ABSTRACT
The receptor tyrosine kinase (RTK) pathway plays an essential role in development and disease by controlling cell proliferation and differentiation. Here, we profile the Drosophila larval brain by single-cell RNA-sequencing and identify Amalgam (Ama), which encodes a cell adhesion protein of the immunoglobulin IgLON family, as regulating the RTK pathway activity during glial cell development. Depletion of Ama reduces cell proliferation, affects glial cell type composition and disrupts the blood–brain barrier (BBB), which leads to hemocyte infiltration and neuronal death. We show that Ama depletion lowers RTK activity by upregulating Sprouty (Sty), a negative regulator of the RTK pathway. Knockdown of Ama blocks oncogenic RTK signaling activation in the Drosophila glioma model and halts malignant transformation. Finally, knockdown of a human ortholog of Ama, LSAMP, results in upregulation of SPROUTY2 in glioblastoma cell lines, suggesting that the relationship between Ama and Sty is conserved.
KEY WORDS: Drosophila, Receptor tyrosine kinase, Blood–brain barrier, Sprouty, Single-cell RNA-sequencing, LSAMP
Summary: Employing single-cell RNA-seq uncovered that Amalgam affects receptor tyrosine kinase signaling and the blood–brain barrier during glia development and in a glioma model in Drosophila.
INTRODUCTION
The receptor tyrosine kinase (RTK) pathway regulates growth, cell proliferation, differentiation and survival, and therefore has a prominent role in development and in cancer (Regad, 2015). Signaling through this pathway is mediated by cell surface receptors that are activated and dimerized upon binding to growth factors and that propagate signals through the RAS-RAF-MEK-ERK axis. The RTK pathway is tightly regulated at multiple levels including inhibition by members of the Sprouty gene family. This type of inhibition, measured by a decrease in ERK phosphorylation, occurs when Sprouty binds to either GRB2 or RAF and disrupts the propagation of the signal (Masoumi-Moghaddam et al., 2014). However, beyond this classical role of binding to different components of the RTK pathway, the mechanism of Sprouty repression and its regulation is not fully understood (Masoumi-Moghaddam et al., 2014). Notably, Sprouty is a potential therapeutic target in many neurological diseases (Hausott and Klimaschewski, 2019) and has been shown to act either as a tumor suppressor or as an oncogene in different cancers (Masoumi-Moghaddam et al., 2014). The latter suggests that the function of Sprouty and its impact on RTK signaling is likely cell type dependent, which further complicates investigation of the role of Sprouty in cancer given high heterogeneity in tumors.
Recent advances in single-cell RNA-sequencing (scRNA-seq) allow characterization of cell diversity at a high resolution and help to dissect cellular heterogeneity. This technology enabled identification of novel biomarkers in cell types as well as the discovery of rare cell subtypes that would have otherwise been missed in bulk RNA-seq (Papalexi and Satija, 2018). Additionally, scRNA-seq has been shown to uncover dynamic spatiotemporal processes such as differentiation (Wagner et al., 2016) and complex cell-to-cell responses following stimuli (Liu and Trapnell, 2016).
Drosophila has proven to be a highly advantageous model to study signaling pathways given its amenability to genetic analysis and remarkable conservation of signal transduction pathways between humans and flies (Chatterjee and Deng, 2019). This is best illustrated by pioneering studies of the RTK signaling in Drosophila (Simon et al., 1991) leading to discovery of several members of the pathway that are preserved across species (Perrimon, 1994). For instance, Sprouty was initially discovered in Drosophila (Hacohen et al., 1998) and it was later found to have a similar role in mammals (Impagnatiello et al., 2001). Drosophila has also been instrumental in studying cancers as several fly tumor models of the lung (Levine and Cagan, 2016), eye (Pagliarini and Xu, 2003), blood (Osman et al., 2009), glia (Read et al., 2009) and colon (Bangi et al., 2016) cancers have recently been established. In the Drosophila third-instar larva, developing glia present an ideal model to study the role of Sprouty as multiple RTK surface receptors have been shown to affect glia proliferation and migration as well as fly locomotion (Read, 2018; Avet-Rochex et al., 2012; Franzdóttir et al., 2009; Ray et al., 2017). Glial cells are also a vital component of the nervous system as they provide support and nutrition to neurons. Moreover, surface glia, which surround the nervous system, form a continuous dynamic membrane called the blood–brain barrier (BBB), which protects neurons from the high solute content of the hemolymph and prevents neurodegeneration (Bainton et al., 2005; Yildirim et al., 2019).
Here, we employed scRNA-seq to identify Amalgam (Ama), a member of the IgLON family encoding a cell adhesion immunoglobulin, as a new regulator of the RTK pathway that acts through sprouty (sty). Depletion of Ama decreases glial cell proliferation, disrupts the BBB and results in a dramatic increase of hemocyte infiltration in the brain. We show that knockdown of Ama increases Sty levels, which reduces RTK signaling pathway in glia during development and in a Drosophila glioma model. Notably, the impact of knockdown of Ama on Sty is conserved in human glioblastoma cell lines, suggesting a functional conservation across species.
RESULTS
Ama is required for glial cell development
RTK signaling has been extensively studied in the Drosophila eye, but primarily during the context of photoreceptor differentiation and less so in the glial cells, called wrapping glia (WG), that envelope axonal projections. This glial cell type differentiates from perineurial glia (PG) as PG are migrating from the brain towards the developing eye disc during the third-instar larval stage. It has been shown that a transient increase in RTK signaling is required during the transition from perineurial to wrapping glia (Franzdóttir et al., 2009).
In order to identify novel genes important in this process, we examined the scRNA-seq dataset of the larval eye imaginal disc (Ariss et al., 2018) to isolate genes that are specifically expressed in the perineurial and wrapping glia. Amalgam (Ama) was found to be one of the top gene markers expressed in glial cells (Fig. 1A) with a higher expression in wrapping glia than in perineurial glia (Fig. S1A,B), which parallels the transient increase in RTK signaling during this transition. Ama is an adhesion protein of the immunoglobulin superfamily (Seeger et al., 1988) and affects axon pathfinding in Drosophila embryos (Fremion et al., 2000). To determine whether Ama is important in glia, we employed the UAS-Gal4 system to knockdown Ama by RNAi using a UAS-AmaRNAi transgene driven by a pan-glial repo-Gal4 driver or a subperineurial glia (SPG) specific moody-Gal4 driver. Depletion of Ama in all glial cells repo-Gal4 resulted in early pupal lethality (∼24 h after pupa formation), indicating that Ama could have an essential role in development. This result was validated with another UAS-AmaRNAi transgene, thus, confirming the specificity of RNAi knockdown (Fig. S1C).
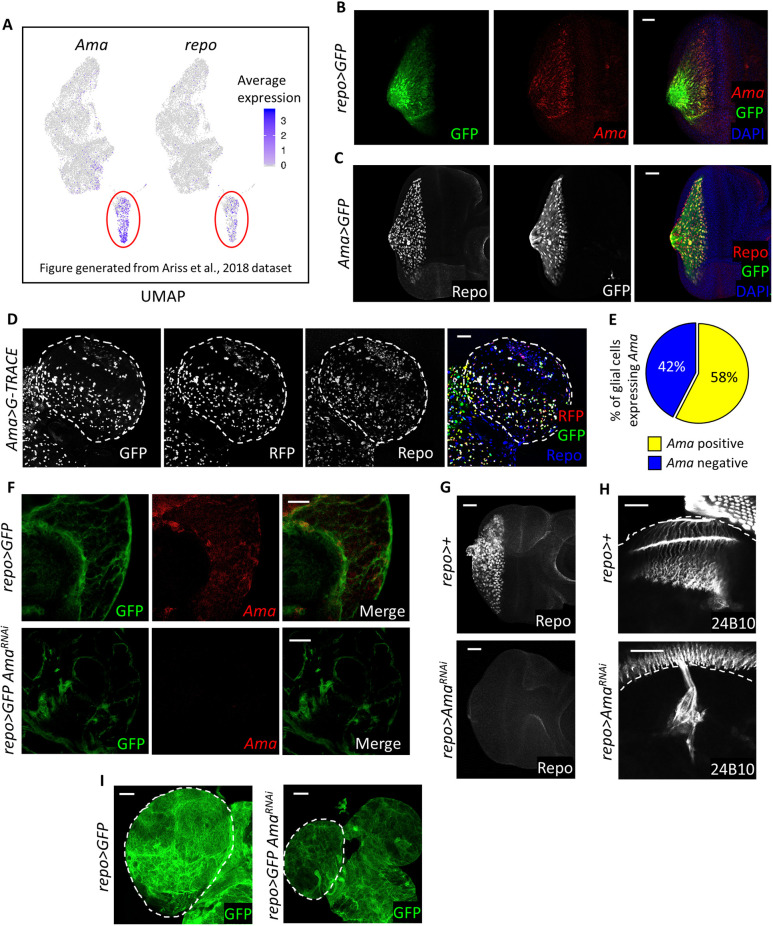
Fig. 1.
Ama is expressed in glia and is required in glial development. (A) Feature plots displaying the expression of genes on the UMAP clusters of a previously published third-instar eye disc scRNA-seq dataset (Ariss et al., 2018), showing that Ama and repo are co-expressed in the same cluster. (B) FISH for Ama mRNA in the eye disc showing expression in glial cells with glial membranes labeled by GFP. Final genotype: repo>mCD8GFP. (C) Repo immunofluorescence in the eye disc displaying expression in Ama-positive cells labeled by GFP. (D) Repo immunofluorescence in the third-instar brain (dashed outline) of Ama>G-TRACE showing the lineage and real-time expression of Ama-Gal4. GFP labels the lineage, whereas RFP represents the real-time expression. (E) Pie chart outlying the percentage of glial cells expressing Ama. Z-stacks of Ama>G-TRACE brains were counted. Data represent means of three experiments (Table S8). (F) FISH for Ama mRNA in the brain showing that there is expression in glial cells, with glial membranes labeled by GFP. Knockdown of Ama in glia results in loss of the FISH signal. Final genotypes: repo>mCD8GFP (top panel) repo>mCD8GFP AmaRNAi (bottom panel). (G) Repo immunofluorescence shows the lack of glia in the eye disc following Ama depletion. (H) Immunofluorescence using 24B10 antibody to label photoreceptor axons indicates defects in axons guidance in repo>AmaRNAi. The dashed lines indicate the outline of the brain. (I) Brains (outlined by dashed lines) are smaller in repo>AmaRNAi than in repo>GFP. GFP labels glial cell membranes. Final genotypes: repo>mCD8GFP (left panel) repo>mCD8GFP AmaRNAi (right panel). Scale bars: 20 µm.
We began the investigation of the function of Ama in glia by confirming its glial-specific expression. repo-Gal4 UAS-mCD8-GFP eye discs, which express GFP in glial cell membranes, were dissected and subjected to a fluorescence in situ hybridization (FISH) protocol using Ama-specific Stellaris probes. As shown in Fig. 1B, Ama transcripts were detected exclusively in GFP-positive glial cells. In a complementary approach, the expression of Ama was examined by immunofluorescence using an Ama-Gal4 enhancer trap line crossed to UAS-GFP, while glial cells were visualized with pan-glial Repo antibody. In agreement with the FISH results, Ama (GFP positive) was detected only in glial cells (Fig. 1C). Notably, the intensity of the GFP signal was higher in wrapping glia than in perineurial glia. Indeed, the more-intense GFP signal colocalized with Cut, a wrapping glia marker that is not expressed in perineurial glia (Fig. S1B). We also noticed that Ama is expressed in a subset of cells of the ventral peripodial membrane epithelium (VPE), as also revealed in the scRNA-seq dataset (Fig. S1D,E). These results suggest that the expression of Ama is primarily restricted to glial cells, with wrapping glia expressing a higher level of Ama than perineurial glia.
