheral blood of patients with EOC compared with healthy controls (Table 2). 17 , 34 , 35 , 36 The levels of miR‐200a and miR‐200c are associated with disease progression, and the level of miR‐200a is shown to be elevated in mucinous and serous carcinomas than in other EOC subtypes. 36 In addition, exosomal miR‐21, ‐100, and ‐320 are upregulated, whereas miR‐16, ‐93, ‐126, and ‐223 are downregulated in the plasma of patients with EOC compared with healthy controls. 17 However, the accuracy of a single miRNA as a diagnostic marker is not sufficient due to histological diversity and individual differences.
TABLE 2.
Clinical utility of miRNAs as biomarkers
| miRNA | Sample type | Methodology | Number of samples | Clinical observation | Ref |
|---|---|---|---|---|---|
| Diagnostic biomarker | |||||
| miR‐21 | Blood (exosome) | Microarray | EOC (n = 106) vs healthy (n = 29) | AUC: 0.740, SN: 61%, SP: 82% | 17 |
| miR‐100 | Blood (exosome) | Microarray | EOC (n = 106) vs healthy (n = 29) | AUC: 0.710, SN: 62%, SP: 73% | 17 |
| miR‐200a | Serum | qRT‐PCR | EOC (n = 28) vs healthy (n = 28) | AUC: 0.675, SN: 85.7%, SP: 35.7% | 35 |
| miR‐200b | Blood (exosome) | Microarray | EOC (n = 106) vs healthy (n = 29) | AUC: 0.868, SN: 64%, SP: 86% | 17 |
| miR‐200b | Serum | qRT‐PCR | EOC (n = 28) vs healthy (n = 28) | AUC: 0.722, SN: 85.7%, SP: 35.7% | 35 |
| miR‐200c | Serum | qRT‐PCR | EOC (n = 28) vs healthy (n = 28) | AUC: 0.727, SN: 71.4%, SP: 57.1% | 35 |
| miR‐320 | Blood (exosome) | Microarray | EOC (n = 106) vs healthy (n = 29) | AUC: 0.658, SN: 56%, SP: 69% | 17 |
| miR‐200b and ‐200c | Serum | qRT‐PCR | EOC (n = 28) vs healthy (n = 28) | AUC: 0.784, SN: 78.6%, SP: 46.4% | 35 |
| miR‐21, ‐362‐5p, and ‐1274a | Plasma | qRT‐PCR | EAOC (n = 14) vs endometriosis (n = 33) | AUC: 0.92, SN: 57%, SP: 91% | 38 |
| A combination of 10 miRNAs a | Serum | Microarray | EOC (n = 333) vs non‐EOC (n = 3713) | AUC: 1.000, SN: 99%, SP: 100% | 37 |
| Prognostic biomarker | |||||
| miR‐200 family | Mixed | Meta‐analysis | Tissue (n = 1353), blood (n = 1269), ascites (n = 22) | HR: 1.207 (1.040‐1.400), P = .013 | 16 |
| miR‐141 | Mixed | Meta‐analysis | Tissue (n = 124), blood (n = 300) | HR: 1.121 (0.960‐1.311), P = .150 | 16 |
| miR‐200a | Mixed | Meta‐analysis | Tissue (n = 456), blood (n = 163) | HR: 1.279 (0.522‐3.132), P = .590 | 16 |
| miR‐200b | Mixed | Meta‐analysis | Tissue (n = 261), blood (n = 370), ascites (n = 22) | HR: 2.306 (1.305‐4.079), P = .004 | 16 |
| miR‐200c | Mixed | Meta‐analysis | Tissue (n = 144), blood (n = 180) | HR: 1.011 (0.512‐1.995), P = .974 | 16 |
| miR‐429 | Mixed | Meta‐analysis | Tissue (n = 368), blood (n = 257) | HR: 1.142 (0.423‐3.085), P = .793 | 16 |
| miR‐181a | FFPE tissue | qRT‐PCR | n = 52 | High expression of miR‐181a is associated with poor PFS and OS | 18 |
| miR‐223 | Tissue | qRT‐PCR | n = 62 | High expression of miR‐223 is associated with poor PFS | 50 |
| miR‐506 | Tissue | TCGA | n = 468 | Low expression of miR‐506 is associated with poor OS | 43 |
| miR‐509‐3p | Tissue | TCGA | n = 477 | Low expression of miR‐509‐3p is associated with poor OS | 44 |
| The chrXq27.3 cluster b | FFPE tissue | Microarray | n = 85 | Low expression of the miRNAs is associated with early relapse | 24 |
| miR‐551b‐3p | FFPE tissue | ISH | n = 145 | High expression of miR‐551b‐3p is associated with poor OS | 19 |
| miR‐622 | Tissue | TCGA | n = 89 | High expression of miR‐662 is associated with poor DFS and OS in tumors with BRCAness | 62 |
Abbreviations: AUC, area under curve; DFS, disease‐free survival; EAOC, endometriosis‐associated ovarian cancer; EOC, epithelial ovarian cancer; FFPE, formalin‐fixed paraffin‐embedded; HR, hazard ratio; ISH, in situ hybridization; OS, overall survival; PFS, progression‐free survival; qRT‐PCR, quantitative reverse transcription‐polymerase chain reaction; Ref, reference; SN, sensitivity; SP, specificity; TCGA, The Cancer Genome Atlas.
