19F-13C spy couple enables NMR visualization of long-lived signals of large RNA structures, folds, and drug binding pockets.
Abstract
RNAs form critical components of biological processes implicated in human diseases, making them attractive for small-molecule therapeutics. Expanding the sites accessible to nuclear magnetic resonance (NMR) spectroscopy will provide atomic-level insights into RNA interactions. Here, we present an efficient strategy to introduce 19F-13C spin pairs into RNA by using a 5-fluorouridine-5′-triphosphate and T7 RNA polymerase–based in vitro transcription. Incorporating the 19F-13C label in two model RNAs produces linewidths that are twice as sharp as the commonly used 1H-13C spin pair. Furthermore, the high sensitivity of the 19F nucleus allows for clear delineation of helical and nonhelical regions as well as GU wobble and Watson-Crick base pairs. Last, the 19F-13C label enables rapid identification of a small-molecule binding pocket within human hepatitis B virus encapsidation signal epsilon (hHBV ε) RNA. We anticipate that the methods described herein will expand the size limitations of RNA NMR and aid with RNA-drug discovery efforts.
INTRODUCTION
RNAs form essential regulators of biological processes and are implicated in human diseases, making them attractive therapeutic targets (1, 2). This extensive functional diversity of RNA derives from its ability to fold into complex three-dimensional (3D) structures. Yet, the number of noncoding RNA sequences far outstrips the number of solved RNA structures deposited in the Protein Data Bank (PDB) necessary for understanding RNA function (3, 4). In comparison to x-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy provides high-resolution structural and dynamic information in solution, making it an ideal biophysical technique to characterize the interactions between target RNAs and small drug-like molecules. Nonetheless, NMR studies of RNA suffer from poor spectral resolution and sensitivity, both of which worsen with increasing molecular weight. In contrast with proteins, which are made up of 20 unique amino acid building blocks, RNAs are composed of only four aromatic residues. These four resonate over a very narrow chemical shift region. At high magnetic field strengths, sizable transverse relaxation rates (R2) cause line broadening and thereby decrease both sensitivity and resolution. These problems are further exacerbated with increasing molecular weight. To overcome these limitations of RNA, novel labeling strategies that expand the number of NMR probes beyond the traditional nonradioactive and stable isotope labels such as hydrogen-1 (1H), phosphorus-31 (31P), carbon-13 (13C), hydrogen-2 (2H), and nitrogen-15 (15N) are needed.
Solution NMR of the magnetically active fluorine-19 (19F) isotope offers clear advantages in the study of RNA structure and conformational changes, which occur upon ligand binding. 19F has high NMR sensitivity (0.83 of 1H) due to a large gyromagnetic ratio that is comparable to 1H (0.94 of 1H), a 100% natural abundance, and ~6 wider chemical shift dispersion than 1H (5, 6). In addition, 19F is also sensitive to changes in its local chemical environment (5, 6). In contrast with other commonly used NMR nuclei (1H/31P/13C/15N), 19F is virtually absent in biological systems, thereby rendering 19F NMR background free. Together, 19F is an attractive probe for incorporation into nucleic acids to study their structure, interactions, and dynamics in solution.
Given its attractive spectroscopic properties, 19F was incorporated into RNA for NMR studies in the 1970s (7–9). Since then, 19F has been successfully incorporated into DNA and RNA oligonucleotides for NMR analysis and used to probe RNA and DNA structure, conformational exchange, and macromolecular interactions (10, 11). Most of these studies were conducted on short oligonucleotides [~30 nucleotides (nt)] prepared by solid-phase synthesis with only a few residues 19F labeled. Even when 2-fluoroadenine (2FA) was incorporated into a 73-nt (~22 kDa) guanine-sensing riboswitch, only 4 of the 16 signals could be assigned. This 2FA study hinted at the limitations of 19F NMR for large RNAs (12). Despite its attractiveness, the application of 19F NMR to study RNA has remained limited because the large 19F chemical shift anisotropy (CSA) contributes substantially to line broadening as a function of increasing molecular weight and polarizing magnetic fields.
To circumvent this limitation, Boeszoermenyi et al. (13) recently showed that direct coupling of 19F to 13C allowed for cancelation of CSA and dipole-dipole (DD) interactions. By incorporating this 19F-13C spin pair into aromatic moieties of proteins and a 16-nt DNA, they showed that a transverse relaxation optimized spectroscopy (TROSY) version of a 19F-13C heteronuclear single-quantum coherence (HSQC) (13) provided improved spectroscopic properties. These exciting results hinted that installing 13C-19F pairs in RNA nucleobases should also lead to improved spectroscopic features.
However, there were no facile methods to readily incorporate 19F-13C spin pairs into RNA. To overcome this technical obstacle of incorporating fluorinated aromatic moieties into RNA, we provide here a straightforward chemoenzymatic synthesis of [5-19F, 5-13C]-uridine 5′-triphosphate (5FUTP) for incorporation into RNA (Fig. 1) using phage T7 RNA polymerase–based in vitro transcription. To showcase its versatility, we transcribed two model RNAs using these labels: the 30-nt (~10-kDa) human immunodeficiency type 2 transactivation response (HIV-2 TAR) element (6, 14) and the 61-nt (~20-kDa) human hepatitis B virus encapsidation signal epsilon (hHBV ε) element (Fig. 1) (15, 16).
Fig. 1. RNA structures and NMR methods described in this study.