Since perineurial glial cells originate in the brain before migrating towards the eye disc, we examined the expression of Ama in the third-instar larval brain using a G-TRACE cell lineage tracing system (Evans et al., 2009). This technique provides a visual representation of temporal gene expression, labeling cells that expressed Ama in the past with GFP, while RFP labels cells that currently express Ama. With the exception of a few cells, Ama was almost exclusively detected in glia, based on the colocalization with Repo (Fig. 1D). Notably, every glial cell that expressed RFP also expressed GFP indicating that Ama remains expressed in the same cell throughout development and is not turned off (Fig. 1D). Individual confocal sections show that Ama is expressed in most types of glial cells but was largely absent in the medulla region of the optic lobe (Fig. S1F). Interestingly, only half of glial cells expressed Ama (Fig. 1E). The glial-specific Ama expression was further confirmed by FISH (Fig. 1F). Importantly, the signal was lost when Ama was depleted by RNAi using repo-Gal4 (Fig. 1F).
To characterize the consequences of glia-specific Ama knockdown, the repo>AmaRNAi eye discs were stained with Repo antibody to visualize glial cells. Strikingly, no glial cells were found in repo>AmaRNAi eye discs (Fig. 1G). Since glial cells play a crucial role in photoreceptor axon guidance (Xie et al., 2014), we examined axons in the brain using a 24B10 antibody. In the wild type, axonal projections were found in the lamina and medulla of the optic lobe; however, axons failed to land in their respective compartments in the brains when glial cells were depleted of Ama (Fig. 1H). Furthermore, there was a severe reduction in the number of glial cells in the repo>AmaRNAi brain and its size of was significantly smaller (Fig. 1I).
Thus, we conclude that Ama is essential for glial cell development. Ama depletion results in severe reduction of glia in the brain and prevents migration of glial cells from the brain towards the eye disc.
Profiling of Ama-depleted glia by scRNA-seq
To precisely characterize the impact of Ama depletion on different glial cell types in the larval brain we employed scRNA-seq. A total of 45 brains across four biological replicates in each of the repo>+ and repo>AmaRNAi larva were dissected and then dissociated into a single-cell suspension to perform scRNA-seq through the Drop-seq technique (Macosko et al., 2015) (Fig. 2A). Following sequencing alignment and digital expression matrix generation, the Seurat v3.0 package (Stuart et al., 2019) was used to normalize and filter out the low-quality cells. In wild type, 16,553 high-quality cells were retained. These cells were grouped into 17 distinct clusters based on similarity in gene expression, and the clustering was visualized through a uniform manifold approximation and projection (UMAP) algorithm. Cells in each cluster are characterized by a specific list of expressed genes or biomarkers (Fig. S2A,B; Tables S1, S2). The neuronal cell clusters were identified based on the neuronal marker elav, whereas the glial clusters were identified by repo expression (Fig. S2A; Table S2). Glial cell clusters were selected for further analysis.
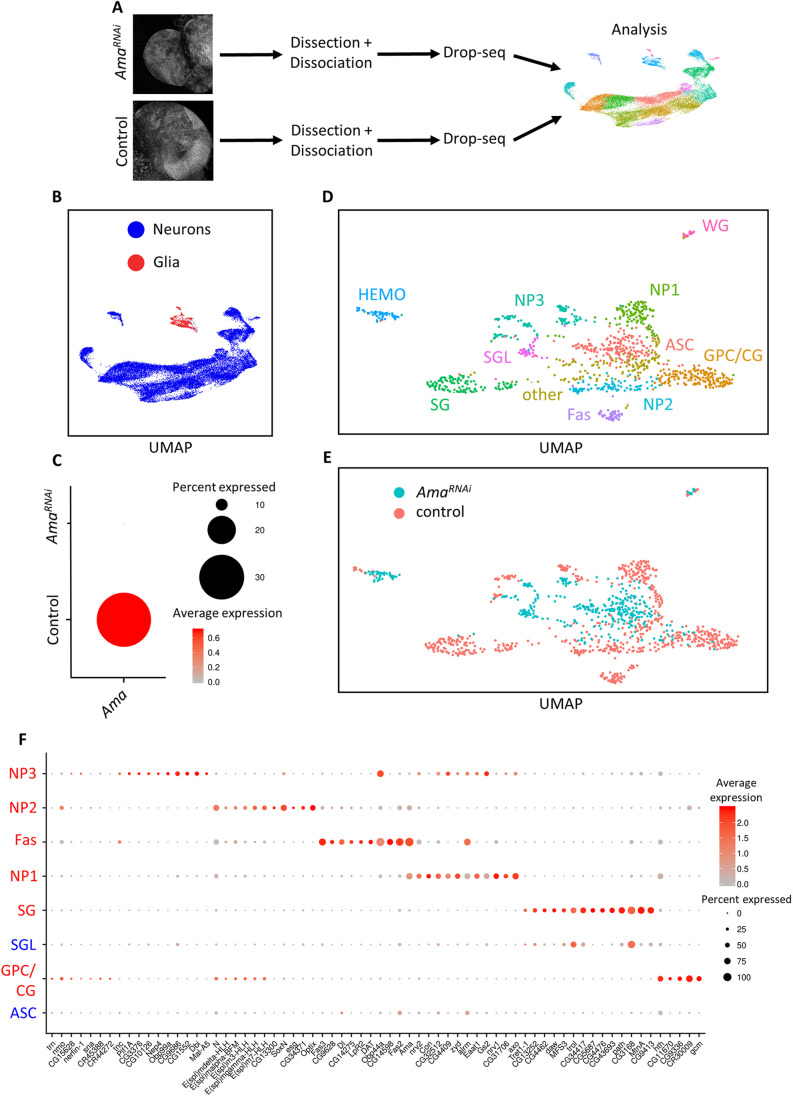
Fig. 2.
scRNA-seq identifies cellular perturbations following Ama knockdown. (A) Illustration of scRNA-seq pipeline. Third-instar larval brains from Control (repo>+) and AmaRNAi (repo>AmaRNAi) were dissected then dissociated into a single-cell suspension. Drop-seq was performed to capture single cells and generate cDNA libraries. Following alignment and generating a single-cell gene expression matrix, the samples were analyzed using Seurat to unbiasedly find cell clusters having distinct gene expression profiles. (B) UMAP of 25,700 cells outlying the neuronal and glial cells from the combined repo>+ (16,553 cells) and repo>AmaRNAi (9147 cells) scRNA-seq brains. (C) Dot plot in repo>+ and repo>AmaRNAi brains displaying the depletion of Ama expression using scRNA-seq. The red color represents the average expression of Ama (average log fold change), whereas the size of the dot represents the percentage of cells expressing Ama. (D) UMAP from the supervised analysis on glial cells from repo>+ and repo>AmaRNAi brains displaying ten distinct clusters. GPC/CG, glia precursor cells/cortex glia; ASC, AmaRNAi specific cluster; SG, surface glia; SGL, surface glia-like; NP1, neuropil glia 1; NP2, neuropil glia 2; NP3=neuropil glia 3; Fas, fasciclin cluster. (E) UMAP from D showing the genotype of each cell. (F) Dot plot displaying the top markers in different clusters from the analysis in D. The red color gradient shows the average expression of the genes (average log fold change), whereas the dot size shows the percentage of cells in the cluster expressing the gene. ASC glia do not share markers of other control glial cluster. SGL share markers with SG cells. Clusters labeled in red are observed in the repo>+ supervised glial analysis. Clusters labeled in blue are additional clusters that appeared after pooling repo>+ cells with repo>AmaRNAi brains. HEMO and WG are excluded as they originate from the hemolymph and the peripheral nervous system, respectively, and not the brain. In this analysis, GPC and CG results were pooled.
There are multiple types of glial cells in the developing third-instar larval brain. Surface glia (SG), consisting of perineurial glia and subperineurial glial cells, are situated at the surface of the third-instar larval brain and form the BBB (Bainton et al., 2005). The barrier protects neurons from degeneration and the acidic environment of hemolymph. There are two types of neuropil-associated glia (NP), astrocyte-like glia (AG) and ensheathing glia (EG), which infiltrate and cover neuropils, respectively, in the central brain and optic lobe (Omoto et al., 2015). Cortex glia (CG) encase neurons and are located between SG and the neuropils (Pereanu et al., 2007). Finally, glia progenitor cells (GPCs) are situated between the lamina neuropil and SG of the optic lobe and give rise to glia in the optic lobe (Yoshida et al., 2005).
The supervised analysis of glial cells selected based on repo expression encompassed 760 cells across nine clusters. The distinct marker list in each cluster was used to assign and label the different glial cell types based on previously published work (Fig. S3A–C; Tables S3 and S4). Thus, cells of GPC, SG, NP, WG cell types were readily identified. Interestingly, cells of the Fasciclin (Fas) cluster express markers of multiple glial cell types. For instance, Fas2 and alrm, which are highly expressed in the Fas cluster, are surface and neuropil glia markers, respectively (DeSalvo et al., 2014; Doherty et al., 2009). Moreover, Ama is also expressed in multiple glial cell types, such as surface glia, neuropil-associated glia and wrapping glia, as well as cortex glia (Fig. S1F), and is a top marker in that Fas cluster (Fig. S3B; Tables S3 and S4). One of the prominent features of the Fas cluster is the expression of cell adhesion proteins (Fas2, Fas3 and Ama), which is consistent with published findings underlying that cell–cell contact is a hallmark of glial cells (DeSalvo et al., 2014; Sasse et al., 2015). Thus, the Fas cluster does not appear to correspond to a specific cell type and therefore may represent a cell state.
Next, we performed Seurat analysis of a combined dataset of 9147 repo>AmaRNAi cells and the 16,553 control cells to characterize glial cells differences between the knockdown and control brains. Cells of non-neuronal clusters were selected as above (Fig. 2B) and used to perform a supervised Seurat analysis. Reassuringly, the expression of Ama was strongly reduced in repo>AmaRNAi cells (Fig. 2C), thus, indicating the efficiency of Ama depletion.
UMAP revealed several major differences in the presentation of Ama-depleted cells (Fig. 2D,E). First, there was a complete loss of the Fas cluster in repo>AmaRNAi cells as it was represented exclusively by wild-type cells. Interestingly, this cluster displays the highest level of Ama expression (Fig. 2D,E). Second, repo>AmaRNAi cells were missing in the SG cluster (Fig. 2D,E). Instead, Ama-depleted cells formed a distinct cluster that displayed the SG markers Tret1-1, troll and CG3168 (Volkenhoff et al., 2015; Ariss et al., 2018) and was therefore labeled SG-like (SGL) (Fig. 2F; Fig. S4A). Third, the UMAP revealed a new Ama knockdown-specific cluster (ASC) (Fig. 2D,E) that expresses repo but none of other known glial cell markers (Fig. 2F; Tables S5 and S6). Instead sprouty (sty) is one of its top markers (Fig. S4B; Table S5). Finally, a hemocyte (HEMO) cluster was identified by the expression of blood cell markers such as Hml and He (Fig. S4B) (Goto et al., 2003; Kurucz et al., 2007), and this predominantly consisted of cells from the repo>AmaRNAi brains (Fig. 2D,E). These results indicate that depletion of Ama affects multiple clusters at the single-cell level.
The BBB is disrupted upon Ama depletion
scRNA-seq profiling revealed that Ama is highly expressed in the Fas cell cluster and that this cluster is lost in repo>AmaRNAi brains. To validate these results in vivo, we selected Fas2, a top marker of the Fas cluster, and examined its expression through immunofluorescence with anti-Fas2 antibody. Glial cells were labeled with membrane-targeted GFP using repo>mCD8-GFP. In the wild-type brain, Fas2 was observed in multiple glial cell types but with strongest expression in surface glia (Fig. 3A). This is consistent with published data showing that Fas2 is predominantly expressed in surface glia (DeSalvo et al., 2014). Notably, there was a striking reduction in Fas2 at the surface of the brain in SG cells of repo>mCD8-GFP AmaRNAi brains (Fig. 3A). Such a defect in surface glia of repo>AmaRNAi is particularly interesting given that SGL cells clustering distinctly from wild-type SG cells on the UMAP plot (Fig. 2D,E) suggesting that the gene expression profile of surface glial cells changes dramatically following Ama knockdown. We therefore decided to investigate the effect of Ama depletion in surface glia.
