Abstract
Physiological equilibrium in the retina depends on coordinated work between rod and cone photoreceptors and can be compromised by the expression of mutant proteins leading to inherited retinal degeneration (IRD). IRD is a diverse group of retinal dystrophies with multifaceted molecular mechanisms that are not fully understood. In this review, we focus on the contribution of chronically activated unfolded protein response (UPR) to inherited retinal pathogenesis, placing special emphasis on studies employing genetically modified animal models. As constitutively active UPR in degenerating retinas may activate pro-apoptotic programs associated with oxidative stress, pro-inflammatory signaling, dysfunctional autophagy, free cytosolic Ca2+ overload, and altered protein synthesis rate in the retina, we focus on the regulatory mechanisms of translational attenuation and approaches to overcoming translational attenuation in degenerating retinas. We also discuss current research on the role of the UPR mediator PERK and its downstream targets in degenerating retinas and highlight the therapeutic benefits of reprogramming PERK signaling in preclinical animal models of IRD. Finally, we describe pharmacological approaches targeting UPR in ocular diseases and consider their potential applications to IRD.
Keywords: Unfolded Protein Response, Retinal Degeneration, Translation, Translational Repressor, Neuroprotection
1. Retinal structure: Rod and cone photoreceptors
The retina is stratified neurological tissue lining the back of the eye that relays visual information to the brain (Bloomfield and Dacheux, 2001; Wassle, 2004; Yarfitz and Hurley, 1994). Vision begins when light enters the eye through the cornea and traverses the entirety of the eye until it strikes visual pigments located in the outer segments (OSs) of photoreceptor cells at the back of the retina (Bloomfield and Dacheux, 2001). There are two types of classical photoreceptors in the retina of vertebrates: rods and cones (Curcio et al., 1987; Wassle, 2004). These two types of cells differ in their morphology, function, retinal structure integrative properties, and biochemical and metabolic characteristics.
Rods are more common than cones; there are approximately 120 million rods but only 5 million cones in the human retina. Rods are active in dim light and account for nighttime (i.e., scotopic) vision, whereas cones are more active at higher lighting intensities and are responsible for daytime (i.e., photopic) and color vision (Schnapf et al., 1988). Rods are more cylindrical, whereas cones are more cone-shaped. Until recently, the entire OS membrane of cones was believed to be continuous with the plasma membrane of the inner segment and enfolded to form a lamellar structure, generating stacks of disk membranes, whereas the rod plasma membrane was believed to surround disk membranes, generating space between disk and plasma membranes. However, studies now show that like cones, rod disks are formed via plasma membrane evagination (Burgoyne et al., 2015; Ding et al., 2015; Volland et al., 2015).
When photoreceptors receive photons, they send signals through other neurons that eventually exit the eye and route to the brain. Rods and cones exhibit different connectivity with other retinal neurons, resulting in differences in their transmission of electrical signals. For example, cone signals exit the retina fairly directly. The transmission process begins as visual pigment molecules in the OS of photoreceptors interact with photons and initiate phototransduction, a complex signaling cascade that converts a light signal into a chemical response (Yarfitz and Hurley, 1994). During phototransduction, cone photoreceptors hyperpolarize, reducing glutamate release at the synapse and thereby depolarizing ON-center and hyperpolarizing OFF-center cone bipolar cells (Bloomfield and Dacheux, 2001; Wassle, 2004; Werblin and Dowling, 1969), likely due to the expression of different types of glutamate receptors (DeVries, 2000; Nomura et al., 1994). Depolarization of ON-center bipolar cells prompts their release of glutamate at the inner plexiform layer, where they synapse with amacrine and ganglion cells.
For rod signals to reach ganglion cells, rod-originating impulses piggyback on cone bipolar signaling; rod bipolar cells send signals to cone bipolar cells through All amacrine cells (Bloomfield and Dacheux, 2001; Masland, 2012; Wassle, 2004). Cone bipolar cells then project this signal to ganglion cells (Bloomfield and Dacheux, 2001; Masland, 2012; Wassle, 2004), which in turn projects to the lateral granular nucleus and higher visual areas in the cortex (Ghodrati et al., 2017). In both the retina and higher brain regions, visual processing depends on the ability of photoreceptors to detect changes in lighting parameters and their capacity to relay information to downstream neural circuitry.
Rods and cones also show differences in cell physiology. Rods have the ability to respond to a single photon in the dark-adapted state (Baylor et al., 1979; Rieke and Baylor, 1998), largely because phototransduction results in much higher gain in rods than in cones. The light sensitivity of rods is proposed to be at least 25–100 times greater than that in cones (Fain and Dowling, 1973; Kefalov, 2012). However, the kinetic response of rods is believed to be five times slower than that in cones (Rushton, 1965), perhaps because rods have slower transduction machinery. For example, reaction product lifetimes are 10–20 times longer in rods than in cones (Morshedian and Fain, 2015). Rods also have about 10 times faster recovery of visual molecules following bleaching than cones, perhaps because the higher rate of pigment thermal activation in cones could lower their sensitivity (Kefalov, 2012). These differences could explain why cones have the remarkable ability to adjust their sensitivity in extremely bright light and remain photosensitive, whereas rods saturate in even moderately bright light and remain nonfunctional during most of the day (Kefalov, 2012).
Rods and cones also have different compositions of visual pigment molecules. The visual pigment in rods is rhodopsin, whereas cones contain three different types of opsins—blue, green, and red—which differ in their absorption maximum efficiency: a wavelength of 420 nm for blue cones, 530 nm for green cones, and 560 nm for red cones. The rhodopsin molecule is composed of a chromophore, 11-cis retinal, and a protein moiety called opsin. The chromophore is covalently linked to the ε-amino group of the lysine residue of opsin and is embedded in the protein moiety (Palczewski et al., 2000). In a mammalian rod, there are about 108 rhodopsin molecules present in the OS, with an overall concentration reaching up to 3 mM (Harosi, 1975).
In addition to biochemical differences in the enzymatic activity, lifespan, and recovery time of major phototransduction molecules between rods and cones, the two photoreceptors are also distinguished by specific expression profiles of genes involved in the phototransduction cascade. For example, in humans, rods and cones have different G-protein receptor kinases (GRKs)—GRK1 in rods and GRK1 and/or GRK7 in cones—whereas mice have only GRK1 in both types of photoreceptors (Chen et al., 2001; Ingram et al., 2016). Mice also have arrestin-1 in rods but arrestin-1 and arrestin-4 in cones (Ingram et al., 2016). Although GTPase-accelerating protein complex proteins are the same in rods and cones, their expression is significantly higher in cones (Zhang et al., 2003b). Rod photoreceptors express α1, β1, and γ1 isoforms of the G protein transducin, whereas cones express α2, β3, and γ8 isoforms. Furthermore, rods and cones have different catalytic subunits of the photoreceptor effector enzyme phosphodiesterase 6 (PDE6), which bind a subunit of activated transducin (in GTP form); rods have PDE6A and PDE6B, whereas cones have two identical PDE6C subunits. Additionally, the inhibitory subunits of PDE6 differ between rods and cones; rods express PDE6G, whereas cones express PDE6H (Ingram et al., 2016).
Rods and cones also show metabolic differences in regard to their sensitivity and demand, partially because the two photoreceptors contain different numbers of mitochondria per cell. For example, in mice, cones have twice the number of mitochondria than rods, whereas in primates, cones contain ~10 times more mitochondria than rods (Perkins et al., 2003). This may explain why cones produce more ATP and are more resistant to metabolic stress and apoptosis than rods (Scarpelli and Craig, 1963). Interestingly, there are also difference in the expression of major metabolic kinases; cones co-express cytosolic creatine kinase isozyme and mitochondrial creatine kinase, which maintain ATP homeostasis, whereas rods do not produce these proteins (Leveillard, 2015).
The retina exhibits the Warburg effect similar to some cancer cells (Leveillard, 2015; Noell, 1952a; Winkler, 1981), suggesting that aerobic glycolysis is a main mode of glucose metabolism and that only a small fraction of glucose-derived pyruvate enters mitochondria (Grenell et al., 2019). Indeed, photoreceptors and their synapses exhibit high capacity for glycolysis, aerobic glycolysis, and oxidative phosphorylation and regulate glucose metabolism through compartment-selective expression of hexokinase-1 and −2 and pyruvate kinase-M1 and -M2 (Rueda et al., 2016). Glycolysis and oxidative phosphorylation are major metabolic pathways that synthesize ATP, and impairments in their signaling not only affect photoreceptor function but can also cause cell death. Previous studies of relationships between energy metabolism and retinal function demonstrate that the two process have interdependent effects (Rueda et al., 2016). For example, studies using an iodoacetate that “kills” the retinal pigment epithelium (RPE), resulting in retinal degeneration, show that inhibition of glycolysis induces the death of rods before cones (Noell, 1951; Wang et al., 2011) and that rods and cones have different sensitivities as measured by electroretinogram (ERG) responses to variations in experimentally-induced cellular glucose concentrations (Dawson et al., 2000). In a monkey model of retinal degeneration, blocking glycolysis with iodoacetate does not induce rapid cell death in cones as it does in rods, suggesting that cones depend on glycolysis to a lesser extent than rods (Noell, 1952b). Consistently, another study showing that diabetic patients have difficulty seeing in the dark suggests that rod function is affected early in the course of diabetes (Bailey and Sparrow, 2001). By contrast, the amplitude of the S cone ERG b-wave is significantly reduced in both non-retinopathic and retinopathic diabetic patients under poor hyperglycemia control (Yamamoto et al., 1996).