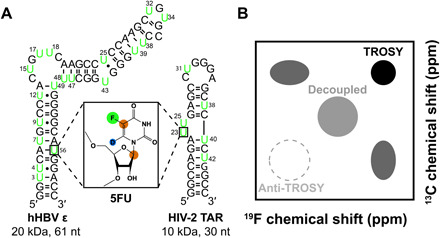
(A) Model RNA systems: HIV-2 TAR (30 nt, 10 kDA) and hHBV ε (61 nt, 20 kDa). Residues highlighted in green are labeled with 19F-13C–5-fluorouridine (5FU) shown in the box. Green circle, 19F; brown circle, 13C; blue circle, 2H. (B) Theoretical 19F,13C spectrum showing the four observable magnetization components of the 19F-13C spin pair as well as the decoupled resonance that has the average chemical shift and linewidths of all four components.
With our new labels, we demonstrate several advantages for RNA NMR studies, including improved resolution and increased sensitivity to ligand binding. We show that a 19F substitution is structurally nonperturbing and has an optimal TROSY effect at readily available magnetic field strength of 600 MHz (1H frequency), in agreement with previous studies (13). Unlike C-H spectra, the resolving power of 19F allows for easy identification of RNA structural elements in helical and nonhelical regions, as well as in wobble GU base-paired regions. With protons substituted with deuterium and depending on the molecular weight of the RNA, the TROSY effect in the 19F-13C pair can reduce the 13C linewidth by a factor >2, compared to a 13C-1H pair, and the 19F-13C label enables detection of a small-molecule binding to a 20-kDa RNA. Thus, our 19F-13C label overcomes several of the limitations in sensitivity and resolution facing RNA NMR studies with the potential to extend the application of solution NMR measurements to larger–molecular weight systems in vivo.
RESULTS
Chemical synthesis of 5-fluorouracil aids enzymatic synthesis of 5-fluorouridine-5′-triphosphate
Given the potential utility but unavailability of 19F-13C spin pairs in aromatic moieties of RNA, we first sought to develop a reliable and scalable method that combined chemical synthesis with enzymatic coupling in almost quantitative yields. This chemoenzymatic approach is a versatile method that combines chemical synthesis of atom-specific labeled nucleobases with commercially available selectively labeled ribose using enzymes from the pentose phosphate pathways (PPPs) (3, 4). To this end, we adapted the method of Santalucia et al. (17) and Kreutz and co-workers (18) and first synthesized the uracil base (U) specifically labeled with 13C at the aromatic C5 and 15N at the N1 and N3 positions (Fig. 1). This synthesis is readily accomplished using unlabeled potassium cyanide, 13C-labeled bromoacetic acid, and 15N-labeled urea. The resulting U was converted to 5-fluorouracil (5FU) by direct fluorination with Selectfluor (19, 20). This strategy allows for efficient and cost-effective synthesis of the 5FU base with high yield of ~63%. In addition, to remove unwanted scalar coupling interactions (14), we selectively deuterated H6 (~95%) using well-established methods (21). Next, using enzymes from the PPP, we coupled 5FU to D-ribose labeled at the C1′ position to give 5FUTP (Fig. 1) (3, 22) with an overall yield of ~50%. This site-specifically labeled 5FUTP was then used for DNA template–directed T7 RNA polymerase–based in vitro transcription with overall yields comparable to those obtained with unmodified nucleotides.
Thermal stabilities of wild-type and 5FU HIV-2 TAR and hHBV ε
Fluorine substitution at uridine C5 is thought to reduce the imino N3 pKa values by about 1.7 to 1.8 units with respect to their protonated analogs (23), leading to extensive line broadening of imino protons in 5FU RNAs (24). To determine if incorporation of 5-fluorouridine alters the folding thermodynamics of our RNAs (Fig. 1), we recorded ultraviolet (UV) thermal melting profiles for both wild-type (WT) and 5FU HIV-2 TAR and hHBV ε (table S1). Both WT and 5FU RNAs showed a single transition in their melting profiles, consistent with unimolecular folding (25). WT and 5FU HIV-2 TAR had melting temperatures within ~1 K of each other (WT: Tm = 355.6 ± 0.5 K; 5FU: Tm = 357.4 ± 0.4 K). Similarly, 5FU hHBV ε had a melting temperature of 327.1 ± 0.1 K, which is within the error of the melting temperature of WT. Together, these results suggest that 5FU does not markedly alter the thermodynamic stability of HIV-2 TAR and hHBV ε, in accordance with previous studies of 5FU RNAs (6, 7, 24).
DD-CSA cross-correlation in 19F-13C 5FU and TROSY effect
The linewidth for aromatic 19F-13C spin pair (Fig. 1B) is expected to become dominated by the CSA mechanism with increasing polarizing magnetic fields (13). To estimate this effect for 5FU, we calculated the chemical shielding tensor (CST) for 19F-13C spin pairs using density functional theory (DFT) methods (tables S2 and S3) (26–29). Using these CST parameters and relaxation theory implemented in the Spinach library (30), we computed the TROSY R2 relaxation rates for the 19F-13C pair of 5FU (13CF and 19FC) and the 13C-1H pair of U (13CH and 1HC) (Fig. 2) assuming isotropic tumbling. The R2 of fluorinated carbon (13CF) TROSY resonance is ~2 times smaller than that of the protonated carbon (13CH) at their respective minima of ~600 and ~950 MHz, respectively, for all molecular weights greater than 5 ns (Fig. 2A). Compared with the decoupled resonance, the R2 of the 13CF TROSY resonance is ~3 times smaller than that of protonated carbon for all molecular weights greater than 5 ns (fig. S1). Although the TROSY effect is quite small for 19F nuclei bonded to 13C (19FC) and for 1H nuclei bonded to 13C (1HC), the R2 of 19FC is three times bigger than that of 1HC (fig. S2). Thus, sensitive, high-resolution NMR spectra for the 19F-13C pair of 5FU in RNAs can be obtained by selective detection of the 13CF TROSY resonance as demonstrated for the 19F-13C pair in aromatic amino acids (13).