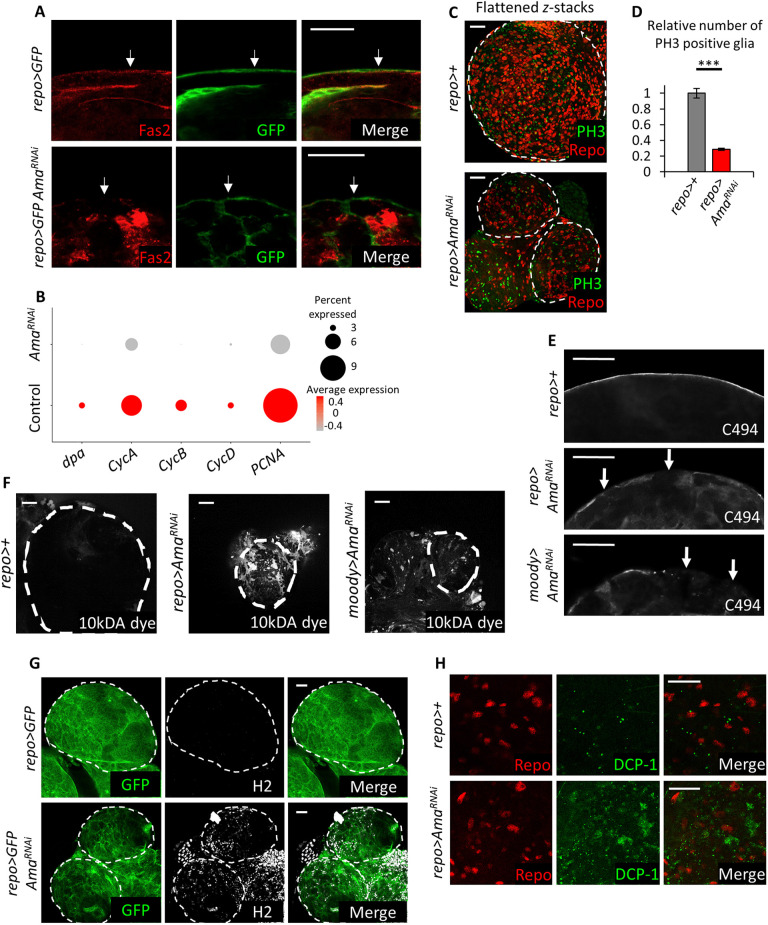
Fig. 3.
Ama depletion decreases glial cell proliferation and disrupts the BBB. (A) Immunofluorescence images showing that there is a decrease in Fas2 in surface glia following Ama knockdown. Final genotype: repo>mCD8GFP (top panel) repo>mCD8GFP AmaRNAi (bottom panel). White arrow points at surface glia. (B) Dot plot indicating a decrease in expression level of cell cycle genes dpa, CycA, CycB, CycD, and PCNA using scRNA-seq in repo>+ and repo>AmaRNAi glia. The red color gradient represents the average expression of the genes (average log fold change), whereas the size of the dot represents the percentage of cells expressing the genes. (C) Flattened z-stacks of repo>+ and repo>AmaRNAi brain immunofluorescence with REPO and PH3 showing that Ama knockdown decreases overall Repo and PH3 colocalization. (D) Quantification of Repo and PH3 colocalization in C reveals a significant reduction in mitotically active glia following Ama depletion. Z-stacks from repo>+ (n=5) and repo>AmaRNAi (n=6) brains were counted and normalized to the average count in repo>+ (set at 1). Data represents the mean±s.e.m. after normalization. ***P<0.001 (Student's t-test; Table S9). (E) C494 immunofluorescence labeling SPG of third-instar larval brains showing discontinuous surface glia membranes following Ama knockdown. White arrows point to discontinuous membranes. (F) Brains labeled with 10 kDa dextran dye indicates that Ama knockdown increases dye penetration in the tissue, implying a damaged BBB. (G) Immunofluorescence using pan hemocyte H2 antibody shows a strong signal in the brain in repo>AmaRNAi. (H) Cleaved DCP-1 immunofluorescence shows a neuronal apoptotic signal in repo>AmaRNAi. Dashed outlines show brain regions. Scale bars: 20 µm.
SG cells proliferate extensively during larval stages (Yildirim et al., 2019) in order to fully cover the rapidly increasing brain mass. This is essential for the maintenance of the BBB, a physical barrier that shields neurons from blood stream solutes and protects them from neurodegeneration (Yeh et al., 2018). The barrier is a vital component of the nervous system and it is damaged in multiple neurodegenerative diseases, such as Parkinson's and Alzheimer's diseases (Sweeney et al., 2018).
In order to determine whether Ama affects glial cell proliferation, the expression of cell cycle genes from the scRNA-seq dataset was examined. In comparison to wild-type controls, repo>AmaRNAi glia strongly downregulate PCNA, CycD, CycB, CycA and dpa (Fig. 3B). Accordingly, there was a striking reduction of glial cells undergoing mitosis as revealed by immunostaining with an anti-PH3 antibody (Fig. 3C,D). These findings suggest that Ama plays a role in glial cell proliferation.
To examine the morphology of SG following Ama depletion, these cells were visualized with the C494 antibody, which labels SG membranes (Fig. S5A). In wild-type brains, C494 reveals a continues membrane layer that encompassed the brain. In contrast, surface glia membranes were discontinuous in repo>AmaRNAi indicating that the brains were not completely covered by SG cells (Fig. 3E). This was confirmed with a different Gal4 driver, moody-Gal4, which is expressed in subperineurial glia (Fig. 3E). Discontinuity of SG membranes is a hallmark of a damaged BBB. To directly confirm this, repo>AmaRNAi and moody>AmaRNAi brains were incubated with a fluorescent dextran dye that only penetrates the brain when the BBB is broken (Bainton et al., 2005). Although no fluorescence signal was detected in control brains, both repo>AmaRNAi and moody>AmaRNAi brains displayed penetration of the fluorescent dye (Fig. 3F). Intriguingly, while analyzing the scRNA-seq dataset we noted an elevated number of hemocytes in the repo>AmaRNAi condition relative to that in the control brain (Fig. 2D,E), which would be in agreement with a defective BBB. This was confirmed by staining with a pan hemocyte antibody (H2) (Kurucz et al., 2007), which showed a dramatic increase of infiltrating hemocytes in repo>AmaRNAi brain lobes (Fig. 3G).
One of the consequences of disruption of BBB is the exposure of neurons to the high concentration of solutes in the hemolymph, which may cause neuronal degeneration and death. To determine whether Ama depletion in glia affects neurons, we examined the expression of apoptotic genes using scRNA-seq dataset. Interestingly, hid, a known pro-apoptotic gene in flies, is highly expressed in repo>AmaRNAi cells but not in the wild-type cells of the neuronal cluster 6 (Fig. S5B–E). Accordingly, there was a significant level of neuronal death in repo>AmaRNAi brains, as revealed by the staining with a cleaved DCP-1 antibody that labels the apoptotic cells (Fig. S5E).
We concluded that Ama has an essential function in glia because its glia-specific depletion severely impairs the BBB, leads to hemocyte infiltration into the brain and extensive neuronal cell death. The latter and severe proliferative defects of Ama-depleted glia may help to explain the small brain size in repo>AmaRNAi animals.
Ama depletion reduces RTK signaling
The other major impact of Ama knockdown is the appearance of the ASC (Fig. 2D,F). Seurat analysis revealed that sty, a general RTK inhibitor (Hacohen et al., 1998), is an ASC top marker (Fig. S4B; Table S5). sty is highly upregulated in Ama-depleted glia cells, while the expression of a downstream effector and a read-out of RTK signaling in glia pointed (pnt) (O'Neill et al., 1994) is reduced (Fig. 4A). Interestingly, RTK signaling through FGFR (Avet-Rochex et al., 2012) and PDGFR (Read, 2018) has been shown to be important for glial cell proliferation in the third-instar brain as well as for glial migration (Franzdóttir et al., 2009). Since Ama-deficient glia proliferate poorly and fail to migrate from brain to the eye disc, we investigated RTK signaling in the repo>AmaRNAi brain.
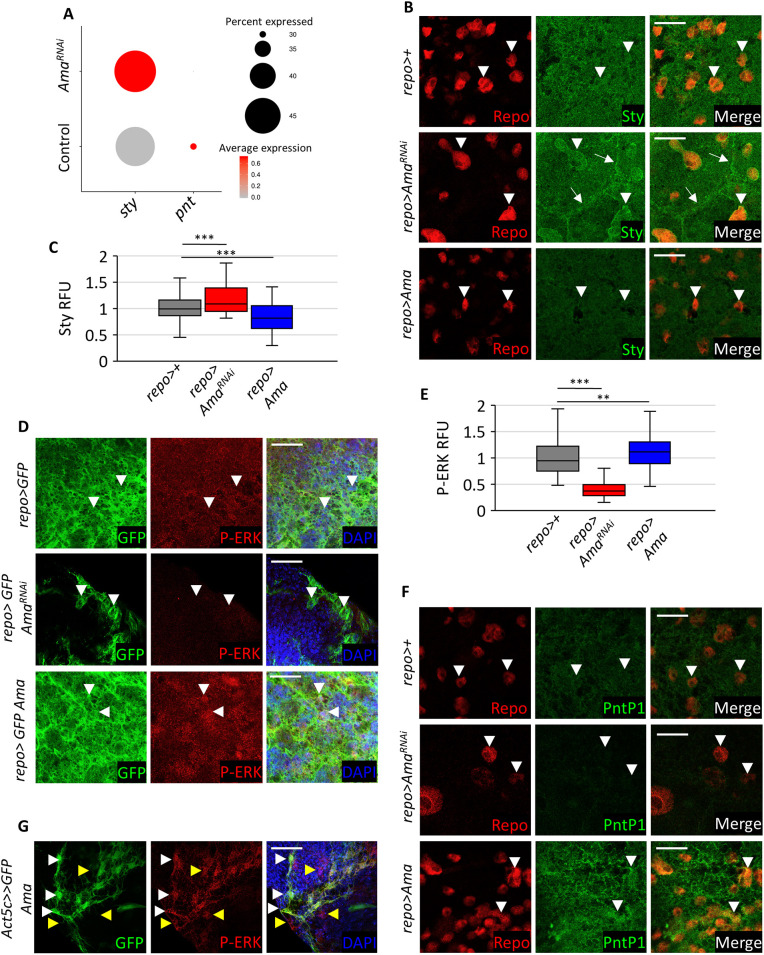
Fig. 4.