Both rods and cones produce reactive oxygen species (ROS) and are sensitive to compromised cellular homeostasis. Increased oxidative stress is associated with varied ocular pathogeneses, including retinitis pigmentosa (RP) (Campochiaro and Mir, 2018; Campochiaro et al., 2015). Under pathological conditions involving rod death leading to high oxygen tissue levels, progressive oxidative damage first affects cones and then other retinal cells. Thus, remaining cones experience an extra burden of production of superoxide radicals, which are powerful damaging agents. The accumulation of these free superoxide radicals overwhelms the cellular antioxidant system and triggers oxidative stress-induced cone cell death. This cell death fate decision occurs through production of peroxynitrites, which results from a reaction of superoxide with nitric oxide (Beckman et al., 1990). Another mode of cone cell death occurs via lack of glucose. Cone glucose uptake depends on rod-derived cone viability factor and insulin signaling (Leveillard, 2015), which may contribute to cone survival during rod photoreceptor degeneration. Altogether, the biochemical, structural, functional, and metabolic differences between rods and cones generate equilibrium in the coordinated work of photoreceptors in the retina. However, no biological system is flawless, and varying factors can result in dying photoreceptors and retinal diseases. Given the differences in light sensitivity and metabolic activity between rods and cones and the activation of cell death pathways as a result of light damage (Wang and Chen, 2014) or metabolic dysfunction (Punzo et al., 2009), it would be interesting to compare activation of the endoplasmic reticulum (ER) stress response of these two photoreceptors in animals expressing different mutant photoreceptor-specific proteins.
2. Retinal degenerative disorders
Many factors can affect vision loss, including inheritance, the environment, aging, and mechanical trauma. Inherited retinal degeneration (IRD) can affect rods (e.g., RP and rod-cone dystrophy) and cones (e.g., cone-rod dystrophy (CORD)), primarily due to mutations in rod- or cone-specific genes. In addition, mutations in genes particularly expressed in RPE cells, such as RPE65, can affect photoreceptors and lead to IRD (e.g., Leber’s congenital amaurosis (LCA)). Environmental factors and aging contribute to age-related macular degeneration (AMD). Physical trauma can cause retinal detachment, which can lead to retinal degeneration. However, cases of retinal detachment as a complication of retinoschisis, uveitis, degenerative myopia, or diabetic retinopathy are also known.
Genetics is a key factor leading to IRD (Birtel et al., 2018; Maeder et al., 2019; Mizobuchi et al., 2019; Tayebi et al., 2019; Tsang and Sharma, 2018; Wu et al., 2018), which is associated with irreversible vision loss and affects an estimated 1 in 2500 individuals (Abisambra et al., 2013). Different classes of genetic retinal dystrophies differ greatly in their etiology and age of onset. About 50% of all inherited human retinal diseases belong to a class of pigmentary retinopathies called RP (Daiger et al., 2013), which is broad heterogenic group of rare genetic disorders characterized by a progressive loss of vision leading to blindness due to degeneration of photoreceptors, primarily rods, in the retina. The prevalence of RP is approximately 1 in 4000 individuals worldwide. The name RP was selected because ophthalmoscopic observation shows an ischemic appearance of the neural retina, with a waxy, pale optic nerve head and brown or black pigmented ‘bone spicules’ in the periphery (Jones and Marc, 2005).
The most common form of RP is rod-cone dystrophy, in which initial night blindness is followed by progressive loss of the peripheral visual field. Cones are maintained for some time and die following rod cell death. Varying modes of RP inheritance include 50–60% autosomal recessive, 30–40% autosomal dominant, and 5–15% X-linked (Bunel et al., 2019). In addition to its mode of inheritance, RP is classified as syndromic or non-syndromic. Examples of syndromic RP are Usher syndrome, which is also associated with deafness, and Bardet Biedl syndrome, which is also associated with obesity and intellectual disability. Non-syndromic RP is more common, accounting for 70–80% of all RP cases (Pfeiffer et al., 2019).
In the clinic, RP patients are usually examined by ERG recordings. In the early stage of the disease, ERG amplitudes are affected more in rods than in cones. However, with RP progression, both rod and cone amplitudes become diminished. Photoreceptor degeneration continues until the retina completely lacks photoreceptors, leaving patients deficient in perceiving light for years to decades of life (Pfeiffer et al., 2019). In addition, RP patients can experience epiretinal membrane formation, atrophy of the RPE, posterior subcapsular cataracts, and cystoid macular edema (Musarella and Macdonald, 2011).
RP patients often become legally blind by their mid-40s. However, the onset and mechanism of photoreceptor deterioration varies widely across RP diseases. Thus, some patients experience vision loss early in life, whereas others may not show signs of retinal degeneration until adulthood (Hartong et al., 2006). As of today, over 60 genes are known to cause non-syndromic RP, and over 20 genes are associated with autosomal dominant RP. Mutations in rhodopsin genes are the most common cause of autosomal dominant RP and are present in 20–30% of cases. For example, the substitution of proline-to-histidine at position 23 (P23H) is the most common mutation found in the United States. Autosomal recessive RP is associated with mutations of at least 35 genes. Although mutations in six genes cause the X-linked form of RP, mutations in the RP GTPase regulator gene account for most cases.
Different from RP, CORD is characterized by deterioration of cones followed by a decline in rod function. Thus, daylight and color vision is affected before night vision in RP patients, whereas CORD often occurs in childhood and manifests as decreased sharpness of vision and increased light sensitivity followed by impaired color vision (i.e., dyschromatopsia), blind spots (i.e., scotomas) in the center of the visual field, and partial side (i.e., peripheral) vision loss (https://ghr.nlm.nih.gov/condition/cone-rod-dystrophy). CORD patients soon develop night blindness and experience a worsening of their peripheral vision, which can limit their independent mobility. A decrease in visual acuity makes reading incredibly difficult, and most affected individuals are legally blind by mid-adulthood. Mutations in at least 10 genes are associated with CORD, which are inherited in an autosomal dominant pattern. Mutations in guanylate cyclase 2D and cone-rod homeobox genes account for about half of these cases. Mutations in the ATP binding cassette subfamily A member 4 protein account for 30–60% of cases.
LCA was first described by Theodore Leber in 1869 (Perrault et al., 1999) and is the most common type of IRD resulting in neonatal blindness (Maeder et al., 2019; Miraldi Utz et al., 2018; Tsang and Sharma, 2018). Currently, there are 18 forms of LCA based on the affected gene (https://omim.org/entry/204000). One is the RPE-specific 65 kDa protein (RPE65), a key player in the visual cycle. RPE65 is responsible for the conversion of all-trans-retinal to 11-cis-retinol during phototransduction. Mutations in this gene affect photoreceptors and result in LCA2. However, mutations in photoreceptor-specific proteins also cause other forms of LCA. Currently, U.S. Food and Drug Administration (FDA)-approved Luxturna is an treatment option for patients with confirmed biallelic RPE65 mutation-associated retinal dystrophy, which can lead to vision loss (https://www.fda.gov/news-events/press-announcements/fda-approves-novel-gene-therapy-treat-patients-rare-form-inherited-vision-loss). LCA is typically inherited in an autosomal recessive pattern (Kumaran et al., 2017; Perrault et al., 1999), meaning that both copies of a gene must be mutant for a patient to present a significant phenotype. In the United States, LCA affects 1–3 per 100,000 newborns, representing ~2% of all cases of inherited retinal dystrophy (Kumaran et al., 2017).
LCA is typically diagnosed within the first few months of life when a parent or doctor notices a lack of visual responsiveness and aberrant eye movements, termed nystagmus (Kumaran et al., 2017). It is common for a fundus image, or clinical image of the retina, to appear normal in an infant with LCA (Weisschuh et al., 2018). However, ERG recordings assessing retinal response to light stimuli reveal that retinal neurons show little or no activity in response to flashes of light (Brecelj and Stirn-Kranjc, 1999; Koenekoop et al., 2002; Kumaran et al., 2017; Perrault et al., 1999; Weisschuh et al., 2018). LCA patients in early adolescence to adulthood typically show several signs and/or symptoms including nystagmus and deep-set eyes (Kumaran et al., 2017; Perrault et al., 1999; Weisschuh et al., 2018). Some LCA patients experience sensitivity to bright light, known as photophobia (Weisschuh et al., 2018). In addition, numerous changes to the retina become apparent in adolescence. In many cases, retinal blood vessels become attenuated, or more constricted, as occurs in RP patients(Ma et al., 2012). It is also common for the retinas of LCA patients to exhibit changes in pigmentation due to alterations in the RPE, which is just superficial to the retina (Kumaran et al., 2017; Weisschuh et al., 2018). Although various physical changes occur in the retinas of LCA patients, vision often remains stable into young adulthood for most patients (Kumaran et al., 2017; Perrault et al., 1999). Patients with LCA typically have visual acuity ranging from 20/200 to complete blindness (Kumaran et al., 2017; Weisschuh et al., 2018). Mutations in as many as 25 genes are known to cause LCA (Kumaran et al., 2017). LCA etiology depends on the mutated gene, as mutations in certain genes can result in differences in the severity of the phenotype. In North America and Europe, mutations in the gene encoding the centrosomal protein of 290 kDa (CEP290) account for 15–20% of cases, which is more than any other gene (Kumaran et al., 2017).
AMD predominantly affects elderly individuals over the age of 50 years. Today, the disease is a leading cause of vision loss worldwide, and its prevalence dramatically increases with aging. Additional risk factors for AMD are the environment, smoking, and nutrition. AMD exists in two forms: dry and wet. The dry form (80–85% of all cases) is associated with accumulation of drusen deposits, creating a distance between the RPE and photoreceptors and a shortening and loss of the photoreceptor OS. The mechanism of retinal pathogenesis is the loss of functional RPE cells, which results in photoreceptor cell death at late stages. Due to the metabolic and phagocytotic dependency of photoreceptors on RPE cells, the loss of retinal integrity results in AMD development. The wet form (15–20% of all cases) originates from neovascularization in the choroid, neural retina, and vitreous, during which leakage of the blood and serum damages ocular tissues. Both wet and dry forms eventually lead to blurriness of vision in the central retinal area if not treated. These symptoms often progress to vision loss that spreads to the periphery of the retina.
Retinal detachment can also induce photoreceptor degeneration. Clinically, patients with retinal detachment often experience photopsias and progressive loss of vision due to damaged photoreceptors and other serious impacts to the retina. Following detachment, OSs degenerate, which is accompanied by the death of retina ganglion cells and glial involvement (Sorensen et al., 2019). For example, Muller cells experience hypertrophy (Liu et al., 2018b) and changes in gene expression (Matsumoto et al., 2018). Together, these events can trigger rewiring and remodeling in degenerating retinas.
