Abstract
Background Colorectal cancer (CRC) is a major public health burden worldwide, and colonoscopy is the most commonly used CRC screening tool. Still, there is variability in adenoma detection rate (ADR) among endoscopists. Recent studies have reported improved ADR using deep learning models trained on videos curated largely from private in-house datasets. Few have focused on the detection of sessile serrated adenomas (SSAs), which are the most challenging target clinically.
Methods We identified 23 colonoscopy videos available in the public domain and for which pathology data were provided, totaling 390 minutes of footage. Expert endoscopists annotated segments of video with adenomatous polyps, from which we captured 509 polyp-positive and 6,875 polyp-free frames. Via data augmentation, we generated 15,270 adenomatous polyp-positive images, of which 2,310 were SSAs, and 20,625 polyp-negative images. We used the CNN AlexNet and fine-tuned its parameters using 90 % of the images, before testing its performance on the remaining 10 % of images unseen by the model.
Results We trained the model on 32,305 images and tested performance on 3,590 images with the same proportion of SSA, non-SSA polyp-positive, and polyp-negative images. The overall accuracy of the model was 0.86, with a sensitivity of 0.73 and a specificity of 0.96. Positive predictive value was 0.93 and negative predictive value was 0.96. The area under the curve was 0.94. SSAs were detected in 93 % of SSA-positive images.
Conclusions Using a relatively small set of publicly-available colonoscopy data, we obtained sizable training and validation sets of endoscopic images using data augmentation, and achieved an excellent performance in adenomatous polyp detection.
Introduction
Colorectal cancer (CRC) is a leading cause of morbidity and mortality in the United States and worldwide, with increasing incidence and mortality rates in many parts of the world 1 2 3 . Screening colonoscopy has been associated with a reduced risk of developing CRC in observational studies, with an up to 60 % reduction in relative risk of incidence and mortality even among patients with negative baseline findings 4 5 6 . The utility of colonoscopy relies on accurate detection of precancerous lesions and subsequent removal via polypectomy; however, clear evidence demonstrates substantial variability in adenoma detection rate (ADR) among endoscopists, with profound implications for patient outcomes 7 8 9 . Importantly, sessile serrated adenomas (SSAs) are recognized as particularly challenging lesions to identify due to their frequently flat morphology and subtle surface features 10 11 12 , which are likely underdiagnosed but overrepresented in development of interval CRC 13 14 .
The application of machine learning is a promising modality to improve endoscopic lesion detection, and many recent studies have demonstrated its successful use in improving ADR, including detection of sessile serrated adenomas (SSAs) 15 . Many of these studies adopted the emerging paradigm of deep learning, where imaging data flow through an intricate network of “neurons” in a non-linear fashion, resulting in highly accurate predictive models capable of polyp detection and localization 16 17 18 19 . However, existing studies have relied on large, curated databases, which can be expensive or laborious to obtain and which are not made available in the public domain. In addition, existing systems are typically trained on large networks requiring significant computational resources. In the present study, we aimed to demonstrate the utility of a relatively small set of publicly available endoscopic videos in training a small convolutional neural network model, which can achieve similar accuracy in detecting adenomatous colon polyps, including challenging cases like SSAs.
Methods
Data curation
Using public video databases (YouTube, VideoGIE, and Vimeo) we identified 23 colonoscopy videos ranging from 1 to 41 minutes in duration, totaling over 390 minutes of footage. All videos were colonoscopy technique teaching videos uploaded by expert endoscopists, and full pathology data were provided in all cases when a polyp was present and polypectomy was performed. All videos were of standard white light colonoscopy. The terms ‘screening colonoscopy’, ‘surveillance colonoscopy’, ‘polypectomy’, ‘endomucosal resection’, ‘adenoma’, ‘sessile serrated adenoma’, and ‘colon polyp’ were used to search for relevant colonoscopy videos on VideoGIE, Vimeo, and Youtube. One expert gastroenterologist (TB), then selected colonoscopy videos which met the following inclusion criteria: 1) videos uploaded by an academic center/physician to VideoGIE, Vimeo, or YouTube; 2) standard-definition or high-definition video quality with at least “adequate” prep quality; and 3) definitive histopathologic diagnosis of removed polyps. Exclusion criteria for colonoscopy videos were: 1) unclear or unconfirmed source/author of colonoscopy video; 2) poor-quality video or prep; and 3) significant pathology not relevant to screening colonoscopy (colitis, active diverticular bleeding). Because each video had the same frame rate of 30 /second, we adapted command line tools from VLC (videolan.org) to capture individual frames from each of the videos. Each video was reviewed by expert gastroenterologists who annotated the segments of videos with polyps and designated the type of each polyp seen (e. g. adenoma, SSA, and other findings such as lipoma and diverticula which were not included in the current study). We specifically captured video frames that contained adenomatous polyps during these annotated segments, and sampled polyp-free frames immediately before and after polyp-containing frames as controls to ensure equal appearance of polypectomy device and other potential confounding elements. Certain videos contained no adenomatous polyps and were also included as controls. A detailed description of each video and each polyp is listed in Supplementary Table 1 . We captured 509 video frames containing adenomatous polyps and 6,875 polyp-free frames which were used in this study.