Ama knockdown decreases RTK signaling. (A) Dot plot showing an increase in sty and a decrease in pnt as determined through scRNA-seq in repo>AmaRNAi relative to repo>+. The red color gradient represents the average expression of sty or pnt (average log fold change), whereas the size of the dot represents the percentage of cells expressing sty or pnt. (B) Sty immunofluorescence in surface glia shows an increase in Sty especially at the membrane in repo>AmaRNAi and a modest decrease in basal Sty in repo>Ama. White arrowheads point to Repo-positive glial cells, whereas white arrows point to SPG membranes. (C) Box plot displaying the Sty relative fluorescence units (RFU) measured in surface glia in each repo>+ (n=70 nuclei, across 5 brains), repo>AmaRNAi (n=84 nuclei, across 8 brains), and repo>Ama (n=162 nuclei, across 9 brains). The RFUs were normalized to the average Sty RFU value in 70 repo>+ nuclei. The line in the center of the box represents the median. The lower and upper box limits are the first and third quartiles, respectively. The Tukey whiskers extend to show the minimum and maximum values outside the first and third quartiles. ***P<0.001 (Student's t-test) to compare each genotype to repo>+ (Table S10). (D) P-ERK immunofluorescence in surface glia shows a decrease in P-ERK signal in repo>AmaRNAi and a modest increase in P-ERK in repo>Ama. White arrowheads point at glia nuclei. Final genotypes: repo>mCD8GFP (top panel), repo>mCD8GFP Ama (middle panel), repo>mCD8GFP AmaRNAi (bottom panel). (E) Box plot displaying the P-ERK RFU measured in surface glia in each repo>+ (n=171 nuclei, across 17 brains), repo>AmaRNAi (n=173 nuclei, across 15 brains), and repo>Ama (n=118 nuclei, across 15 brains). The RFUs were normalized to the average P-ERK RFU value in 171 repo>+ nuclei. The line in the center of the box represents the median. The lower and upper box limits are the first and third quartiles, respectively. The Tukey whiskers extend to show the minimum and maximum values outside the first and third quartiles. **P<0.01, ***P<0.001 (Student's t-test) to compare each genotype to repo>+ (Table S11). (F) Immunofluorescence images in SG using PntP1 antibodies reveals that there is a decrease in signal in repo>AmaRNAi and a modest increase in PntP1 in repo>Ama. White arrowheads point at Repo-positive glia nuclei. Final genotypes: repo>mCD8GFP (top panel), repo>mCD8GFP Ama (middle panel), repo>mCD8GFP AmaRNAi (bottom panel). (G) P-ERK immunofluorescence in a clone of cells using FLP-Out to overexpress Ama. GFP labels PG cells in the clone on the surface of the brain that over express Ama. White arrowheads point at cell autonomous increase of P-ERK. Yellow arrowheads point at cell non-autonomous increase of P-ERK. Scale bars: 20 µm.
We began by examining the Sty expression in surface glia cells of repo>AmaRNAi brains by immunofluorescence using a Sty antibody that has been previously validated (Hacohen et al., 1998). In the control, Sty is present at a low level uniformly throughout the confocal plane. In Ama-depleted glia, Sty was markedly elevated especially at plasma and nuclear membranes, and this result was confirmed with two different UAS-AmaRNAi lines (Fig. 4B,C; Fig. S6A). Conversely, Sty staining was moderately reduced when Ama was overexpressed using a repo-Gal4 driver (Fig. 4B,C). To determine whether changes in Sty levels in Ama depleted glia are consequential, we examined the phospho-ERK (P-ERK) staining, a commonly used marker of RTK signaling, in surface glia. Strikingly, the P-ERK signal was largely lost in repo>AmaRNAi while overexpression of Ama resulted in a modest increase in P-ERK (Fig. 4D,E). Another readout of RTK signaling, Pointed (PntP1), was strongly reduced in Ama-depleted glia and slightly elevated in repo>Ama (Fig. 4F), which is in agreement with scRNA-seq data (Fig. 4A).
Since Ama is found at the membrane and is also secreted (Fremion et al., 2000), we asked whether it could exert a non-cell-autonomous effect on P-ERK. Clones of glia cells overexpressing Ama were generated by heat shock in a hs-FLP Act≫Ama GFP line, stained with P-ERK and distinguished by GFP. Notably, an elevated P-ERK signal was observed within the clone of cells expressing Ama (GFP positive) as well as in cells adjacent to the clonal boundary (Fig. 4G). This suggests that Ama can affect RTK signaling in non-cell-autonomous manner. Additionally, ectopic expression of Ama in the posterior of the eye disc resulted in elevated P-ERK and a rough eye phenotype (Fig. S7A–C), while clonal analysis demonstrates a non-cell-autonomous effect (Fig. S7D). We also note that an increase in Sty levels occur in PG and SPG even when Ama is depleted using an SPG driver (Fig. S6A). This also suggests that the effect of Ama on Sty can occur in a non-cell-autonomous manner, which may, in turn, explain the non-cell-autonomous impact of Ama on RTK activity described above.
These findings suggest that Ama affects RTK in different cell types and can act in a non-cell-autonomous manner. This is consistent with previous biochemical studies of Ama showing that it is present at the plasma membrane and is also secreted (Fremion et al., 2000).
Ama affects RTK signaling through Sty
As described above, repo>AmaRNAi animals have reduced glial cell proliferation, a small brain size and defects in glial migration. Previous studies highlight the importance of RTK signaling in proliferation and migration of glial cells (Avet-Rochex et al., 2012; Read, 2018; Franzdóttir et al., 2009). Our results show that a major consequence of Ama knockdown is upregulation of Sty, a negative regulator of the RTK pathway, and reduced RTK signaling. These observations suggest a simple model where a high level of Sty in Ama depleted cells lowers RTK signaling in these cells and may account for the Ama phenotype.
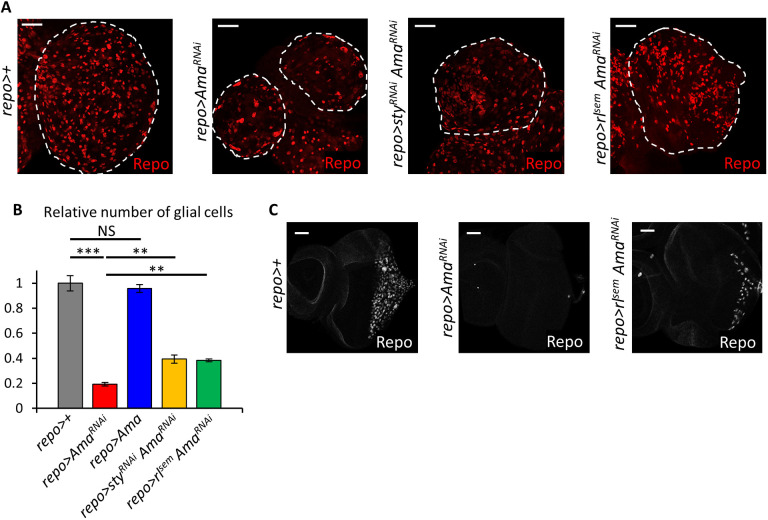
This model was tested in two ways. First, we asked whether upregulation of Sty is an important event in Ama-depleted glia. This was determined by a genetic test in which Sty was downregulated by RNAi in glial cells, after which brains of repo>styRNAiAmaRNAi and repo>AmaRNAi were compared. The efficiency of sty RNAi was confirmed by staining with the Sty antibody, and showed that Sty was no longer upregulated in repo>styRNAiAmaRNAi (Fig. S8A). Downregulation of Sty partially rescued the small brain size of repo>AmaRNAi as the repo>styRNAi AmaRNAi brains were larger in size than in repo>AmaRNAi brains (Fig. 5A). Accordingly, cell counting showed a significant increase in the number of glial cells in repo>styRNAi AmaRNAi compared to the number in repo>AmaRNAi (Fig. 5B). However, Sty depletion was insufficient to rescue the discontinuous SPG membranes and neuronal cell death, as there was no significant difference in the C494 and DCP-1 staining, respectively, between repo>AmaRNAi and repo>styRNAi AmaRNAi conditions (Fig. S8B,C). However, we cannot completely exclude the possibility that Sty is involved in the effect of Ama on the BBB.
Fig. 5.
Sty knockdown largely rescues the phenotype of Ama depletion. (A) Repo immunofluorescence of third-instar larval brains show a partial increase in size in repo>styRNAi AmaRNAi, and repo>rlsem AmaRNAi relative to that in repo>AmaRNAi. Dashed outlines show brain regions. (B) Quantification of Repo in brains in A reveal that sty knockdown or overexpressing activated ERK in Ama-depleted brains partially rescues the number of glia repo>AmaRNAi. Z-stacks from repo>+ (n=4), repo>AmaRNAi (n=4), repo>Ama (n=3), repo>AmaRNAi (n=4), repo>styRNAi AmaRNAi (n=4), repo>rlsem AmaRNAi (n=4) brains were counted and values normalized to the average count in repo>+ (set at 1). Data represents the mean±s.e.m. after normalization. **P<0.01; ***P<0.001; NS, not significant (Student's t-test) to compare each genotype to repo>+ (Table S12). (C) Repo immunofluorescence of eye discs indicates that overexpression of activated ERK in Ama-depleted brains partially rescues glial cell migration in repo>AmaRNAi eye discs. Scale bars: 20 µm.
As another test of the model, we asked whether Erk [also known as Rolled (Rl) in flies], which is downstream of Sty and therefore insensitive to Sty upregulation, rescues the phenotype of Ama depletion. To do this, a constitutively active Erk transgene (UAS-rlsem) was overexpressed in Ama-depleted glial cells. As shown in Fig. 5A, brains of repo>rlsem AmaRNAi animals were significantly larger than those of repo>AmaRNAi and there was a significant increase in glial cell number relative to repo>AmaRNAi (Fig. 5B). Interestingly, glial migration on the eye disc was partially rescued in repo>rlsem AmaRNAi (Fig. 5C), further underscoring the importance of RTK and ERK in this process.
From these results, we conclude that the phenotype of glial-specific Ama knockdown is, at least partially, caused by reduced RTK signaling given that elevating RTK signaling through expression of activated ERK largely rescues glial cell migration and the small brain size in repo>AmaRNAi. Our results suggest that Ama is upstream of ERK and acts through Sty to alter RTK signaling. Thus, upregulation of Sty is functionally important in Ama-depleted cells.
Ama knockdown suppresses neoplasia in a Drosophila glioma model
One of the best cancer models developed in Drosophila is a glioma model in which co-activation of the EGFR-Ras and PI3K pathways faithfully recapitulates many hallmarks of human glioma (Furnari et al., 2007). In the glioma fly model, co-expression of dEGFRλ and dp110CAAX transgenes in glial cells gives rise to highly proliferative, transplantable and invasive neoplastic tumor-like growth (Read et al., 2009). Since Ama is important in regulation of normal RTK signaling during development, we asked whether the role of Ama is conserved in the oncogenic context of the glioma model.