Recently, retinal remodeling was proposed to be a consequence of photoreceptor degeneration, with three common phases across different forms of retinal degeneration (Pfeiffer et al., 2019). In phase 1, degeneration triggered by stress begins to launch neural reprogramming and glial responses. In phase 2, the progressive loss of remaining photoreceptors, particularly cones, induces remodeling. This stage persists as long as remnant cones are viable. Phase 3 starts upon the complete loss of photoreceptors. At this point in degeneration, retinal disintegration is complete, leading to neurite outgrowth and widespread cell death (Pfeiffer et al., 2019). Despite progress toward understanding variations in retinal pathogenesis, the question of whether the molecular mechanisms contributing to photoreceptor cell death at the same phase of retinal degeneration are similar regardless of the etiology of retinal degeneration remains to be addressed.
3. Animal models of retinal degeneration
Transgenic, knock-out (KO), and knock-in animal models enable scientific investigation of the pathogenesis of photoreceptor degeneration in RP, CORD, LCA, AMD, and retinal detachment. Due to the focus of this review, we have highlighted animal models with retinal dystrophies mimicking RP and LCA in humans that have been used by our laboratory to unravel the molecular mechanisms of retinal degeneration; however, many more models of RP and LCA exist.
Rhodopsin is an essential 39 kDa protein for phototransduction. It belongs to a class of G protein-coupled receptors consisting of scotopsin and the covalently bound cofactor retinal. Embedded in the lipid bilayer of the disk’s membranes, rhodopsin forms three general domains: intracellular, transmembrane, and extracellular. Its structure consists of an extracellular N-terminal segment, seven transmembrane segments (TM1–7), and an intracellular C-terminus tail. Three segments link TM regions on the extracellular surface (E1–3), and another three segments link TM regions on the cytoplasmic surface (C1–3) in addition to a final cytoplasmic segment (C4). The rhodopsin protein (RHO) efficiently absorbs green-blue light and appears reddish-purple, giving it the name “visual pigment”.
Rhodopsin undergoes a few conformational changes, including Meta II state, which involves isomerization of retinol into its active all-trans-retinal conformation. Meta II activates the transducin protein, the α subunit of which activates cGMP PDE. In the “dark” state, cGMP directly activates cation channels, resulting in a net depolarization of rods. This depolarization leads to the release of glutamate, which hyperpolarizes some surrounding cells and depolarizes others. In the “light” state, the transducin α subunit activates PDE, which hydrolyses cGMP into GMP, decreasing cation channel activity and causing hyperpolarization of rods. This reduces the release of glutamate by rods, which results in the sensation of light. Notably, rods exhibit a significant extent of signal amplification. In vitro electrophysiological experiments with intact rod photoreceptors indicate that a single photon activates 16 G protein molecules in the mouse and ~60 in the frog (Arshavsky and Burns, 2014). This further results in hydrolysis of ~2000 and ~72 000 molecules of cGMP, respectively, representing the total degree of biochemical amplification of the phototransduction cascade in these species. The return of rods to a resting state occurs through RHO deactivation, closure of cGMP channels, inactivation of transduction, and a decrease in intracellular Ca2+, triggering intracellular proteins to activate guanylate cyclase and restore the level of cGMP. Given the pivotal role of RHO in vision signaling, it is worth mentioning that varied mutations in the rhodopsin gene correlate with congenital retinopathies like RP and congenital stationary night blindness (Toledo et al., 2011).
P23H RHO mutation exhibits characteristics of both types of dominant mutation: toxic gain-of-function and dominant negative (Mendes and Cheetham, 2008; Wilson and Wensel, 2003). Currently, a few animal models express aberrant P23H RHO, including transgenic mice, rats, and frogs as well as knock-in mice. P23H transgenic rats express a mouse RHO transgene with a defective N-terminal domain. These rats were developed to manifest different rates of retinal degeneration, from relatively slow-progressing (line 3) to moderate (line 1) retinal degeneration (Bryda and LaVail, 2019). We previously investigated the molecular mechanism of photoreceptor deterioration in P23H line 3 rats (Gorbatyuk et al., 2012a; Gorbatyuk et al., 2010; Sizova et al., 2014). At postnatal day (P)30, these rats show only a 20% reduction in outer nuclear layer thickness as compared with wild-type (i.e., albino background) rats, whereas P23H line 1 rats show a 50% reduction. In these rats, scotopic ERG responses generally follow degeneration curves that closely mimic those of outer nuclear layer thickness degeneration (https://view.officeapps.live.com/op/view.aspx?src=http%3A%2F%2Fophthalmology.ucsf.edu%2Fwp-content%2Fuploads%2FLaVail-RD-Rat-Model-Resource-063011.docx).
Another model is P23H transgenic mice, which express a human P23H RHO transgene that leads to relatively slow retinal degeneration. At P30, these mice already show a marked reduction in scotopic ERG amplitudes and thinning of the outer nuclear layer, which further proceeds with age (Mao et al., 2012). At P90, these mice show a 30% loss of photoreceptors accompanied by diminishing scotopic ERG responses (Chiang et al., 2016).
P23H RHO knock-in mice are proposed to more closely mimic the genetic background and pathological progression of human autosomal dominant RP with P23H opsin mutation (Sakami et al., 2014), as they have photoreceptors that undergo rapid degeneration after P14. In this model, P23H RHO is improperly glycosylated and expressed at a level 1–10% of that of wild-type opsin (Sakami et al., 2011). Moreover, expression of P23H RHO disrupts rod disk formation, resulting in greater functional deficits in rods than in cones. At P40, these mice show a ~50% decline in rod photoreceptor function measured by scotopic ERG responses as compared with wild-type mice. In addition, these mice exhibit a strong intraretinal gradient of degeneration, with a greater degree of degeneration in the inferior retina, similar to that observed in humans (Olsson et al., 1992; Sakami et al., 2011). By contrast, P23H RHO transgenic rats show greater involvement of the superior retina (LaVail et al., 2000).
Another animal model expressing aberrant rhodopsin is T17M RHO mice. The T17M (tyrosine-to-methionine at position 17) mutation is located close to the rhodopsin glycosylation site (N2–15) on the N-terminal and likely affects protein folding due to lack of glycosylation (Murray et al., 2015). These mice develop severe retinopathy (Choudhury et al., 2013; Choudhury et al., 2014; Kunte et al., 2012; Nashine et al., 2013). At P30, scotopic ERG amplitudes are diminished by over 60%, and this decline in retinal function is tightly associated with photoreceptor loss (Nashine et al., 2013). Interestingly, photopic ERG amplitudes are also reduced at P30, resulting in massive cone loss at P90 (Rana et al., 2017). These findings indicate that these mice exhibit rapid retinal degeneration, which makes rescue of deficits challenging. Moreover, T17M RHO mouse retinal function and morphology change dramatically after light exposure (Krebs et al., 2009); 3 hours of illumination results in 50% recovery of scotopic ERG amplitudes, whereas up to 6 hours of illumination results in normal photopic responses. Interestingly, extensive light exposure can damage photoreceptor cells via activation of the ER stress response (Nakanishi et al., 2013). Therefore, it not surprising that cell death in these mice peaks 1 day after exposure, and outer nuclear layer thickness declines between 1 and 5 days (Krebs et al., 2009).
rd1 and rd10 mice harbor different mutations in PDEβ subunits and transmit retinal degeneration through an autosomal recessive mode. In rd1 mice, the affected allele in the Pde6b gene carries a nonsense mutation resulting from viral insertion in intron 1 or nonsense mutation in exon 7 C→A transversion in codon 347 (Dong et al., 2017). These mice show early onset of retinal degeneration; the outer retina starts deteriorating at P8, which rapidly proceeds to the complete loss of rods by P36 (Carter-Dawson et al., 1978). However, cones degenerate at a slower pace in these mice. At P17, ~75% of cones exist in the retina, whereas at 18 months of age, 1.5% of cones are still present. Photoreceptor cell death occurs through via activation of apoptosis-inducing factor, calpains, and caspases and elevation of Ca2+ concentration (Johnson et al., 2005; Sanges et al., 2006). In rd10 mice, spontaneous mutation in the PDE gene also leads to development of autosomal recessive RP. Retinal degeneration in these mice starts at P18 and peaks around P25 with significant rod degeneration, although cones also eventually deteriorate. At P30, these mice still show light responses as assessed by ERG mimicking the typical progression of RP in humans. The overall changes observed in these mice are similar to those found in rd1 mice except that decay in retinal integrity and function occurs at a slower pace.
An animal model of rapid retinal degeneration is BXD24/TyJ-CEP290rd16/J (rd16) mice, which have a spontaneous mutation in Cep290 that was discovered in the BXD24 inbred strain at Jackson Laboratories (Chang et al., 2006). Since their discovery, rd16 mice have been well characterized (Chang et al., 2006; Craige et al., 2010; Drivas et al., 2013; Mookherjee et al., 2018; Rachel et al., 2015; Starr et al., 2018; Subramanian et al., 2014; Wu et al., 2017; Zhang et al., 2018). Chang et al. discovered that 897 base pairs corresponding to exons 35–39 of the Cep290 gene are deleted in rd16 mice (Chang et al., 2006). This deletion corresponds to amino acids 1599–1897 that contain region M, the microtubule binding region or myosin tail homology domain (Chang et al., 2006; Mookherjee et al., 2018; Rachel et al., 2015). rd16 mice display rapid degeneration of the retina from as early as P18, but the gross morphology of kidneys and the brain, which are known to be susceptible to CEP290-associated ciliopathies, surprisingly remain unaffected (Chang et al., 2006; Subramanian et al., 2014). This suggests that the region deleted in Cep290 encodes a domain that plays a specific role in retinal photoreceptors (Chang et al., 2006). Although rods degenerate rapidly in rd16 mice as demonstrated by a low but detectable scotopic ERG response (Chang et al., 2006; Mookherjee et al., 2018; Murga-Zamalloa et al., 2011; Subramanian et al., 2014; Zhang et al., 2018), cones are still present in the retina until at least P60 despite a near complete loss of photopic response at P18 (Chang et al., 2006). The remainder of the retina appears to be mainly unaffected, with no loss of rows in the inner nuclear layer or ganglion cell layer until at least P60 (Chang et al., 2006). Photoreceptors in rd16 mice display uncharacteristic ciliogenesis and form a shorter than normal OS (Chang et al., 2006; Rachel et al., 2015), different from CEP290 KO mice that do not form an OS (Rachel et al., 2015). In these mice, the 9+0 microtubule doublet of the axoneme of cilia appears relatively normal, but the microtubule ring has an irregular shape (Rachel et al., 2015). As expected, multiple proteins, including those that typically reside in the transition zone or OS, are mislocalized to the photoreceptor inner segment (Chang et al., 2006; Mookherjee et al., 2018; Murga-Zamalloa et al., 2011; Subramanian et al., 2014). For example, RHO and arrestin are mislocalized to the inner segment of rd16 mouse photoreceptors (Chang et al., 2006). Thus, we found that the level of RHO protein is significantly diminished (Starr et al., 2019) and mislocalized in rd16 mice, consistent with a lower level of total protein synthesis in photoreceptors (Starr et al., 2018). A few ciliary proteins that interact with CEP290 show aberrant interactions in rd16 mice, indicating that CEP290 is at least partially responsible for the localization of proteins to the connecting cilium (Chang et al., 2006; Murga-Zamalloa et al., 2011; Subramanian et al., 2014). However, the exact role of CEP290 in trafficking is not clear, and whether OS proteins are mislocalized in rd16 mouse photoreceptors due to improper transport or the lack of a proper OS remains elusive.
