Abstract
Amyotrophic lateral sclerosis (ALS) is a multifactorial and progressive neurodegenerative disease of unknown etiology. Due to ALS’s unpredictable onset and progression rate, the search for biomarkers that allow the detection and tracking of its development and therapeutic efficacy would be of significant medical value. Considering that alterations of energy supply are one of ALS’s main hallmarks and that a correlation has been established between gene expression in human brain tissue and peripheral blood mononuclear cells (PBMCs), the present work investigates whether changes in mitochondrial function could be used to monitor ALS. To achieve this goal, PBMCs from ALS patients and control subjects were used; blood sampling is a quite non-invasive method and is cost-effective. Different parameters were evaluated, namely cytosolic calcium levels, mitochondrial membrane potential, oxidative stress, and metabolic compounds levels, as well as mitochondrial dynamics and degradation. Altogether, we observed lower mitochondrial calcium uptake/retention, mitochondria depolarization, and redox homeostasis deregulation, in addition to a decrease in critical metabolic genes, a diminishment in mitochondrial biogenesis, and an augmentation in mitochondrial fission and autophagy-related gene expression. All of these changes can contribute to the decreased ATP and pyruvate levels observed in ALS PBMCs. Our data indicate that PBMCs from ALS patients show a significant mitochondrial dysfunction, resembling several findings from ALS’ neural cells/models, which could be exploited as a powerful tool in ALS research. Our findings can also guide future studies on new pharmacological interventions for ALS since assessments of brain samples are challenging and represent a relevant limited strategy.
Graphical abstract
Electronic supplementary material
The online version of this article (10.1007/s12035-020-02059-1) contains supplementary material, which is available to authorized users.
Keywords: Amyotrophic lateral sclerosis, Peripheral blood mononuclear cells, Patients, Mitochondrial depolarization, Mitochondrial fission, Biogenesis
Introduction
Amyotrophic lateral sclerosis (ALS) is a fatal disorder characterized by a specific and progressive degeneration of upper and lower motor neurons followed by muscular atrophy [1–9]. Although several pathological mechanisms have been postulated to explain ALS genesis and progression, this matter is still under debate and goes beyond genetic alterations; mutations (~ 24 genes) account for 68% of familial cases of ALS (fALS) and for just 11% of sporadic ALS (sALS) [10]. In this respect, mitochondrial dysfunction has assumed greater importance, since alteration of energy supply is one of the main features of ALS [11–16]. Nevertheless, the importance of mitochondria is not restricted to adenosine triphosphate (ATP) production [17]; the “metabolic organelle” is also involved in many cellular processes, namely, calcium homeostasis, cellular growth, cellular differentiation, cell death, antioxidant metabolism, and axonal transport [18–21].
Unfortunately, there is no cure or effective therapy for ALS [22], and the diagnosis often occurs in the late stages of the disease and usually accompanies a 50% loss of motor neurons [16]. Therefore, the development of strategies to allow ALS detection and development tracking would be of significant medical value. Toward this end, attention has been focused on potential biological components as biomarkers, which may enable not only a reliable diagnosis but also a predictable follow-up and efficacy assessments of therapeutic interventions [23]. In the last decades, several ALS-related biological biomarkers have been recorded [24–33], some of which are from blood [34–38]. Interestingly, biomarkers from blood samples are receiving increased attention and have even contributed to the evaluation of oxidative stress in ALS patients treated with riluzole (Rilutec®) [39].
Thus, considering that mitochondrial deregulation is associated with ALS and that correlation between gene expression in human brain tissue and peripheral blood mononuclear cells (PBMCs) has already been established [40], the present work investigates whether changes in mitochondrial function could be used to monitor ALS. To achieve this goal, PBMCs from ALS patients and control subjects were used; blood sampling is a fairly non-invasive method and is cost-effective. Different parameters were evaluated in this study, namely, cytosolic calcium levels, mitochondrial membrane potential, oxidative stress, and metabolic compound levels, in addition to gene expression associated with mitochondrial metabolism, dynamics, and degradation.
In PBMCs from ALS patients, we observed an overall lower mitochondrial calcium uptake/retention, mitochondrial depolarization, and redox homeostasis unbalance. Moreover, ALS PBMCs showed a significant decrease in critical metabolic genes and a diminishment in mitochondrial biogenesis and content. These findings seem to occur in parallel to an increase in mitochondrial fission and autophagy-related gene expression, despite a reduction in mitochondrial degradation signaling. Accordingly, ALS cells present a decrease in energy-producing metabolic compounds, such as ATP production and pyruvate, corroborating the fact that mitochondrial deregulation can lead to an energetic deficiency in peripheral cells as an ultimate consequence of the disorder. Thus, mitochondrial function evaluation in PBMCs could be a valuable strategy to detect ALS, as well as to assess its progression and therapy efficacy.
Material and Methods
Subjects
The work presented was a hospital-based case-control study, in which patients and control subjects were recruited at the Neurology Department from Irmandade da Santa Casa de Misericórdia de São Paulo (ISCMSP). We got access to 14 ALS individuals. According to El Escorial, clinical diagnosis of ALS individuals was performed based on the presence of clinical signs, which requires upper and lower motor symptoms and a history of progressive disability [41]. At ISCMSP, physicians classified ALS patients in familial when subjects mention that their relatives (first or second degree) are clinically affected—without genetically testing them. Because of the patients’ physical conditions, named for delicate veins, sampling has been carried out in 10–11 affected individuals. Importantly, blood samples were collected in two different time points; the first one (blood sampling 1, BS1) was executed soon after medical evaluation; the second blood sampling (BS2) was collected in 6 patients after 3 months of BS1.
The inclusion criteria of control subjects were (i) no clinical signs of any neuromotor disorder (e.g., frontotemporal dementia, FTD), (ii) no family members with ALS, and (iii) no metabolic disorder. The exclusion criteria were as follows: (i) present any pathology associated to ALS, such as respiratory diseases, difficulty in swallowing, and cognitive disorders, (ii) have comorbidities, namely diabetes and hypertension, (iii) show acute and chronic inflammatory disorders, and (iv) have smoking habits. Control subjects were age- and gender-matched.
Sampling and Human Peripheral Blood Mononuclear Cell Isolation
Blood from all participants was collected in heparinized tubes (8 mL/each). Following this procedure, blood was transferred to tubes containing Ficoll® (Paque Plus GE Healthcare, 17-1440-02), previously diluted in phosphate buffer (1:1) (PBS, mM: 137 NaCl, 2.7 KCl, 1.8 KH2PO4, 10 Na2HPO4, 2H2O, pH 7.4). Soon after, samples were centrifuged at 3000 rpm for 20 min (min) at 18 °C. PBMC-containing layer (a cloudy ring) was then transferred to a new centrifuge tube. The serum, located in the top layer of the gradient, was aliquoted (400 μL was maintained at room temperature for further use—in the culture medium). The collected cells were diluted in 45 mL of sterile PBS and centrifuged at 2000 rpm for 10 min at 18 °C. Pellets were cultured, as described below.
Peripheral Blood Mononuclear Cell Culture
The final pellet containing PBMCs was resuspended in RPMI 1640 medium supplemented with 10% (v/v) serum; each participant’s PBMCs were cultured in its serum. Each cell suspension was transferred to four wells of a six-well plate (2 mL of medium per well). Cells were formerly kept at 37 °C and 5% CO2 for 18–24 h [42]. Shortly, all the desired experiments were performed.
Evaluation of Mitochondrial Functional Parameters
To check mitochondrial functional parameters, different experiments were conducted. To analyze calcium homeostasis, 3 × 105 cells were allocated into a well (in a 96-well plate). At that point, cells were loaded for 1 h at 37 °C with Fluo-4-AM (10 μM), a cytosolic calcium indicator, in microscopy medium (mM: 120 NaCl, 3.5 KCl, 0.4 KH2PO4, 5 NaCOH3, 1.2 NaSO4, 20 HEPES, and glucose; pH 7.4) supplemented with 1 mM of calcium chloride (CaCl2) in the presence of Pluronic F-127™ (20%), a nonionic detergent which facilitates the fluorescent probe entry into the cells [43–47]. Experiments were performed using the spectrofluorometer Spectramaxi3™ (485-nm excitation; 525-nm emission).
To investigate, indirectly, mitochondrial membrane potential (ΔΨm), cells (3 × 105/well in the 96-well plate) were incubated with tetramethylhydrodamine ethyl ester (TMRE) (500 nM, 1 h) at 37 °C, in microscopy medium [43–46, 48–53]. TMRE is a fluorescent cationic indicator that accumulates preferentially into negative mitochondria (showing mitochondrial polarization). The experiments were also run on the plate reader Spectramaxi3™ (540-nm excitation; 590-nm emission).
The oxidative stress level was evaluated after 3 × 105 cells were transferred to each well of a 96-well plate and incubated with carboxylated diclohydro-fluorescein reagent (CM-H2DCF-DA) (20 μM, 1 h, at 37 °C) in microscopy medium [43–46, 48, 53]. CM-H2DCF-DA is used to detect the generation of reactive oxygen intermediates and passively diffuse into the cytosol, where it is cleaved by esterases forming DCFH. DCFH is oxidized to the green fluorescent compound 2,7-dichlorofluorescein (DCF). Thus, the higher is the level of reactive oxygen species (ROS) within the cell, the higher is the fluorescence signal. The experiments were also performed on Spectramaxi3™ (495-nm excitation; 520-nm emission).
All experiments were acquired for 10 min. In the first 5 min, the basal fluorescence was measured; after that, another 5 min was recorded to measure fluorocarbonyl cyanide phenylhydrazone (FCCP, 5 μM) effect, a mitochondrial protonophore used as an internal control. The baseline fluorescence was considered the mean of the last 5–8 points of the initial reading (before FCCP), while the FCCP outcome was calculated as the mean of the first 5–8 readings after FCCP [43–46, 48, 53]. All experiments were performed in duplicate, and data were represented as the percentage of the control group.
Expression of Mitochondrial Function–Related Genes
The expression of genes related to mitochondrial metabolism, dynamic, and degradation was evaluated by real-time PCR (qRT-PCR) [51–53] in 7500 Real-Time PCR Instrument (ThermoFisher Scientific). Briefly, messenger RNA (mRNA) of about 100,000 cells was extracted with Purezol (BioRad) according to the manufacturer’s protocol. The complementary DNA (cDNA) was obtained using 1 μg mRNA, and the iScript DNA Synthesis Kit (Biorad) (total volume of 20 μL). For each qRT-PCR reaction, performed with the SsoAdvanced™ Universal SYBR®Green Supermix kit (BioRad), 200 ng of cDNA and 300 nM of primer (Foward and Reverse) were used. The template was performed with an initial cycle at 95.0 °C for 30 s, followed by 45 cycles of 95 °C (15 s) and 60 °C (30 s).
The expression of the following genes was evaluated (primers sequences are in the Supplementary Material): Nuclear respiratory factor 1 (NRF1) and Nuclear factor (erythroid-derived2)-like 2 (NFE2L2), to evaluate nuclear-encoded electron transport chain subunits transcription and antioxidant defense; Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α); mitochondrial transcription factor A (TFAM) and tRNAleu, to investigate mitochondrial biogenesis and content (tRNAleu is within an area of mtDNA that is rarely deleted and has few polymorphisms); Dynamin related protein 1 (DNM1L) and Mitochondrial fission 1 (FIS-1), to verify mitochondrial dynamics; PTEN-induced putative kinase protein 1 (PINK1) and E3 ubiquitin-protein ligase parkin (PARKIN), to investigate mitochondrial degradation signaling; and Beclin 1 (BECN1), Microtubule-associated proteins 1A/1B light chain 3 (LC3) and Sequestosome-1 (SQSTM1), to check changes in autophagy [54–70]. Gene expression was determined with 2−ΔΔCT [71].
Measurement of Adenine Nucleotides and Metabolic Compounds
Afterward, we evaluated the levels of bioproducts related to energy metabolism, namely ATP, adenosine diphosphate ADP, pyruvate, and lactate. Then, total protein extraction was performed as described previously [46; 50-53]. Briefly, cells were resuspended in Ripa buffer (mM: 20 Tris, 100 NaCl, 2 EDTA, 2 EGTA, 1% Triton X-100, pH 7.0) supplemented with MS-SAFE (protease and phosphatase inhibitor) (Sigma-Aldrich) and were frozen-thaw placed in liquid nitrogen (frozen) and hot water bath (37 °C) for three times. Samples were then sonicated for 3 min in a Sonicator Bath to be then centrifuged for 10 min at 1400 rpm. The final homogenate was considered the total fraction. We quantified protein levels, and we standardize 50 μg to be added in all reactions. All energetic compounds were measured using commercially available kits following (Abcam, ab83355, ab83359, ab65342, ab65330).