Supplementary Table 1. Detailed description of 23 publicly available videos used in the current study.
| Video # | Total Video Length | Pathological Findings | Captured Video Length With Pathology (i. e. polyp) |
| 1 | 59 s | Large adenoma | 59 s |
| 2 | 27m59 s | Adenoma | 212 s |
| 3 | 11m33 s | Polyp – unknown type Lipoma Adenoma |
110 s |
| 4 | 30m09 s | Polyp – unknown type | 0 s |
| 5 | 16m14 s | Normal | 0 s |
| 6 | 14m27 s | Normal | 0 s |
| 7 | 09m07 s | Normal | 0 s |
| 8 | 03m55 s | Normal | 0 s |
| 9 | 13m45 s | Normal | 0 s |
| 10 | 30m25 s | Adenoma (3 unique) | 53 s |
| 11 | 40m37 s | Normal | 0 s |
| 12 | 36m08 s | Adenoma SSA |
93 s |
| 13 | 41m13 s | Adenoma (2 unique) SSA Small polyp (not removed) Sigmoid diverticuli |
78 s |
| 14 | 01m50 s | Large adenoma | 14 s |
| 15 | 01m46 s | Normal | 0 s |
| 16 | 19m14 s | Normal | 0 s |
| 17 | 20m00 s | Adenoma (3 unique) Diverticuli |
58 s |
| 18 | 18m14 s | Adenoma (3 unique) Prior right colon resection |
80 s |
| 19 | 12m43 s | Normal | 0 s |
| 20 | 25m17 s | Adenoma (4 unique) | 131 s |
| 21 | 02m30 s | Adenoma with polypectomy | 0 s |
| 22 | 02m00 s | Adenoma | 34 s |
| 23 | 01m29 s | SSA | 5 s |
SSA,
Data pre-processing
To overcome the limitations of small sample size and prevent overfitting, we applied data augmentation techniques, including translation, reflections, color palette transformations, cropping, rotations, and Gaussian blurring on each image (frame), as was performed and benchmarked previously 20 21 . We excluded images that were significantly blurred as detected by the variance of Laplacian filter edge detection introduced in Huertas et al. 22 . This procedure generated 15,720 adenomatous polyp-positive images, of which 2,310 were SSA, and 20,625 images without adenomatous polyps.
Deep learning framework
We adopted a transfer learning approach and fine-tuned a pretrained convolutional neural network (CNN), AlexNet, which is one of the earliest deep learning frameworks to significantly improve object recognition 20 . AlexNet consists of five convolutional layers, three of which are by max-pooling layers, and three fully connected layers. As such, it is significantly smaller than many modern CNNs, which we believed to achieve a reasonable balance between discriminatory power and computational efficiency. To adapt the AlexNet architecture for our polyp detection application, we created our model using parameters trained on ImageNet data hosted on the Caffe deep learning framework 23 , and fed our pre-processed images with labels to fine-tune these parameters. Specifically, using Caffe, we trained this model on 90 % of the images, applying dropout, batch normalization, and initialization/learning rate adjustment similarly to methods utilized by Krizhevsky et al. 20 , which are considered standard techniques to improve generalizability of CNNs. We tested the performance of the model on the remaining 10 % of the images unseen by the program during training. Training and validation datasets contain images from different polyps.