miR‐320a, ‐665, ‐1275, ‐3184‐5p, ‐3185, ‐3195, ‐4459, 4640‐5p, ‐6076, and ‐6717‐5p.
miR‐506, ‐507 ‐508‐3p, ‐509‐3p, ‐509‐5p, ‐513a‐5p, ‐513b, and ‐514.
To improve the accuracy of screening, combination models using several miRNAs have been proposed (Figure 1C). According to a large‐scale study performed using 4046 serum samples, a diagnostic model based on 10 miRNAs can distinguish patients with EOC and healthy controls with a high efficiency and accuracy (sensitivity: 99%; specificity: 100%). 37 The 10 miRNAs obtained in the study are miR‐320a, ‐665, ‐1275, ‐3184‐5p, ‐3185, ‐3195, ‐4459, 4640‐5p, ‐6076, and ‐6717‐5p. Regarding histological differences, a combination of plasma miR‐21, ‐191, and ‐1975 can distinguish patients with endometriosis‐associated carcinoma and serous carcinoma, whereas miR‐21, ‐362‐5p, and ‐1274a can distinguish patients with endometriosis and endometriosis‐associated carcinoma. 38 Therefore, an appropriate combination of circulating miRNAs will be a potential diagnostic marker in the near future. However, there are several challenges in this research field. Firstly, circulating miRNAs can be influenced not only by the existence of specific cancer but also by presence of many other conditions, such as benign diseases, medication, stress, and any other individual factors. Secondly, depending on the experimental methods, such as next‐generation sequencing or microarray, optimal combination, and cut‐off value can be varied. Therefore, to efficiently screen EOC patients, further optimization is required.
4. ROLES OF miRNAS IN REGULATING THERAPY RESISTANCE IN EOC
4.1. Conventional chemotherapy
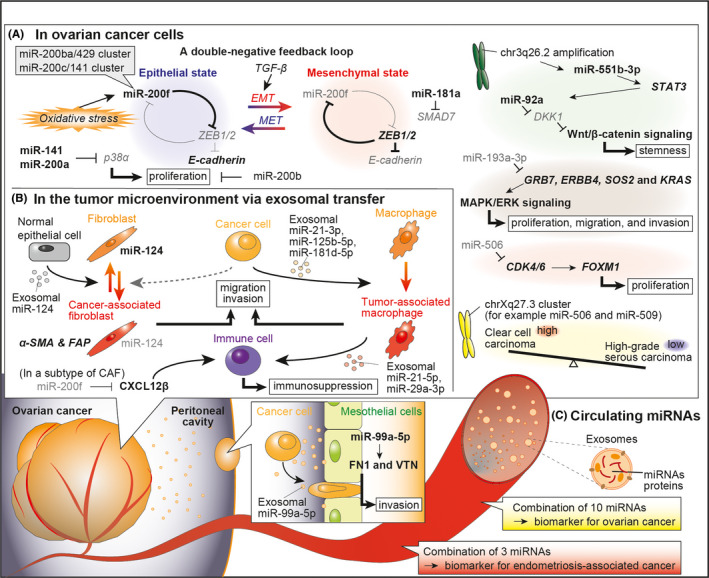
Over several decades, taxane‐ and platinum‐based chemotherapy has been the standard treatment for EOC. 2 Downregulation of miR‐200f confers paclitaxel resistance, whereas taxane‐resistant cell lines with EMT phenotype show decreased expression of miR‐200f (Figure 2A). 12 , 39 Thus, EMT phenotype is associated with taxane resistance. Inversely, upregulation of miR‐200f and miR‐591 enhances paclitaxel sensitivity by targeting ZEB1. 12 , 40 Conversely, the functions of miR‐200f in platinum resistance are reversed. Overexpression of miR‐141 enhances cisplatin resistance by regulating KEAP1, and miR‐141 or miR‐200c confer further carboplatin resistance in multidrug‐resistant cells (Figure 2B). 39 , 41 Therefore, it can be concluded that the EMT phenotype does not always contribute to drug resistance.