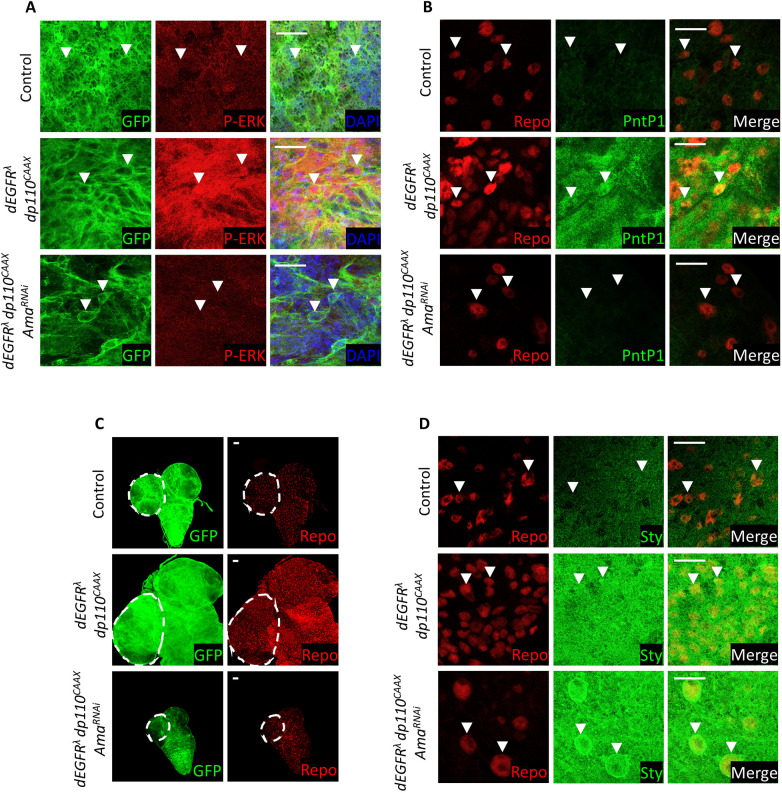
As expected, glial-specific expression of dEGFRλ and dp110CAAX resulted in the activation of EGFR-Ras pathway, which was visualized by a dramatic increase in the P-ERK and PntP1 staining relative to the control (Fig. 6A,B) (Read et al., 2009). This led to excessive glia proliferation and enlargement of the brain (Fig. 6C) (Read et al., 2009). Remarkably, concomitant depletion of Ama blocked hyperactivation of the pathway as the levels of P-ERK and PntP1 were reduced below the wild-type control (Fig. 6A,B). Accordingly, Ama depletion reduced the abnormally large brain size of repo>dEGFRλ dp110CAAX (Fig. 6C). Since upregulation of Sty is a key event of Ama downregulation, we compared Sty expression between repo>dEGFRλ dp110CAAX and repo>dEGFRλ dp110CAAX Ama. In repo>dEGFRλ dp110CAAX brains, Sty was uniformly upregulated throughout the glial plane (Fig. 6D). This is expected as Sty was shown to be directly regulated by EGFR signaling (Butchar et al., 2012) and fine-tunes RTK signaling (Rubin et al., 2003). Strikingly, in repo>dEGFRλ dp110CAAX AmaRNAi glial cells, Sty was upregulated to a much higher level that was especially evident at the nuclear membrane (Fig. 6D). Thus, Ama depletion results in increase of Sty, reduces RTK activation caused by activated EGFR and blocks neoplasia in a glioma fly model.
Fig. 6.
Ama depletion suppresses neoplastic growth in the Drosophila glioma model. (A) P-ERK immunofluorescence in surface glia reveals that knockdown of Ama in the Drosophila glioma model causes a striking decrease in P-ERK. White arrowheads point at glial cells. (B) PntP1 immunofluorescence in surface glia indicates that there is a drastic decrease in PntP1 following Ama depletion in the Drosophila glioma model. White arrowheads point at Repo-positive glial cells. (C) Repo immunofluorescence in surface glia shows that Ama depletion in a Drosophila glioma model drastically decreases the brain size and number of glia. GFP labels glial cell membranes. Dashed outlines show brain regions. (D) Immunofluorescence reveals that there is an increase in Sty levels following Ama knockdown in the Drosophila glioma model in surface glia. White arrowheads point at Repo-positive glial cells. Scale bars: 20 µm. Final genotypes in A–D: repo>mCD8GFP (top panel), repo>dEGFRλ dp110CAAX mCD8GFP (middle panel), repo>dEGFRλ dp110CAAX mCD8GFP AmaRNAi (bottom panel).
Knockdown of the Ama ortholog LSAMP increases SPROUTY2 levels in human glioblastoma cell lines
Ama has several human orthologs that belong to the IgLON immunoglobulin cell adhesion family (https://flybase.org/; Thurmond et al., 2019), with one member of this family LSAMP showing the highest sequence similarity. Interestingly, IgLONs family members are expressed in astrocyte glia and have been shown to promote astrocyte proliferation through FGF signaling (Sugimoto et al., 2012). The experiments described above suggest that upregulation of Sty is the major consequence of Ama depletion in glia during normal development and in the glioma model. Therefore, we asked whether the relationship between Ama and Sty is conserved in human cells.
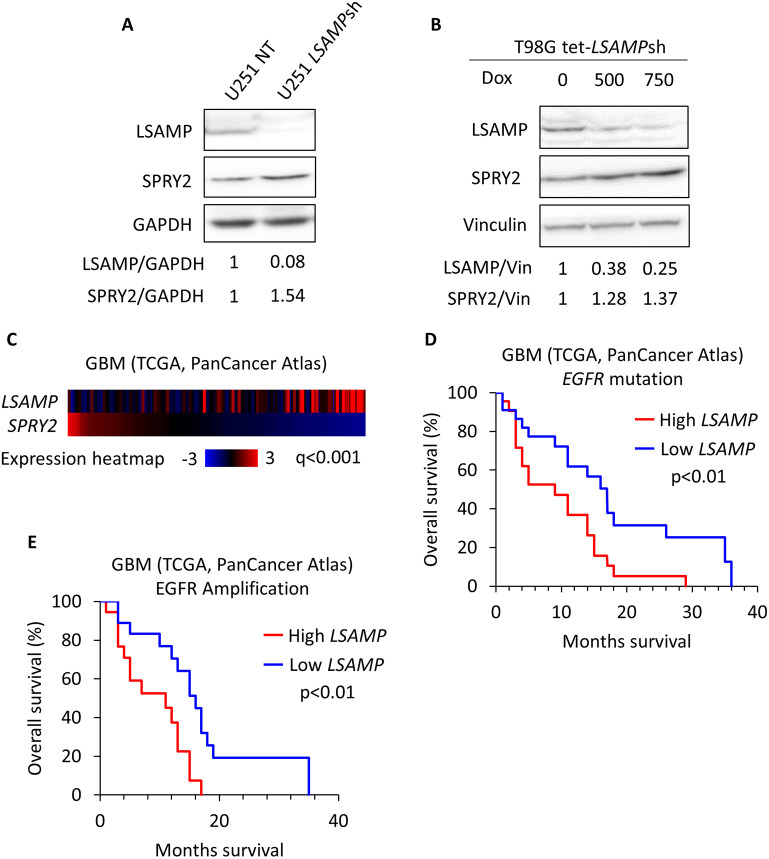
To address this question, we selected two glioblastoma cell lines, U251 and T98G, and examined the expression of SPROUTY2 (SPRY2) following LSAMP knockdown by western blotting. Endogenous LSAMP was detected in U251 and T98G glioblastoma cell lines by western blotting (Fig. 7A,B). U251 cells expressing LSAMP shRNA were generated using a lentiviral construct, and western blotting confirmed that LSAMP was successfully knocked down by the LSAMPsh (Fig. 7A). A doxycycline (Dox)-inducible knockdown system was generated in the T98G cell line, since conventional lentiviral shRNA resulted in reduced cell viability. A dose-dependent decrease in LSAMP was observed with increasing concentration of Dox (Fig. 7B). Having established the efficiency of LSAMP knockdown, we examined SPRY2 levels. As shown in Fig. 7A,B, in both U251 and T98G cell lines, LSAMPsh expression led to an increase in SPRY2 expression, as revealed by western blotting.
Fig. 7.
The relationship between Ama and Sprouty is conserved in human glioblastoma cell lines. (A) Immunoblot showing LSAMP, SPRY2 and GAPDH levels in U251 cells stably expressing non-targeting shRNA (NT) or LSAMP shRNA (LSAMPsh). Knockdown of LSAMP increases SPRY2, with relative densitometry levels for each target labeled below. (B) Immunoblot showing LSAMP, SPRY2 and vinculin levels in T98G cells expressing a doxycycline-inducible shRNA. Cells were treated for 3 days with the corresponding amounts of doxycycline (ng/ml in 0.15% DMSO) before harvesting. Dose-dependent knockdown of LSAMP gradually increases SPRY2. Relative densitometry levels for each target are labeled below. Results in A and B are representative of three independent experiments. (C) Heatmap displaying mRNA expression z-score (RNA Seq V2 RSEM) of LSAMP and SPRY2 in each row in GBM human patients (TCGA PanCancer Atlas) showing a significant inverse correlation between those genes. Each column represents a tumor tissue sample. The q-value was generated from a two-sample t-test. (D) Kaplan–Meier plot revealing that GBM patients (TCGA PanCancer Atlas) with EGFR mutations have a significant decrease in survivability with high levels of LSAMP mRNA. The P-value was generated from a logrank test. (E) Kaplan–Meier plot of GBM patients with EGFR amplifications reveals that patients with high LSAMP mRNA have a significant decrease in survivability. The P-value was generated using a logrank test.
To explore the relationship between LSAMP and SPRY2 in human glioblastoma, we analyzed the publicly available glioblastoma multiforme (GBM) TCGA PanCancer Atlas database (cBioportal.org). We found a significant inverse correlation in mRNA levels between LSAMP and SPRY2 (Fig. 7C). GBM patients with EGFR mutations and amplifications were selected to perform Kaplan–Meier survival analyses based on LSAMP expression. The results revealed a significant decrease in survival in patients with high expression of LSAMP (Fig. 7D,E). These findings suggest that the relationship between Ama and Sty is conserved in human glioblastoma cell lines.
DISCUSSION
The RTK signaling pathway is critical in a plethora of glial cell functions, such as migration, differentiation, proliferation, neurodegeneration and locomotion (Yildirim et al., 2019; Ray et al., 2017). Here, we identify Ama as a regulator of RTK signaling in glial cells and uncover an essential function of Ama in the maintenance of the BBB. Our study underscores the power of scRNA-seq profiling to explore a knockdown phenotype and led to the identification of Sty as a major Ama target during regulation of RTK signaling.
Profiling cells by scRNA-seq identified distinct changes in gene expression profiles across multiple glial cell types in Ama-depleted brain that otherwise would have not been possible using conventional approaches. First, we find that Ama depletion affects gene expression in surface glia, which in turn leads to a disruption in the BBB. Second, the increase in hemocyte numbers in repo>AmaRNAi scRNA-seq dataset enabled the discovery of infiltrating blood cells in Ama-depleted brains. Third, the increase in Sty and decrease in PntP1 levels following Ama knockdown in glia guided us to explore the impact of Ama in RTK signaling.
scRNA-seq identifies and clusters cells based on similarity in gene expression profile to uncover the cellular heterogeneity in a tissue. In this study, scRNA-seq of the normal fly brains identified all the glial cell types in addition to a Fas cluster that appeared to encompass multiple glial cell types. This cell cluster displayed high expression levels of cell adhesion proteins, which is a hallmark of glial cells (DeSalvo et al., 2014; Sasse et al., 2015), while also supporting the observation that scRNA-seq not only clusters cells by type but also by similar biological features (Ariss et al., 2018). This underlies the robustness of scRNA-seq, since it uncovers complex cellular dynamics related to certain stimuli and continuous temporal differentiation processes (Liu and Trapnell, 2016), as well as the spatial arrangement of cells (Wagner et al., 2016). Notably, the cellular perturbations in clustering that we observed following Ama depletion were validated experimentally by genetic analysis, immunofluorescence and other assays. This type of single-cell data validation is essential as it addresses the concern of a batch effect between scRNA-seq samples (Haghverdi et al., 2018). Thus, our work highlights the power of scRNA-seq to profile a knockdown or mutant phenotype.
Our results revealed that Ama is critical for maintaining the BBB, as its depletion results in discontinuous SG membranes, suggesting a lack of tight junctions or organization that leads to disruption of the barrier (Babatz et al., 2018). This in turn exposes the larval brains to the high potassium content of the hemolymph, which damages neurons (Yildirim et al., 2019). Ama knockdown decreases overall glial cell proliferation, which can affect SG cells and the BBB in two additional ways. First, lack of proliferation in PG cells can potentially affect the secretion of metabolites, which is important to prevent neurodegeneration (Volkenhoff et al., 2015). Second, Ama depletion can alter SPG growth by reducing endoreplication and endomitosis, as evident by reduced expression of cell cycle genes in repo>AmaRNAi, and, therefore, hinder the ability of SPG to accommodate the growing brain during late larval stages (Unhavaithaya and Orr-Weaver, 2012).