4. Molecular mechanisms of retinal degeneration
IRD has varied etiologies due to causative mutations in genes. Despite this fact, the molecular mechanisms promoting advanced-stage RP and photoreceptor cell death could be similar. In this regard, apoptosis has been proposed as a general pathway of photoreceptor cell death in retinal degeneration involving rod defects (Chang et al., 1993; Gregory and Bird, 1995; Reme et al., 1998). However, the causative role of apoptosis as a mechanism underlying all rod degenerative diseases has been questioned. For example, cells that normally die by apoptosis execute cell death by necrosis if the normal pathway is blocked (Kunchithapautham and Rohrer, 2007a) and that multiple rescue paradigms targeting all active cellular pathways should be considered until a common upstream initiator of a given photoreceptor dystrophy cannot be found (Kunchithapautham and Rohrer, 2007b). To demonstrate the connection between apoptosis and photoreceptor cell death in T17M RHO transgenic mice, our laboratory conducted an experiment in which we ablated the UPR mediator, C/EBP homologous protein (CHOP), also known for its pro-apoptotic properties in the mouse retina (Nashine et al., 2013). The results were surprising, as CHOP ablation did not rescue degenerating retinas. However, later two independent groups also came to the same conclusion; ablation of pro-apoptotic CHOP does not prevent loss of visual function or photoreceptor cell death in another IRD model, P23H RHO mice (Adekeye et al., 2014; Chiang et al., 2016). Altogether, these results suggest that photoreceptor cell death occurs through either the activation of multiple upstream pro-apoptotic pathways or another pro-death signaling pathway.
Alternatively, programmed necrosis (i.e., necroptosis) was proposed as mode of photoreceptor cell death when investigators observed that photoreceptors can execute necrosis in a programmed manner and that the immunogenic nature of this process favors suicide through a caspase-independent approach (Arango-Gonzalez et al., 2014; Murakami et al., 2012; Sato et al., 2013; Viringipurampeer et al., 2019). Since its discovery, necroptosis has become an attractive therapeutic target for retinal degenerative diseases. However, the study of necroptotic cell death has primarily focused on cones. Deficiency in receptor-interacting protein kinase 3 (RIP3), which contributes to inflammation by regulating programmed necrosis, preserves cones in rd10 mice despite their sustained rod degeneration (Murakami et al., 2012). This study also reports that necrotic mechanisms engaging RIP3 may be crucial to cone cell death in IRD, implicating this molecule as a potential target for treating central and peripheral vision loss in patients with RP. Another study in a pde6cw59 mutant zebrafish model of RP reveals that the underlying mechanism of cone cell death occurs through necroptosis, whereas rods degenerate via a caspase-dependent mechanism (Viringipurampeer et al., 2014). Thus, knocking down RIP3 in pde6cw59 mutant zebrafish rescues dying cones by preventing ROS generation, leading to improved cone adaptation and upregulation of PDE6A and PDE6B expression. In this study, degenerating rods were not responsive to inhibition of RIP1/3 activity and showed positive immunostaining for caspase 3.
In further investigations of the RIP kinase pathway, mice lacking interphotoreceptor retinoid-binding protein were found to show a 3-fold elevation of RIP1 and RIP3 kinases and 10-fold elevation of TNFα receptor 1, a membrane death receptor that triggers both programmed apoptosis and necrosis (Sato et al., 2013). In this study, investigators found that overall TNF-RIP-mediated necrosis strongly contributes to photoreceptor cell death (Sato et al., 2013) and that pharmacological inhibition of RIP1 kinase significantly prevents both cone and rod cell death. Although the results of this study are generally consistent with previous findings, the proposed mode of rod cell death is inconsistent with the role of RIP kinases in rod deterioration and should be further investigated. Interestingly, our study with T17M RHO mice also shows a 2-fold elevation in TNF receptor-associated factor 2, which is a part of the RIP1 kinase complex that promotes apoptosis in the retina (Choudhury et al., 2013).
As cone degeneration usually follows mutation-induced rod cell death in RP, a few hypotheses concerning the specific pathways triggering cone cell death were recently proposed. One hypothesis is that dying rods secrete toxic substances transmitted through gap junctions, which serve as intercellular communication pathways between rods and cones (Smith et al., 1986), and thereby affects cone viability (Ripps, 2002). Migration of activated microglia and inflammation during retinal degeneration is another potential mechanism of the demise of cones in affected retinas (Peng et al., 2014). In one study, degenerating retinas in rd10 mice were treated with minocycline, a pharmacological inhibitor of microglia activation, showing that both rods and cones are responsive to microglia activation and survive via anti-inflammatory and anti-apoptotic mechanisms. However, given the nature of pharmacological treatment, it would be difficult to draw a firm conclusion about the major factor contributing to cone viability. Therefore, it remains to be determined whether cone survival is a result of rod survival or a direct benefit of treatment. Interestingly, in rd1 mice, microglia-regulated photoreceptor survival also occurs by necroptosis as well as activation of RIP1 and RIP3 kinase and the toll-like receptor 4 (TLR4) pathway. A study conducted in our laboratory with T17M RHO mice shows that deficits in TNFα, an activator of the TNF receptor-RIP kinase pathway, dramatically increases photopic ERG amplitudes and the number of surviving cones, suggesting a role of this signaling pathway in cone survival (Rana et al., 2017). However, as in the above-mentioned study in which microglia were pharmacologically inhibited (Peng et al., 2014), we could not conclude that cone viability was a direct result of TNFα deficiency. Therefore, future investigations should identify whether autocrine versus paracrine regulation of TNFα expression affects cone survival in TNFα-deficient T17M RHO mice.
Another factor affecting cone survival is rod-derived cone viability factor, the level of which dramatically reduces upon the loss of rods (Rana et al., 2017). Cone survival is also affected by oxidative stress, which leads to modifications in DNA, proteins, and lipids, as well as exposure to high oxygen content due to a loss of rods (Campochiaro and Mir, 2018). Together, however, the existing literature reveals a gap in our knowledge of how rods and cones die during retinal degeneration and whether the major mode of cell death can switch based on the duration of the disease and its causative factors. Many studies with various animal models of retinal degeneration propose multiple signaling pathways triggering apoptosis. The list includes, but is not limited to, oxidative stress, aberrant autophagy, Ca2+ dysfunction, the inflammatory response, and the ER stress response. Although these processes could be involved in degenerating retinas by themselves, results from our laboratory suggest that activation of these processes could result from sustained activation of the ER stress response or unfolded protein response (UPR), which is the focus of the current review.
5. ER and UPR
5.1. ER
The ER is a eukaryotic organelle composed of a complex membranous network of branching tubules and flattened sacs. The ER has multiple functions including protein maturation, sorting and degradation, and lipid biosynthesis (Aviram and Schuldiner, 2017; Schwarz and Blower, 2016). The ER is also involved in carbohydrate metabolism due to the fact that glucose-6 phosphatase is an ER membrane protein (Burchell et al., 1994). Furthermore, the ER controls Ca2+ homeostasis in cells by buffering free Ca2+ (Wakai and Fissore, 2013).
In eukaryotic cells, the largest targeting destinations for nascent proteins include the ER, mitochondria, nucleus, lysosomes, and peroxisomes. Nearly all proteins destined for the ER, Golgi apparatus, cell membrane, or lysosomes are synthesized on the ER membrane and transported inside the ER through a process known as co-translational ER translocation (Aviram and Schuldiner, 2017). Inside the ER, proteins undergo various post-translational modifications that aid their folding into three-dimensional, or tertiary, structures (Shrimal et al., 2015). Through their catalytic activity, glycosylating enzymes (Bause and Lehle, 1979) and oxidoreductases (Noiva et al., 1991) help immature proteins achieve proper folding. Also, inside the ER are protein chaperones that stabilize proteins while they correctly fold (Garcia-Huerta et al., 2016; Sun et al., 2019). The ionic environment in the ER is optimally suited for chaperones and enzymes participating in protein folding (Garcia-Huerta et al., 2016). In fact, the ER is the primary site of Ca2+ storage in cells, with a Ca2+ concentration 1,000–10,000 times higher than that in cytosol (Bollimuntha et al., 2017; Bygrave and Benedetti, 1996; Glaser et al., 2018; Samtleben et al., 2013) depending on the measurement method and cell type (Bygrave and Benedetti, 1996). After proteins correctly fold, they are trafficked to their destination (Liu et al., 2018a). The ER has a complex surveillance system known as the UPR that detects misfolded proteins, and any terminally misfolded proteins are trafficked out of the ER to be degraded through a process called ER-associated degradation (ERAD) (McCracken and Brodsky, 1996; Wu and Rapoport, 2018).