FIGURE 2.
Involvement of miRNAs in therapeutic resistance
In addition, the expression levels of miR‐9, miR‐506, and miR‐509‐3p are associated with a better response to platinum‐containing chemotherapy in patients with EOC. 42 , 43 , 44 miR‐9 mediates the downregulation of BRCA1 and improves cisplatin sensitivity. 42 Moreover, miR‐506 and miR‐509‐3p enhance cisplatin sensitivity by targeting RAD51, which is an important gene for DNA repair. 43 , 44 Downregulation of PARP1 by miR‐216b also reverses cisplatin resistance in EOC cells. 45 Hence, downregulation of these miRNAs targeting DNA repair‐related genes is important for cisplatin resistance. Moreover, activation of the Wnt/β‐catenin signaling pathway is involved in platinum resistance, and the pathway is shown to be activated by miR‐126‐5p but suppressed by miR‐195‐5p. 46 , 47
Furthermore, exosomal transfer of miRNAs may also lead to development of chemoresistance. TAMs receive miR‐1246 abundant exosomes derived from paclitaxel‐resistant EOC cells, and miR‐1246 further promotes paclitaxel resistance. 48 In contrast, EOC cells receive miR‐21 abundant exosomes from cancer‐associated adipocytes and fibroblasts, resulting in paclitaxel resistance. 49 Moreover, in hypoxic conditions, miR‐223 enriched TAM‐derived exosomes promote cisplatin resistance in EOC cells via the PTEN‐PI3K/AKT pathway. 50
Therefore, various miRNA‐related mechanisms are involved in chemoresistance, and combination of different chemotherapies is advisable considering this the aspect of miRNAs.
4.2. Anti‐VEGF therapy
As cancer progresses, the center of cancer tissues become hypoxic, and cancer cells promote angiogenesis. EOC tissues are known to represent high levels of vascular endothelial growth factor A (VEGFA). 47 Therefore, bevacizumab, a monoclonal antibody for all VEGFA isoforms, suppresses angiogenic processes, resulting in prolonged progression‐free survival in patients with EOC. 3 However, its clinical benefits are usually transient.
Anti‐angiogenic miR‐125b, ‐195‐5p, and ‐199a are downregulated in EOC, and these miRNAs target HIF1A and VEGFA (Figure 2C). 47 , 51 Conversely, low expression of angiogenic miR‐378 is associated with prolonged survival due to progression‐free effects observed due to bevacizumab in EOC patients. 52 Moreover, exosomes also contribute to angiogenesis, and EOC‐derived miR‐141‐3p, miR‐205, and adipocyte‐derived miR‐21 promote vascularization of endothelial cells. 53 , 54 , 55 Conversely, miR‐200f inhibits angiogenesis by modulating the expression of interleukin‐8 and CXCL1 in cancer cells and directly affecting endothelial cells. 56 In addition, miR‐484 modulates tumor vasculature by targeting VEGFB and VEGFR2, and it alters the chemosensitivity of EOC cells in vivo but not in vitro. 57
It is suggested that, not only VEGFA, but also activation of alternative angiogenic pathways is associated with bevacizumab tolerance. 57 , 58 Therefore, intervention with these miRNAs and their targets may reverse bevacizumab resistance.
4.3. Poly ADP ribose polymerase (PARP) inhibitors (PARPi)
PARP1 and PARP2 proteins mediate the repair of single‐stranded DNA breaks, whereas BRCA1 and BRCA2 proteins play critical roles in the repair of double‐stranded DNA breaks called homologous recombination repair. 59 PARPi are the first clinically approved drugs designed to exploit synthetic lethality and show a higher response to patients with germline BRCA1/2 mutations or acquired BRCAness phenotype. 4 , 59
Based on the concept of synthetic lethality, miR‐182 overexpressing breast cancer cells are hypersensitive to PARPi by targeting BRCA1 (Figure 2D). 60 However, miR‐182 also has functions related to aggressive behaviors by regulating multiple tumor suppressor genes. 61 Therefore, BRCA1 targeting therapy seems ambivalent. Moreover, miR‐506 and miR‐509‐3p enhance the efficacy of PARPi as well as cisplatin by targeting RAD51. 43 , 44 However, despite of the presence of BRCA‐mutation, resistance to PARPi can occur. High expression of miR‐622 is associated with poor survival in EOC with BRCA1 mutation, and miR‐622 also induces PARPi resistance by downregulating the Ku complex during the cell cycle in BRCA1‐mutant in vitro. 62 Conversely, miR‐200c reversed PARPi resistance by targeting NRP1 which is overexpressed in PARPi‐resistant cells. 63 Therefore, miRNAs are also powerful modulators of PARPi response.