Statistical Analysis
Results are expressed as mean ± standard deviation (SD) of the number of independent experiments indicated in figure legends. The fluorescence was normalized to 1 to assemble graphic lines, and data are represented in relation to the control group (in percentage). Gene expression was determined with 2−ΔΔCT (normalizing values to actin expression in relation to the control group), and results regarding the bioenergetics compounds are also represented concerning the control group (in percentage). The graphs were put together using the GraphPad Prism 6 program (GraphPad Prism Version 6.0), and the same program was used to execute the statistical analysis. We performed one-way ANOVA followed by post hoc Bonferroni and Student’s t test to achieve statistical comparisons between groups. It was considered statistically different p < 0.05.
Results
Subjects Characteristics and Socio-Demographic Data
All outcomes regarding demographic and clinical data are shown in Table 1. Information concerning age, gender, time with the disease, current medication, first motor symptoms, and El Escorial are listed. Our work included 12 control subjects and 14 patients, which were evaluated according to the El Escorial criteria [41]. It is essential to mention that the small sample size was due to several subjects in the Neurology Department presenting motor neuron disorder that was not classified as ALS. In Table 1, we also indicated the familial cases among ALS patients and the presence of other neurodegenerative diseases. Data about age and gender of control subjects are also presented.
Table 1.
Demographic characteristics of ALS patients and control subject at the time of blood sampling at Irmandade da Santa Casa de Misericórdia de São Paulo (ISCMSP). In total, 12 control individuals and 14 patients were recruited
| ALS patients | Male (7)/female (7) | Age (mean), 52 | |||
|---|---|---|---|---|---|
| Control subjects | Male (6)/female (6) | Age (mean), 49 | |||
| No. | Age | Duration | Medication | First motor signals | El Escorial |
| 1 | 63 | 9 years | Riluzole | Left hemisphere | Definite1 |
| 2 | 45 | 3 years | Riluzole | Right superior | Definite |
| 3 | 70 | 3 years | Riluzole | Right superior | Definite2 |
| 4 | 56 | 2 years | Riluzole | Left hemisphere | Definite |
| 5 | 57 | 1 year | Riluzole | Left lower | Probable2 |
| 6 | 46 | 12 years | Riluzole | Left lower | Probable |
| 7 | 33 | 2 years | Riluzole | Right superior | Definite |
| 8 | 55 | 5 years | Riluzole | Lower | Definite |
| 9 | 44 | 4 years | Riluzole | Superior | Possible |
| 10 | 51 | 7 years | Riluzole | Right superior | Definite3 |
| 11 | 59 | 5 years | Riluzole | Bulbar | Definite |
| 12 | 56 | 4 years | Riluzole | Left lower | Definite |
| 13 | 40 | 6 years | Riluzole | Bulbar | Probable |
| 14 | 57 | 3 years | Riluzole | Right superior | Definite |
1ALS patient with multiple sclerosis
2ALS patients with motor diseases cases in relatives
3fALS (familiar case)
Of the 14 ALS subjects in this study, 78.5% of them have presented with the disease for 6 years or less (1–6 years of ALS duration). Only 21.5% of patients have had ALS for 7 years or more (one subject has presented with the disease for 12 years). Despite such a range, the duration of ALS seems not to be correlated with either the diagnosis of fALS or with different outcomes related to mitochondrial functional parameters and gene expression, once the results of those patients were under the group’s mean (see below). Importantly, all individuals evaluated were Caucasian. As mentioned previously [9], we believe that ethnicity is crucial to the interpretation of results, since metabolism can be modulated, at least partially, by polymorphisms that might be ethnically dependent [9].
The mean age of ALS individuals in our study is 52 years, while in the controls is 49 years. Moreover, there are 7 ALS female patients and seven males; the same proportion is found in the control group. Curiously, just one patient has fALS, meaning that 92.86% has sALS; the mother, uncle, and two cousins of the fALS patient are also affected.
Furthermore, none of ALS patients smokes, and the most predominant first motor symptoms are the upper or lower limbs (known as the spinal form)—42.85% of each; just 14.3% of patients presented bulbar symptoms first (Table 1). The treatment used by all patients of this cohort is riluzole, the most used medication in ALS [72].
Depolarized Mitochondria and Disturbances in Calcium Handling in ALS PBMCs
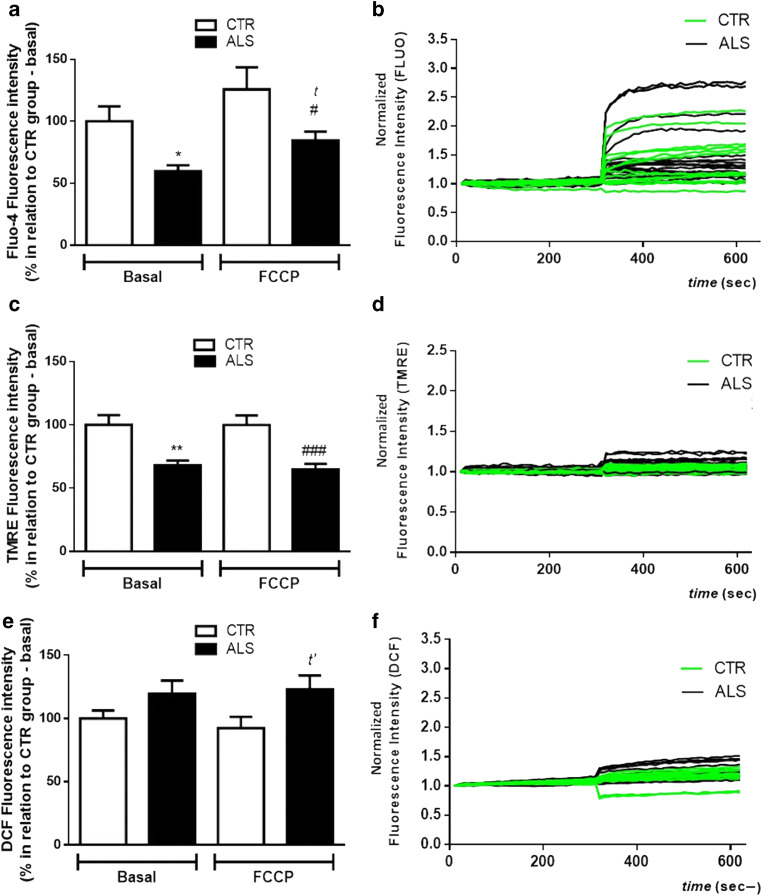
Considering the importance of mitochondria function in keeping calcium handling, and because modifications in the transport of electron and the proton motive force can interfere with ATP production [73, 74], we evaluated the cytosolic Ca2+ level in PBMCs from ALS and control subjects (Fig. 1a, b). As we can notice, there is a significant reduction in the Fluo-4 fluorescence in the ALS group, suggesting a decrease in cytosolic calcium in the patient’s cells. Curiously, in the presence of FCCP (5 μM), there is also a diminishment in the cytosolic calcium in ALS PBMCs.
Fig. 1.
Mitochondrial depolarization and changes in calcium handling in PBMCs from ALS patients. PBMCs (300,000 cells/well—in a 96-well plate) were loaded with different fluorescent probes for 1 h at 37 °C before mitochondrial evaluation. a The histogram represents Fluo-4 fluorescence (10 μM) before and after stimulation with FCCP (5 μM). b Representative graphic line of normalized Fluo-4 fluorescence intensity in CTR/control (green lines) and ALS (black lines) samples. c Representative histogram of TMRE fluorescence (500 nM) before and after stimulation with FCCP (5 μM). d Representative graphic line of normalized TMRE fluorescence intensity of different samples in CTR/control (green lines) and ALS (black lines) samples. e The histogram represents the level of H2DCF-DA fluorescence (20 μM) before and after stimulation with FCCP (5 μM). f Representative graphic line of DCF normalized fluorescence intensity (values normalized to 1) in CTR/control (green lines) and ALS (black lines) samples. Data is represented by mean ± SD, and the results were normalized as a percentage of CTR/control group (basal fluorescence) (ALS group: N = 11; control group: N = 10, in duplicate). Statistical analysis was performed using one-way ANOVA followed by the post hoc Bonferroni and Student’s t test. It was considered significant when p < 0.05; *p < 0.05 and **p < 0.01, in relation control/CTR group (basal fluorescence); tp < 0.05, in relation to ALS group (basal fluorescence); t’p < 0.05, #p < 0.05, and ###p < 0.001, in relation control/CTR group in the presence of FCCP
As mitochondrial calcium buffering is influenced by ΔΨm (and vice versa), PBMCs were also loaded with TMRE. As shown in Fig. 1c, d, PBMCs from ALS patients have a significantly lower TMRE fluorescence than the control group (less TMRE in the cytosol). After the stimulation with FCCP, TMRE signal is also significantly decreased in ALS PBMCs. This data suggests that mitochondria from ALS patients are depolarized and, therefore, present less TMRE (and calcium) retention capability.
Since mitochondria are one of ROS’s primary sources, we evaluated the oxidative stress level in PBMCs from both ALS and control groups (Fig. 1e, f). Although no change is observed in basal DCF fluorescence between ALS and control PBMCs, there is a significant augmentation in DCF fluorescence after FCCP in ALS PBMCs when compared with controls’ cells.
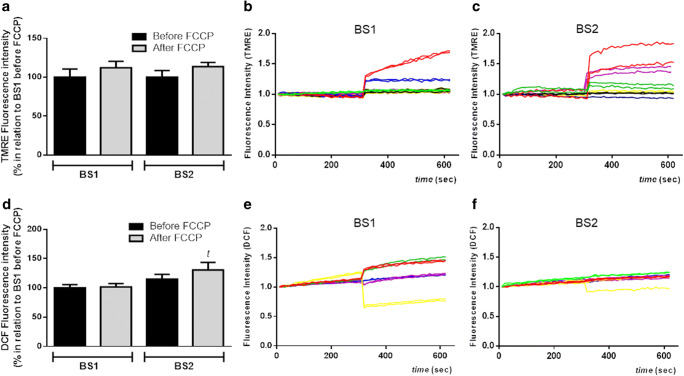
To test if ALS patients present changes in mitochondrial functional parameters with time, named mitochondrial membrane potential and oxidative stress, samples were re-evaluated in 6 patients after 3 months of the first blood sampling (BS1). As shown in Fig. 2a, c, there is no difference regarding TMRE fluorescence between groups (BS1 and BS2), neither before nor after FCCP. Although we do not observe any change in basal DCF fluorescence with time, the DCF signal is significantly higher in the ALS group in BS2 in comparison with that in BS1 after FCCP (Fig. 2d–f).
Fig. 2.