Fig. 2. Theoretical R2 values for the TROSY components for 19F-13C of 5-fluorouracil (13CF) or 1H-13C of uracil (13CH).
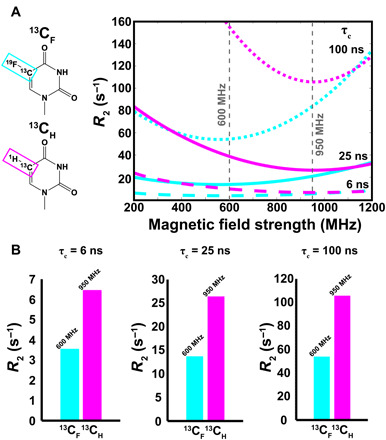
(A) Theoretical curves showing the expected R2 values for the TROSY component of 13CF (cyan) and 13CH (magenta) as a function of magnetic field strength (relative to 1H Larmor frequency) for τc = 6 ns (dashed line), 25 ns (solid line), and 100 ns (dotted line) at 25°C. (B) Theoretical R2 values taken at the commercially available magnetic field strength closest to the maximum TROSY effect (13CH = 950 MHz; 13CF = 600 MHz) for τc = 6, 25, and 100 ns at 25°C.
19F-13C TROSY on 5FU HIV-2 TAR and hHBV ε
To validate these theoretical TROSY predictions experimentally, we adapted the 1H-15N TROSY experiment (3, 31, 32) to perform a 19F-13C TROSY experiment on a ~10-kDa 5FU HIV-2 TAR and on a ~20-kDa 5FU hHBV ε RNAs (Fig. 3). Because of hardware limitations, we could only run experiments that start with and end on the magnetization of 19F, with the 13C frequency encoded in the indirect dimension. That is, we used the so-called 19F-detected out-and-back method, rather than the more sensitive 19F-excited out-and-stay 13C-detected experiment (13). We collected spectra for each of the four components (Fig. 1B) of the 19F-13C (1H-13C) correlations for both 5FU (WT) HIV-2 TAR and hHBV ε (figs. S3 to S6).
Fig. 3. TROSY spectra of RNA systems.
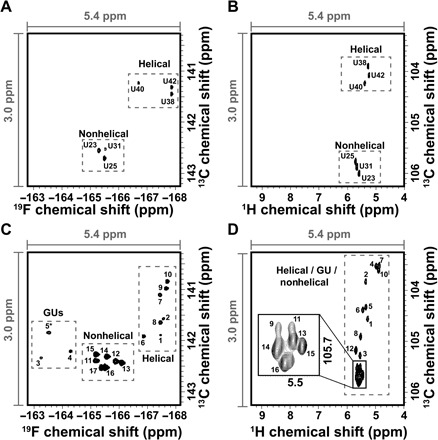
(A) 19F-13C TROSY of 5FU HIV-2 TAR. (B) 1H-13C TROSY of WT HIV-2 TAR. (C) 19F-13C TROSY of 5FU hHBV ε. (D) 1H-13C TROSY of WT hHBV ε. The assignments of 5FU and WT TAR-2 are indicated, as well as the arbitrary peak numbers for 5FU and WT hHBV ε. The same window size was used in all four spectra to aid in comparison. Gray dashed boxes indicate signals from helical, GU, and nonhelical regions. For (D), the black box indicates a zoom-in view of poorly resolved signals.
Both HIV-2 TAR and hHBV ε show ~6-fold improvement in chemical shift dispersion of 19F compared with 1H and similar dispersion in 13C (Fig. 3). All six correlations of HIV-2 TAR are well resolved for both 1H-13C and 19F-13C correlations and are in agreement with previously published 1H-19F and 1H-13C RNA spectra (6, 24, 33). Nonetheless, even for this small RNA, the 19F-13C spin pair markedly improves the spectral resolution. 5FU HIV-2 TAR shows a chemical shift dispersion of 2.6 parts per million (ppm) in the 19F dimension and only 0.5 ppm in the 1H dimension for WT (Fig. 3, A and B). Replacing 1H with 19F at C5 results in a slight reduction in chemical shift dispersion along the 13C dimension from 2.1 to 1.5 ppm, although this effect is much smaller than the gain in resolution for 19F over 1H (Fig. 3, A and B). Similarly, the 19F resonances of 5FU hHBV ε are spread over 4.5 ppm, whereas the WT 1H signals resonate over a narrow 0.8-ppm window. This represents 5.7 times better dispersion (Fig. 3, C and D). Again, substitution of 1H with 19F at C5 results in a reduction in chemical shift dispersion of 2.3 to 1.7 ppm along the 13C dimension for hHBV ε (Fig. 3, C and D). Of the anticipated 18 signals for hHBV ε, 16 are resolved for WT and 17 for 5FU. Together, these results demonstrate the marked gain in resolution afforded by the 19F-13C spin pair in 5FU RNAs compared with the 1H-13C spin pair in WT.