5.2. UPR
The UPR is a series of signaling cascades originating in the ER that are initiated in response to ER stress (Kohno et al., 1993; Partaledis and Berlin, 1993; Robinson et al., 1993). The UPR can become active following insults to the ER, many of which result in the misfolding and retention of proteins in the ER. For example, stresses such as glucose deprivation (Doerrler and Lehrman, 1999) and Ca2+ irregularities (Hojmann Larsen et al., 2001; Li et al., 1997) are well-described triggers of ER stress signaling (Wang and Kaufman, 2016). Glucose deprivation or fluctuating Ca2+ concentrations can result in the accumulation of misfolded proteins in the ER due to the vital role of these components in protein folding. Glucose is essential for protein glycosylation; without it, proteins cannot properly fold or exit the ER (Doerrler and Lehrman, 1999). Ca2+ is found in the catalytic site of many ER enzymes that assist in protein folding (Appenzeller-Herzog and Simmen, 2016). A reduction in the protein folding capacity of the ER results in the accumulation of misfolded proteins and activation of the ER stress response. In addition, genetic predispositions, such as those involved in IRD, can also result in activation of the UPR (Athanasiou et al., 2017b; Bhootada et al., 2015; Bhootada et al., 2016; DeLuca et al., 2016; Rana et al., 2014).
Following an insult to the ER, the UPR initiates the activation of three ER transmembrane enzymes: inositol-requiring enzyme 1 (IRE1), activating transcription factor 6 (ATF6), and protein kinase R-like endoplasmic reticulum kinase (PERK) (Figure 1). The widely accepted model for UPR signaling initiation involves a dynamic interaction between glucose-regulated protein 78 (GRP78), an ER-resident protein chaperone, and the three ER enzymes (Oikawa et al., 2009; Pobre et al., 2019; Walter and Ron, 2011; Wang and Kaufman, 2016). In unstressed conditions, GRP78, also known as binding immunoglobulin protein, physically interacts with the luminal domains of the three UPR enzymes, keeping them in an inactive state (Bertolotti et al., 2000; Oikawa et al., 2009). Upon ER stress, misfolded proteins accumulate in the ER lumen, and as GRP78 has a higher affinity for misfolded proteins than for enzymes, it dissociates from PERK, ATF6, and IRE1 and associates with the misfolded proteins. Now unbound, the three UPR enzymes are free to activate the ER stress response by triggering PERK, ATF4, and the IRE1 UPR arms (Bertolotti et al., 2000) (Figure 1).
Figure 1.
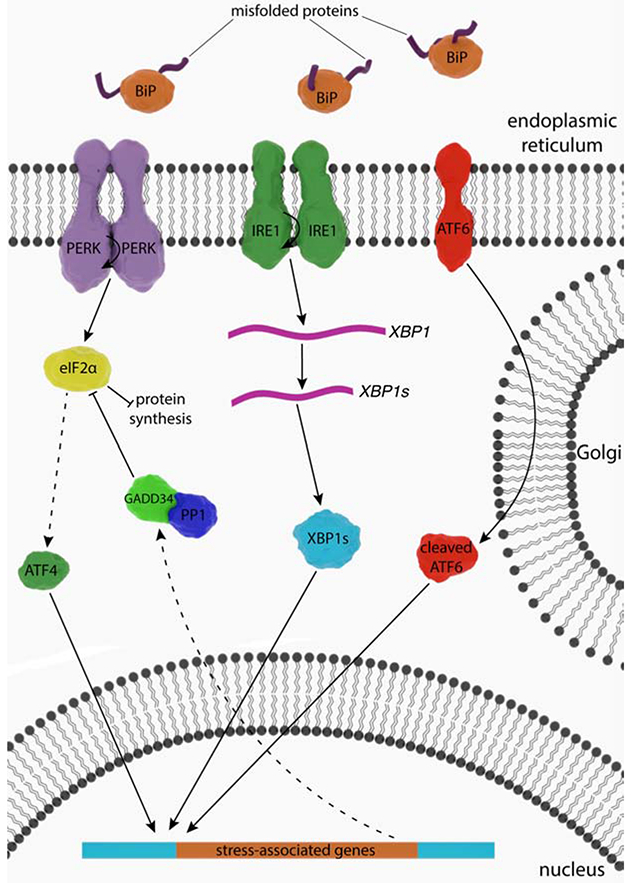
The UPR. Schematic depicting signaling through the three ER transmembrane proteins—PERK, ATF6 and IRE—during ER stress. BiP dissociation results in activation of all three membrane proteins. PERK attenuates protein synthesis through phosphorylation of eIF2α. IRE1 and ATF6 proteins activate specialized transcriptional programs to help alleviate ER stress. IRE1 phosphorylation results in unconventional splicing of XBP1 mRNA, which promotes transcription of stress-induced genes.
5.2.1. ATF6
When GRP78 dissociates from ATF6, the ~90-kDa ATF6 travels to the Golgi body, where it is cleaved by site 1 and 2 proteases (Haze et al., 1999; Shen et al., 2002; Shen et al., 2005). This leaves a soluble ~50-kDa transcription factor that migrates to the nucleus (Haze et al., 1999) and promotes the transcription of dozens of genes (Walter and Ron, 2011; Wang and Kaufman, 2016). Most proteins whose genes are targeted following ATF6 cleavage are protein chaperones that assist in protein folding, proteins involved in Ca2+ homeostasis, or proteins involved in ERAD (Wang and Kaufman, 2016; Yoshida et al., 2000). A recent study shows that ATF6 plays a key role in homeostatic adaptation to ER stress, whereas IRE1α and PERK are expendable in this process (Vitale et al., 2019). Another study reports that mutations in ATF6 lead to achromatopsia, an autosomal recessive disorder characterized by cone photoreceptor dysfunction (Chiang et al., 2017).
5.2.2. IRE1
Following the departure of GRP78 from the luminal domain of IRE1 unbound, IRE1 enzymes dimerize, forming a catalytically active homodimer that becomes active by autophosphorylation (Ali et al., 2011; Credle et al., 2005). The cytosolic portion of IRE1 has two catalytic domains: a kinase and an endoribonuclease (Ali et al., 2011; Tirasophon et al., 2000; Yoshida et al., 2001). Following ER stress, an activated IRE1 dimer alternatively splices RNAs via its endoribonuclease activity (Walter and Ron, 2011), such as that encoding XBP1 (Yoshida et al., 2001). Interestingly, transcription of XBP1 is promoted by cleaved ATF6 (Walter and Ron, 2011), demonstrating crosstalk between these two arms of the UPR. When translated, spliced XBP1 is a highly active transcription factor for genes encoding chaperones and proteins involved in ERAD (Walter and Ron, 2011; Yoshida et al., 2001). IRE1 also has the ability to phosphorylate targets such as JNK, which can activate MAPK signaling (Urano et al., 2000). A recent study shows that IRE1α activity is repressed by the binding of endogenous BiP to the lumen domain of IRE1α and promotes IRE1α monomerization (Amin-Wetzel et al., 2019). Furthermore, enforced loading of BiP on IRE1 results in repression of the UPR in CHO cells.
5.2.3. PERK
Similar to IRE1 activation, PERK is activated in response to dissociation of GRP78 and dimerizes to form an active homodimer (Cui et al., 2011; Liu et al., 2000). PERK is a ~125 kDa kinase with multiple domains (Carrara et al., 2015; Cui et al., 2011; Walter and Ron, 2011; Wang et al., 2018) and a structure similar to that of other two-lobed membrane kinases. The luminal domain of PERK consists of its smaller N-terminal lobe (Carrara et al., 2015; Wang et al., 2018), whereas a larger C-terminal lobe constitutes its cytosolic domain (Cui et al., 2011) and contains its catalytic site (Cui et al., 2011). Together with IRE1, PERK switches BiP from its chaperone cycle into an ER stress sensor cycle by inhibiting ATPase stimulation upon the binding of BiP to the luminal domains of UPR mediators (Kopp et al., 2019).
Recent research has focused on understanding the role of PERK in the induction of ER stress. One study shows that mammalian PERK interacts with misfolded proteins at a region of its luminal domain, meaning that upon induction of ER stress, PERK itself may act as a chaperone that binds exposed hydrophobic regions of misfolded polypeptides and initiates its dimerization and subsequent activation (Wang et al., 2018), which directly argues against canonical UPR activation. Furthermore, this study provides evidence that PERK activation is independent of its interaction with GRP78, as cells expressing PERK lacking a luminal domain do not exhibit higher than normal PERK signaling but are still susceptible to induction of ER stress following treatment with UPR-inducing compounds.
Following dimerization, PERK becomes activated by phosphorylation at Thr982 through a process known as trans-autophosphorylation, meaning each PERK monomer phosphorylates the other (Su et al., 2008; Walter and Ron, 2011). PERK is also known as eukaryotic translation initiation factor 2-alpha kinase 3 (EIF2AK3), named after its first described and primary role of interacting with and phosphorylating the α subunit of the eukaryotic translation initiation factor 2 (eIF2α) complex (Shi et al., 1998; Taniuchi et al., 2016). PERK is also required for the formation of ER-plasma membrane associations, which is important for regulating Ca2+ stores in the ER (van Vliet et al., 2017). As the name EIF2AK3 suggests, PERK is a kinase that phosphorylates eIF2α (Taniuchi et al., 2016). To fully appreciate the impact of eIF2α phosphorylation on cells, we must first understand the role of the protein under normal conditions. The following section discusses the role of eIF2 in normal and stressed conditions.
5.2.4. eIF2 and the integrated stress response
eIF2 is a heterotrimeric translation factor important for the initiation of protein synthesis in eukaryotes. The eIF2 complex consists of three subunits—α, β, and γ—each of which has distinct roles (Alone and Dever, 2006; Barrieux and Rosenfeld, 1977, 1978). The role of eIF2 in translation initiation involves the ternary complex (TC), which consists of eIF2, Met-tRNA, and guanosine tri-phosphate (GTP). In forming the TC, the initiator Met-tRNA is brought to a start codon on an mRNA in a process that depends on GTP being bound to the eIF2γ subunit (Barrieux and Rosenfeld, 1977, 1978; Hinnebusch, 2017; Hinnebusch and Lorsch, 2012; Sonenberg and Hinnebusch, 2009) (Figure 2). The TC joins the 40S ribosomal subunit with other initiation factors, and the pre-initiation complex (PIC) scans the mRNA until a start codon is located. When the start codon binds the anti-codon on Met-tRNA, GTP is hydrolyzed by eIF2, and the TC, which now contains guanosine diphosphate (GDP), exits the PIC and must be reactivated to begin another round of initiation (Hinnebusch, 2017; Hinnebusch and Lorsch, 2012). GDP must be exchanged by a nucleotide exchange factor for eIF2, with EIF2B being a guanine exchange factor that binds eIF2β to exchange GTP for GDP on eIF2γ (Konieczny and Safer, 1983). When this exchange occurs, eIF2 is free to participate in translation initiation (Hinnebusch, 2017).