Changes in mitochondrial membrane potential and oxidative stress level in PBMCs from ALS subjects after first blood sampling (BS1) and 3 months later (BS2). PBMCs (300,000 cells/well—in a 96-well plate) were loaded with a–c TMRE (500 nM) and d–f DCF (20 μM) for 1 h at 37 °C. a Representative histogram of TMRE fluorescence before and after stimulation with FCCP (5 μM). b Representative graphic line of BS1 normalized TMRE fluorescence intensity. c Representative graphic line of BS2 normalized TMRE fluorescence intensity. d Representative histogram of DCF fluorescence before and after stimulation with FCCP (5 μM). e Representative graphic line of BS1 normalized DCF fluorescence intensity (values normalized to one). f Representative graphic line of BS2 normalized DCF fluorescence intensity (values normalized to one) (patient 1 blue, patient 2 pink, patient 3 red, patient 4 yellow, patient 5 green, patient 6 black). Data is represented by mean ± SD, and the results were normalized as a percentage of BS1 group before FCCP (basal) (N = 6, in duplicate). Statistical analysis was performed using one-way ANOVA followed by the post hoc Bonferroni and Student’s t test. It was considered significant when p < 0.05; tp < 0.05, in relation to BS1 after FCCP
Decreased Antioxidant Defense and Metabolic Gene Expression in ALS PBMCs
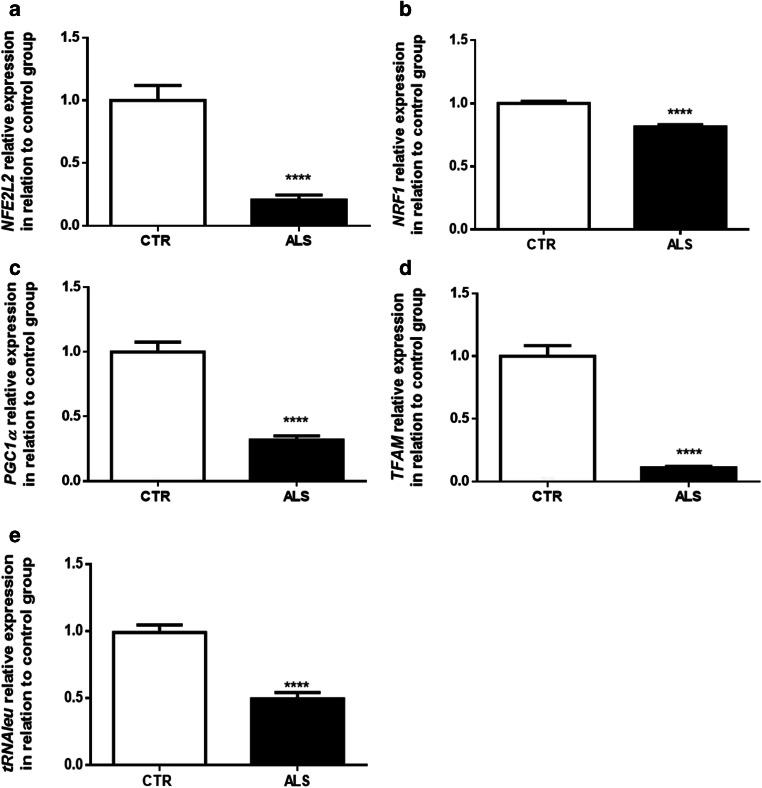
Because oxidative stress is increased in peripheral cells from ALS subjects, we further investigated the expression of NFE2L2, a transcription factor related to the synthesis of antioxidant enzymes [57, 75–77]. As shown in Fig. 3a, there is a massive reduction in NFE2L2 in ALS cells, indicating that ALS PBMCs have a decreased antioxidant defense [78–80].
Fig. 3.
Diminishment in the expression of genes related to mitochondrial metabolism, biogenesis, and content in ALS PBMCs. Relative expression (2−ΔΔCT) of NFE2L2 (a), NRF1 (b), PGC-1α (c), TFAM (d), and tRNAleu (e) in relation to actin and control group. Data are represented by mean ± SD (N = 5, in duplicates) and the results expressed in 2−ΔΔCT. Statistical analysis was performed using Student’s t test. It was considered significant when p < 0.05; ****p < 0.0001, in relation to CTR/control group
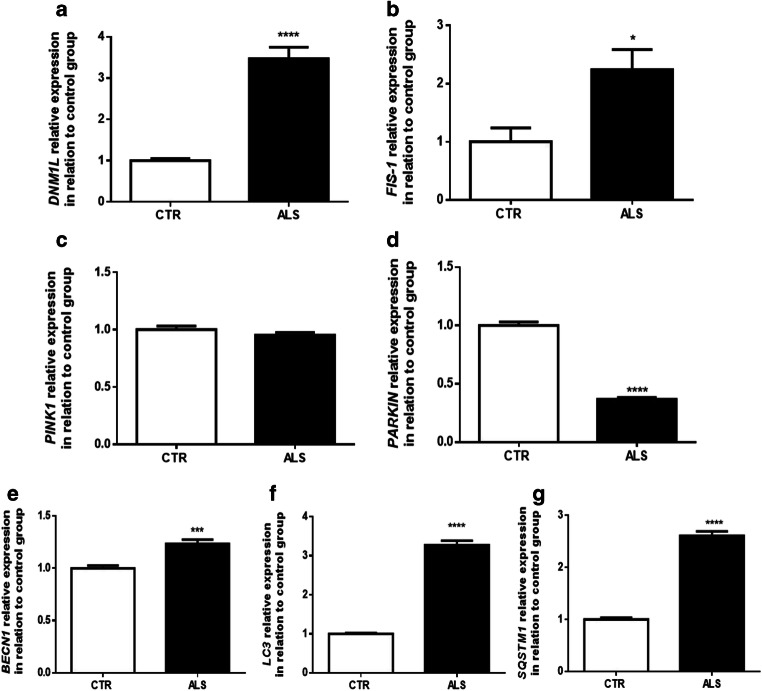
Regarding the expression NRF1, responsible for regulating several nuclear-encoded electron transport chain proteins [70, 81, 82], there is a significant diminishment in ALS cells compared with taht in controls (Fig. 3b).
Reduced Mitochondrial Biogenesis and Content in PBMCs from ALS Patients
To evaluate changes in mitochondrial biogenesis and content, we assessed PGC-1α, TFAM, and tRNAleu expression in PBMCs [59, 65, 66, 68, 83–92]. As shown in Fig. 3c, there is a significant decrease in PGC-1α expression in ALS cells. Corroborating this data, we demonstrated that ALS PBMCs also present a significant decrease in TFAM and tRNAleu expression (Fig. 3d, e).
Increased Mitochondrial Fission and Autophagy Gene Expression in ALS Patients’ Cells
Knowing that mitochondrial membrane potential and biogenesis are related to modifications in mitochondrial dynamics [93, 94], we investigated the expression of DNM1L and FIS-1 in PBMCs; both genes are linked to mitochondrial fission [95–97]. We observe a significant enrichment in both gene expressions in ALS patients’ PBMCs compared with that in the controls, indicating an augmentation in organelle fission (Fig. 4a, b).
Fig. 4.
Increase in mitochondrial fission genes expression and diminishment in the expression of mitochondrial degradation genes, despite an augmentation in autophagy, in PMBCs from ALS patients. Relative expression of DNM1L (a) and FIS-1 (b) (genes linked to mitochondrial fission), PINK (c) and PARKIN (d) (genes related to mitochondrial target degradation through autophagy), and BECLIN (e), LC3 (f), and SQSTM1 (g) (genes of autophagy pathway) in relation to actin and control group. Data in graphs are the mean ± SD (N = 5, in duplicates) and the results expressed in 2−ΔΔCT. Statistical analysis was performed using Student’s t test. It was considered significant when p < 0.05; *p < 0.05, ***p < 0.001, ****p < 0.0001, in relation to CTR/control group
In order to understand if the augmentation in fission machinery could lead to an increase in mitophagy [98, 99], we analyzed the expression of both PINK1 (responsible to preserve of mitochondrial functioning and integrity) and PARKIN (recruited by PINK1 for the degradation of damaged mitochondria) [100, 101]. There is no significant difference in the expression of PINK1 between ALS and control groups (Fig. 4c). However, the expression of PARKIN is significantly lower in PBMCs from ALS patients (Fig. 4d). This outcome indicates that despite mitochondrial function is deregulated, fewer mitochondria seems to be degraded.
To evaluate whether ALS PBMCs present changes in autophagy-related gene expression, we examined BECN1 (essential for nucleation/expansion of autophagosome), LC3 (responsible for autophagosome membrane formation), and SQSTM1 (in charge of the recruitment of specific cargo) [102–107]. It is possible to observe that there is a significant increment in the expression of all genes tested in ALS patients’ cells when compared with controls’ PBMCs (Fig. 4e–g). This data indicates that despite lower levels of PARKIN in ALS patients’ PBMCs, the autophagy pathway seems to be activated.
Changes in Bioenergetics Metabolic Compounds in PMBCs from ALS Patients
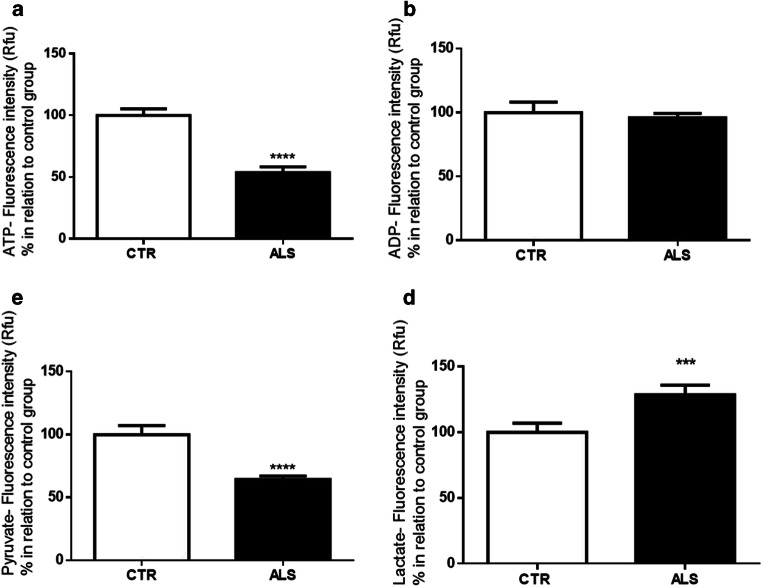
As we observed changes in mitochondrial function in ALS’ PBMCs, besides deviations in the expression of genes related to mitochondrial metabolism, biogenesis, and fission, we investigated whether the bioproducts related to energy metabolisms, such as ATP, ADP, pyruvate, and lactate, could vary. As observed in Fig. 5a, there is a significant decrease in ATP levels in PBMCs from ALS patients compared with control individuals; no modification is observed regarding ADP levels (Fig. 5b). As we can notice in Fig. 5c, the pyruvate level significantly dropped in ALS PBMCs, while the level of lactate is significantly elevated (Fig. 5d). This outcome suggests that pyruvate may be converted into lactate to produce energy since ATP synthesis, once oxidative phosphorylation might be jeopardized because of mitochondrial deregulation (Fig. 6).
Fig. 5.
Changes in bioenergetics metabolic compounds (ATP, ADP, pyruvate, lactate) in PMBCs from ALS patients. a ATP, b ADP, c pyruvate, and d lactate levels. Data in graphs are the mean ± SD (N = 3, in duplicates), and the results were normalized as a percentage of the control group. Statistical analysis was performed using Student’s t test. It was considered significant when p < 0.05; ***p < 0.001 and ****p < 0.0001, in relation to CTR/control group
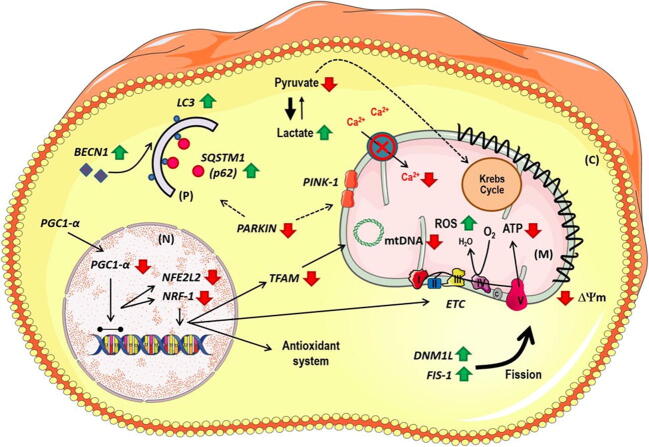
Fig. 6.
Schematic representation of mitochondrial deregulation observed in peripheral blood mononuclear cells from ALS patients. Cells from ALS patients present mitochondrial depolarization and diminishment in mitochondrial calcium uptake and/or retention, changes in redox homeostasis, and a decrease in metabolism biogenesis–related gene expression, namely, PGC-1α, NFE2L, NRF1, TFAM, and tRNAleu. Moreover, PBMCs from ALS individuals show an augmentation in mitochondrial fission (represented by an increase in DNM1L and FIS-1 expression) and a reduction in mitochondrial degradation signaling since there is a significant decrease in PARKIN expression. Notwithstanding, there is a significant increase in autophagy-related genes, as BECLIN, LC3, and SQSTM1. All these changes can further contribute to the decreased levels of ATP production and pyruvate in ALS PBMCs, and the augmented lactate levels observed in patients’ cells
Discussion
In this manuscript, we showed that PBMCs from ALS patients present a lower mitochondrial calcium uptake/retention, mitochondrial depolarization, and redox homeostasis unbalance, together with a significant decrease in vital metabolic gene expression and a diminishment in mitochondrial biogenesis and content. These outcomes seem to occur in parallel with an increase in mitochondrial fission and autophagy-related gene expression, despite a reduction in mitochondrial degradation signaling demonstrated by PINK and PARKIN expression. Accordingly, ALS cells present a decrease in high energy–producing metabolic compounds, corroborating the fact that mitochondrial deregulation can lead to an energetic deficiency in peripheral cells as an ultimate consequence of the disorder. Thus, mitochondrial function evaluation in PBMCs could be a valuable strategy to detect ALS, as well as to assess its progression and therapy efficacy.