In addition to this considerable gain in resolution, 19F-13C labeling confers favorable 13CF TROSY linewidths. We compared the relative linewidths for both RNAs, which we assume to be Lorentzian (Figs. 4 and 5). For 5FU HIV-2 TAR, the 13CF TROSY linewidths were 1.5 times sharper on average than the anti-TROSY components, with a range of 1.3 to 1.7 (Fig. 4A). For WT HIV-2 TAR, the 13CH TROSY component was 3.7-fold narrower than the anti-TROSY component (range, 1.6 to 8.7) (Fig. 4B). Similarly, for 5FU HBV ε, the 13CF TROSY linewidths were 2.2-fold narrower than the anti-TROSY ones over a range of 1.5 to 3.3 (Fig. 4C). For WT HBV ε, only 5 of the 16 13CH anti-TROSY signals were observed and were 2.6 times broader than the TROSY resonances (range, 2.0 to 3.3) (Fig. 4D). As predicted from our simulations (Fig. 2), the 13CF TROSY component relaxes ~2 times slower than the 13CH TROSY component in both HIV-2 TAR and hHBV ε. The 19FC TROSY linewidths for 5FU HIV-2 TAR and 5FU HBV ε were 1.4 (range, 1.3 to 1.6) and 1.6 (range, 1.1 to 2.5) times narrower than the anti-TROSY components, respectively (Fig. 5, A and C). For both WT HIV-2 and WT HBV ε, the 1HC TROSY and anti-TROSY linewidths were comparable (Fig. 5, B and D). Consistent with our simulations, the 19FC TROSY linewidth is ~2-fold larger than that of the 1HC component for both RNAs (fig. S3). Again, this is in line with the poor performance of 19F NMR experiments due to the large CSA-induced relaxation. Thus, the incorporation of the 13C label mitigates the deleterious relaxation of the 19F nuclei within a 19F-13C spin pair. However, even for medium-sized RNAs ~20 kDa, 19F TROSY detection of the 19F-13C spin pair still outperforms that for a 1H-13C spin pair. Therefore, to reap the maximum benefits of this label, it is advantageous to monitor the 13C nuclei rather than the 19F nuclei. We anticipate that the 19F-13C TROSY effect will continue to scale with molecular weight for RNAs as was seen recently with proteins (13) and our simulations.
Fig. 4. Measured 13C linewidths for 5FU and WT HIV-2 TAR and hHBV ε.
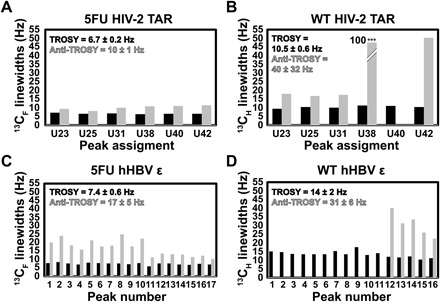
Quantification of TROSY (black) and anti-TROSY (gray) (A) 13CF and (B) 13CH linewidths for HIV-2 TAR. Note that U40 was not observed in the anti-TROSY spectrum of WT HIV-2 TAR (B). In addition, the anti-TROSY component of U38 in (B) was 97 Hz and truncated to fit in the plot. Quantification of TROSY (black) and anti-TROSY (gray) (C) 13CF and (D) 13CH linewidths for hHBV ε. Note that peaks 1 through 11 in WT hHBV ε were not observed in the anti-TROSY spectrum (D). The average ± SD in Hz is shown for the TROSY and anti-TROSY components in each plot. Peak numbers and assignments are given in Fig. 3.
Fig. 5. Measured 19F and 1H linewidths for 5FU and WT HIV-2 TAR and hHBV ε.
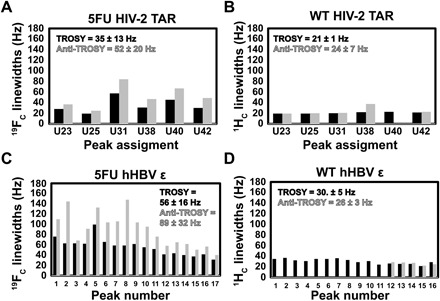
Quantification of TROSY (black) and anti-TROSY (gray) (A) 19FC and (B) 1HC linewidths for HIV-2 TAR. Quantification of TROSY (black) and anti-TROSY (gray) (C) 19FC and (D) 1HC linewidths for hHBV ε. The average ± SD in Hz is shown for the TROSY and anti-TROSY components in each plot. Peak numbers and assignments are given in Fig. 3.
19F chemical shifts reveal RNA structural equilibria
In addition to these gains in resolution and favorable linewidths, previous work suggested the 19F chemical shifts serve as sensitive markers of RNA secondary structure (10, 11). For example, GU wobble base pairs are deshielded and shifted by ~4.5 ppm to lower fields compared with AUs within Watson-Crick geometries (34). On the basis of these earlier observations, we hypothesized that 19F-13C correlations of HIV-2 TAR and hHBV ε can be grouped on the basis of whether or not they are in helical, nonhelical, or GU base-paired regions of the RNA. As a positive control, we note that nonhelical U23, U25, and U31 in 5FU HIV-2 TAR resonate around ~−165.5 ppm in 19F and ~142.5 ppm in 13C (Fig. 3A). On the other hand, the helical residues U38, U40, and U42 of 5FU HIV-2 TAR are centered around ~−167.5 ppm in 19F and ~141.5 ppm in 13C in line with previous observations for 19F-1H samples of HIV-2 TAR (6) and tRNA (34). Comparison of the equivalent 1H-13C spectra shown in Fig. 3B indicates that even though helical residues cannot be distinguished from nonhelical residues in the 1H dimension, nonhelical residues can be differentiated from helical base pairs in the 13C dimension for a 1H-13C spin pair.