Figure 2.
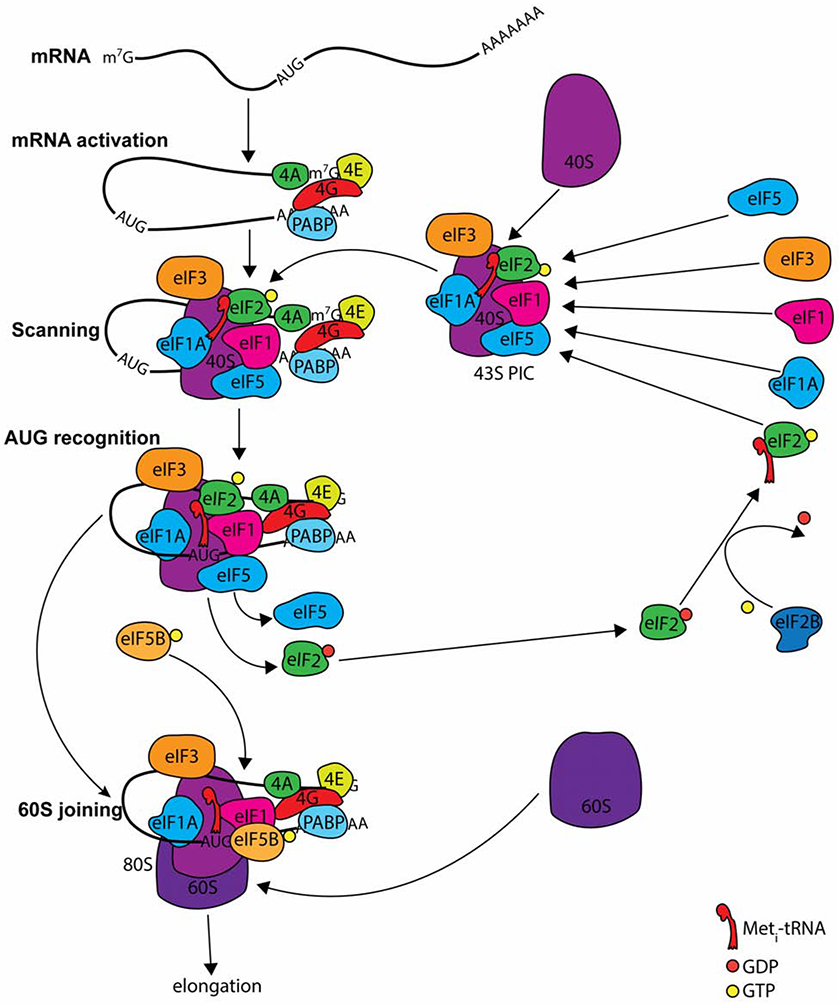
Depiction of translation initiation. Many factors are involved in protein synthesis regulation. Step 1: Formation of the 43S PIC when Met-tRNAiMet is delivered by eIF2 to the P site of the 40S ribosomal subunit. Step 2: Recruitment of the 43S complex to the 5' end of the mRNA by eIF3 and eIF4 factors. mRNAs are activated by the eIF4F complex (4E, 4G, and 4A) and Poly A binding protein (PABP). Multiple factors (eIFs 1, 1A, 2, 3, 5) join the 40S ribosome to generate the 43S PIC. eIF2 brings the initiator Met-tRNA to the complex. Step 3: Scanning of the 5' UTR and recognition of the AUG codon. The complex scans while eIF1 and 1A keep the complex inactive. When the AUG is found, GTP on eIF2 is hydrolyzed in a process mediated by eIF5, and eIF2-GDP leaves the complex. eIF2-GDP must be reactivated to eIF2-GTP by eIF2B for eIF2 to begin another round of initiation. Step 4: Assembly of the 80S ribosome. eIF5B promotes joining of the 60S ribosome to form the first peptidyl bond, which is followed by elongation. Adapted from Hinnebusch AG, Trends in Biochemical Sciences, 2017.
Numerous cellular stresses can increase eIF2α phosphorylation (Taniuchi et al., 2016). In addition to PERK, three kinases (i.e., heme-regulated kinase, protein kinase R, and general control nonderepressible 2) in vertebrates phosphorylate eIF2α at serine 51 in response to stress (Taniuchi et al., 2016). eIF2α phosphorylation prevents canonical translation initiation resulting from failed guanine nucleotide exchange between eIF2 and EIF2B (Bogorad et al., 2018). This is because EIF2B has a higher affinity for phosphorylated eIF2α than it does for eIF2β; therefore, what is typically a transient complex between EIF2B and eIF2β becomes a stable complex of EIF2B and phosphorylated eIF2α. This is thought to result in the entrapment of EIF2B, the rate-limiting component of TC formation, which prevents canonical translation initiation (Bogorad et al., 2017). When general translation comes to a halt, synthesis of most proteins is inhibited; however, translation of some stress-related mRNAs are upregulated under these conditions. One of these mRNAs encodes ATF4. Interestingly, translation of ATF4 mRNA occurs in the presence of an active TC (Ross et al., 2018), arguing against central model ATF4 activation.
5.2.5. ATF4
Exactly how ATF4 mRNA escapes canonical regulation is unknown but involves the existence of upstream open-reading frames (uORFs) in ATF4. The general structure of mRNAs with uORFs consists of one or more regions upstream from the primary ORF region of the mRNA that is converted to a stable protein (Kearse and Wilusz, 2017). Although a type of mRNA modification, N6-methyladenosine, regulates ATF4 mRNA (Zhou et al., 2018), how this modification modulates translation has not been determined, leaving the exact mechanism of stress-induced upregulation of ATF4 unknown.
ATF4 promotes transcription of several genes involved in various cellular processes ranging from development to apoptosis (Bhootada et al., 2016; Fusakio et al., 2016; Pitale et al., 2017; Zhao et al., 2015b). The most studied targets of ATF4 are those that are promoted after cellular stressors that result in elevated phosphorylated eIF2α levels (Pitale et al., 2017; Taniuchi et al., 2016). Together with the phosphorylation of eIF2α and the subsequent halt in translation, this alternative translational program is known as the integrative stress response (ISR) (Taniuchi et al., 2016). Growth arrest and DNA damage-inducible 34 (GADD34), CHOP, and tribbles homolog 3 (TRB3) are a few examples of proteins whose genes are targeted by ATF4 during stress (Fusakio et al., 2016; Gorbatyuk and Gorbatyuk, 2013). Interestingly, ATF4 regulates the transcription of 254 downstream genes, one of which is another molecule involved in 5’ cap dependent translation—eukaryotic translation initiation factor 4E-binding protein 1, a translational repressor (Han et al., 2013). Studies in our laboratory with T17M RHO (Bhootada et al., 2016) and rd16 mice deficient in ATF4 show that reductions in ATF4 significantly retard retinal degeneration (Figure 3). For example, ATF4 deficiency in rd16 retinas delays photoreceptor cell death.
Figure 3.
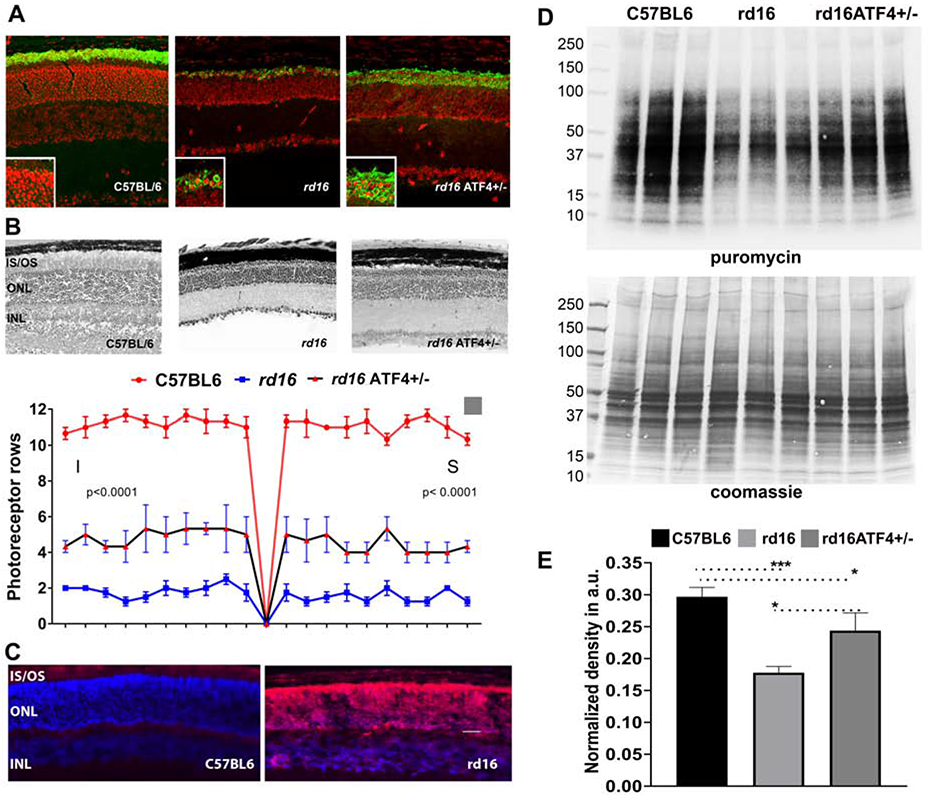
Molecular mechanisms of retinopathy in rd16 mice. Immunohistochemical analysis of C57BL6, rd16, and rd16 ATF4+/− retinas show mislocalization of rhodopsin in rd16 retinas at P18 (unpublished data). Detection of rhodopsin (green) was performed using primary anti-rhodopsin antibody ID4. Propidium iodide was used to stain retinal cell nuclei (red) (A). Delay of photoreceptor cell death in rd16 ATF4+/− retinas. H&E-stained retina sections are shown in the upper panel, and counts of photoreceptor rows are shown in the bottom panel (B). P15 rd16 retinas show significant accumulation of adenylyl cyclase (red) in the ONL. Based on the mechanism proposed by Nakao et al., this mislocalization could contribute to the retinal pathophysiology of rd16 mice (C). Study of the molecular mechanisms responsible for the delay of retinal degeneration in rd16 ATF4+/− retinas demonstrates enhanced synthesis of proteins in general (D) and rhodopsin in particular (A). Protein synthesis was assessed by the SUnSET method (Starr et al., 2019). C57BL6, rd16, and rd16 ATF4+/− mice were intraperitoneally injected with puromycin at a dosage of 0.04 μmol/g body mass. After 30 min of puromycin injection, retinas were harvested. Retinal proteins with incorporated puromycin were extracted in RIPA buffer, and their concentrations were estimated. Protein (60 μg) was separated by SDS-PAGE and transferred to a PVDF membrane, which was then probed with anti-puromycin antibody. A secondary antibody specific for IgG2a was also applied. As a loading control, membranes were stained with Coomassie blue after washing with distilled water (D). The relative densities of entire lanes were measured using ImageJ software. Puromycin densities were normalized to their corresponding Coomassie densities (E).