Statistical analysis
All statistical analyses were performed using the R programming language version 3.3. We calculated the probability of an image having a polyp using a normalized softmax output from the neural network classifier, and used 0.5 as the threshold to assign predicted class (polyp-containing vs polyp-free), which were then compared with physician-labeled ground truth. We performed sensitivity, specificity, and predictive value analyses using epiR package version 1.0, with default assumptions of normality to calculate confidence intervals. We computed the area under the ROC curve (AUC) using the standard pROC package version 1.8, where 10,000 stratified bootstrap replicates were drawn to calculate confidence intervals. To investigate the limitations of our model, especially those containing SSAs, we manually examined mis-classified images (including all 17 SSA-containing false-negative images and 80 false-positive images), and presented those which are representative.
Results
We repurposed the AlexNet CNN model 20 and implemented it for colonoscopy-specific image recognition, using 32,305 endoscopic images for training (90 %) and 3,590 for testing (10 %). Training and testing images came from different sets of polyps. We balanced our dataset such that 43 % of images were polyp-positive, and 6.4 % were SSA-positive (15 % of polyp-positive images were SSA-positive), in both training and validation sets.
The model achieved an overall binary classification (presence of adenomatous polyps) accuracy of 0.86, with a sensitivity of 0.73 (95 % confidence interval [0.71, 0.75]), and a specificity of 0.96 (95 % confidence interval [0.95, 0.97]). Positive predictive value was 0.93 (95 % confidence interval [0.92, 0.95]), and negative predictive value was 0.83 (95 % confidence interval [0.81, 0.84]) ( Fig. 1 and Table 1 ). The area under the receiver operating characteristic curve (AUC) was 0.94 (95 % confidence interval [0.9401, 0.9445] based on 10,000 stratified bootstrap replicates ( Fig. 2 ).
Fig. 1 .
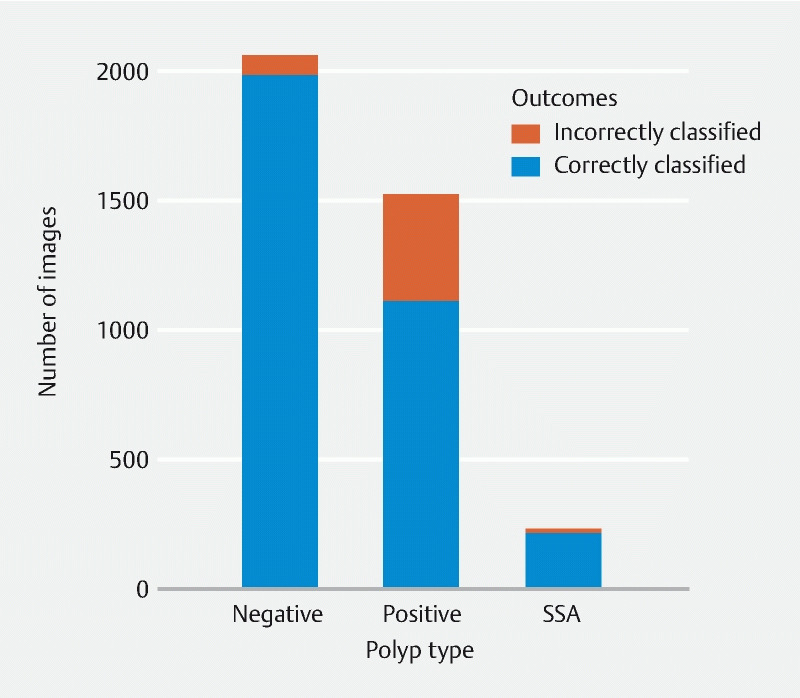
Validation result by polyp type, using a validation set of N = 3,590 images. Detailed performance metrics are in Table 1 . This model achieved a high specificity (0.96) and accurately detected sessile serrated adenomas (SSAs).
Table 1. Detailed model performance on a validation set of 3,590 images.
| Predicted Condition | ||||
| Total N = 3,590 | Predicted Positive | Predictive Negative | ||
| True Condition | Condition Positive | Overall | 1,114 | 413 |
| SSA | 214 | 17 | ||
| Condition Negative | 80 | 1,983 | ||
SSA, sessile serrated adenoma.
Fig. 2.