An intensification in mitochondrial calcium influx is described as the primary process linked to cell death and excitotoxicity [108–113]. Hence, we evaluated calcium homeostasis in PBMCs from control and ALS subjects. As we show in Fig. 1a, there is a significant decrease in the basal fluorescence of Fluo-4 in the ALS group, along with a lower fluorescence in ALS PBMCs after stimulation with FCCP. Such data is in agreement with the literature and indicates that probably less mitochondria are buffering the ion, which could lead to cellular dysfunction [114–116].
Because calcium homeostasis is necessary for keeping the mitochondrial membrane potential [117, 118], and mitochondria membrane potential plays a direct role in calcium handling [119], we indirectly evaluated such parameters using the fluorophore TMRE [120]. As shown in Fig. 1c, there is a significant decrease in the basal TMRE fluorescence in patients’ cells. Because TMRE is a cationic indicator (accumulates in hyperpolarized mitochondria), we hypothesized that if mitochondria are depolarized, as indicated by Fluo-4 experiments, TMRE fluorescence would not vary in the presence of FCCP (mitochondria would not internalize TMRE). Indeed, as we conjectured, after FCCP, TMRE fluorescence was still significantly lower in the ALS group when compared with that in the control. Similarly, other studies also showed a decrease in ΔΨm [47, 121, 122]. Thus, the decreased capability of ALS PBMC mitochondria in uptake and/or retaining calcium might be related to the organelle’s depolarization. These results reinforce the need to identify biomarkers using both blood samples and mitochondrial functional parameters as a target.
As calcium homeostasis and ΔΨm interfere with the electron transport chain and, therefore, oxidative stress levels [123, 124], we investigated general ROS formation in PBMCs. Indeed, physiologically, mitochondria are one of ROS’s major sources, together with several mechanisms, but ROS are a key factor in the etiology of various neurodegenerative pathologies since its levels are associated with neuronal degeneration [36, 125–129]. The results demonstrate that there is more ROS in ALS PBMCs than in the control group in both basal (although not significant) and post-FCCP conditions (Fig. 1e). Such findings might be due to an impaired antioxidant system. Indeed, it was already demonstrated a significant diminishment in antioxidant defense markers in patients with ALS, including reduced glutathione in the motor cortex of ALS patients. Nonetheless, the results are somehow inconsistent due to the heterogeneity of patients [129]. Importantly, our data is in accordance with literature that also shows a small increase in oxidative stress levels in ALS PBMCs at the baseline and a significant increase after FCCP [130–132]. Moreover, and corroborating our findings, a clinical study using serum and whole blood from 10 ALS patients demonstrated an increment in malondialdehyde and 8-hydroxy-2′-deoxyguanosine levels [36].
Since ALS is a progressive degenerative disorder, we also investigated whether PBMCs from ALS patients present modification in mitochondrial functional parameters with time. Unfortunately, the amount of blood in the BS2 was lower than in the BS1, a reflection of the fragile patients’ veins. Consequently, we evaluated membrane potential (Fig. 2a) and oxidative stress level (Fig. 2b). As shown, only the DCF signal, after the addition of FCCP, is significantly higher in the BS2 group in relation to the BS1. This result suggests that patients become more susceptible with time to toxic stimuli, such as FCCP. The increase in the fluorescence could be due to a slight decrease in basal DCF fluorescence in CTR group, an impaired antioxidant system, and a further leak of electron from the mitochondrial complexes chain due to the stimulation of electron’s transport through the mitochondrial complexes chain induced by FCCP. It is also important to mention that DCF is capable of providing data about general ROS synthesis, and no specific measurements of ROS were performed (superoxide, hydrogen peroxide, MDA, or HNE). For this reason, more studies are necessary to investigate whether other mitochondrial parameters related to oxidative stress modulation change with ALS progression. It is essential to mention that the lower number of patients reassessed was due to patients missing scheduled medical appointments because of motor difficulties. Although only six patients were re-evaluated, and the period between BSs was 3 months, we did observe a significant increase in oxidative stress.
Due to the increased ROS level in ALS PBMCs, we decided to investigate NFE2L2 expression [57], which is implicated in cellular detoxifying systems [78–80]. We observed a large diminishment in NFE2L2 expression (more than 50%) in ALS cells (Fig. 3a). The increased levels of ROS could have been related to a failure of NFE2L2 in translocating to the nucleus, reducing the transcription of secondary factors of the antioxidant cascade [79, 133]. Interestingly, in previous work with PBMCs, the expression of NFE2L2 had no changes in relation to controls, but ALS patients were 10 years older than in our study [13], which could indicate a time-dependent NFE2L2 expression. In agreement with our outcome, the expression of NFE2L2 was declined in humans with ALS and in the SOD1-G93A model [134, 135]. Of importance, NFE2L2 overexpression was associated with more prolonged survival and neuroprotection in the SOD1-G93A mouse model [134, 136].
Considering that mitochondrial function can vary due to changes in mitochondrial protein content, including in mitochondrial complexes, we further investigated the expression of NRF1. Our study demonstrated a significant decrease in NRF1 expression in ALS PBMCs (Fig. 3b), a finding that corroborates previous data in the literature in which a reduction in NRF1 expression in the spinal cord and muscles from ALS patients and SOD1-G93A animal models was shown [135, 137]. Our data then suggest that mitochondria from ALS PBMCs can present less mitochondrial complex inhibiting, consequently, mitochondrial oxidative phosphorylation system (OXPHOS).
Given that there is a significant reduction in both NFE2L2 and NRF1 expressions in PBMCs from ALS patients, and that PGC-1α regulates both, we investigated mitochondrial biogenesis in our cellular model. Importantly, PGC-1α leads to an increase in mitochondrial mass and number, stimulating the division of pre-existing mitochondria in order to sustain energetic cellular status [55, 138]. As we can observe in Fig. 3c, there is a significant decrease in PGC-1α expression in the ALS group. This evidence aligns with a diminished expression of PGC-1α in the spinal cord of SOD1-G93A mice [139], and in muscle and motor cortex of human sporadic ALS [135]. Intriguingly, low PGC-1α expression is related to lower mitochondrial activity and to a worsened regulation of reactive oxygen species [54, 55], and fewer PGC-1α seems to be associated with a decrease in mitochondrial activity [140] and an increase in muscle degeneration in a different animal model of ALS [141, 142]. Therefore, decreasing PGC-1α may contribute to minor ROS protection [143–145]. These findings seem to corroborate our data so far and can also be associated with changes in the basal level of cytosolic Fluo-4 and TMRE.
To corroborate the reduction in mitochondrial biogenesis, we also evaluated TFAM expression, a mitochondrial transcription factor regulated by PGC-1α responsible for the transcription of mtDNA genes and bioenergetic function [146–148]. We detected significant mitigation in TFAM expression in the ALS group (Fig. 3d). Curiously, a drop in TFAM expression was associated with an augmentation of oxidative damage [149]. In line with our data, a reduction in TFAM was also observed in the spinal cord and motor cortex of human sporadic ALS [135]. We also observed a decrease in tRNAleu expression (Fig. 3e), a unique sequence in mtDNA, suggesting fewer copies of mtDNA in ALS cells [150]. A reduced mtDNA copy number was also described on neurons and the spinal cords of ALS patients [148, 151]. Therefore, even with a limited sample size, we do consider that our results represent a true argument in favor of an augmentation in mitochondrial biogenesis, as four different approaches were assessed.
Cellular death among degenerative disorders is commonly associated with an increase of mitochondrial fission [152]. Indeed, fission can decrease energy production, encourage oxidative stress, lead to mtDNA deletion, and impair calcium buffering [153]. Remarkably, mitochondrial fission can be stimulated by oxidative stress [154–156]. Because we observed a diminishment in mitochondrial biogenesis and in mtDNA copy number, as well as changes in calcium homeostasis, we also evaluated DNM1L and FIS-1 expression in PBMCs from ALS and control groups. As shown in Fig. 4a, b, there is a significant increase in the expression of both genes in patients’ cells. Significantly, and corroborating our findings, an increase in fission, including changes in DNM1L and FIS-1, was already reported in neurons from fALS patients [157, 158], in ALS lymphoblastoid human cells [159], in ALS patient–derived fibroblasts [96], in cultured motor neurons expressing SOD1 mutant [96], and in animal models of ALS [154, 157, 160]. Thus, so far, our data implies that mitochondrial functional parameters and dynamics should be further investigated as a biomarker, even though we had few samples and no further techniques to explore such processes, namely, Western blot or immunofluorescence.
Taking into consideration that changes in DNM1L and FIS-1 expression, together with mitochondrial depolarization and biogenesis modification, are related to selective mitochondrial degradation [56, 161], we hypothesized that mitophagy, the major machinery to eliminate dysfunctional mitochondria [162], could also be dysfunctional in PBMCs from ALS subjects. Subsequently, we investigated in PBMCs the expression of PINK-1 and PARKIN, genes related to mitophagy [99, 163–165]. It is important to point out, however, that although PARKIN is associated with mitophagy [166], its translocation to deregulated mitochondria is one of the earlier steps in mitochondrial control quality [167]. As shown in Fig. 4c, there is no difference in PINK-1 between groups, but there is a significant decrease in PARKIN in ALS PBMCs. Supporting our results, Lagier-Tourenne, Stribl, and their colleagues also observed a diminishment in PARKIN in samples from autopsied brains of ALS patients and from hTDP-43A315T animals [168, 169]. Therefore, our data suggest that the ubiquitination of target proteins enrolled in autophagy machinery could be impaired in PBMCs from individuals with ALS.
To investigate whether autophagy genes would be changed in PBMCs from ALS patients, we verified the expression of BECN1, LC3, and SQSTM1 (Fig. 4e–g). We showed a significant augmentation in all three genes in ALS cells. In this sense, an increase in autophagy-related gene expression could be a compensatory mechanism in an attempt to clear out unwanted organelles. Our data is supported by the literature, in which transgenic models have increased BECN1, LC3, and SQSTM1 expression [170–175]. It should be noted, still, that in respect to autophagy, our goal in verifying BECN1, LC3, and SQSTM1 expressions was to identify a possible alteration in this mechanism (in general), rather than examine the functionality/blockage of the whole pathway (initiation, formation, and maturation). To investigate autophagy in greater depth, various methodologies including fluorescence and high-content imaging, flow cytometry, and luminescence detection could be performed [102, 107]. Therefore, future studies should also look for autophagy markers (end-points) in ALS cohorts.
Since we observed changes in organelles’ functional parameters, specifically calcium handling, membrane potential, and oxidative stress level, together with modification in the expression of genes related to mitochondrial dynamics and degradation in ALS cells, we additionally evaluated the level of energy metabolites (Fig. 5). Besides the decreased ATP levels observed in PBMCs from ALS patients (Fig. 5a), there was also a reduction in the pyruvate status (Fig. 5c) and an increased level of lactate (Fig. 5d). Our findings seem to be in accordance with the literature that shows a diminishment in ATP levels [176–178], denoting that mitochondrial deregulation is leading to a decrease in energy reservoir. Moreover, a reduced level of pyruvate was demonstrated in SOD1-G93A mice [179, 180], as well as an increment in lactate in the same rodent model, in ALS homogenate from sporadic patients, and in G39-neuroblastoma spinal cord mutant cells [181–183]. The lactate outcomes are consistent with an augmentation in the aerobic glycolysis, the so-called Warburg effect that has been noted in ALS [184]. In fact, lactate augmentation accounts for a compensatory glycolytic response to favor ATP synthesis [185]. Although the synthesis of ATP through lactate is not efficient and is less cost-effective than through OXPHOS, metabolic modifications might be an attempt to restore energy homeostasis [186] since ATP depletion contributes to ALS progression [187]. Furthermore, the accumulation of lactate can be a strategy to convert NADH (reduced form of nicotinamide adenine dinucleotide) into NAD+ (oxidized form of nicotinamide adenine dinucleotide), reverting reductive stress in order to compensate for an oxidative phosphorylation deficiency [186, 188].
To date, the most common medication used by ALS patients is RilutekTM (riluzole) [189–191], known as a wide-spectrum medication [9, 39, 192–195] and showing different effects in later stages of the disease [192, 196]. Therefore, one might think that the data presented would point toward the effect of riluzole. However, it is important to mention that our findings are in opposition to riluzole’s influence described so far [192, 196]. Thus, it can be assumed that at least in part, the results presented herein are not riluzole-dependent.