In conclusion, because various targets of miRNAs are involved in treatment resistance, miRNAs can act as potential therapeutic targets.
5. NOVEL FINDINGS OF miRNAS IN NON‐EPITHELIAL MALIGNANT OVARIAN NEOPLASMS
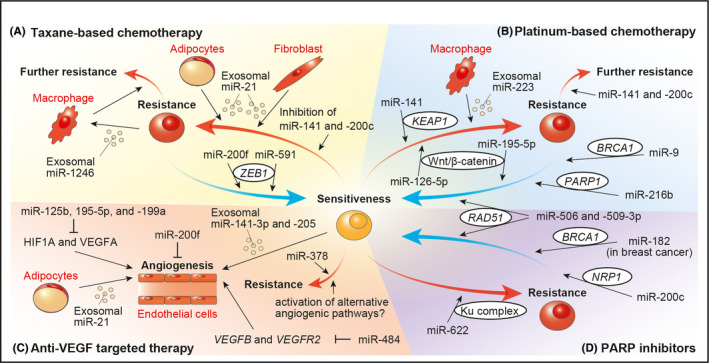
Germ cell tumors (GCTs) are frequently diagnosed in childhood and adolescent age, and are derived from primordial germ cells, embryonic precursors of egg and sperm (Figure 3A). 64 According to miRNA microarrays, miR‐371‐373 and miR‐302 clusters are overexpressed in malignant GCTs compared with nonmalignant tissues, regardless of histological subtype. 65 , 66 In particular, these clusters show even higher overexpression in yolk sac tumors, whereas miR‐146b‐5p, ‐155, and ‐182 are overexpressed in germinomas. 66 , 67 Members of the miR‐302 cluster and miR‐200f, having high expression in yolk sac tumors, are also associated with the TGF‐β/BMP signaling pathway. 67 Moreover, differentially expressed mRNAs and proteins are identified in GCTs, and elucidation of their differentiation processes is expected. 64
FIGURE 3.
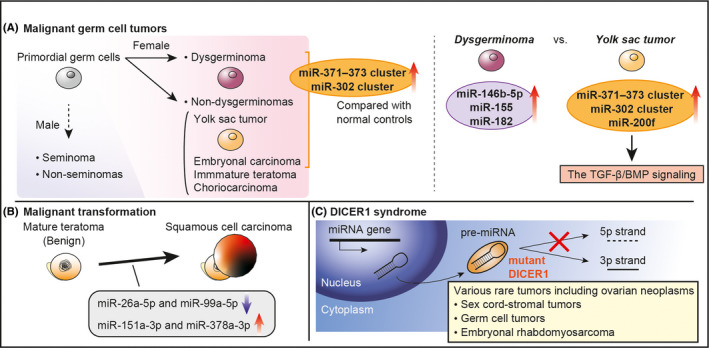
miRNAs in non‐epithelial ovarian malignancies
Mature teratoma is a benign GCT. However, secondary malignant transformation can occur in postmenopausal women, and its clinical presentation is far from that of other GCTs. 68 Comprehensive miRNA sequencing revealed that 4 miRNAs (miR‐26a‐5p, ‐99a‐5p, ‐151a‐3p and ‐378a‐3p) are dysregulated in this malignancy compared with mature teratoma or normal ovary, suggesting that these miRNAs might be involved in its carcinogenesis (Figure 3B). 69
Sex cord‐stromal tumors are frequently associated with DICER1 syndrome, a hereditary cancer predisposition syndrome characterized by the deleterious germline DICER1. 70 , 71 DICER protein is involved in cleavage of precursor miRNAs by recognizing the 5′‐end of RNA, and DICER1‐deficient cells results in loss of mature 5p miRNA strands (Figure 3C). 7 , 8 , 72 , 73 Therefore, the major alteration in miRNA expression causes a variety of rare cancers such as pleuropulmonary blastoma, neuroblastoma, and thyroid carcinoma. 74 GCTs and ovarian embryonal rhabdomyosarcoma are also recognized as DICER1‐associated tumors. 70 , 71 , 75 This syndrome clearly shows the importance of miRNAs in carcinogenesis.