The conventional way to determine the intactness of the BBB function is by labeling brains with a fluorescent dextran dye. If the barrier is permissible to large molecules, such as the dye, then the BBB is considered to be broken. Here, we present an alternative approach to monitor a disruption in the BBB by measuring the increase in infiltrating hemocytes in larval brains both by scRNA-seq and through staining with a hemocyte-specific antibody. Although SG protect neurons from the hemolymph (Yeh et al., 2018), penetration of hemocytes into the brain through the damaged BBB has not been previously reported. Whether infiltrating hemocytes have a role in inflicting the damage in the brain is unknown but raises the possibility that they might have a function in that context.
Ama and Lachesin are part of the IgLON family, with Lachesin also shown to be required for the BBB maintenance (Strigini et al., 2006). Since IgLONs are also expressed in the BBB in mammals, this suggests that the immunoglobin superfamily may indeed also have an evolutionarily conserved BBB function in humans (Kubick et al., 2018). These findings highlight an important role of IgLONs in neurodegenerative diseases that result in BBB breakdown (Sweeney et al., 2018).
Through scRNA-seq and measuring P-ERK and PntP1 levels, we found a drastic reduction in the level of RTK signaling in Ama-depleted brains. We suggest that Ama regulates the RTK pathway since its depletion increases Sty levels, a general inhibitor of the pathway, and, conversely, overexpression of Ama has an opposite effect. Sty in repo>AmaRNAi brains predominantly localizes to the nuclear and plasma membranes. Intriguingly, in mammalian cells, SPRY2 localization to the membrane has been shown to be crucial for its phosphorylation and inhibitory effect (Hanafusa et al., 2002). Our results therefore suggest that, while depletion of Ama increases Sty levels, there might additionally be an effect in the cellular localization or post-translational modification of Sty (Jarvis et al., 2006). Genetic experiments indicate that Sty is the key target of Ama and that activated ERK can partially rescue the repo>AmaRNAi phenotype, whereas activated EGFR in the glioma fly model cannot. These epistatic interactions support the model that Ama, acts through Sty, and is downstream of EGFR but upstream of ERK in the RTK signaling pathway (Fig. 8).
Fig. 8.
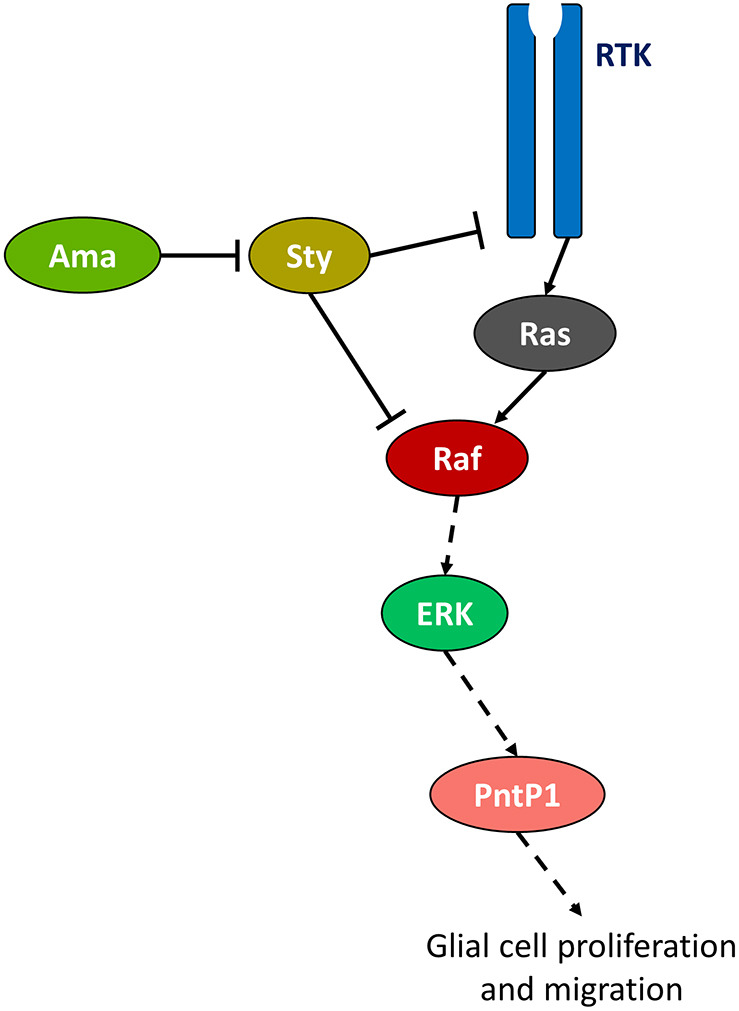
Ama affects RTK signaling. Illustration of the molecular mechanism of Ama acting through Sty to inhibit RTK signaling.
Although knockdown of Sty in Ama-depleted brains partially rescues the glial cell numbers and brain size, it fails to suppress the neuronal apoptosis in repo>AmaRNAi brains. Since Ama can act non-cell-autonomously, we cannot exclude the possibility that Ama might also affect neurons non-cell-autonomously and, therefore, the expression of Sty in glial cells fails to rescue neurons. This is noteworthy since LSAMP, the human counterpart of Ama, was reported to control neurite growth (Akeel et al., 2011). The precise mechanism of how Ama affects Sty is unknown and requires further investigation. However, SPRY2 and EGFR in human cell lines have been shown to compete with c-Cbl (a ubiquitin ligase) binding. In this context, the ubiquitin ligase attenuates the inhibitory effect of SPRY2 and vice versa (Rubin et al., 2003). Since Ama and Sty localize to cell membranes, one possible explanation is that they may interact with each other, whereby loss of Ama releases Sty to bind to RTK receptors and promotes inhibition of the signaling pathway.
Finally, we show that the role of Ama in controlling Sty levels is conserved in human glioblastoma (GBM) cell lines. Additionally, GBM patients with EGFR mutations display a significant increase in survival when they have low expression levels of LSAMP, suggesting a potential role for the IgLON family member in this cancer. Previous work has shown that LSAMP either promotes growth or acts as a tumor suppressor (Kresse et al., 2009). Intriguingly, SPRY2 inhibits or activates RTK signaling based on the context and cell type and also acts either as an oncogene or tumor suppressor (Masoumi-Moghaddam et al., 2014). Our results suggest a potential connection between LSAMP and SPRY2 that may help to explain the role of LSAMP in tumorigenesis and, thus, point to LSAMP as a potential therapeutic target.
MATERIALS AND METHODS
Fly stocks
Drosophila stocks and crosses were kept at 25°C using standard cornmeal agar medium unless mentioned otherwise. Stocks were obtained from the Bloomington Drosophila Stock Center (BDSC) the Kyoto Drosophila Genetic Resource Center (DGRC), and the Vienna Drosophila Resource Center (VDRC).
The stocks used were: (1) w[1118]; P{w[+m*]=GAL4}repo/TM3, Sb[1] (BDSC P{GAL4}repo); (2) y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00297}attP2 (BDSC HMS00297, UAS-AmaRNAi); (3) y[1] v[1]; P{y[+t7.7] v[+t1.8]=UAS-GFP.VALIUM10}attP2 (BDSC); (4) w[*]; P{y[+t7.7] w[+mC]=10XUAS-IVS-mCD8::GFP}attP2 (BDSC); (5) y[*] w[*]; P{w[+mW.hs]=GawB}Ama[NP1297] / TM6, P{w[-]=UAS-lacZ.UW23-1}UW23-1 (Kyoto DGRC); (6) moody-Gal4 (gift from Vicki Losick, Biology Department, Boston College, USA); (7) UAS-dEGFRλ, UAS-dp110CAAX; repo-Gal4, UAS-mCD8-GFP/TM6C, Tb, tub-Gal80 (gift from Renee Read, Department of Pharmacology and Chemical Biology, Emory University School of Medicine, USA); (8) repo-Gal4, UAS-mCD8-GFP/TM6C, Tb (gift from Renee Read); (9) UAS-styRNAi (VDRC shRNA330208); (10) y[1] w[*]; P{w[+mC]=UAS-rl[Sem].S}2 (BDSC); (11) w[1118]; P{y[+t7.7] w[+mC]=GMR57E08-GAL4}attP2 (BDSC); (12) y[1] w[*]; P{y[+t7.7] w[+mC]=E(spl)mdelta-HLH-GFP.FPTB}attP40 (BDSC); (13) hs-FLP; act5c-FRT-stop-FRT-Gal4, UAS-GFP (gift from Teresa Orenic, Biological Sciences Department, University of Illinois at Chicago, USA); (14) UAS-AmaRNAi (VDRC GD22945).
The moody>AmaRNAi (VDRC GD22945) cross was performed at 25°C for 2 days then the vial was transferred to 30°C for the rest of development.
All the Ama knockdown experiments were performed using BDSC HMS00297 unless the VDRC GD22945 UAS-AmaRNAi line is mentioned in figure and figure legend.
Ama transgene
A UAS-Ama C-terminal FLAG-HA tagged clone UFO01101 having a white gene selectable marker and an attB site was used from the Universal Proteomics Resource and part of the Berkeley Drosophila Genome Project (BDGP) (https://www.fruitfly.org/EST/proteomics.shtml). This clone was received from the Drosophila Genomics Resource Center (DGRC) (https://dgrc.bio.indiana.edu/Home) stock number 1621050.
The PhiC31 transformation services in BestGene (http://www.thebestgene.com/) were performed on the y[1] w[1118]; PBac{y[+]-attP-9A}VK00023 line (BDSC 9741) with an estimated CytoSite 70A2 (chromosome 3) using the UAS-Ama construct above.
Fluorescence in situ hybridization
A total of 38 RNA probes labeled with Quasar 670 fluorophores were generated with the Stellaris custom probe designer v4.2 that target the Ama-RA coding sequence (oligonucleotide length=18; min. spacing length=2; masking level=5) (https://biosearchassets.blob.core.windows.net/assets/bti_custom_stellaris_drosophila_protocol.pdf).
Third-instar larval brains were dissected and the Stellaris imaginal disc fluorescence in situ hybridization protocol was followed (https://biosearchassets.blob.core.windows.net/assets/bti_custom_stellaris_drosophila_protocol.pdf). Samples were imaged using a Zeiss Confocal microscope.
Immunofluorescence
Third-instar larval brains were dissected in 1× PBS then fixed for 30 min in 4% formaldehyde in 1× PBS. The brains were permeabilized and washed twice with 0.3% PBST (1× PBS and 0.3% Triton X-100) for 10 min. Primary antibodies were incubated overnight in 10% normal donkey serum (NDS; Jackson Immunoresearch). The samples were washed three times on the second day with 0.1% PBST (1× PBS and 0.1% Triton X-100) and incubated with the fluorescently labeled secondary antibody in 10% NDS for 1 h. Brains were then washed five times with 0.1% PBST and mounted on a glass slide using FluorSave (EMD Millipore).
Wandering third-instar larval eye discs were dissected in 1× PBS and fixed in 4% formaldehyde in 1× PBS for 15 min, permeabilized in 0.3% PBST two times for 10 min and then incubated with antibodies overnight at 4°C in 1× PBS plus 10% NDS and 0.1% Triton X-100 blocking serum. The following day, samples were washed in 0.1% PBST three times for 5 min. Samples were then incubated with appropriate fluorescently labeled secondary antibodies (Jackson Immunoresearch) for 1 h in 1× PBS plus 10% NDS and 0.1% Triton X-100 followed by 4′,6-diamidino-2-phenylindole (DAPI) for 5 min. Finally, samples were washed five times for 5 min then mounted in FluorSave (EMD Millipore) on glass slides. All steps were carried out at room temperature and with gentle rocking, unless specified otherwise. Whenever fluorescence images have been compared, they were obtained with the same acquisition and display settings. The slides were imaged using Zeiss Confocal microscope.