5.2.6. GADD34
GADD34, first identified as a pro-apoptotic member of the GADD family of proteins that suppress cell growth and division (Zhan et al., 1994), is best known for its role as a phosphatase regulatory subunit responsible for initiating the dephosphorylation of eIF2α by bringing protein phosphatase 1 (PP1) to eIF2α and thereby promoting translational recovery (Harding et al., 2009; Kojima et al., 2003; Novoa et al., 2001). To this end, GADD34 provides the ISR with a feedback mechanism that restores translational homeostasis. Although ATF4 acts on the GADD34 promoter to restore protein synthesis rates (Ma and Hendershot, 2003), c-Jun can also upregulate transcription of GADD34 under DNA damage (Haneda et al., 2004) and proteotoxic stress associated with Alzheimer’s disease (AD) (Xu et al., 2015). It is well known that GADD34 levels correlate with apoptosis in multiple cell types (Farook et al., 2013; Gu et al., 2014; Honjo et al., 2015; Kang et al., 2019; Zhan et al., 1994). For example, Farook et al. show that the pro-apoptotic role of GADD34 could stem from its inhibition of AKT phosphorylation (Farook et al., 2013). Recently, our laboratory demonstrated that Gadd34 ablation in rd16 mouse retinas reduces apoptosis and elevates phosphorylated AKT, suggesting that GADD34 has a powerful impact on cell fate decision (Starr and Gorbatyuk, 2019). We proposed that Gadd34 ablation could selectively regulate translation of anti-apoptotic proteins in degenerating retinas, which could be due to a 5′-cap independent or internal ribosome entry site (IRES) mechanism (Starr and Gorbatyuk, 2019).
In addition, GADD34 interacts with TRAF6 and inhibits TRAF6-mediated polyubiquitination of AKT, a process that primes AKT for subsequent phosphorylation. Also, GADD34 interacts with and initiates PP1-mediated dephosphorylation of SMAD7 (Shi et al., 2004). However, GADD34 is proposed to play a pro-survival role apart from its contribution to restoring protein synthesis. For example, GADD34 is important for the integrity of the Golgi apparatus, as its deficiency causes Golgi bodies to become fractionated in hyperosmotic corneal cells (Krokowski et al., 2017). Golgi fractionation also lessens the membrane translocation of receptors in these cells, suggesting that the role of GADD34 may be more complicated than previously thought. In fact, GADD34/PP1 alters the metabolic states of cells by acting on hundreds of proteins (Dedigama-Arachchige et al., 2018). Therefore, recent research is allowing us to grasp the full breadth of roles played by GADD34. For example, studies from our laboratory indicate that the role of GADD34 in the retinal degeneration of rd16 mice is multifaceted and that in addition to regulating gliosis and apoptosis in degenerating retinas, GADD34 controls apoptotic photoreceptor cell death (Starr and Gorbatyuk, 2019).
5.2.7. CHOP and TRB3
CHOP (also known as GADD153) is a pro-apoptotic molecule that promotes the transcription of several stress-related genes in response to stress (Bartlett et al., 1992; Ron and Habener, 1992; Wang et al., 1996). By forming heterodimers with ATF4, some transcription factors whose genes are ATF4 targets can alter the activity of ATF4. For example, CHOP forms heterodimers with ATF4 (Talukder et al., 2002) and thus could mediate the transcription of tribbles homolog 3 protein (TRB3) (Ohoka et al., 2005). In turn, the activity of ATF4 can be regulated by TRB3 (Jousse et al., 2007). In addition, the migration of CEP290 to the nucleus elevates ATF4 activity (Sayer et al., 2006). The role of TRB3 in the progression of RP is currently under investigation in our laboratory.
5.3. ERAD
Approximately 30% of newly synthesized proteins in eukaryotes undergo degradation within minutes of formation, likely due to unsuccessful folding or failure in multimer assembly (Greene and McElvaney, 2010). After multiple unsuccessful attempts to assist proteins in folding, they are redirected for degradation. Thus, ERAD regulates protein levels in response to cellular demands. This quality control process generally consists of four distinct steps: substrate recognition, retro-translocation across the ER membrane, substrate polyubiquitination, and proteasomal degradation (Dupzyk and Tsai, 2016). ER-resident chaperones recognize misfolded proteins and target them to re-translocation machinery. These misfolded proteins are identified by improper glycosylation patterns or inaccurate disulfide bonds that can result in aberrant folding due to exposure of hydrophobic spots or oligomer formation. Recently, we demonstrated that expression of non-glycosylated rhodopsin in the retina leads to a retinal degenerative disorder involving OS malformation, similar to that observed in mice with the human T17M RHO transgene (Murray et al., 2015).
A few ER-resident enzymes, such as protein disulfide isomerase, BiP/GRP78/HSPA5, ER class I mannosidase, ER degradation-enhancing mannosidase-like 1/3, and osteosarcoma amplified 9/XTP3, carry out the initial step of the ERAD pathway. In subsequent steps, identified proteins translocate from the ER into the cytosol. The ER transmembrane proteins Hrd1, sel-1 homolog 1, and Derlins assist in this process. For example, sel-1 homolog 1 binds a misfolded protein and passes it to Hrd1, which prepares the targeted protein for transport across the ER membrane. Hrd1, a self-regulating protein, is capable of translocating targeted proteins through both its structural properties and autoubiquitination function. Hrd1 contains a catalytic really interesting new gene (RING)-finger domain facing the cytosol. In the presence of the ubiquitin-conjugating enzyme UBC7, this domain exhibits in vitro ubiquitination activity for Lys48-specific polyubiquitin linkage (Kikkert et al., 2004). Polyubiquitination of Hrd1 prevents sliding a client protein back to the ER lumen, thereby providing release into the cytosol. Thus, in the next step, this process is catalyzed by cytosolic chaperones, and polyubiquitinated substrates are directed to the proteasome for degradation. Delivery to the proteasome is provided by a recently discovered holdase complex that consists of Bcl2-associated athanogene 6, ubiquitin-like protein 4A, transmembrane domain recognition complex 35, and small glutamine-rich tetratricopeptide repeat-containing protein alpha, which prevents the aggregation of misfolded proteins in the cytosol.
Recent studies have examined the role of ERAD in general and Hrd1 in particular in retinal degeneration, including the role of members of the ER membrane complex that physically interact with ERAD components (e.g., UbAC2, Derlin1/2) and thus influence ERAD (Christianson et al., 2011). A point mutation in the ER membrane complex EMC1 gene has been proposed to cause retinal dystrophy in humans (Abu-Safieh et al., 2013). However, it is worth mentioning that, apart from its hypothetical role in human retinal dystrophy, the role of EMC1 has not been validated by in vivo studies. Another study shows that EMC3, EMC5, and EMC6 regulate the biogenesis and trafficking of rhodopsin protein in Drosophila and mice, implying a link between ERAD and the accumulation of misfolded rhodopsin (Xiong et al., 2019). ERAD induction leads to ubiquitination and induces rapid degradation of P23H rhodopsin in vivo (Chiang et al., 2015). In addition, Hrd1 overexpression suppresses the late onset of retinal degeneration in a Drosophila model of autosomal dominant RP (Kang and Ryoo, 2009). Together, these findings suggest that the proper function of ERAD is critical for photoreceptor homeostasis and that enhancing ERAD could have beneficial effects in degenerating retinas.
5.4. Interplay between UPR and other signaling pathways
A broad spectrum of signaling pathways are regulated by the UPR, include autophagy, oxidative stress, Ca2+-mediated cell death, and inflammation.
5.4.1. UPR and autophagy
Autophagy is a catabolic process that removes unnecessary or dysfunctional cellular components. It exists in three forms: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Boya et al., 2016). Each pathway degrades substances by lysosomal enzymes. Macroautophagy affords a substantial portion of cellular protection under normal conditions. This process destroys targeted dysfunctional cytoplasmic components within the autophagosome. After incorporation of a substance, an autophagosome fuses with a lysosome to form an autolysosome that degrades incorporated content. Unlike macroautophagy, microautophagy is mediated by direct lysosomal engulfment of cytoplasmic dysfunctional content after it becomes trapped in lysosomes through membrane invagination (Sahu et al., 2011). This process plays an important role in membrane protein turnover and glycogen delivery into the lysosome. CMA usually refers to the chaperone-dependent targeting of soluble cytosolic proteins that are destined for further degradation in lysosomes (Kaushik and Cuervo, 2012). The most distinguishing feature of this pathway is the high specificity of the process. Heat shock protein 70 recognizes cytosolic proteins and targets them for CMA based on a specific amino acid sequence. Also, CMA contributes to several other cellular functions, including recycling the amino acids of degraded proteins and cellular metabolism (Kaushik and Cuervo, 2012).