Therefore, research on miRNAs in non‐epithelial tumors is currently in progress, and further studies are desired with a view to identify novel therapeutic targets.
6. FUTURE PERSPECTIVES AND CONCLUSIONS
In EOC, there are no effective screening methods, and treatment for advanced/recurrent disease still remains challenging. Because miRNAs play various roles in cancer progression and treatment resistance, clinical application of miRNAs is highly expected. Firstly, due to the stability of miRNAs in the body fluid, circulating miRNAs can be promising non‐invasive biomarkers (Figure 4). To improve screening effectiveness, the development of a detection method and target miRNAs is essential to advance this field forward. Recently, deep learning computational frameworks has enabled the utilization of enormous multi‐omics data, and it provides more robust prognosis prediction models. 76 Moreover, it aids in accurate target prediction of miRNAs and accelerates miRNA research. 77 Secondly, miRNAs have the potential to be novel therapeutic targets and agents. To perform miRNA replacement therapy, it is essential to develop a suitable delivery system with high specificity for targeting cell and protection them from RNA degradation. 78 In cancer‐bearing mouse model, intravenous or peritoneal administration of miRNA or anti‐miRNA treatment showed therapeutic effects. 19 , 44 , 56 , 61 In addition, an acceptable toxicity of TargomiRs, which are minicells loaded with miR‐16‐based mimic miRNA, was shown in a phase 1 study of patients with malignant pleural mesothelioma. 79 Hence, miRNA replacement therapy can be a novel attractive therapy for different malignancies, and it is necessary to study how to carry the specific miRNA depending on the type of cancer.
FIGURE 4.
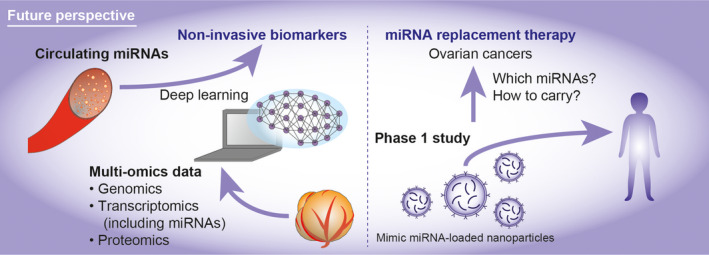
Future perspectives
In conclusion, miRNAs are powerful mediators that alter the tumor microenvironment. The application of miRNAs in the clinical setting is advancing, and research on miRNAs continues to receive much attention.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Yoshida K, Yokoi A, Kato T, Ochiya T, Yamamoto Y. The clinical impact of intra‐ and extracellular miRNAs in ovarian cancer. Cancer Sci. 2020;111:3435–3444. 10.1111/cas.14599
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393:1240‐1253. [DOI] [PubMed] [Google Scholar]
- 3. Perren TJ, Swart AM, Pfisterer J, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365:2484‐2496. [DOI] [PubMed] [Google Scholar]
- 4. Pujade‐Lauraine E, Ledermann JA, Selle F, et al. Olaparib tablets as maintenance therapy in patients with platinum‐sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT‐Ov21): a double‐blind, randomised, placebo‐controlled, phase 3 trial. Lancet Oncol. 2017;18:1274‐1284. [DOI] [PubMed] [Google Scholar]
- 5. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell. 1993;75:843‐854. [DOI] [PubMed] [Google Scholar]
- 6. Kozomara A, Birgaoanu M, Griffiths‐Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155‐D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281‐297. [DOI] [PubMed] [Google Scholar]
- 8. Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126‐139. [DOI] [PubMed] [Google Scholar]
- 9. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412‐9420. [PubMed] [Google Scholar]
- 10. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654‐659. [DOI] [PubMed] [Google Scholar]
- 11. Yokoi A, Yoshioka Y, Yamamoto Y, et al. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun. 2017;8:14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mateescu B, Batista L, Cardon M, et al. miR‐141 and miR‐200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627‐1635. [DOI] [PubMed] [Google Scholar]
- 13. Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Can Res. 2007;67:8699‐8707. [DOI] [PubMed] [Google Scholar]
- 14. Bendoraite A, Knouf EC, Garg KS, et al. Regulation of miR‐200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial‐to‐epithelial transition. Gynecol Oncol. 2010;116:117‐125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial‐mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69‐84. [DOI] [PubMed] [Google Scholar]
- 16. Huang GL, Sun J, Lu Y, et al. MiR‐200 family and cancer: From a meta‐analysis view. Mol Aspects Med. 2019;70:57‐71. [DOI] [PubMed] [Google Scholar]