Primary antibodies were used: anti-Repo: DSHB 8D13 (1:50), anti-cleaved Drosophila Dcp-1 (Cell Signaling Asp216; 1:500), anti-GFP (FITC) (Abcam ab6662; 1:1000), anti-P-ERK (Sigma M8159; 1:200), anti-Sty-C-terminal (a gift from Mark Krasnow, Biochemistry Department, Stanford University, USA; 1:900), anti-P-Glycoprotein, C494 (recommended by Roland Bainton, Department of Anesthesia and Perioperative Care, University of California, San Francisco, USA; ThermoFisher Scientific MA1-26529 1:100), anti-Fas2 (DSHB 1D4; 1:50), anti-PH3 (Millipore Sigma 06-570, 1:1000), 24B10 antibody (DSHB; 1:100), anti-PntP1 (gift from James Skeath, Department of Genetics, Washington University, USA; 1:200), H2 antibody (gift from István Ando, Institute of Genetics, Hungarian Academy of Sciences, Hungary; 1:100).
Heat-shock treatment
To generate third-instar larvae with clones of cells that overexpress Ama, heat shock was induced 72 h after egg deposition at 37°C for 1 h then embryos were transferred to 25°C. Third-instar larvae were collected around 55 h after heat-shock for immunostaining.
Dextran labeling
Larval brains were dissected and rinsed twice with 1× PBS then incubated for 10 to 15 min in 2.5 mM of Tetramethylrhodamine-labeled 10 kDa dextran dye (ThermoFisher D1816) in 1× PBS. The brains were rinsed again twice with 1× PBS and mounted on a glass slide using FluorSave (EMD Millipore). A Zeiss Confocal microscope was quickly used to image the slides.
Immunofluorescence quantification
The Sty and P-ERK fluorescence intensities were measured after selecting individual glial cell nuclei in multiple brains using the imageJ software (NIH). The average Sty (70 nuclei) and P-ERK (171 nuclei) intensities in the control repo>+ glia nuclei were determined and compared to that in the repo>AmaRNAi and repo>Ama glia nuclei to measure the relative fluorescent intensities of each immunostaining.
Tissue dissociation and Drop-seq
The dissociation protocol uses both collagenase (Sigma C9891) resuspended in 10× Krebs-Ringer with Ca2+ solution and 10× trypsin-EDTA (Sigma 59418C). The EDTA partially inhibits the collagenase to minimize tissue damage. Ten to fifteen third-instar brains were dissected in 30 min in 1× PBS on ice then transferred to a 1.5 ml microcentrifuge tube and spun down at 5000 rpm (∼2000 g) 4°C for 5 min. The brains were washed with Rinaldini solution and spundown at 5000 rpm (∼2000 g) 4°C for 5 min. While washing, the pellet remained carefully submerged to prevent the tissues from being pipetted up and sticking to the pipette tip. A digestion mix consisting of 1× trypsin and 2.5 mg/ml collagenase in Rinaldini's solution (500 μl) was added to resuspend the pelleted tissues. The microcentrifuge tube was oriented horizontally (to maximize the mechanical digestion and dissociation mix movement in the tube) on a shaker at 225 rpm for 45 min to an hour. The microcentrifuge tube was flicked every 10 min during the digestion step. The tube was spun down at 5000 rpm (∼2000 g) 4°C for 5 min before and between each following step: rinsing with 1× PBS with 0.01% BSA, rinsing with 1× PBS and resuspending cells in 60–100 μl 1× PBS. This protocol results in single cells and less than 5% clumps. The Drop-seq protocol was performed on the single-cell suspension as previously described (Ariss et al., 2018).
scRNA-seq alignment
Drop-seq data was analyzed using the pipeline wrapped in DropSeqPipe (version 0.5) (https://github.com/Hoohm/dropSeqPipe). This is based on Drop-seq core computational pipeline described in ‘Drop-seq_Alignment_Cookbook.pdf’ version 2.0.0 on 9-28-18 (http://mccarrolllab.com/dropseq/) and the Drop-seq tools (version-2.3.0) was used to process the single-cell RNA-seq Illumina paired end raw sequences (FastQ files). STAR aligner was used to align the raw sequences against the Drosophila melanogaster genome version BDGP6 (Ensembl gene model version 90). The quality of reads and mapping were examined with the FastQC program (v0.11.8) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and poor quality reads (below 20) were eliminated before mapping.
The digital gene expression (DGE) was generated using the Drop-seq alignment protocol. The number of cells extracted from aligned BAM file is based on the knee plot, which extracts the number of reads per cell, then plotting the cumulative distribution of reads and selecting the knee of the distribution.
Cell clustering and single-cell analysis
The DGE files were subjected to Seurat (version 3.0.0) single-cell analysis to generate computational figures, such as dot plots, feature plots, UMAP and violin plots in R software (version 3.5.3). The standard Seurat workflow was followed, and we filtered the low-quality cells and outlier cells with a high gene number (min. gene cutoff=200 and max. gene cutoff=3000). The first 20 principle components were selected to run the UMAP with a resolution of 0.5 for the analyses of brains and a resolution 1.2 for the supervised glial cell analyses. The repo>+, repo>AmaRNAi combined supervised glia analysis was performed using the cell barcodes listed in Table S7. The population labeled as ‘other’ display high levels of ribosomal genes and, therefore, considered to represent low quality cells.
Human cell lines
U251 cells were gifted by Nissim Hay (Department of Biochemistry & Molecular Genetics, University of Illinois at Chicago, USA) and T98G cells were gifted by Elizaveta Benevolenskaya (Department of Biochemistry & Molecular Genetics, University of Illinois at Chicago, USA). 293FT cells were acquired from Thermo Fisher. Both GBM cell lines were grown in MEM (Corning 10-010-CV) and 293FT were grown in DMEM (Corning 10-017-CV). All media were supplemented with 10% FBS (Gemini) and 1% penicillin-streptomycin (Corning 30-002-Cl). U251 cells were transduced with non-targeting (NT) or LSAMP shRNAs that were stably expressed. For immunoblotting experiments, 800,000 cells were split onto 6 cm plates and harvested 24 h later. T98G cells were transduced with a doxycycline-inducible shRNA targeting LSAMP. Following transduction, T98G cells were grown in medium with 10% tet-free FBS (Gemini). For immunoblotting experiments, 300,000 cells were plated onto 6 cm plates with the corresponding amount of doxycycline (sigma) added and grown for 72 h before harvesting. All conditions had 0.15% DMSO. Results from replicates are available upon request.
Plasmids and lentiviral production and transduction
Stably expressed shRNA plasmids, plko.1 empty vector and non-target (NT), were purchased from Sigma. The pTIP doxycycline inducible shRNA plasmid was a gift from Marcus Peter (xxx dept., institute, country xxx). The LSAMP-specific shRNA cloned into each plasmid was: 5′-CCGGCAAGTTTACTTGATCGTACAA-3′. For lentiviral production, 6×106 293FT cells were reverse transfected with 9 μg Virapower (Thermo Fisher) and 3 μg of the corresponding shRNA vector in 36 μl of Lipofectamine. The medium was changed 16 h after transfection, and 6 ml of viral particles was collected twice before filtering through a 0.45 μm filter. Cells were transduced with 2 ml of viral supernatant diluted in 2 ml of complete medium supplemented with 8 μg/ml of polybrene (Sigma 107689). 48 h after transduction, cells were selected in 1 μg/ml of puromycin (ACROS organics 227420100) for 5 days. (Putzbach et al., 2017).
Immunoblotting
At the indicated time point, cells were washed twice with cold PBS on ice. Then, 100 μl of 1× RIPA (Cell Signaling #9806S), supplemented with phosphatase and protease inhibitor tablets (Peirce A32957 and A32953), was added to each plate for 5 min before scraping and collecting into 1.5 ml Eppendorf tubes. Lysates were vortexed on high for 10 s every 10 min for 30 min before centrifuging at 13,000 g for 10 min. The lysate supernatant was collected, and the protein concentration was determined with the Bradford Method (Bio-Rad). A total of 20–30 μg of protein was loaded for each sample and ran via standard electrophoresis on a 10% polyacrylamide gel and transferred for 75 min at 115 v onto PVDF membranes. Membranes were then blocked in 5% milk Tris-buffered saline with 0.1% Tween 20 (TBS-T) with for 1 h. Primary antibodies were incubated on a shaker over night at 4°C in 2.5% milk in TBS-T. Membranes were washed 3× in TBS-T before the corresponding secondary antibody (1:3000) was added for 1 h at room temperature. Membranes were washed 3× in TBS-T before developing on an Azure cSeries with chemiluminescence. Primary antibodies used were against: LSAMP (Abcam ab64427; 1:500), SPRY2 (Millipore #07-524; 1:1000), GAPDH (Cell Signaling D16H11; 1:5000) and vinculin (Sigma V9131; 1:5000). The relative densitometries were determined using Image Studio Lite Ver 5.2.
TCGA – cBioportal
Glioblastoma TCGA PanCancer Atlas patient samples were selected in cBioportal (https://www.cbioportal.org/) (Gao et al., 2013; Cerami et al., 2012). The LSAMP and SPRY2 heatmap was generated using the ‘OncoPrint’ tab in cBioportal.
Using the ‘Custom Selection’ in cBioportal, GBM patients with EGFR mutations were split into two groups based on the median LSAMP expression (LSAMP high versus LSAMP low) in the ‘Groups’ tab. The survivability was then plotted. The same was achieved using GBM patients with EGFR amplifications that were split into two groups based on the first and fourth quartiles in LSAMP expression (LSAMP high versus LSAMP low).
Supplementary Material
Acknowledgements
The authors are grateful to Renee Read, Vicki Losick, and Teresa Orenic for sharing Drosophila stocks as well as Roland Bainton and Mark Krasnow, James Skeath, and István Ando for recommending and sharing antibodies and Elizaveta Benevolenskaya for sharing cell lines. We thank the Developmental Studies Hybridoma Bank for the antibodies and Bloomington Stock Center at Indiana University for Drosophila stocks.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.M.A., M.V.F.; Software: A.B.M.M.K.I.; Formal analysis: A.B.M.M.K.I., M.M.A.; Investigation: M.M.A., A.R.T.; Writing - original draft: M.M.A.; Writing - review & editing: M.V.F.; Visualization: M.M.A.; Supervision: N.H., M.V.F.; Project administration: M.V.F.; Funding acquisition: A.R.T., N.H., M.V.F.
Funding
This work is funded by the National Institutes of Health grants R35GM131707 to M.V.F., R01AG016927, R01CA090764, R01CA206167 to N.H. and F30CA225058 to A.R.T.; the U.S. Department of Veterans Affairs grants BX000733 and IK6BX004602 to N.H. Deposited in PMC for release after 12 months.