The 17 19F-13C resolved correlations of 5FU hHBV ε show similar clustering as 5FU HIV-2 TAR (Fig. 3C). For instance, the six most intense signals are centered around ~−165.5 ppm in 19F and ~142.5 ppm in 13C where the nonhelical signals of HIV-2 TAR are located. On the basis of the secondary structure of hHBV ε (Fig. 1A), these six intense peaks belong to the six nonhelical uridines (U15, U17, U18, U32, U34, and U43) (Fig. 3C). A seventh peak is also seen in this region, most likely due to U48 or U49, both of which flank the bulge region. The weaker peaks are from the helical portions of hHBV ε because these signals located at ~−167.5 ppm in 19F and ~141.5 ppm in 13C resonate in the same region as the helical signals from 5FU HIV-2 TAR (6). HIV-2 TAR contains only Watson-Crick base pairs, and so, signals in this region of the hHBV ε spectrum correspond to AUs (U3, U7, U38, U39, U47, U48, U49, and U56). Of the eight anticipated peaks belonging to helical residues, only seven are observed, further suggesting that U48 or U49 may fray and resonate within the nonhelical region. Unlike HIV-2 TAR, hHBV ε has four noncanonical GU wobble base pairs embedded within helical regions. The three signals resonating in a distinct region centered at ~−163.5 ppm in 19F and ~142.0 ppm in 13C are from the four GUs (U4, U9, U12, and U25). This is in line with previous observations of GU base pairs in tRNA (34). Peak 5 (Fig. 3C) is most likely two GUs that are overlapped. Again, comparison of the equivalent 1H-13C spectra shown in Fig. 3D indicates that even though helical residues can be distinguished from nonhelical residues, nonhelical residues cannot be differentiated from GU base pairs for a 1H-13C spin pair. Thus, the spectroscopic discrimination of helical and nonhelical regions as well as GU wobble and Watson-Crick base pairs in RNA structures becomes possible with the high sensitivity of 19F to the local chemical environment of a 19F-13C spin pair. This distinguishing feature is not readily available for a 1H-13C spin pair.
19F chemical shift perturbation enables facile identification of site-specific RNA binders
Ligand-based (35) and protein-observed (36) 19F NMR screening methods are important for identifying small drug-like molecules that act as protein inhibitors. Although most work to date has focused on proteins, recent work suggests that RNAs also contain specific binding pockets that could be easily distinguished and targeted with small molecules (1, 2). hHBV ε is at the center of the viral replication cycle since the first two residues in its internal bulge are used by the virus to initiate synthesis of the minus-strand DNA. Thus, targeting this RNA structure will notably expand the repertoire of HBV drug targets beyond the current focus on viral proteins (37). Given 19F chemical shifts serve as sensitive markers of RNA secondary structure, we reasoned that 19F-13C spectroscopy will likely pinpoint loop over helical region binders. Rather satisfyingly, we found a small molecule that specifically binds a subset of nonhelical residues in 5FU hHBV ε (Fig. 6). Overlay of the full spectra of 5FU hHBV ε with and without the small-molecule shows chemical shift perturbations (CSPs) (38) predominantly confined to nonhelical regions (Fig. 6). Within the nonhelical residues, only four of the seven signals shift with the addition of the small molecule, which suggests selectivity for certain nonhelical residues over others (Fig. 6). We propose a model whereby our small molecule binds hHBV ε in the 6-nt bulge formed between C14 and C19, but not anywhere else in the RNA. The minor CSPs seen in the helical portion of the 5FU hHBV spectra are from U residues flanking the 6-nt bulge, specifically U47, U48, and U49. Last, the CSP seen in the GU portion is from U12, which also flanks our proposed binding pocket.
Fig. 6. Small-molecule binding to 5FU hHBV ε.
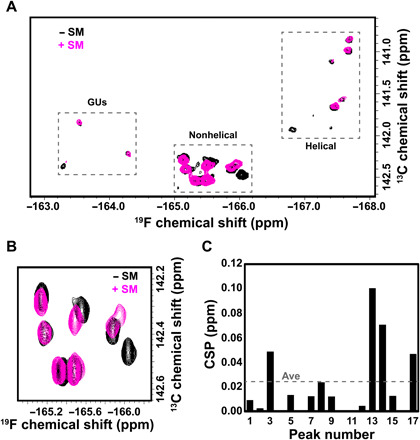
(A) Overlay of 19F-13C-TROSY spectra for hHBV ε without (black) and with small molecule (SM, magenta). (B) Zoom-in of nonhelical residues showing chemical shift perturbations (CSPs) upon addition of SM. (C) Quantification of the CSPs upon addition of SM. The average (Ave) CSP is shown as a dashed line.
DISCUSSION
19F is an attractive spectroscopic probe to study biomolecular structure, interactions, and dynamics in solution. Nonetheless, a number of obstacles must be overcome for it to become widely useful. First, we must be able to easily install the label into any biopolymer. While incorporation of fluorinated aromatic amino acids and nucleobases into proteins and nucleic acids is usually not a technical challenge, until now, synthesis of carbon-labeled and fluorinated nucleobase to create a 19F-13C spin pair has been problematic for RNA. Here, we present a facile strategy to incorporate 19F-13C 5-fluorouridine into RNA using in vitro transcription for characterization of small-molecule binding interactions by NMR. Our protocol to prepare 19F-13C 5-fluorouridine-5′-triphosphate (5FUTP) involves chemically synthesizing 5FU and then enzymatically coupling it to 13C-labeled D-ribose. Our synthetic strategy can be generalized to selectively place labels in the pyrimidine nucleobase at either 15N1, 15N3, 13C2, 13C4, 13C5, or 13C6 or any combinations thereof, and then enzymatically couple ribose labeled at either 13C1′, 13C2′, 13C3′, 13C4′, or 13C5′ or any of the preceding ribose combinations to the base. The resulting isotopically enriched 5FUTP is then readily incorporated into any desired RNA using DNA template–directed T7 RNA polymerase–based in vitro transcription. This enzymatic approach, unlike solid-phase RNA synthesis, is not limited to RNAs less than 70 nt or to nucleotides made of labeled nucleobase coupled to unlabeled ribose. Although fluorine substitution at C5 in pyrimidines strongly affects the shielding of the nearby H6, it has little effect on the anomeric H1′ chemical shifts (24). We therefore anticipate that our unique strategy that combines ribose 13C1′ label with 19F-13C uracil should allow the transfer of assignments from unmodified RNAs to 5-fluoropyrimidine–substituted RNAs made with our labels.