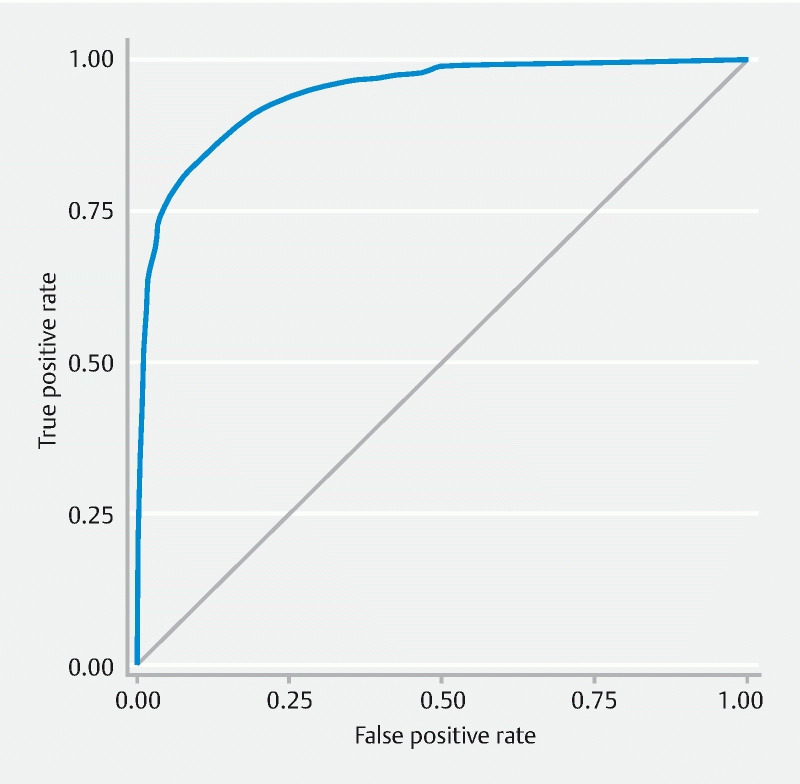
Receiver operating characteristic (ROC) curve of the overall model. The area under the ROC curve (AUC) is 0.94.
When examining the activation patterns of different CNN layers, we observed a sequential enrichment of higher-level features (e. g. edges, light intensity, vascular patterns, Fig. 3 ), suggesting that our resulting model was not strongly influenced by noises in the training database of images derived from publicly available videos, and was thus less likely to overfit. This was also supported by the observation that the accuracy in the validation set tracked with that in the training set fairly well throughout training epochs.
Fig. 3 .
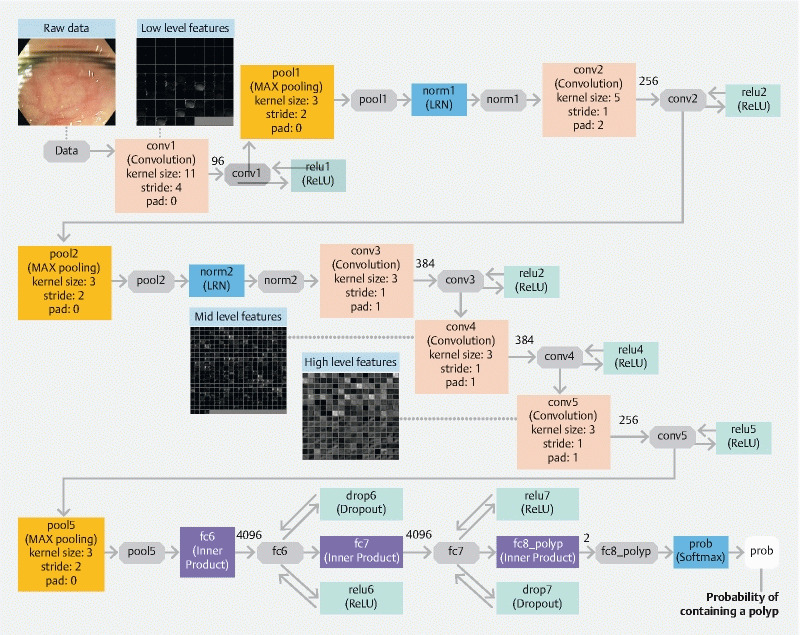
Architecture of the underlying convolutional neural network (“AlexNet”) with an exemplary image showing the activation of three convolutional layers. As raw image data passes through the CNN model, higher-level features of the polyp (bottom of the image) begin to emerge.