- 17. Pan C, Stevic I, Muller V, et al. Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol Oncol. 2018;12:1935‐1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parikh A, Lee C, Joseph P, et al. microRNA‐181a has a critical role in ovarian cancer progression through the regulation of the epithelial‐mesenchymal transition. Nat Commun. 2014;5:2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chaluvally‐Raghavan P, Jeong KJ, Pradeep S, et al. Direct upregulation of STAT3 by MicroRNA‐551b‐3p deregulates growth and metastasis of ovarian cancer. Cell Rep. 2016;15:1493‐1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen MW, Yang ST, Chien MH, et al. The STAT3‐miRNA‐92‐Wnt signaling pathway regulates spheroid formation and malignant progression in ovarian cancer. Can Res. 2017;77:1955‐1967. [DOI] [PubMed] [Google Scholar]
- 21. Liu G, Sun Y, Ji P, et al. MiR‐506 suppresses proliferation and induces senescence by directly targeting the CDK4/6‐FOXM1 axis in ovarian cancer. J Pathol. 2014;233:308‐318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen K, Liu MX, Mak CS, et al. Methylation‐associated silencing of miR‐193a‐3p promotes ovarian cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways. Theranostics. 2018;8:423‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vilming Elgaaen B, Olstad OK, Haug KB, et al. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR‐200c‐3p as a prognostic marker. BMC Cancer. 2014;14:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bagnoli M, De Cecco L, Granata A, et al. Identification of a chrXq27.3 microRNA cluster associated with early relapse in advanced stage ovarian cancer patients. Oncotarget. 2011;2:1265‐1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y, Cai H, Chen S, Sun D, Zhang D, He Y. Exosomal transfer of miR‐124 inhibits normal fibroblasts to cancer‐associated fibroblasts transition by targeting sphingosine kinase 1 in ovarian cancer. J Cell Biochem. 2019;120:13187‐13201. [DOI] [PubMed] [Google Scholar]
- 26. Givel AM, Kieffer Y, Scholer‐Dahirel A, et al. miR200‐regulated CXCL12β promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat Commun. 2018;9:1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen X, Zhou J, Li X, Wang X, Lin Y, Wang X. Exosomes derived from hypoxic epithelial ovarian cancer cells deliver microRNAs to macrophages and elicit a tumor‐promoted phenotype. Cancer Lett. 2018;435:80‐91. [DOI] [PubMed] [Google Scholar]
- 28. Zhou J, Li X, Wu X, et al. Exosomes Released from Tumor‐Associated Macrophages Transfer miRNAs That Induce a Treg/Th17 Cell Imbalance in Epithelial Ovarian Cancer. Cancer Immunol Res. 2018;6:1578‐1592. [DOI] [PubMed] [Google Scholar]
- 29. Vaksman O, Trope C, Davidson B, Reich R. Exosome‐derived miRNAs and ovarian carcinoma progression. Carcinogenesis. 2014;35:2113‐2120. [DOI] [PubMed] [Google Scholar]
- 30. Yoshimura A, Sawada K, Nakamura K, et al. Exosomal miR‐99a‐5p is elevated in sera of ovarian cancer patients and promotes cancer cell invasion by increasing fibronectin and vitronectin expression in neighboring peritoneal mesothelial cells. BMC Cancer. 2018;18:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clarke‐Pearson DL. Clinical practice. Screening for ovarian cancer. The New England journal of medicine. 2009;361:170‐177. [DOI] [PubMed] [Google Scholar]
- 32. Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997‐1006. [DOI] [PubMed] [Google Scholar]
- 33. Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087‐2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taylor DD, Gercel‐Taylor C. MicroRNA signatures of tumor‐derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13‐21. [DOI] [PubMed] [Google Scholar]
- 35. Kan CW, Hahn MA, Gard GB, et al. Elevated levels of circulating microRNA‐200 family members correlate with serous epithelial ovarian cancer. BMC Cancer. 2012;12:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zuberi M, Mir R, Das J, et al. Expression of serum miR‐200a, miR‐200b, and miR‐200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin Transl Oncol. 2015;17:779‐787. [DOI] [PubMed] [Google Scholar]