Second, because of van der Waals radii comparable to that of 1H, 19F is considered minimally perturbing when incorporated into biopolymers (24). Although fluorine substitution in 5FU RNAs leads to sizeable line broadening of the imino protons, thermal melting analysis indicates that the 5FU RNAs are thermodynamically equivalent to the nonfluorinated RNAs (6, 7, 24). In future work, it will be important to systematically investigate the effect of fluorine substitution not only on thermodynamic stability but also on folding kinetics of RNAs. Insights derived from solving, at high-resolution, the 3D structures of fluorinated and nonfluorinated RNA could potentially guide the use of these spin pairs to spy on the biological processes within the cell.
Third, despite its huge potential, nucleic acid observed 19F (NOF) NMR has remained underused because the large 19F CSA induces severe line broadening at high molecular weights and magnetic fields. Using DFT calculations of CST parameters, we show that an optimal 19F-13C TROSY enhancement occurs at 600-MHz 1H frequency to enable slow relaxation of 13C bonded to 19F. Our RNAs show an enhanced 19F-13C TROSY effect with increasing molecular weight and 13C linewidths that are twice as sharp as seen with traditional 1H-13C spin pairs. Thus, nucleobase 19F -13C TROSY will expand the applicability of RNA NMR beyond the ~30-nt (~10-kDa) average.
Fourth, the RNA secondary structure is made up of segments of nucleotides that are either base paired or not. The arrangements of base-paired with unpaired regions can leave distinct NMR chemical shift signatures that can provide low-resolution structural information with minimum expenditure of time and cost. For example, the H5 of a pyrimidine is sensitive to the nature of the residue that comes before it within a triplet of canonical Watson-Crick AU and GC base pairs. When the A in a central UA base pair is substituted by a G, the H5 resonance shifts downfield because of the formation of the GU base pair. Yet, an analysis of the commonly used 1H-13C probes fails to unambiguously separate nonhelical residues from helical ones (39). In contrast, the 19F-13C labels resonate in distinct chemical shift regions based on their secondary structure. For instance, nonhelical residues resonate in spectral regions distinct from helical ones, which are further separated into GU wobble and AU Watson-Crick base-paired regions. The ability to differentiate between different structural features in an RNA simply based on chemical shifts removes the need for the time-consuming and laborious process of resonance assignment.
Given the ubiquity and functional importance of GU wobble base pairs (40) in all kingdoms of life (41), the ability to easily distinguish GU from canonical GC and AU base pairs has several important implications. For instance, in the minor groove, a GU base pair presents a distinctive exocyclic amino group that is unpaired and the U’s C1′ atom rotates counterclockwise compared with the C’s C1′ atom in a canonical GC base pair. This region serves as an important site for protein-RNA interactions. Similarly, in the major groove, G N7 and O6 together with U O4 create an area of intense negative electrostatic potential conducive for binding divalent metal ions. Furthermore, all canonical Watson-Crick base pairs are circumscribed by ~10.6-Å diameters formed by a line connecting their C1′-C1′ centers. These ribose-connected centers are superimposable with almost perfect alignment. In contrast, a GU base pair is misaligned counterclockwise by a residual twist of +14°, and an UG base pair is misaligned clockwise by a residual twist of −11° (42). That is, the GU base pair is not isosteric with canonical Watson-Crick pairs. Rather, these wobble base pairs either overtwist or undertwist the RNA double helix. 19F-13C labels might aid in elucidating the structural and dynamic basis of these twists depending on the identity of the base pairs neighboring the wobble pair. We, therefore, anticipate that our new label could potentially open up avenues for probing GU wobble pairs in various structural contexts outlined above, such as 19F-13C–labeled RNA-protein interactions and metalloribozyme-ion interactions.
In summary, the labeling technologies presented here open the door for characterizing the structure, dynamics, and interactions of RNA, RNA-RNA, RNA-DNA, RNA-protein, and RNA-drug complexes in vitro and in vivo for complexes as large as 100 kDa or higher with the appropriate fluorine NMR hardware. This 19F -13C labeling approach will also enable correlating chemical shift−structure relationships to aid chemical shift–centered probing of RNA structure, dynamics, and interactions. We envision that the 19F-13C spin pair, by providing a clear demarcation of RNA structural elements, may facilitate the discovery and identification of small drug-like molecules that target RNA binding pockets in vitro and in vivo.
MATERIALS AND METHODS
The full description of Materials and Methods can be found in the Supplementary Materials. A brief summary is provided here.
Chemoenzymatic synthesis of 5FUTPs
[5-19F, 5-13C, 6-2H]– and [5-19F, 5-13C, 6-2H, 1,3-15 N2]–5FU were synthesized from unlabeled potassium cyanide, 13C-labeled bromoacetic acid, and 15N-labeled urea as described elsewhere (3, 17, 18, 24). The resulting uracil was converted to 5FU by direct fluorination with Selectfluor and deuteration (19–21). [1′,5-13C2, 5-19F, 6-2H]–5FUTP and [1′,5-13C2, 5-19F, 6-2H, 1,3-15 N2]–5FUTP were synthesized using PPP enzymes (3, 4, 6, 22, 24, 43).
RNA in vitro transcription
All RNAs were prepared by in vitro transcription and purified as previously described (3, 4). RNA concentrations were approximated by UV absorbance using extinction coefficients of 387.5 mM−1 cm−1 for HIV-2 TAR and 768.3 mM−1 cm−1 for hHBV ε. All RNA concentrations were >0.5 mM (~0.3 ml) in Shigemi NMR tubes.