With respect to SSA polyps, among the 231 images in the validation set, the model obtained an accuracy of 0.93. Upon further examination of SSA images which were either correctly or incorrectly classified, we observed that a few incorrectly-classified images came from frames of video with significant motion-induced blurring ( Fig. 4 ). On the other hand, we also investigated polyp-free images that were incorrectly classified as polyp-containing (false positives). These images commonly demonstrated evidence of motion-induce blurring ( Fig. 5 ), suggesting that rapid motions can present challenges to accurately detect polyps using our current model.
Fig. 4 .
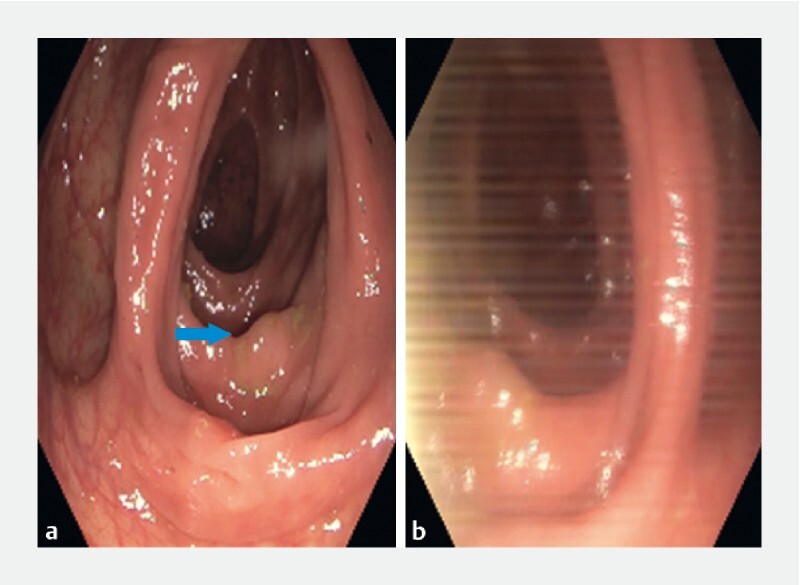
Example images of correctly classified (a) and incorrectly classified (b) sessile serrated adenoma (SSA) images. These two images come from the same video (#12) but the image on the right has motion-induced blurring.
Fig. 5 .
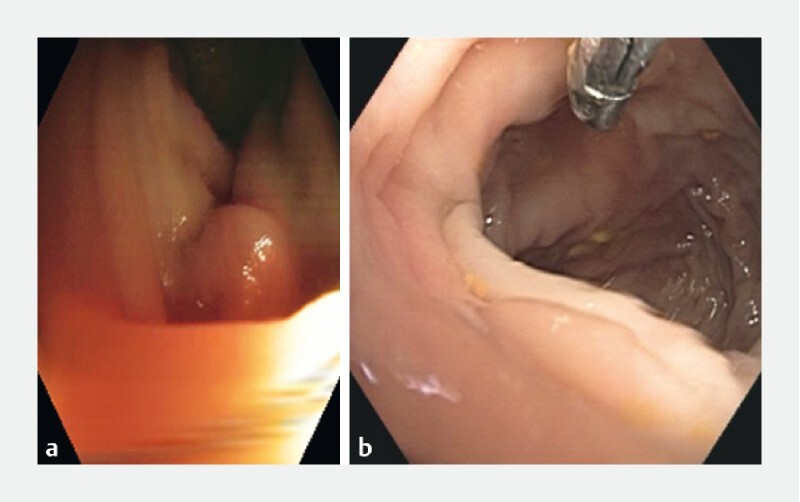
Example images of adenomatous polyp-free images incorrectly classified as polyp-containing (false positive). Both a and b are misclassified as polyp-free and are examples of motion-induced blurring, which represents the most common feature of these false positives.
Discussion
Artificial intelligence (AI) has profoundly impacted biomedical research and many aspects of clinical care, and gastroenterology is poised to benefit tremendously from the rapidly expanding repertoire of analytical tools and rich imaging data from patients 15 16 . In this context, we have shown that using a relatively small set of publicly-available colonoscopy videos and subsequently applying data augmentation and transfer learning of a simple convolutional neural network (CNN), it is possible to identify the presence of colorectal adenomas based on images captured from colonoscopy videos with high accuracy. Accurate detection of SSAs as observed in the current study is particularly promising, because they are difficult to identify with estimated miss rate ranging from 15 % to 41 % 24 25 26 . Because CNN architecture has been shown to excel in image object recognition tasks, we were encouraged to observe the remarkable improvement in SSA detection, resulting in higher accuracy than other types of adenomatous polyps, likely due to the intrinsic but subtle consistency of SSA morphology not easily appreciated by humans.