Altogether, this work brings new insights into how ALS affects peripheral blood cells and how mitochondria deregulation plays a role in this hallmark. Specifically, it seems that PBMC mitochondria from ALS patients become depolarized, losing its ability to uptake and/or retain calcium from cytosol, contributing to the increase in oxidative stress and decrease in NFE2L2 expression. ALS PBMCs also present lower mitochondrial content, which may further corroborate the decreased cytosolic levels of Fluo-4 and TMRE observed. Notably, as mentioned, despite fewer mitochondria, the ROS level is slightly higher in ALS cells. Moreover, ALS PBMCs present an unblemished decrease in metabolic intermediates, indicating that, ultimately, the energetic cellular homeostasis is jeopardized. Importantly, the collapse of mitochondrial membrane potential, indicated by TMRE fluorescence, and high levels of calcium and ROS are triggered by mitochondrial degradation through mitophagy. Oddly, ALS PBMCs present a decrease in PARKIN, which is needed to target malfunctioning mitochondria and recruit autophagy proteins. Eventually, the accumulation of dysfunctional organelles could contribute to a worsening ALS scenario.
It should be noted that our sample size is small, as several subjects in the Neurology Department presented motor neuron disorders that were not classified as ALS. Nevertheless, not only should the sample size be increased in future studies but other time points could also be checked. Although the outcomes presented here seem not to be riluzole-dependent, additional studies should be carried out in order to investigate these biases. Moreover, the lack of knowledge regarding secondary mechanisms related to the disease progression represents a struggle for the academic community, especially because diagnosis and prognosis are mainly based on physical exams [23]. Unfortunately, this is a matter of debate that affects all studies performed with ALS patients [9].
It is important to emphasize, though, that we established a parallel between the outcomes presented with findings in (i) ALS animal models, (ii) ALS lymphocytes (established culture bought from cell banks), (iii) neuroblastomas expressing mutant proteins, (iv) iPSC derived-neurons from ALS patients with specific mutations, and (v) human brain samples. Therefore, the novelty of our work is as follows. (1) At once, we could show changes observed in a variety of ALS models and human tissues in the same ALS patients’ cell type, PBMCs. (2) ALS patients from our study were 93% sALS subjects. (3) Mitochondrial functional parameters and gene expression evaluated in our work seem not to be correlated with the diagnosis of fALS—results from the only one fALS individual were under the ALS group’s mean. (4) All the parameters showed were measured in only 8 mL of blood (sampled once). (5) It was used an only primary culture of PBMCs from ALS patients along with the study, and no cellular transformation was performed in these cells.
Curiously, in the literature, very few studies reassess cellular mechanisms in ALS patients’ samples along with the progression of the disorder. To date, to our knowledge, these works focus on clinical evaluation, epidemiology, oximetry, respiratory failure, and genetic counseling. Hence, we can say that we are one of the groups that considered this fact; we could match our results regarding NFE2L2 expression with patients 10 years older than those in our group. Such finding is auspicious, since the values differ, showing a change in this transcription factor over time. Considering that we evaluated distinct parameters investigated in many models and human samples and that such changes are observed in fALS and sALS, the assessed factors might be a significant biomarker to study disease progression in both familial and sporadic ALS patients.
Conclusion
Overall, our results indicate that PBMCs from ALS patients present a significant mitochondrial dysfunction that could be used as a powerful tool in investigating ALS disclosure and improvement. Importantly, the usage of PBMCs in biomarker research was already validated, and our findings can further contribute to future findings concerning new pharmacological strategies against ALS.
Electronic Supplementary Material
(PDF 286 kb)
Acknowledgments
We thank all the technicians from the Department of Physiological Science of Santa Casa de São Paulo School of Medical Science and the staff of the Neurology Department from Irmandade da Santa Casa de Misericórdia de São Paulo for helping us in this study.
Abbreviations
- ΔΨm
Mitochondrial membrane potential
- ALS
Amyotrophic lateral sclerosis
- ADP
Adenosine diphosphate
- ATP
Adenosine triphosphate
- BECN1
Beclin 1
- BS1
Blood sampling 1
- BS2
Blood sampling 2
- CaCl2
Calcium chloride
- CM-H2DCF-DA
Carboxylated diclohydro-fluorescein reagent
- DNM1L
Dynamin-related protein 1
- fALS
Familial cases of ALS
- FCCP
Fluorocarbonyl cyanide phenylhydrazone
- FIS-1
Mitochondrial fission 1
- FTD
Frontotemporal dementia
- iPSC
Induced pluripotent stem cells
- ISCMSP
Irmandade da Santa Casa de Misericórdia de São Paulo
- LC3
Microtubule-associated proteins 1A/1B light chain 3
- Min
Minutes
- mRNA
Messenger RNA
- mtDNA
Mitochondrial DNA
- NAD+
Oxidized form of nicotinamide adenine dinucleotide
- NADH
Reduced form of nicotinamide adenine dinucleotide
- NFE2L2
Nuclear factor (erythroid-derived2)-like 2
- NRF1
Nuclear respiratory factor 1
- PARKIN
E3 ubiquitin-protein ligase parkin
- PBMCs
Peripheral blood mononuclear cells
- PDH
Pyruvate dehydrogenase complex
- PGC-1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PINK1
PTEN-induced putative kinase protein 1
- qRT-PCR
Real-time PCR
- ROS
Reactive oxygen species
- sALS
Sporadic cases of ALS
- SD
Standard deviation
- SQSTM1
Sequestosome-1
- TFAM
Mitochondrial transcription factor A
- TMRE
Tetramethylhydrodamine ethyl ester
- Δ
Difference between the fluorescence after FCCP and the basal fluorescence intensity
Authors’ Contributions
BGA performed experiments regarding mitochondrial function (calcium homeostasis, mitochondrial membrane potential, and ROS levels, in addition to qRT-PCR); she also performed data analysis and wrote this manuscript. LFSS and JLBT performed the ATP, ADP, and pyruvate and lactate assays. AS performed some of PBMC culture assay. BCOV was responsible for blood sampling, and clinical and medical records data. MDB contributed by performing qRT-PCR analysis. TRR designed the study, wrote the protocols, and revised the manuscript. All authors approved the final version of the manuscript.
Funding Information
This work was supported by São Paulo Research Foundation (FAPESP) (2015/02041-1) and Fundação de Amparo à Pesquisa of Santa Casa de São Paulo School of Medical Science (FCMSCSP) (2017–2019; 2018–2020). BGA was supported by FAPESP/IC (ref. 2018/09084-6), LFSS had Master’s fellowships from FAPESP (ref. 2015/25595-2); now, LFSS is supported by CAPES (PhD fellowship). JLBT had PIBIC fellowship and AS was supported by PROSUC/CAPES. MDB was supported by CAPES and FAPESP/Master (2016/12039-7).
Data Availability
All data generated or analyzed during this study are included in the manuscript and the supplementary files.
Compliance with Ethical Standards
Ethics Approval and Consent to Participate
The study was approved by the Institutional Human Research Ethics Committee from Santa Casa de São Paulo, protocol number 1.923.109, following the Brazilian laws, the principles of the Declaration of Helsinki, and the Guideline for Good Clinical Practices. Each participant provided written consent.
Consent to Publication
Each participant provided written consent.
Competing Interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344(22):1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Seetharaman SV, Prudencio M, Karch C, et al. Immature copper-zinc superoxide dismutase and familial amyotrophic lateral sclerosis. Exp Biol Med. 2009;234(10):1140–1154. doi: 10.3181/0903-MR-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Alessandro G, Calcagno E, Tartari S, et al. Glutamate and glutathione interplay in a motor neuronal model of amyotrophic lateral sclerosis reveals altered energy metabolism. Neurobiol Dis. 2011;43(2):346–355. doi: 10.1016/j.nbd.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Al-Chalabi A, Jones A, Troakes C, et al. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012;124(3):339–352. doi: 10.1007/s00401-012-1022-4. [DOI] [PubMed] [Google Scholar]
- 5.Saberi S, Stauffer JE, Schulte DJ, et al. Neuropathology of amyotrophic lateral sclerosis and its variants. Neurol Clin. 2015;33(4):855–876. doi: 10.1016/j.ncl.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarei S, Carr K, Reiley L, et al. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol. 2015;6:171. doi: 10.4103/2152-7806.169561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozzo F, Mirra A, Carrì MT. Oxidative stress and mitochondrial damage in the pathogenesis of ALS: new perspectives. Neurosci Lett. 2017;636:3–8. doi: 10.1016/j.neulet.2016.04.065. [DOI] [PubMed] [Google Scholar]
- 8.Vandoorne T, De Bock K, Van Den Bosch L. Energy metabolism in ALS: an underappreciated opportunity? Acta Neuropathol. 2018;135(4):489–509. doi: 10.1007/s00401-018-1835-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brito MD, da Silva GFG, Tilieri EM, et al. Metabolic alteration and amyotrophic lateral sclerosis outcome: a systematic review. Front Neurol. 2019;10:1205. doi: 10.3389/fneur.2019.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripps ME, Huntley GW, Hof PR, et al. Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 1995;92:689–693. doi: 10.1073/pnas.92.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi P, Gal J, Kwinter DM, et al. Mitochondrial dysfunction in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2010;1802:45–51. doi: 10.1016/j.bbadis.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladd AC, Keeney PM, Govind MM, et al. Mitochondrial oxidative phosphorylation transcriptome alterations in human amyotrophic lateral sclerosis spinal cord and blood. NeuroMolecular Med. 2014;16(4):714–726. doi: 10.1007/s12017-014-8321-y. [DOI] [PubMed] [Google Scholar]
- 14.Allen SP, Rajan S, Duffy L, et al. Superoxide dismutase 1 mutation in a cellular model of amyotrophic lateral sclerosis shifts energy generation from oxidative phosphorylation to glycolysis. Neurobiol Aging. 2014;35(6):1499–1509. doi: 10.1016/j.neurobiolaging.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Carrì MT, Valle C, Bozzo F, et al. Oxidative stress and mitochondrial damage: importance in non-SOD1 ALS. Front Neurosci. 2015;9:41. doi: 10.3389/fncel.2015.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laferriere F, Polymenidou M (2015) Advances and challenges in understanding the multifaceted pathogenesis of amyotrophic lateral sclerosis. Swiss Med Wkly 145(0506). 10.4414/smw.2015.14054 [DOI] [PubMed]
- 17.Kadenbach B (2012) Introduction to mitochondrial oxidative phosphorylation. Adv Exp Med Biol Springer, New York, NY 1–11. 10.1007/978-1-4614-3573-0_1. [DOI] [PubMed]
- 18.Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxid Redox Signal. 2013;8:240–242. doi: 10.1089/ars.2013.5255. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz TL. Mitochondrial trafficking in neurons. Cold Spring Harb Perspect Biol. 2013;5(6):a011304. doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osellame LD, Duchen MR. Quality control gone wrong: mitochondria, lysosomal storage disorders and neurodegeneration. B J Pharmacol. 2014;171(8):1958–1972. doi: 10.1111/bph.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A, Kukreti R, Saso L, et al. Oxidative stress: a key modulator in neurodegenerative. Dis Mol. 2019;24(8):1583. doi: 10.3390/molecules24081583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieira FG, Ping Q, Moreno AJ, et al. Guanabenz treatment accelerates disease in a mutant SOD1 mouse model of ALS. PLoS One. 2015;10(8):e0135570. doi: 10.1371/journal.pone.0135570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shook SJ, Pioro EP. Racing against the clock: recognizing, differentiating, diagnosing, and referring the amyotrophic lateral sclerosis patient. Ann Neurol. 2009;65(S1):S10–S16. doi: 10.1002/ana.21545. [DOI] [PubMed] [Google Scholar]
- 24.Paillisse C, Lacomblez L, Dib M, et al. Prognostic factors for survival in amyotrophic lateral sclerosis patients treated with riluzole. Amyotroph Lateral Scler Other Motor Neuron Disord. 2005;6(1):37–44. doi: 10.1080/14660820510027035. [DOI] [PubMed] [Google Scholar]