- 37. Yokoi A, Matsuzaki J, Yamamoto Y, et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9:4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suryawanshi S, Vlad AM, Lin HM, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis‐associated ovarian cancer. Clin Cancer Res. 2013;19:1213‐1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brozovic A, Duran GE, Wang YC, Francisco EB, Sikic BI. The miR‐200 family differentially regulates sensitivity to paclitaxel and carboplatin in human ovarian carcinoma OVCAR‐3 and MES‐OV cells. Mol Oncol. 2015;9:1678‐1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huh JH, Kim TH, Kim K, et al. Dysregulation of miR‐106a and miR‐591 confers paclitaxel resistance to ovarian cancer. Br J Cancer. 2013;109:452‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Jaarsveld MTM, Helleman J, Boersma AWM, et al. miR‐141 regulates KEAP1 and modulates cisplatin sensitivity in ovarian cancer cells. Oncogene. 2013;32:4284‐4293. [DOI] [PubMed] [Google Scholar]
- 42. Sun C, Li N, Yang Z, et al. miR‐9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J Natl Cancer Inst. 2013;105:1750‐1758. [DOI] [PubMed] [Google Scholar]
- 43. Liu G, Yang D, Rupaimoole R, et al. Augmentation of response to chemotherapy by microRNA‐506 through regulation of RAD51 in serous ovarian cancers. J Natl Cancer Inst. 2015;107:djv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun C, Cao W, Qiu C, et al. MiR‐509‐3 augments the synthetic lethality of PARPi by regulating HR repair in PDX model of HGSOC. J Hematol Oncol. 2020;13:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y, Niu Z, Lin X, Tian Y. MiR‐216b increases cisplatin sensitivity in ovarian cancer cells by targeting PARP1. Cancer Gene Ther. 2017;24:208‐214. [DOI] [PubMed] [Google Scholar]
- 46. Wu G, Cao L, Zhu J, et al. Loss of RBMS3 Confers Platinum Resistance in Epithelial Ovarian Cancer via Activation of miR‐126‐5p/β‐catenin/CBP signaling. Clin Cancer Res. 2019;25:1022‐1035. [DOI] [PubMed] [Google Scholar]
- 47. Dai J, Wei R, Zhang P, Kong B. Overexpression of microRNA‐195‐5p reduces cisplatin resistance and angiogenesis in ovarian cancer by inhibiting the PSAT1‐dependent GSK3beta/beta‐catenin signaling pathway. J Transl Med. 2019;17:190. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Kanlikilicer P, Bayraktar R, Denizli M, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p‐gp/M2‐type macrophage axis in ovarian cancer. EBioMedicine. 2018;38:100‐112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Au Yeung CL, Co NN, Tsuruga T, et al. Exosomal transfer of stroma‐derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhu X, Shen H, Yin X, et al. Macrophages derived exosomes deliver miR‐223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J Exp Clin Cancer Res. 2019;38:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He J, Jing Y, Li W, et al. Roles and mechanism of miR‐199a and miR‐125b in tumor angiogenesis. PLoS One. 2013;8:e56647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chan JK, Kiet TK, Blansit K, et al. MiR‐378 as a biomarker for response to anti‐angiogenic treatment in ovarian cancer. Gynecol Oncol. 2014;133:568‐574. [DOI] [PubMed] [Google Scholar]
- 53. He L, Zhu W, Chen Q, et al. Ovarian cancer cell‐secreted exosomal miR‐205 promotes metastasis by inducing angiogenesis. Theranostics. 2019;9:8206‐8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Masoumi‐Dehghi S, Babashah S, Sadeghizadeh M. microRNA‐141‐3p‐containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK/STAT3 and NF‐kappaB signaling pathways. J Cell Commun Signal. 2020;14:233‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. An Y, Zhao J, Nie F, et al. Exosomes from adipose‐derived stem cells (ADSCs) overexpressing miR‐21 promote vascularization of endothelial cells. Sci Rep. 2019;9:12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pecot CV, Rupaimoole R, Yang D, et al. Tumour angiogenesis regulation by the miR‐200 family. Nat Commun. 2013;4:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vecchione A, Belletti B, Lovat F, et al. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc Natl Acad Sci USA. 2013;110:9845‐9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jayson GC, Kerbel R, Ellis LM, Harris AL. Antiangiogenic therapy in oncology: current status and future directions. Lancet. 2016;388:518‐529. [DOI] [PubMed] [Google Scholar]