The present study is meaningful in addressing two of the common challenges in the application of AI-assisted colonoscopy: first, existing studies 17 18 19 using computer-aided detection and/or deep learning models typically require large database of carefully selected and annotated images which are often difficult to obtain, and require extensive investment in computational resources in addition to the time needed for clinicians to curate these data. Second, datasets collected at a single medical center in a research environment may be homogeneous with site-specific settings, and it is unclear whether these studies can directly generalize to other institutions. We have developed a computational pipeline that can harness the heterogeneous collection of colonoscopy videos in the public domain, which by data augmentation and standard CNN training techniques achieved accurate polyp detection with no evidence of overfitting. In addition, our model was trained on images directly captured from colonoscopy videos with very high specificity, suggesting that it is potentially suitable for clinical use as a real-time computer-aided diagnostic tool.
Compared to some other studies based on deep learning (e. g. 17 18 ) or other machine learning modalities (e. g. 27 ), our sensitivity is lower (0.73 compared to > 0.90) but specificity is higher (0.96 compared to < 0.80), potentially due to our polyp-free images coming from a wider set of clinical scenarios spanning all of the videos. This study is also limited by the small number of videos and images used in both training and testing of the resulting model, potentially contributing to the lower sensitivity of the study and making it challenging to further dissect the factors resulting in missed detections and false positive results (in addition to motion-induced blurring). We acknowledge that these limitations can be addressed by curating more colonoscopy videos due to the scalability of our pipeline.
Conclusion
While prospectively collected videos from expert centers has been a typical approach in developing AI algorithms for gastrointestinal endoscopy, our work shows that publicly available video samples are sufficient to develop a viable algorithm for polyp detection. The next important step will be to determine if an algorithm developed from data in the public domain can function equally well if applied to high-quality prospectively collected videos from endoscopy centers and in the setting of live clinical care. This proof-of-concept study has shown the potential for using publicly available endoscopy resources to formulate computational models that can accurately detect adenomatous lesions that would otherwise be easily missed by clinicians. By further fine-tuning and characterizing the current model with training and testing on larger datasets, we believe this is the first step to establish a safe, universal, and easy-to-use AI platform to improve adenoma detection and patient outcomes.
Importantly, progress in the field of computer-aided polyp detection has generally been limited to groups with direct access to proprietary data sets of colonoscopy videos and images obtained through IRB-approved research protocols. Our study is the first to apply these techniques using endoscopic videos already available in the public domain, the number and quality of which are only expected to increase markedly over time. Our important next steps would be to compare images and videos obtained from the public domain to those captured from endoscopy centers and in the setting of live clinical care to further establish the unique benefits of this modality. Along with the rapid ongoing progress in machine learning and AI capabilities, leveraging publicly available data may help accelerate progress in the field of computer-aided polyp detection.
Footnotes
Competing interests Tyler Berzin has served as a consultant for Wision AI, Fujifilm and Medtronic. The remaining authors have no conflicts of interest to disclose.