- 25.Dupuis L, di Scala F, Rene F, et al. Up-regulation of mitochondrial uncoupling protein 3 reveals an early muscular metabolic defect in amyotrophic lateral sclerosis. FASEB J. 2003;17(14):2091–2093. doi: 10.1096/fj.02-1182fje. [DOI] [PubMed] [Google Scholar]
- 26.Chiò A, Logroscino G, Hardiman O, et al. Prognostic factors in ALS: a critical review. Amyotroph Lateral Scler. 2009;10(5–6):310–323. doi: 10.3109/17482960802566824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorst J, Kühnlein P, Hendrich C, et al. Patients with elevated triglyceride and cholesterol serum levels have a prolonged survival in amyotrophic lateral sclerosis. J Neurol. 2011;258(4):613–617. doi: 10.1007/s00415-010-5805-z. [DOI] [PubMed] [Google Scholar]
- 28.Sutedja NA, Van Der Schouw YT, Fischer K, et al. Beneficial vascular risk profile is associated with amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82(6):638–642. doi: 10.1136/jnnp.2010.236752. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda Y, Sciarretta S, Nagarajan N, et al. New insights into the role of mitochondrial dynamics and autophagy during oxidative stress and aging in the heart. Oxidative Med Cell Longev. 2014;2014:210934. doi: 10.1155/2014/210934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paganoni S, Zhang M, Zárate AQ, et al. Uric acid levels predict survival in men with amyotrophic lateral sclerosis. J Neurol. 2012;259(9):1923–1928. doi: 10.1007/s00415-012-6440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang YM, Gupta SK, Kim KJ, et al. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Stem Cells. 2013;12(6):713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiò A, Battistini S, Calvo A, et al. Genetic counselling in ALS: facts, uncertainties and clinical suggestions. J Neurol Neurosurg Psychiatry. 2014;85(5):478–485. doi: 10.1136/jnnp-2013-305546. [DOI] [PubMed] [Google Scholar]
- 33.Creemers H, Grupstra H, Nollet F, et al. Prognostic factors for the course of functional status of patients with ALS: a systematic review. J Neurol. 2015;262(6):1407–1423. doi: 10.1007/s00415-014-7564-8. [DOI] [PubMed] [Google Scholar]
- 34.Cereda C, Leoni E, Milani P, et al. Altered intracellular localization of SOD1 in leukocytes from patients with sporadic amyotrophic lateral sclerosis. PLoS One. 2013;8(10):e75916. doi: 10.1371/journal.pone.0075916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu CH, Macdonald-Wallis C, Gray E, et al. Neurofilament light chain: a prognostic biomarker in amyotrophic lateral sclerosis. J Neurol. 2015;84(22):2247–2257. doi: 10.1212/WNL.0000000000001642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blasco H, Garcon G, Patin F, et al. Panel of oxidative stress and inflammatory biomarkers in ALS: a pilot study. Can J Neurol Sci. 2017;44(1):90–95. doi: 10.1017/cjn.2016.284. [DOI] [PubMed] [Google Scholar]
- 37.Coppedè F, Stoccoro A, Mosca L, et al. Increase in DNA methylation in patients with amyotrophic lateral sclerosis carriers of not fully penetrant SOD1 mutations. Amyotroph Lateral Scler Frontotemporal Degener. 2018;19(1–2):93–101. doi: 10.1080/21678421.2017.1367401. [DOI] [PubMed] [Google Scholar]
- 38.Verber NS, Shepheard SR, Sassani M, et al. Biomarkers in motor neuron disease: a state of the art review. Front Neurol. 2019;10:291. doi: 10.3389/fneur.2019.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin D, Thompson MA, Nadler JV. The neuroprotective agent riluzole inhibits release of glutamate and aspartate from slices of hippocampal area CA1. Eur J Pharmacol. 1993;250(3):473–476. doi: 10.1016/0014-2999(93)90037-I. [DOI] [PubMed] [Google Scholar]
- 40.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(3):261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 41.Ludolph A, Drory V, Hardiman O, et al. A revision of the El Escorial criteria - 2015. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16(5–6):291–292. doi: 10.3109/21678421.2015.1049183. [DOI] [PubMed] [Google Scholar]
- 42.Mota SI, Costa RO, Ferreira IL, et al. Oxidative stress involving changes in Nrf2 and ER stress in early stages of Alzheimer’s disease. Biochim Biophys Acta. 2015;1852(7):1428–1441. doi: 10.1016/j.bbadis.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 43.Carvalho ACP, Sharpe J, Rosenstock TR, et al. Bax affects intracellular Ca 2+ stores and induces Ca 2+ wave propagation. Cell Death Differ. 2004;11(12):1265. doi: 10.1038/sj.cdd.4401508. [DOI] [PubMed] [Google Scholar]
- 44.Rosenstock TR, Carvalho ACP, Jurkiewicz A, et al. Mitochondrial calcium, oxidative stress and apoptosis in a neurodegenerative disease model induced by 3-nitropropionic acid. J Neurochem. 2004;88:1220–1228. doi: 10.1046/j.1471-4159.2003.02250.x1220. [DOI] [PubMed] [Google Scholar]
- 45.Rosenstock TR, Bertoncini CRA, Teles AV, et al. Glutamate-induced alterations in Ca2+ signaling are modulated by mitochondrial Ca2+ handling capacity in brain slices of R6/1 transgenic mice. Eur J Neurosci. 2010;32(1):60–70. doi: 10.1111/j.1460-9568.2010.07268.x. [DOI] [PubMed] [Google Scholar]
- 46.Naia L, Ferreira IL, Cunha-Oliveira T, et al. Activation of IGF-1 and insulin signaling pathways ameliorate mitochondrial function and energy metabolism in Huntington’s disease human lymphoblasts. Mol Neurobiol. 2015;51(1):331–348. doi: 10.1007/s12035-014-8735-4. [DOI] [PubMed] [Google Scholar]
- 47.Onesto EC, Colombrita V, Gumina MO, et al. Gene-specific mitochondria dysfunctions in human TARDBP and C9ORF72 fibroblasts. Acta Neuropathol Commun. 2016;4(1):47. doi: 10.1186/s40478-016-0316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribeiro M, Rosenstock TR, Cunha-Oliveira T, et al. Glutathione redox cycle dysregulation in Huntington’s disease knock-in striatal cells. Free Radic Biol Med. 2012;53(10):1857–1867. doi: 10.1016/j.freeradbiomed.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Ribeiro M, Rosenstock TR, Oliveira AM, et al. Insulin and IGF-1 improve mitochondrial function in a PI-3K/Akt-dependent manner and reduce mitochondrial generation of reactive oxygen species in Huntington’s disease knock-in striatal cells. Free Radic Biol Med. 2014;74:129–144. doi: 10.1016/j.freeradbiomed.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 50.Naia L, Ribeiro M, Rodrigues J, et al. Insulin and IGF-1 regularize energy metabolites in neural cells expressing full-length mutant huntingtin. Neuropeptides. 2016;58:73–81. doi: 10.1016/j.npep.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 51.Naia L, Cunha-Oliveira T, Rodrigues J, et al. Histone deacetylase inhibitors protect against pyruvate dehydrogenase dysfunction in Huntington’s disease. J Neurosci. 2017;37(10):2776–2794. doi: 10.1523/JNEUROSCI.2006-14.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Naia L, Rosenstock TR, Oliveira AM, et al. Comparative mitochondrial-based protective effects of resveratrol and nicotinamide in Huntington’s disease models. Mol Neurobiol. 2017;54(7):5385–5399. doi: 10.1007/s12035-016-0048-3. [DOI] [PubMed] [Google Scholar]
- 53.E Silva LFS, Brito MD, Yuzawa JMC, et al. Mitochondrial dysfunction and changes in high-energy compounds in different cellular models associated to hypoxia: implication to schizophrenia. Sci Rep. 2019;9(1):18049. doi: 10.1038/s41598-019-53605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27(7):728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 55.Weydt P, Pineda VV, Torrence AE, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1α in Huntington’s disease neurodegeneration. Cell Metab. 2006;4(5):349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 56.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Copple IM, Goldring CE, Kitteringham NR, et al. The Nrf2–Keap1 defence pathway: role in protection against drug-induced toxicity. Toxicology. 2008;246(1):24–33. doi: 10.1016/j.tox.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 58.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 59.Handschin C. The biology of PGC-1α and its therapeutic potential. Trends Pharmacol Sci. 2009;30(6):322–329. doi: 10.1016/j.tips.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 60.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 61.Rubinsztein DC, Cuervo AM, Ravikumar B, et al. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 62.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5(5):713–716. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osellame LD, Blacker TS, Duchen MR. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab. 2012;26(6):711–723. doi: 10.1016/j.beem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venegas V, Halberg MC. Measurement of mitochondrial DNA copy number. Methods Mol Biol. 2012;837:327–335. doi: 10.1007/978-1-61779-504-6_22. [DOI] [PubMed] [Google Scholar]
- 65.Choi J, Chandrasekaran K, Inoue T, et al. PGC-1α regulation of mitochondrial degeneration in experimental diabetic neuropathy. Neurobiol Dis. 2014;64:118–130. doi: 10.1016/j.nbd.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weydt P, Soyal SM, Landwehrmeyer GB, et al. A single nucleotide polymorphism in the coding region of PGC-1α is a male-specific modifier of Huntington disease age-at-onset in a large European cohort. BMC Neurol. 2014;14(1):1. doi: 10.1186/1471-2377-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Rydzewski N, Hider A, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol. 2016;18(4):404. doi: 10.1038/ncb3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bayer H, Lang K, Buck E, et al. ALS-causing mutations differentially affect PGC-1α expression and function in the brain vs. peripheral tissues. Neurobiol Dis. 2017;97:36–45. doi: 10.1016/j.nbd.2016. [DOI] [PubMed] [Google Scholar]
- 69.King GA, Hashemi Shabestari M, Taris KKH, et al. Acetylation and phosphorylation of human TFAM regulate TFAM–DNA interactions via contrasting mechanisms. Nucleic Acids Res. 2018;46(7):3633–3642. doi: 10.1093/nar/gky204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kiyama T, Chen CK, Wang SW, et al. Essential roles of mitochondrial biogenesis regulator Nrf1 in retinal development and homeostasis. Mol Neurodegener. 2018;13(1):56. doi: 10.1186/s13024-018-0287-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak K, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.Ceccanti M, Onesti E, Rubino A, et al. Modulation of human corticospinal excitability by paired associative stimulation in patients with amyotrophic lateral sclerosis and effects of Riluzole. Brain Stimul. 2018;11(4):775–781. doi: 10.1016/j.brs.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 73.Crompton M, Barksby E, Johnson N, et al. Mitochondrial intermembrane junctional complexes and their involvement in cell death. Biochimie. 2002;84(2–3):143–152. doi: 10.1016/S0300-9084(02)01368-8. [DOI] [PubMed] [Google Scholar]
- 74.Gnaiger E. Mitochondrial pathways and respiratory control. An introduction to OXPHOS analysis Mitochondr. Physiol Network. 4. 2007. pp. 19–80. [Google Scholar]
- 75.Huang H, Tu R, Liu F, et al. Effects of resveratrol on Nrf2 signal pathway of chronic lead-exposed mouse brain tissue. Wei Sheng Yan Jiu. 2015;44(6):954–958. [PubMed] [Google Scholar]
- 76.Chen S, Meng XF, Zhang C. Role of NADPH oxidase-mediated reactive oxygen species in podocyte injury. Biomed Res Int. 2013;2013:839761. doi: 10.1155/2013/839761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dinkova-Kostova AT, Kazantsev AG. Activation of Nrf2 signaling as a common treatment of neurodegenerative diseases. Neurodegener Dis. 2017;7(2):97–100. doi: 10.2217/nmt-2017-0011. [DOI] [PubMed] [Google Scholar]
- 78.Liu T, Knowlton AA. Repetitive ROS injury leads to inactivation of NRF2-induced antioxidant defense and mitochondrial dysfunction in ischemic heart failure. FASEB J. 2016;31(1_supplement):1080.19-1080.19. doi: 10.1096/fasebj.31.1_supplement.1080.19. [DOI] [Google Scholar]
- 79.Morales Pantoja IE, Hu CL, Perrone-Bizzozero NI, et al. Nrf2-dysregulation correlates with reduced synthesis and low glutathione levels in experimental autoimmune encephalomyelitis. J Neurochem. 2016;139(4):640–650. doi: 10.1111/jnc.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang RY, Liu LH, Liu H, et al. Nrf2 protects against diabetic dysfunction of endothelial progenitor cells via regulating cell senescence. Int J Mol Med. 2018;42(3):1327–1340. doi: 10.3892/ijmm.2018.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang WT, Chen HI, Chiou RJ, et al. A novel function of transcription factor α-Pal/NRF-1: Increasing neurite outgrowth. Biochem Biophys Res Commun. 2005;334(1):199–206. doi: 10.1016/j.bbrc.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 82.Wang CH, Wang CC, Wei YH. Mitochondrial dysfunction in insulin insensitivity: implication of mitochondrial role in type 2 diabetes. Ann N Y Acad Sci. 2010;1201(1):157–165. doi: 10.1111/j.1749-6632.2010.05625.x. [DOI] [PubMed] [Google Scholar]