- 59. Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152‐1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moskwa P, Buffa FM, Pan Y, et al. miR‐182‐mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41:210‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu X, Ayub B, Liu Z, et al. Anti‐miR182 reduces ovarian cancer burden, invasion, and metastasis: an in vivo study in orthotopic xenografts of nude mice. Mol Cancer Ther. 2014;13:1729‐1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Choi YE, Meghani K, Brault ME, et al. Platinum and PARP inhibitor resistance due to overexpression of MicroRNA‐622 in BRCA1‐mutant ovarian cancer. Cell Rep. 2016;14:429‐439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vescarelli E, Gerini G, Megiorni F, et al. MiR‐200c sensitizes Olaparib‐resistant ovarian cancer cells by targeting Neuropilin 1. J Exp Clin Cancer Res. 2020;39:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kraggerud SM, Hoei‐Hansen CE, Alagaratnam S, et al. Molecular characteristics of malignant ovarian germ cell tumors and comparison with testicular counterparts: implications for pathogenesis. Endocr Rev. 2013;34:339‐376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Palmer RD, Murray MJ, Saini HK, et al. Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Can Res. 2010;70:2911‐2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Murray MJ, Saini HK, van Dongen S, et al. The 2 most common histological subtypes of malignant germ cell tumour are distinguished by global microRNA profiles, associated with differential transcription factor expression. Mol Cancer. 2010;9:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fustino N, Rakheja D, Ateek CS, Neumann JC, Amatruda JF. Bone morphogenetic protein signalling activity distinguishes histological subsets of paediatric germ cell tumours. Int J Androl. 2011;34:e218‐e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hackethal A, Brueggmann D, Bohlmann MK, Franke FE, Tinneberg HR, Munstedt K. Squamous‐cell carcinoma in mature cystic teratoma of the ovary: systematic review and analysis of published data. Lancet Oncol. 2008;9:1173‐1180. [DOI] [PubMed] [Google Scholar]
- 69. Yoshida K, Yokoi A, Kagawa T, et al. Unique miRNA profiling of squamous cell carcinoma arising from ovarian mature teratoma: comprehensive miRNA sequence analysis of its molecular background. Carcinogenesis. 2019;40:1435‐1444. [DOI] [PubMed] [Google Scholar]
- 70. Heravi‐Moussavi A, Anglesio MS, Cheng SW, et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366:234‐242. [DOI] [PubMed] [Google Scholar]
- 71. Witkowski L, Mattina J, Schonberger S, et al. DICER1 hotspot mutations in non‐epithelial gonadal tumours. Br J Cancer. 2013;109:2744‐2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Park JE, Heo I, Tian Y, et al. Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature. 2011;475:201‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Anglesio MS, Wang Y, Yang W, et al. Cancer‐associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse‐strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol. 2013;229:400‐409. [DOI] [PubMed] [Google Scholar]
- 74. Slade I, Bacchelli C, Davies H, et al. DICER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet. 2011;48:273‐278. [DOI] [PubMed] [Google Scholar]
- 75. McCluggage WG, Apellaniz‐Ruiz M, Chong AL, et al. Embryonal Rhabdomyosarcoma of the Ovary and Fallopian Tube: Rare Neoplasms Associated With Germline and Somatic DICER1 Mutations. Am J Surg Pathol. 2020;44:738‐747. [DOI] [PubMed] [Google Scholar]
- 76. Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning‐Based Multi‐Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248‐1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wen M, Cong P, Zhang Z, Lu H, Li T. DeepMirTar: a deep‐learning approach for predicting human miRNA targets. Bioinformatics. 2018;34:3781‐3787. [DOI] [PubMed] [Google Scholar]
- 78. Ofek P, Tiram G, Satchi‐Fainaro R. Angiogenesis regulation by nanocarriers bearing RNA interference. Adv Drug Deliv Rev. 2017;119:3‐19. [DOI] [PubMed] [Google Scholar]
- 79. van Zandwijk N, Pavlakis N, Kao SC, et al. Safety and activity of microRNA‐loaded minicells in patients with recurrent malignant pleural mesothelioma: a first‐in‐man, phase 1, open‐label, dose‐escalation study. Lancet Oncol. 2017;18:1386‐1396. [DOI] [PubMed] [Google Scholar]