References
- 1.Arnold M, Sierra M S, Laversanne M et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Brody H. Colorectal cancer. Nature. 2015;521:S1. doi: 10.1038/521S1a. [DOI] [PubMed] [Google Scholar]
- 3.Center M M, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2009;18:1688–1694. doi: 10.1158/1055-9965.EPI-09-0090. [DOI] [PubMed] [Google Scholar]
- 4.Kahi C J. Imperiale TF, Juliar BE et al. Effect of screening colonoscopy on colorectal cancer incidence and mortality. Clin Gastroenterol Hepatol. 2009;7:770–775. doi: 10.1016/j.cgh.2008.12.030. [DOI] [PubMed] [Google Scholar]
- 5.Pan J, Xin L, Ma Y-F et al. Colonoscopy reduces colorectal cancer incidence and mortality in patients with non-malignant findings: a meta-analysis. Am J Gastroenterol. 2016;111:355–365. doi: 10.1038/ajg.2015.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacob B J, Moineddin R, Sutradhar R et al. Effect of colonoscopy on colorectal cancer incidence and mortality: an instrumental variable analysis. Gastrointest. Endosc. 2012;76:355–640. doi: 10.1016/j.gie.2012.03.247. [DOI] [PubMed] [Google Scholar]
- 7.Ahn S B, Han D S, Bae J H et al. The miss rate for colorectal adenoma determined by quality-adjusted: back-to-back colonoscopies. Gut Liver. 2012;6:64–70. doi: 10.5009/gnl.2012.6.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corley D A, Levin T R, Doubeni C A. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:1298–1306. doi: 10.1056/NEJMoa1309086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C-L, Huang Z-P, Chen K et al. Adenoma miss rate determined by very shortly repeated colonoscopy: Retrospective analysis of data from a single tertiary medical center in China. Medicine (Baltimore) 2018;97:e12297. doi: 10.1097/MD.0000000000012297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rex D K, Ahnen D J, Baron J A.Serrated lesions of the colorectum: review and recommendations from an expert panel Am J Gastroenterol 20121071315–1329.; quiz 1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouwens M W, van Herwaarden Y J, Winkens B et al. Endoscopic characterization of sessile serrated adenomas/polyps with and without dysplasia. Endoscopy. 2014;46:225–235. doi: 10.1055/s-0034-1364936. [DOI] [PubMed] [Google Scholar]
- 12.Ma M X, Bourke M J. Sessile serrated adenomas: how to detect: characterize and resect. Gut Liver. 2017;11:747–760. doi: 10.5009/gnl16523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettington M, Walker N, Rosty C et al. Critical appraisal of the diagnosis of the sessile serrated adenoma. Am J Surg Pathol. 2014;38:158–166. doi: 10.1097/PAS.0000000000000103. [DOI] [PubMed] [Google Scholar]
- 14.Benedict M, Galvao Neto A, Zhang X. Interval colorectal carcinoma: An unsolved debate. World J. Gastroenterol. 2014;21:12735–12741. doi: 10.3748/wjg.v21.i45.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alagappan M, Brown J RG, Mori Y et al. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J Gastrointest Endosc. 2018;10:239–249. doi: 10.4253/wjge.v10.i10.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Topol E J. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- 17.Chen P-J, Lin M-C, Lai M-J et al. Accurate Classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology. 2018;154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Misawa M, Kudo S-E, Mori Y et al. Artificial intelligence-assisted polyp detection for colonoscopy: initial experience. Gastroenterology. 2018;154:2027–2.029E6. doi: 10.1053/j.gastro.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Wang P, Berzin T M, Glissen Brown J R et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813–1819. doi: 10.1136/gutjnl-2018-317500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krizhevsky A, Sutskever I, Hinton G E. Curran Associates: Inc; 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 21.Perez L, Wang J. The effectiveness of data augmentation in image classification using deep learning. arXiv:1712.04621 [cs.CV] 2017.
- 22.Huertas A, Medioni G. Detection of intensity changes with subpixel accuracy using Laplacian-Gaussian masks. IEEE Trans. Pattern Anal Mach Intell PAMI. 1986;8:651–664. doi: 10.1109/tpami.1986.4767838. [DOI] [PubMed] [Google Scholar]
- 23.Jia Y, Shelhamer E, Donahue J Caffe: convolutional architecture for fast feature embedding. arXiv:1408.5093 [cs.CV] 2014.
- 24.Obuch J C, Pigott C M, Ahnen D J. Sessile serrated polyps: detection: eradication: and prevention of the evil twin. Curr Treat Options Gastroenterol. 2015;13:156–170. doi: 10.1007/s11938-015-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim N H, Jung Y S, Jeong W S et al. Miss rate of colorectal neoplastic polyps and risk factors for missed polyps in consecutive colonoscopies. Intest Res. 2017;15:411–418. doi: 10.5217/ir.2017.15.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajaratnam R, Purchiaroni F, Wilson A et al. Mo1075 high levels of presumed polyp miss rate at 1 and 3 years following index screening colonoscopy: no room for complacency. Gastrointest Endosc. 2017;85:AB417. [Google Scholar]
- 27.Mori Y, Kudo S-E, Misawa M et al. Real-Time use of artificial intelligence in identification of diminutive polyps during colonoscopy: a prospective study. Ann Intern Med. 2018;169:357–366. doi: 10.7326/M18-0249. [DOI] [PubMed] [Google Scholar]