- 83.Larsson NG, Wang J, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintance and embryogenesis in mice. Nat Genet. 1998;18(3):231. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 84.Rapaport D, Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J Cell Biol. 1999;146(2):321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reyes A, Mezzina M, Gadaleta G. Human mitochondrial transcription factor A (mtTFA): gene structure and characterization of related pseudogenes. Gene. 2002;291(1–2):223–232. doi: 10.1016/S0378-1119(02)00600-5. [DOI] [PubMed] [Google Scholar]
- 86.Facucho-Oliveira JM, Alderson J, Spikings EC, et al. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120(22):4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- 87.Weydt P, Soyal SM, Gellera C, et al. The gene coding for PGC-1α modifies age at onset in Huntington’s disease. Mol Neurodegener. 2009;4(1):3. doi: 10.1186/1750-1326-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taherzadeh-Fard E, Saft C, Akkad DA, et al. PGC-1alpha downstream transcription factors NRF-1 and TFAM are genetic modifiers of Huntington disease. Mol Neurodegener. 2011;6(1):32. doi: 10.1186/1750-1326-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lezza AM. Mitochondrial transcription factor A (TFAM): one actor for different roles. Front Biol. 2012;7(1):30–39. doi: 10.1007/s11515-011-1175-x. [DOI] [Google Scholar]
- 90.Ngo HB, Lovely GA, Phillips R, et al. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Novin MG, Allahveisi A, Noruzinia M, et al. The relationship between transcript expression levels of nuclear encoded (TFAM, NRF1) and mitochondrial encoded (MT-CO1) genes in single human oocytes during oocyte maturation. Balkan J Med Genet. 2015;18(1):39–46. doi: 10.1515/bjmg-2015-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Novin MG, Noruzinia M, Allahveisi A, et al. Comparison of mitochondrial-related transcriptional levels of TFAM, NRF1 and MT-CO1 genes in single human oocytes at various stages of the oocyte maturation. Iran Biomed J. 2015;19(1):23. doi: 10.6091/ibj.1400.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sebastián D, Zorzano A. Mitochondrial dynamics and metabolic homeostasis. Curr Opin Physiol. 2018;3:34–40. doi: 10.1016/j.cophys.2018.02.006. [DOI] [Google Scholar]
- 94.Yu J, Li Y, Liu X, et al. Mitochondrial dynamics modulation as a critical contribution for Shenmai injection in attenuating hypoxia/reoxygenation injury. J Ethnopharmacol. 2019;37:9–19. doi: 10.1016/j.jep.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 95.Losón OC, Song Z, Chen H, et al. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Joshi AU, Saw NL, Vogel H, et al. Inhibition of Drp1/Fis1 interaction slows progression of amyotrophic lateral sclerosis. EMBO Mol Med. 2018;10(3):e8166. doi: 10.15252/emmm.201708166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manczak M, Kandimalla R, Yin X, et al. Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity. Hum Mol Genet. 2018;28(2):177–199. doi: 10.1093/hmg/ddy335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hamacher-Brady A, Brady NR. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci. 2016;73(4):775–795. doi: 10.1007/s00018-015-2087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Williams JA, Ding WX. Mechanisms, pathophysiological roles and methods for analyzing mitophagy–recent insights. Biol Chem. 2018;399(2):147–178. doi: 10.1515/hsz-2017-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhuang N, Li L, Chen S, et al. PINK1-dependent phosphorylation of PINK1 and Parkin is essential for mitochondrial quality control. Cell Death Dis. 2016;7(12):e2501. doi: 10.1038/cddis.2016.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Truban D, Hou X, Caulfield TR, et al. PINK1, Parkin, and mitochondrial quality control: what can we learn about Parkinson’s disease pathobiology? J Parkinsons Dis. 2017;7(1):13–29. doi: 10.3233/JPD-160989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seranova E, Palhegyi AM, Verma S, et al. Human induced pluripotent stem cell models of neurodegenerative disorders for studying the biomedical implications of autophagy. J Mol Biol. 2020;432(8):2754–2798. doi: 10.1016/j.jmb.2020.01.024. [DOI] [PubMed] [Google Scholar]
- 103.Sarkar S, Korolchuk VI, Renna M, et al. Complex inhibitory effects of nitric oxide on autophagy. Cell Mol. 2011;43(1):19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014;16(6):495. doi: 10.1016/j.jim.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 105.Chiang WC, Wei Y, Kuo YC, et al. High-throughput screens to identify autophagy inducers that function by disrupting Beclin 1/Bcl-2 binding. ACS Chem Biol. 2018;13(8):2247–2260. doi: 10.1021/acschembio.8b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kraft LJ, Dowler J, Manral P, et al. Size, organization, and dynamics of soluble SQSTM1 and LC3-SQSTM1 complexes in living cells. Autophagy. 2016;12(9):1660–1674. doi: 10.1080/15548627.2016.1199299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seranova E, Ward C, Chipara M, et al. In vitro screening platforms for identifying autophagy modulators in mammalian cells. Autophagy. 2019;1880:389–428. doi: 10.1007/978-1-4939-8873-0_26. [DOI] [PubMed] [Google Scholar]
- 108.Brookes PS. Mitochondrial nitric oxide synthase. Mitochondrion. 2004;3(4):187–204. doi: 10.1016/j.mito.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 109.Corona JC, Tapia R. Ca2+-permeable AMPA receptors and intracellular Ca2+ determine motoneuron vulnerability in rat spinal cord in vivo. Neuropharmacol. 2007;52(5):1219–1228. doi: 10.1016/j.neuropharm.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 110.Kawamata H, Manfredi G. Mitochondrial dysfunction and intracellular calcium dysregulation in ALS. Mech Ageing Dev. 2010;131(7–8):517–526. doi: 10.1016/j.mad.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jaiswal MK. Calcium, mitochondria, and the pathogenesis of ALS: the good, the bad, and the ugly. Front Cell Neurosci. 2013;7:199. doi: 10.3389/fncel.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith EF, Shaw PJ, De Vos KJ. The role of mitochondria in amyotrophic lateral sclerosis. Neurosci Lett. 2017;S0304-3940(17):30544–3054X. doi: 10.1016/j.neulet.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 113.Verma M, Wills ZP, Chu CT. Excitatory dendritic mitochondrial calcium toxicity: implications for Parkinson’s and other neurodegenerative diseases. Front Neurosci. 2018;12:523. doi: 10.3389/fnins.2018.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jaiswal MK, Zech WD, Goos M, et al. Impairment of mitochondrial calcium handling in a mtSOD1 cell culture model of motoneuron disease. BMC Neurosci. 2009;10:64. doi: 10.1186/1471-2202-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tadić V, Adam A, Goldhammer N, et al. Investigation of mitochondrial calcium uniporter role in embryonic and adult motor neurons from G93AhSOD1 mice. Neurobiol Aging. 2019;75:209–222. doi: 10.1016/j.neurobiolaging.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 116.Von Lewinski F, Fuchs J, Vanselow BK, et al. Low Ca2+ buffering in hypoglossal motoneurons of mutant SOD1 (G93A) mice. Neurosci Lett. 2008;445(3):224–228. doi: 10.1016/j.neulet.2008.08.084. [DOI] [PubMed] [Google Scholar]
- 117.Abdallah Y, Kasseckert SA, Iraqi W, et al. Interplay between Ca2+ cycling and mitochondrial permeability transition pores promotes reperfusion-induced injury of cardiac myocytes. J Cell Mol Med. 2011;15(11):2478–2485. doi: 10.1111/j.1582-4934.2010.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fernandez-Sanz C, De la Fuente S, Sheu SS. Mitochondrial Ca2+ concentrations in live cells: quantification methods and discrepancies. FEBS Lett. 2019;593(13):1528–1541. doi: 10.1002/1873-3468.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ly JD, Grubb DR, Lawen A. The mitochondrial membrane potential (Δψ m) in apoptosis; an update. Apoptosis. 2003;8(2):115–128. doi: 10.1023/A:10229451. [DOI] [PubMed] [Google Scholar]
- 120.Chazotte B. Labeling mitochondria with TMRM or TMRE. Cold Spring Harb Protoc. 2011;2011(7):pdb-prot5641. doi: 10.1101/pdb.prot5641. [DOI] [PubMed] [Google Scholar]
- 121.Cassina P, Cassina A, Pehar M, et al. Mitochondrial dysfunction in SOD1G93A-bearing astrocytes promotes motor neuron degeneration: prevention by mitochondrial-targeted antioxidants. J Neurosci. 2008;28(16):4115–4122. doi: 10.1523/JNEUROSCI.5308-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shrivastava M, Vivekanandhan S, Pati U, et al. Mitochondrial perturbance and execution of apoptosis in platelet mitochondria of patients with amyotrophic lateral sclerosis. Int J Neurosci. 2011;121(3):149–158. doi: 10.3109/00207454.2010.537416. [DOI] [PubMed] [Google Scholar]
- 123.Heath PR, Shaw PJ. Update on the glutamatergic neurotransmitter system and the role of excitotoxicity in amyotrophic lateral sclerosis. Muscle Nerve. 2002;26(4):438–458. doi: 10.1002/mus.10186. [DOI] [PubMed] [Google Scholar]
- 124.Zhou H, Wang J, Hu S, et al. BI1 alleviates cardiac microvascular ischemia-reperfusion injury via modifying mitochondrial fission and inhibiting XO/ROS/F-actin pathways. J Cell Physiol. 2019;234(4):5056–5069. doi: 10.1002/jcp.27308. [DOI] [PubMed] [Google Scholar]
- 125.Halliwell B. Oxygen and nitrogen are pro-carcinogens. Damage to DNA by reactive oxygen, chlorine and nitrogen species: measurement, mechanism and the effects of nutrition. Mutation Research/Genetic Toxic Environment Mutagen. 1999;443(1–2):37–52. doi: 10.1016/S1383-5742(99)00009-5. [DOI] [PubMed] [Google Scholar]
- 126.Barber SC, Shaw PJ. Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic Biol Med. 2010;48(5):629–641. doi: 10.1016/j.freeradbiomed.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 127.D’Amico E, Factor-Litvak P, Santella RM, et al. Clinical perspective on oxidative stress in sporadic amyotrophic lateral sclerosis. Free Radic Biol Med. 2013;65:509–527. doi: 10.1016/j.freeradbiomed.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bond L, Bernhardt K, Madria P, et al. A metadata analysis of oxidative stress etiology in preclinical amyotrophic lateral sclerosis: benefits of antioxidant therapy. Front Neurosci. 2018;12:10. doi: 10.3389/fnins.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carrera-Juliá S, Moreno ML, Barrios C, et al. Antioxidant alternatives in the treatment of amyotrophic lateral sclerosis: a comprehensive review. Front Physiol. 2020;11:63. doi: 10.3389/fphys.2020.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Curti D, Malaspina A, Facchetti G, et al. Amyotrophic lateral sclerosis: oxidative energy metabolism and calcium homeostasis in peripheral blood lymphocytes. Neurol. 1996;47(4):1060–1064. doi: 10.1212/WNL.47.4.1060. [DOI] [PubMed] [Google Scholar]
- 131.Oteiza PI, Uchitel OD, Carrasquedo F, et al. Evaluation of antioxidants, protein, and lipid oxidation products in blood from sporadic amiotrophic lateral sclerosis patients. Neurol Res. 1997;22(4):535–539. doi: 10.1023/a:1027384432715. [DOI] [PubMed] [Google Scholar]
- 132.Bonnefont-Rousselot D, Lacomblez L, Jaudon MC, et al. Blood oxidative stress in amyotrophic lateral sclerosis. J Neurol Sci. 2000;178(1):57–62. doi: 10.1016/s0022-510x(00)00365-8. [DOI] [PubMed] [Google Scholar]
- 133.Tebay LE, Robertson H, Durant ST, et al. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med. 2015;88(Pt B):108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vargas MR, Pehar M, Cassina P, et al. Fibroblast growth factor-1 induces heme oxygenase-1 via nuclear factor erythroid 2-related factor 2 (Nrf2) in spinal cord astrocytes consequences for motor neuron survival. J Biol Chem. 2005;280(27):25571–25579. doi: 10.1074/jbc.M501920200. [DOI] [PubMed] [Google Scholar]
- 135.Thau N, Knippenberg S, Körner S, et al. Decreased mRNA expression of PGC-1α and PGC-1α-regulated factors in the SOD1G93A ALS mouse model and in human sporadic ALS. J Neuropathol Exp Neurol. 2012;71(12):1064–1074. doi: 10.1097/NEN.0b013e318275df4b. [DOI] [PubMed] [Google Scholar]