Thermal melt analysis
We collected thermal melting profiles for both WT and 5FU-substituted HIV-2 TAR and hHBV ε as previously described (24, 25).
Electronic structure calculations
Calculations were carried out on 1-methyl-uracil and 5-fluoro-1-methyl uracil using optimized geometries (26–28). All calculations used the Gaussian-16 program (29). Details are provided in the Supplementary Materials.
Solution NMR spectroscopy
All 19F-13C TROSY spectra were collected at 298 K using a Bruker 600 MHz Avance III spectrometer equipped with TXI (triple resonance inverse) and BBI (broad band inverse) probes. All data were processed with Bruker’s Topspin 4.0.7 software. 1H chemical shifts were internally referenced to DSS (0.00 ppm), with the 13C chemical shifts referenced indirectly using the gyromagnetic ratios of 13C/1H (44). The 19F chemical shifts were internally referenced to trifluoroacetic acid (−75.51 ppm) (45). Experiments showing each component of the 1H/19F-13C correlations were adapted from a sensitivity- and gradient-enhanced 1H-15N TROSY used for proteins (31).
Supplementary Material
Acknowledgments
We thank P. Deshong, J. Kahn, L.-X. Wang, and P. Y. Zavalij (University of Maryland) and H. Arthanari (Harvard University) for the helpful comments. We thank S. Bentz and D. Oh for help in preparing samples for thermal melt analysis, and M. Svirydava for help in analyzing samples by mass spectrometry. Funding: We thank the National Science Foundation (DBI1040158 to T.K.D. for NMR instrumentation) and the NIH (U54AI50470 to T.K.D. and D.A.C.) for support. Author contributions: T.K.D.: conceptualization. T.K.D. and O.B.B.: implementation of the project and manuscript preparation. G.Z., B.C., K.M.T., and T.K.D.: synthesis of 5FU. O.B.: synthesis of 5FUTP, RNA synthesis, and thermal melt analysis. T.K.D., K.M.T., B.C., and O.B.B.: TROSY measurements. O.B.B.: small-molecule titration. D.A.C.: DFT calculations. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/41/eabc6572/DC1
References and Notes
- 1.Warner K. D., Hajdin C. E., Weeks K. M., Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 17, 547–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donlic A., Hargrove A. E., Targeting RNA in Mammalian Systems with Small Molecules. WIREs RNA 9, e1477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarado L. J., LeBlanc R. M., Longhini A. P., Keane S. C., Jain N., Yildiz Z. F., Tolbert B. S., D’Souza V. M., Summers M. F., Kreutz C., Dayie T. K., Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. Chembiochem 15, 1573–1577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longhini A. P., LeBlanc R. M., Becette O., Salguero C., Wunderlich C. H., Johnson B. A., D’Souza V. M., Kreutz C., Dayie T. K., Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 44, e52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rastinejad F., Evilia C., Lu P., Studies of nucleic acids and their protein interactions by 19F NMR. Methods Enzymol. 261, 560–575 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Hennig M., Scott L. G., Sperling E., Bermel W., Williamson J. R., Synthesis of 5-fluoropyrimidine nucleotides as sensitive NMR probes of RNA structure. J. Am. Chem. Soc. 129, 14911–14921 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Horowitz J., Ching-Nan O., Ishaq M., Ofengand J., Bierbaum J., Isolation and partial characterization of Escherichia coli valine transfer RNA with uridine and uridine-derived residues replaced by 5-fluorouridine. J. Mol. Biol. 88, 301–312 (1974). [DOI] [PubMed] [Google Scholar]
- 8.Horowitz J., Ofendgan J., Daniel W. Jr., Cohn M., 19F nuclear magnetic resonance of 5-fluorouridine-substituted tRNA1Val from Escherichia coli. J. Biol. Chem. 252, 4418–4420 (1977). [PubMed] [Google Scholar]
- 9.Marshall A. G., Smith J. L., Nuclear Spin-Labeled Nucleic Acids. 1. 19F Nuclear Magnetic Resonance of Escherichia coli 5-Fluorouracil-5S-RNA. J. Am. Chem. Soc. 99, 635–636 (1977). [DOI] [PubMed] [Google Scholar]
- 10.Cobb S. L., Murphy C. D., 19F NMR applications in chemical biology. J. Fluor. Chem. 130, 132–143 (2009). [Google Scholar]
- 11.Guo F., Li Q., Zhou C., Synthesis and biological applications of fluoro-modified nucleic acids. Org. Biomol. Chem. 15, 9552–9565 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Sochor F., Silvers R., Müller D., Richter C., Fürtig B., Schwalbe H., 19F -labeling of the adenine H2-site to study large RNAs by NMR spectroscopy. J. Biomol. NMR 64, 63–74 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Boeszoermenyi A., Chhabra S., Dubey A., Radeva D. L., Burdzhiev N. T., Chanev C. D., Petrov O. I., Gelev V. M., Zhang M., Anklin C., Kovacs H., Wagner G., Kuprov I., Takeuchi K., Arthanari H., Aromatic 19F-13C TROSY: a background-free approach to probe biomolecular structure, function, and dynamics. Nat. Methods 16, 333–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennig M., Munzarová M. L., Bermel W., Scott L. G., Sklenář V., Williamson J. R., Measurement of Long-Range 1H−19F Scalar Coupling Constants and Their Glycosidic Torsion Dependence in 5-Fluoropyrimidine-Substituted RNA. J. Am. Chem. Soc. 128, 5851–5858 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knaus T., Nassal M., The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 21, 3967–3975 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flodell S., Petersen M., Girard F., Zdunek J., Kidd-Ljunggren K., Schleucher J., Wijmenga S., Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 34, 4449–4457 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santalucia J. Jr., Shen L. X., Cai Z., Lewis H., Tinoco I. Jr., Synthesis and NMR of RNA with selective isotopic enrichment in the bases. Nucleic Acids Res. 23, 4913–4921 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wunderlich C. H., Spitzer R., Santner T., Fauster K., Tollinger M., Kreutz C., Synthesis of (6-C-13)Pyrimidine Nucleotides as Spin-Labels for RNA Dynamics. J. Am. Chem. Soc. 134, 7558–7569 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Lal G. S., Pastore W., Pesaresi R., A Convenient Synthesis of 5-Fluoropyrimidines Using 1-(Chloromethyl)-4-fluoro-1,4-diazabicyclo[2.2.2]octane Bis(tetrafluoroborate)-SELECTFLUOR Reagent. J. Organomet. Chem. 60, 7340–7342 (2005). [Google Scholar]