Data availability
All raw sequencing data are deposited in the Gene Expression Omnibus (GEO) database under accession code GSE147688.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.250837.supplemental
References
- Akeel M., McNamee C. J., Youssef S. and Moss D. (2011). DIgLONs inhibit initiation of neurite outgrowth from forebrain neurons via an IgLON-containing receptor complex. Brain Res. 1374, 27-35. 10.1016/j.brainres.2010.12.028 [DOI] [PubMed] [Google Scholar]
- Ariss M. M., Islam A. B. M. M. K., Critcher M., Zappia M. P. and Frolov M. V. (2018). Single cell RNA-sequencing identifies a metabolic aspect of apoptosis in Rbf mutant. Nat. Commun. 9, 1-13. 10.1038/s41467-018-07540-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avet-Rochex A., Kaul A. K., Gatt A. P., McNeill H. and Bateman J. M. (2012). Concerted control of gliogenesis by InR/TOR and FGF signalling in the Drosophila post-embryonic brain. Development 139, 2763-2772. 10.1242/dev.074179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babatz F., Naffin E. and Klämbt C. (2018). The Drosophila blood-brain barrier adapts to cell growth by unfolding of pre-existing septate junctions. Dev. Cell 47, 697-710.e3. 10.1016/j.devcel.2018.10.002 [DOI] [PubMed] [Google Scholar]
- Bainton R. J., Tsai L. T.-Y., Schwabe T., DeSalvo M., Gaul U. and Heberlein U. (2005). moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell 123, 145-156. 10.1016/j.cell.2005.07.029 [DOI] [PubMed] [Google Scholar]
- Bangi E., Murgia C., Teague A. G. S., Sansom O. J. and Cagan R. L. (2016). Functional exploration of colorectal cancer genomes using Drosophila. Nat. Commun. 7, 13615 10.1038/ncomms13615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchar J. P., Cain D., Manivannan S. N., McCue A. D., Bonanno L., Halula S., Truesdell S., Austin C. L., Jacobsen T. L. and Simcox A. (2012). New negative feedback regulators of Egfr signaling in Drosophila. Genetics 191, 1213-1226. 10.1534/genetics.112.141093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E. et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401-404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D. and Deng W. M. (2019). Drosophila Model in Cancer: An Introduction. Adv Exp. Med. Biol. 1167, 1-14. 10.1007/978-3-030-23629-8_1 [DOI]
- DeSalvo M. K., Hindle S. J., Rusan Z. M., Orng S., Eddison M., Halliwill K. and Bainton R. J. (2014). The Drosophila surface glia transcriptome: evolutionary conserved blood-brain barrier processes. Front. Neurosci. 8, 346 10.3389/fnins.2014.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J., Logan M. A., Taşdemir Ö. E. and Freeman M. R. (2009). Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 29, 4768-4781. 10.1523/JNEUROSCI.5951-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C. J., Olson J. M., Ngo K. T., Kim E., Lee N. E., Kuoy E., Patananan A. N., Sitz D., Tran P., Do M. T. et al. (2009). G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 6, 603 10.1038/nmeth.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzdóttir S. R., Engelen D., Yuva-Aydemir Y., Schmidt I., Aho A. and Klämbt C. (2009). Switch in FGF signalling initiates glial differentiation in the Drosophila eye. Nature 460, 758-761. 10.1038/nature08167 [DOI] [PubMed] [Google Scholar]
- Fremion F., Darboux I., Diano M., Hipeau–Jacquotte R., Seeger M. A. and Piovant M. (2000). Amalgam is a ligand for the transmembrane receptor neurotactin and is required for neurotactin–mediated cell adhesion and axon fasciculation in Drosophila. EMBO J. 19, 4463-4472. 10.1093/emboj/19.17.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari F. B., Fenton T., Bachoo R. M., Mukasa A., Stommel J. M., Stegh A., Hahn W. C., Ligon K. L., Louis D. N., Brennan C. et al. (2007). Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 21, 2683-2710. 10.1101/gad.1596707 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A., Kadowaki T. and Kitagawa Y. (2003). Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Dev. Biol. 264, 582-591. 10.1016/j.ydbio.2003.06.001 [DOI] [PubMed] [Google Scholar]
- Hacohen N., Kramer S., Sutherland D., Hiromi Y. and Krasnow M. A. (1998). sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92, 253-263. 10.1016/S0092-8674(00)80919-8 [DOI] [PubMed] [Google Scholar]
- Haghverdi L., Lun A. T. L., Morgan M. D. and Marioni J. C. (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421-427. 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa H., Torii S., Yasunaga T. and Nishida E. (2002). Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 4, 850-858. 10.1038/ncb867 [DOI] [PubMed] [Google Scholar]
- Hausott B. and Klimaschewski L. (2019). Sprouty2—A novel therapeutic target in the nervous system? Mol. Neurobiol. 56, 3897-3903. 10.1007/s12035-018-1338-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagnatiello M.-A., Weitzer S., Gannon G., Compagni A., Cotten M. and Christofori G. (2001). Mammalian sprouty-1 and-2 are membrane-anchored phosphoprotein inhibitors of growth factor signaling in endothelial cells. J. Cell Biol. 152, 1087-1098. 10.1083/jcb.152.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis L. A., Toering S. J., Simon M. A., Krasnow M. A. and Smith-Bolton R. K. (2006). Sprouty proteins are in vivo targets of Corkscrew/SHP-2 tyrosine phosphatases. Development 133, 1133-1142. 10.1242/dev.02255 [DOI] [PubMed] [Google Scholar]
- Kresse S. H., Ohnstad H. O., Paulsen E. B., Bjerkehagen B., Szuhai K., Serra M., Schaefer K.-L., Myklebost O. and Meza-Zepeda L. A. (2009). LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer 48, 679-693. 10.1002/gcc.20675 [DOI] [PubMed] [Google Scholar]
- Kubick N., Brösamle D. and Mickael M.-E. (2018). Molecular evolution and functional divergence of the IgLON family. Evol. Bioinform. 14, 1176934318775081 10.1177/1176934318775081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz É., Váczi B., Márkus R., Laurinyecz B., Vilmos P., Zsámboki J., Csorba K., Gateff E., Hultmark D. and Andó I. (2007). Definition of Drosophila hemocyte subsets by cell-type specific antigens. Acta Biol. Hung. 58, 95-111. 10.1556/ABiol.58.2007.Suppl.8 [DOI] [PubMed] [Google Scholar]
- Levine B. D. and Cagan R. L. (2016). Drosophila lung cancer models identify trametinib plus statin as candidate therapeutic. Cell Rep. 14, 1477-1487. 10.1016/j.celrep.2015.12.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. and Trapnell C. (2016). Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Research 5, 182 10.12688/f1000research.7223.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M. et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202-1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoumi-Moghaddam S., Amini A. and Morris D. L. (2014). The developing story of Sprouty and cancer. Cancer Metastasis Rev. 33, 695-720. 10.1007/s10555-014-9497-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto J. J., Yogi P. and Hartenstein V. (2015). Origin and development of neuropil glia of the Drosophila larval and adult brain: two distinct glial populations derived from separate progenitors. Dev. Biol. 404, 2-20. 10.1016/j.ydbio.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. M., Rebay I., Tjian R. and Rubin G. M. (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137-147. 10.1016/0092-8674(94)90580-0 [DOI] [PubMed] [Google Scholar]
- Osman D., Gobert V., Ponthan F., Heidenreich O., Haenlin M. and Waltzer L. (2009). A Drosophila model identifies calpains as modulators of the human leukemogenic fusion protein AML1-ETO. Proc. Natl Acad. Sci. USA 106, 12043-12048. 10.1073/pnas.0902449106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini R. A. and Xu T. (2003). A genetic screen in Drosophila for metastatic behavior. Science 302, 1227-1231. 10.1126/science.1088474 [DOI] [PubMed] [Google Scholar]
- Papalexi E. and Satija R. (2018). Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 18, 35 10.1038/nri.2017.76 [DOI] [PubMed] [Google Scholar]
- Pereanu W., Spindler S., Cruz L. and Hartenstein V. (2007). Tracheal development in the Drosophila brain is constrained by glial cells. Dev. Biol. 302, 169-180. 10.1016/j.ydbio.2006.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N. (1994). Signalling pathways initiated by receptor protein tyrosine kinases in Drosophila. Curr. Opin. Cell Biol. 6, 260-266. 10.1016/0955-0674(94)90145-7 [DOI] [PubMed] [Google Scholar]
- Putzbach W., Gao Q. Q., Patel M., Van Dongen S., Haluck-Kangas A., Sarshad A. A., Bartom E. T., Kim K.-Y. A., Scholtens D. M., Hafner M. et al. (2017). Many si/shRNAs can kill cancer cells by targeting multiple survival genes through an off-target mechanism. eLife 6, e29702 10.7554/eLife.29702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Speese S. D. and Logan M. A. (2017). Glial Draper rescues Aβ toxicity in a Drosophila model of Alzheimer's Disease. J. Neurosci. 37, 11881-11893. 10.1523/JNEUROSCI.0862-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R. D. (2018). Pvr receptor tyrosine kinase signaling promotes post-embryonic morphogenesis, and survival of glia and neural progenitor cells in Drosophila. Development 145, dev164285 10.1242/dev.164285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R. D., Cavenee W. K., Furnari F. B. and Thomas J. B. (2009). A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 5, e1000374 10.1371/journal.pgen.1000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T. (2015). Targeting RTK signaling pathways in cancer. Cancers 7, 1758-1784. 10.3390/cancers7030860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C., Litvak V., Medvedovsky H., Zwang Y., Lev S. and Yarden Y. (2003). Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr. Biol. 13, 297-307. 10.1016/S0960-9822(03)00053-8 [DOI] [PubMed] [Google Scholar]
- Sasse S., Neuert H. and Klämbt C. (2015). Differentiation of Drosophila glial cells. Wiley Interdiscipl. Rev. Dev. Biol. 4, 623-636. 10.1002/wdev.198 [DOI] [PubMed] [Google Scholar]
- Seeger M. A., Haffley L. and Kaufman T. C. (1988). Characterization of amalgam: a member of the immunoglobulin superfamily from Drosophila. Cell 55, 589-600. 10.1016/0092-8674(88)90217-6 [DOI] [PubMed] [Google Scholar]
- Simon M. A., Bowtell D. D., Dodson G. S., Laverty T. R. and Rubin G. M. (1991). Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67, 701-716. 10.1016/0092-8674(91)90065-7 [DOI] [PubMed] [Google Scholar]
- Strigini M., Cantera R., Morin X., Bastiani M. J., Bate M. and Karagogeos D. (2006). The IgLON protein Lachesin is required for the blood–brain barrier in Drosophila. Mol. Cell. Neurosci. 32, 91-101. 10.1016/j.mcn.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P. and Satija R. (2019). Comprehensive integration of single-cell data. Cell 177, 1888-1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto C., Morita S. and Miyata S. (2012). Overexpression of IgLON cell adhesion molecules changes proliferation and cell size of cortical astrocytes. Cell Biochem. Funct. 30, 400-405. 10.1002/cbf.2813 [DOI] [PubMed] [Google Scholar]
- Sweeney M. D., Sagare A. P. and Zlokovic B. V. (2018). Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133 10.1038/nrneurol.2017.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S., Marygold S. J., Matthews B. B., Millburn G., Antonazzo G., Trovisco V. et al. (2019). FlyBase 2.0: the next generation. Nucleic Acids Res. 47, D759-D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unhavaithaya Y. and Orr-Weaver T. L. (2012). Polyploidization of glia in neural development links tissue growth to blood–brain barrier integrity. Genes Dev. 26, 31-36. 10.1101/gad.177436.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkenhoff A., Weiler A., Letzel M., Stehling M., Klämbt C. and Schirmeier S. (2015). Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab. 22, 437-447. 10.1016/j.cmet.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Wagner A., Regev A. and Yosef N. (2016). Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 34, 1145 10.1038/nbt.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Gilbert M., Petley-Ragan L. and Auld V. J. (2014). Loss of focal adhesions in glia disrupts both glial and photoreceptor axon migration in the Drosophila visual system. Development 141, 3072-3083. 10.1242/dev.101972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh P.-A., Liu Y.-H., Chu W.-C., Liu J.-Y. and Sun Y. H. (2018). Glial expression of disease-associated poly-glutamine proteins impairs the blood–brain barrier in Drosophila. Hum. Mol. Genet. 27, 2546-2562. 10.1093/hmg/ddy160 [DOI] [PubMed] [Google Scholar]
- Yildirim K., Petri J., Kottmeier R. and Klämbt C. (2019). Drosophila glia: few cell types and many conserved functions. Glia 67, 5-26. 10.1002/glia.23459 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Soustelle L., Giangrande A., Umetsu D., Murakami S., Yasugi T., Awasaki T., Ito K., Sato M. and Tabata T. (2005). DPP signaling controls development of the lamina glia required for retinal axon targeting in the visual system of Drosophila. Development 132, 4587-4598. 10.1242/dev.02040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.