- 136.Guo Y, Zhang Y, Wen D, et al. The modest impact of transcription factor Nrf2 on the course of disease in an ALS animal model. Lab Investig. 2013;93(7):825. doi: 10.1038/labinvest.2013.73. [DOI] [PubMed] [Google Scholar]
- 137.Russell AP, Wada S, Vergani L, et al. Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol Dis. 2013;49:107. doi: 10.1016/j.nbd.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 138.Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1α. Cardiovasc Res. 2008;79(2):208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 139.Palomo GM, Granatiero V, Kawamata H, et al. Parkin is a disease modifier in the mutant SOD1 mouse model of ALS. EMBO Mol Med. 2018;10(10):e8888. doi: 10.15252/emmm.201808888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sandri M, Lin J, Handschin C, et al. PGC-1α protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci. 2006;103(44):16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 142.Qi Y, Yin X, Wang S, et al. PGC-1 silencing compounds the perturbation of mitochondrial function caused by mutant SOD1 in skeletal muscle of ALS mouse model. Front Aging Neurosci. 2015;7:204. doi: 10.3389/fnagi.2015.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Finkel T. Cell biology: a clean energy programme. Nat Res. 2006;444(7116):151. doi: 10.1038/444151a. [DOI] [PubMed] [Google Scholar]
- 144.St-Pierre J, Drori S, Uldry M, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 145.Chen X, Xue H, Fang W, et al. Adropin protects against liver injury in nonalcoholic steatohepatitis via the Nrf2 mediated antioxidant capacity. Redox Biol. 2019;21:101068. doi: 10.1016/j.redox.2018.101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91(4):1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem Sci. 2007;32(3):111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 148.Ladd AC, Brohawn DG, Thomas RR, et al. RNA-seq analyses reveal that cervical spinal cords and anterior motor neurons from amyotrophic lateral sclerosis subjects show reduced expression of mitochondrial DNA-encoded respiratory genes, and rhTFAM may correct this respiratory deficiency. Brain Res. 2017;1667:74–83. doi: 10.1016/j.brainres.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 149.Hayashi Y, Yoshida M, Yamato M, et al. Reverse of age-dependent memory impairment and mitochondrial DNA damage in microglia by an overexpression of human mitochondrial transcription factor in mice. J Neurosci. 2008;28(34):8624–8634. doi: 10.1523/JNEUROSCI.1957-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Florentz C, Sohm B, Tryoen-Toth P, et al. Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci. 2003;60(7):1356–1375. doi: 10.1007/s00018-003-2343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Keeney PM, Bennett JP. ALS spinal neurons show varied and reduced mtDNA gene copy numbers and increased mtDNA gene deletions. Mol Neurodegener. 2010;5(1):21. doi: 10.1186/1750-1326-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reddy PH. Mitochondrial dysfunction in aging and Alzheimer’s disease: strategies to protect neurons. Antioxid Redox Signal. 2007;9(10):1647–1658. doi: 10.1089/ars.2007.1754. [DOI] [PubMed] [Google Scholar]
- 153.Knott AB, Perkins G, Schwarzenbacher R, et al. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9(7):505. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou Q, Zhu L, Qiu W, et al. Nicotinamide riboside enhances mitochondrial proteostasis and adult neurogenesis through activation of mitochondrial unfolded protein response signaling in the brain of ALS SOD1G93A mice. Int J Biol Sci. 2020;16(2):284–297. doi: 10.7150/ijbs.38487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jahani-Asl A, Cheung EC, Neuspiel M, et al. Mitofusin 2 protects cerebellar granule neurons against injury-induced cell death. J Biol Chem. 2007;282(33):23788–23798. doi: 10.1074/jbc.M703812200. [DOI] [PubMed] [Google Scholar]
- 156.Gao J, Wang L, Liu J, et al. Abnormalities of mitochondrial dynamics in neurodegenerative diseases. Antioxid. 2017;6(2):25. doi: 10.3390/antiox6020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang W, Li L, Lin WL, et al. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum Mol Genet. 2013;22(23):4706–4719. doi: 10.1093/hmg/ddt319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Deng J, Yang M, Chen Y, et al. FUS interacts with HSP60 to promote mitochondrial damage. PLoS Genet. 2015;11(9):e1005357. doi: 10.1371/journal.pgen.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pansarasa O, Bordoni M, Drufuca L, et al. Lymphoblastoid cell lines as a model to understand amyotrophic lateral sclerosis disease mechanisms. Dis Model Mech. 2018;11(3):dmm031625. doi: 10.1242/dmm.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu W, Yamashita T, Tian F, et al. Mitochondrial fusion and fission proteins expression dynamically change in a murine model of amyotrophic lateral sclerosis. Curr Neurovasc Res. 2013;10(3):222–230. doi: 10.2174/15672026113109990060. [DOI] [PubMed] [Google Scholar]
- 161.Reddy PH, Reddy TP, Manczak M, et al. Dynamin-related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res Rev. 2011;67(1–2):103–118. doi: 10.1016/j.brainresrev.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Geisler S, Holmström KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 164.Okatsu K, Saisho K, Shimanuki M, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15(8):887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lim GG, Chua DS, Basil AH, et al. Cytosolic PTEN-induced putative kinase 1 is stabilized by the NF-κB pathway and promotes non-selective mitophagy. J Biol Chem. 2015;290(27):16882–16893. doi: 10.1096/fasebj.31.1_supplement.1080.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Narendra D, Kane LA, Hauser DN, et al. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6(8):1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stribl C, Samara A, Trümbach D, et al. Mitochondrial dysfunction and decrease in body weight of a transgenic knock-in mouse model for TDP-43. J Biol Chem. 2014;289(15):10769–10784. doi: 10.1074/jbc.M113.515940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morimoto N, Nagai M, Ohta Y, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 171.Li L, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4(3):290–293. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- 172.Sasaki S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2011;70(5):349–359. doi: 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- 173.Zhang X, Li L, Chen S, et al. Rapamycin treatment augments motor neuron degeneration in SOD1G93A mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7(4):412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 174.Nassif M, Valenzuela V, Rojas-Rivera D, et al. Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy. 2014;10(7):1256–1271. doi: 10.4161/auto.28784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mitsui S, Otomo A, Nozaki M, et al. Systemic overexpression of SQSTM1/p62 accelerates disease onset in a SOD1 H46R-expressing ALS mouse model. Mol Brain. 2018;11(1):30. doi: 10.1186/s13041-018-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rizzardini M, Mangolini A, Lupi M, et al. Low levels of ALS-linked Cu/Zn superoxide dismutase increase the production of reactive oxygen species and cause mitochondrial damage and death in motor neuron-like cells. J Neurol Sci. 2005;232(1–2):95–103. doi: 10.1016/j.jns.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 177.Browne SE, Yang L, DiMauro JP, et al. Bioenergetic abnormalities in discrete cerebral motor pathways presage spinal cord pathology in the G93A SOD1 mouse model of ALS. Neurobiol Dis. 2006;22(3):599–610. doi: 10.1016/j.nbd.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 178.Ehinger JK, Morota S, Hansson MJ, et al. Mitochondrial dysfunction in blood cells from amyotrophic lateral sclerosis patients. J Neurol. 2015;262(6):1493–1503. doi: 10.1007/s00415-015-7737-0. [DOI] [PubMed] [Google Scholar]
- 179.Dobrowolny G, Lepore E, Martini M, et al. Metabolic changes associated with muscle expression of SOD1G93A. Front Physiol. 2018;9:831. doi: 10.3389/fphys.2018.00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tefera TW, Bartlett K, Tran SS, et al. Impaired pentose phosphate pathway in the spinal cord of the hSOD1 G93A mouse model of amyotrophic lateral sclerosis. Mol Neurobiol. 2019;56(8):5844–5855. doi: 10.1007/s12035-019-1485-6. [DOI] [PubMed] [Google Scholar]
- 181.Mali Y, Zisapels N. Gain of interaction of ALS-linked G93A superoxide dismutase with cytosolic malate dehydrogenase. Neurobiol Dis. 2008;32(1):133–141. doi: 10.1016/j.nbd.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 182.Valbuena GN, Rizzardini M, Cimini S, et al. Metabolomic analysis reveals increased aerobic glycolysis and amino acid deficit in a cellular model of amyotrophic lateral sclerosis. Mol Neurobiol. 2016;53(4):2222–2240. doi: 10.1007/s12035-015-9165-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Veyrat-Durebex C, Bris C, Codron P, et al. Metabo-lipidomics of fibroblasts and mitochondrial-endoplasmic reticulum extracts from ALS patients shows alterations in purine, pyrimidine, energetic, and phospholipid metabolisms. Mol Neurobiol. 2019;56(8):5780–5791. doi: 10.1007/s12035-019-1484-7. [DOI] [PubMed] [Google Scholar]
- 184.Vallée A, Lecarpentier Y, Guillevin R, et al. Aerobic glycolysis in amyotrophic lateral sclerosis and Huntington’s disease. Rev Neurosci. 2018;29(5):547–555. doi: 10.1515/revneuro-2017-0075. [DOI] [PubMed] [Google Scholar]
- 185.Josefsen K, Nielsen SM, Campos A, et al. Reduced gluconeogenesis and lactate clearance in Huntington’s disease. Neurobiol Dis. 2010;40(3):656–662. doi: 10.1016/j.nbd.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 186.Ravera S, Torazza C, Bonifacino T, et al. Altered glucose catabolism in the presynaptic and perisynaptic compartments of SOD1G93A mouse spinal cord and motor cortex indicates that mitochondria are the site of bioenergetic imbalance in ALS. J Neurochem. 2019;151(3):336–350. doi: 10.1111/jnc.14819. [DOI] [PubMed] [Google Scholar]
- 187.Vallée A. Aerobic glycolysis activation through canonical WNT/β-catenin pathway in ALS. Med Sci (Paris) 2018;34(4):326–330. doi: 10.1051/medsci/20183404013. [DOI] [PubMed] [Google Scholar]
- 188.Harlan BA, Killoy KM, Pehar M, et al. Evaluation of the NAD+ biosynthetic pathway in ALS patients and effect of modulating NAD+ levels in hSOD1-linked ALS mouse models. Exp Neurol. 2020;327:113219. doi: 10.1016/j.expneurol.2020.113219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bensimon G, Lacomblez L, Meininger VA. Controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N Engl J Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 190.Lacomblez L, Bensimon G, Leigh PN, et al. Doseranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic Lateral Sclerosis/Riluzole Study Group II. Lancet. 1996;347:1425–1431. doi: 10.1016/S0140-6736(96)91680-3. [DOI] [PubMed] [Google Scholar]
- 191.Tripathi VB, Al-Chalabi A. Molecular insights and therapeutic targets in amyotrophic lateral sclerosis. CNS Neurol Disord Drug Targets. 2008;7:11–19. doi: 10.2174/187152708783885110. [DOI] [PubMed] [Google Scholar]
- 192.Li RW, Deng Y, Pham HN, et al. Riluzole protects against skeletal muscle ischaemia-reperfusion injury in a porcine model. Injury. 2020;51(2):178–184. doi: 10.1016/j.injury.2019.12.030. [DOI] [PubMed] [Google Scholar]
- 193.Umemiya M, Berger AJ. Inhibition by riluzole of glycinergic postsynaptic currents in rat hypoglossal motoneurones. Br J Pharmacol. 1995;116:3227–3230. doi: 10.1111/j.1476-5381.1995.tb15128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zona C, Cavalcanti S, De Sarro G, et al. Kainate-induced currents in rat cortical neurons in culture are modulated by riluzole. Synapse. 2002;43:244–251. doi: 10.1002/syn.10040. [DOI] [PubMed] [Google Scholar]
- 195.Fumagalli E, Funicello M, Rauen T, et al. Riluzole enhances the activity of glutamate transporters GLAST, GLT1 and EAAC1. Eur J Pharmacol. 2008;578:171–176. doi: 10.1016/j.ejphar.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 196.Jaiswal MK. Riluzole and edaravone: a tale of two amyotrophic lateral sclerosis drugs. Med Res Rev. 2019;39:733–748. doi: 10.1002/med.21528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 286 kb)
Data Availability Statement
All data generated or analyzed during this study are included in the manuscript and the supplementary files.