- 20.Rangwala H. S., Giraldes J. W., Gurvich V. J., Synthesis and purification of [2-13C]-5-fluorouracil. J. Label. Compd. Radiopharm. 54, 340–343 (2011). [Google Scholar]
- 21.Cushley R. J., Lipsky S. R., Reactions of 5-fluorouracil derivatives with sodium deuteroxide. Tetrahedron Lett. 52, 5393–5396 (1968). [Google Scholar]
- 22.Alvarado L. J., Longhini A. P., LeBlanc R. M., Chen B., Kreutz C., Dayie T. K., Chemo-enzymatic synthesis of selectively 13C/15N-labeled RNA for NMR structural and dynamics studies. Methods Enzymol. 549, 133–162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bevilacqua P. C., Brown T. S., Nakano S. I., Yajima R., Catalytic Roles for Proton Transfer and Protonation in Ribozymes. Biopolymers 73, 90–109 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Scott L. G., Hennig M., 19F-site-specific-labeled nucleotides for nucleic acid structural analysis by NMR. Methods Enzymol. 566, 59–87 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Mergny J. L., Lacroix L., UV melting of G-quadruplexes. Curr. Protoc. Nucleic Acid Chem. 37, 17.1.1–17.1.15 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Handy N. C., Cohen A. J., Left-right correlation energy. Mol. Phys. 99, 403–412 (2001). [Google Scholar]
- 27.Adamo C., Barone V., Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 110, 6158–6170 (1999). [Google Scholar]
- 28.Jensen F., Segmented contracted basis sets optimized for nuclear magnetic shielding. J. Chem. Theory Comput. 11, 132–138 (2015). [DOI] [PubMed] [Google Scholar]
- 29.M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. Montgomery, J. A., J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, D. J. Fox, Gaussian 16, Revision A.03 (2016).
- 30.Kuprov I., Wagner-Rundell N., Hore P. J., Bloch-Redfield-Wangsness theory engine implementation using symbolic processing software. J. Magn. Reson. 184, 196–206 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Weigelt J., Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 120, 10778–10779 (1998). [Google Scholar]
- 32.Pervushin K., Riek R., Wider G., Wüthrich K., Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 94, 12366–12371 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlomagno T., Amata I., Williamson J. R., Hennig M., NMR assignments of HIV-2 TAR RNA. Biomol. NMR Assign. 2, 167–169 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Chu W., Horowitz J., 19F NMR of 5-fluorouracil-substituted transfer RNA transcribed in vitro: resonance assignment of fluorouracil-guanine base pairs. Nucleic Acids Res. 17, 7241–7252 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dalvit C., Vulpetti A., Ligand-Based Fluorine NMR Screening: Principles and Applications in Drug Discovery Projects. J. Med. Chem. 62, 2218–2244 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Divakaran A., Kirberger S. E., Pomerantz W. C. K., SAR by (Protein-Observed) 19F NMR. Acc. Chem. Res. 52, 3407–3418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gill U. S., Kennedy P. T. F., The impact of currently licensed therapies on viral and immune responses in chronic hepatitis B: Considerations for future novel therapeutics. J. Viral Hepat. 26, 4–15 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Williamson M. P., Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 73, 1–16 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Barton S., Heng X., Johnson B. A., Summers M. F., Database proton NMR chemical shifts for RNA signal assignment and validation. J. Biomol. NMR 55, 33–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crick F. H., Codon--anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555 (1966). [DOI] [PubMed] [Google Scholar]
- 41.Varani G., McClain W. H., The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1, 18–23 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ananth P., Goldsmith G., Yathindra N., An innate twist between Crick’s wobble and Watson-Crick base pairs. RNA 19, 1038–1053 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arthur P. K., Alvarado L. J., Dayie T. K., Expression, purification and analysis of the activity of enzymes from the pentose phosphate pathway. Protein Expr. Purif. 76, 229–237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aeschbacher T., Schubert M., Allain F. H. T., A procedure to validate and correct the 13C chemical shift calibration of RNA datasets. J. Biomol. NMR 52, 179–190 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Rosenau C. P., Jelier B. J., Gossert A. D., Togni A., Exposing the Origins of Irreproducibility in Fluorine NMR Spectroscopy. Angew. Chem. Int. Ed. 57, 9528–9533 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Rovnyak D., Frueh D. P., Sastry M., Sun Z. Y. J., Stern A. S., Hoch J. C., Wagner G., Accelerated acquisition of high resolution triple-resonance spectra using non-uniform sampling and maximum entropy reconstruction. J. Magn. Reson. 170, 15–21 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/41/eabc6572/DC1