Under certain conditions, such as a low nutrition supply for an extended period of time, the UPR can use cellular autophagy to generate energy through lysosomal degradation (Dash et al., 2019). Specifically, activation of the UPR PERK-peIF2α and IRE1 arms trigger autophagy (Senft and Ronai, 2015). The PERK-eIF2α pathway is essential for autophagy induction after stress initiated by tunicamycin (TM) treatment (B'Chir et al., 2013). Indeed, ATF4 and CHOP transcriptionally regulate multiple ATG genes. However, despite the overall role of PERK arm mediators in autophagy signaling, their specific contributions to regulating autophagy genes may not be the same. For example, one study reports that ATF4 and PERK play distinct roles in autophagy (Luhr et al., 2019); ATF4 controls transcription and is essential for autophagosome formation, whereas PERK is involved in the post-sequestration step of autophagy. Interestingly, the over-expression of ATF4 in T17M RHO mouse retinas is associated with defective autophagy as evidenced by p62 and LC3-II levels (Bhootada et al., 2016).
The other UPR mediator, IRE1, is also implicated in the activation of autophagy (Senft and Ronai, 2015). TNF receptor-associated factor 2-dependent activation of IRE1 and c-Jun N-terminal kinase (JNK) cause Bcl-2 phosphorylation, leading to dissociation of Beclin-1 and activation of the phosphoinositide-3-kinase (PI3K) complex, thereby activating autophagy (Deegan et al., 2013). By contrast, another study concludes that under TM-induced UPR activation, IRE1 acts as a negative regulator of autophagy (Luhr et al., 2019), indicating that the role of IRE1 remains to be clarified. Although current literature suggests a link between UPR activation and regulation of autophagy, additional studies investigating the transcriptional or post-translational regulation of autophagy-associated genes via UPR mediators are necessary to unravel the precise mechanisms.
5.4.2. UPR and oxidative stress
Under normal conditions, the ER carries out oxidative protein folding and has limited antioxidant activity due to a balance between reduced and oxidized protein stages. Protein folding is associated with disulfide bond formation that is coupled with ROS formation in the ER (Malhotra and Kaufman, 2007). ER-resident oxidoreductase protein disulfide isomerases together with ER oxidoreductase 1 catalyze disulfide bond formation (Pollard et al., 1998), leading to the generation of hydrogen peroxide. Therefore, disulfide bond generation during folding requires a stable redox environment maintained by several buffering factors such as glutathione (GSH), ascorbic acid, and flavin nucleotides. For example, GSH reacts with non-native disulfide bonds formed as a result of oxidative stress or enzyme-catalyzed oxidation, allowing misfolded proteins to fold again. In addition, detoxification of ROS occurs with the help of reduced GSH, leading to a depletion of oxidized GSSG (Ushioda et al., 2008). The ratio of reduced-to-oxidized GSH/GSSG often serves as an indicator of oxidative stress within cells.
The adaptive stage of the UPR can not only reduce the folding demand but also trigger a compensative transcriptional program activating genes encoding antioxidant factors. Antioxidant control is associated with the PERK and IRE1 pathways (Harding et al., 2003). In particular, ATF4 is an essential factor for GSH synthesis (Dickhout et al., 2012; Huggins et al., 2015). In addition to ATF4, the PERK arm controls the activation of a potent oxidative stress regulator and transcription factor involved in the antioxidant response: nuclear factor erythroid 2-related factor 2 (NRF2). By enhancing NRF2, PERK may be a critical survival factor in P23H RHO retinas (Comitato et al., 2019; Zhang et al., 2019).
In addition, activation of the UPR IRE1/XBP1 axis stimulates the hexosamine biosynthetic pathway and the production of UDP-N-acetylglucosamine, a crucial component for stress-induced O-GlcNAc modification and cellular defense against ROS (Vincenz and Hartl, 2014). The IRE1 arm regulates oxidative stress by activating apoptosis signal regulating kinase 1, which is associated with mitochondrial ROS production through its inhibition of the mitochondrial electron transport chain by downstream cJun (Urano et al., 2000; Win et al., 2014). Our study of T17M RHO retinas with chronic UPR activation also shows an increase in ROS production, confirming a link between the UPR and oxidative stress in degenerating retinas (Bhootada et al., 2016).
5.4.3. UPR and Ca2+ homeostasis
The ER is an essential organelle for Ca2+ storage. Photoreceptors show marked variations in Ca2+ concentrations depending on their cellular compartments and demands. These compartments are associated with varied functions: transduction, transcription, translation, metabolism, and synaptic transmission. This results in higher Ca2+ influx in the OS and lower Ca2+ influx in the cell body and synaptic terminal. Types of Ca2+ channels in photoreceptors include cyclic nucleotide-gated channels in the OS, which are regulated by the phototransduction cascade, and voltage-gated Ca2+ channels in the cell body and synaptic terminal, which are regulated by light-induced cell membrane hyperpolarization. Store-operated channels in the plasma membrane; sarco/ER Ca2+-ATPase (SERCA) transporters, IP3 receptors, and ryanodine receptors in the ER; and voltage-dependent anion channels in IS mitochondria also contribute to the regulation of intracellular Ca2+ in photoreceptors (Butler et al., 2017; Shinde et al., 2016). For example, an important function of Grp78 is to keep Ca2+ within the ER lumen to control its free cytosolic form. Other ER-resident proteins include calreticulin and Bax inhibitor-1 protein. In addition, free Ca2+ is controlled by M calpain and calcineurin in the cytoplasm. Free intracellular Ca2+ can trigger several cellular pathways including mitochondria homeostasis, Ca2+ channels, and calmodulin signaling and eventually lead to apoptosis.
Aberrant intracellular Ca2+ signaling is found in a variety of neurodegenerative diseases including AD, Huntington disease, and Parkinson’s disease (PD). Ca2+ homeostasis can be disturbed by leakage of Ca2+ from the ER, with inositol-1,4,5-trisphosphate receptors (IP3R1) and ryanodine receptors being two major intracellular Ca2+ channels releasing ER Ca2+ stores. Thus, mice lacking IP3R1 or carrying spontaneous mutations show severe ataxic behavior (Cheung et al., 2008; Joshi et al., 2016). By recording a single channel, researchers have demonstrated the stimulatory effect of IP3R1 in a mouse model of a familial form of AD, showing exaggerated cellular Ca2+ signaling in the presence of agonist stimulation or enhanced low-level Ca2+ signaling in untreated cells (Cheung et al., 2008). Another study suggests the importance of SERCA pumps, which are responsible for Ca2+ uptake from the cytoplasm, in neurodegeneration (Rahate et al., 2019). Our study shows that during autosomal dominant RP progression in P23H-3 and S334ter-RHO rat retinas, free cytosolic Ca2+ is elevated in photoreceptors and is accompanied by increased calpastatin, VDAC, calreticulin, and BI-1 (Shinde et al., 2016). Interestingly, a study using rd1 mice reported a decrease in calpastatin (Paquet-Durand et al., 2006). In addition, we found increased expression of IP3R and SERCA2b mRNA and protein in degenerating photoreceptors and show that UPR activation is responsible for the increased calpain and JNK activity that, together with CDK5, result in cleavage of caspase 3 (Shinde et al., 2016).
5.4.4. UPR and inflammation
TNFα, interleukin (IL)-1b, and IL-6 are hallmarks of degenerating retinas. The UPR can promote pro-inflammatory cytokine production via activation of NF-kB, IRF-3, JNK, and JAK/STAT signaling (Beisel et al., 2017; Martinon et al., 2010; Meares et al., 2014). For example, TLR2 and TLR4 induce ER stress and activate IRE1a→spliced XBP1, resulting in production of IL-6 and TNF.
Neuroinflammation is also modulated through the NLR family pyrin domain-containing-3 (NLRP3) inflammasome. A multimeric protein complex composed of NLRP3, ASC adaptor, and activated caspase 1 mediates the proteolytic activation of IL-1b and IL-18 and promotes a type of inflammatory cell death referred to as pyroptosis (Labzin et al., 2018). The interplay between the UPR and inflammasome is carried out through the IRE1 UPR arm; the RNase domain of IRE1 regulates expression of thioredoxin-interacting protein (TXNIP), the activator of NLRP3, by degrading a miR-17 that destabilizes TXNIP (Lerner et al., 2012). Meantime, the PERK pathway is associated with production of IL-6 and the chemokines CCL2 and CCL20, which promote microglial activation (Guthrie et al., 2016; Meares et al., 2014). In spinal cord injury, ATF4 deficiency diminishes microglial activation and alters expression of IL-6, TNFα, and IL-1b (Hughes and Mallucci, 2019; Onate et al., 2016; Valenzuela et al., 2012).
Multiple studies show inflammation in the retina of animals with IRD. For example, rd10 mice exhibit strong upregulation of the microglia-specific genes Cx3cr1, Aif1, Irf8, and C1qc before the onset of neurodegeneration at P12 (Zhao et al., 2015a). Another study with rd1 mice demonstrates accumulation of Iba1, IL-4, and TGFβ cytokines in the retina at P14 (A et al., 2019). One study shows that NLRP3 inflammasome activation drives bystander cone cell death in a P23H rhodopsin model of retinal degeneration (Viringipurampeer et al., 2016). Another study with canine models of early-onset (rcd1, xlpra2, and erd) and late-onset (xpra1) RP shows that the inflammasome components Nlrp3, Casp1, Asc, Il-1b, and Il-1ra are present in the retina and participate in retinal degeneration (Appelbaum et al., 2017). Our group observed upregulation of pro-inflammatory cytokines in the retinas of T17M RHO and Ter349Glu mice, which show increased levels of TNFα, IL-1β, p65, monocyte chemoattractant protein-1, and IBA1 (Rana et al., 2014). Moreover, a variety of proinflammatory cytokines and chemokines, including monocyte chemotactic protein-1, are increased in the aqueous humor and vitreous fluid in RP patients as compared with control individuals (Yoshida et al., 2013). These findings lead to the conclusion that chronic inflammation may underlie the pathogenesis of RP, suggesting interventions for ocular inflammatory reactions as potential treatments for RP patients. We recently provided evidence that ATF4 and CHOP mediators of UPR PERK signaling could be responsible for elevated pro-inflammatory IL-1β cytokines in autosomal dominant RP retinas (Rana et al., 2017; Rana et al., 2014).
5.4.5. UPR and mitochondrial dysfunction
The ER and outer mitochondrial membrane are in tight contact, with merged areas known as mitochondria-associated ER membranes (MAMs). In degenerating neurons, the association between the ER and mitochondria is stronger in the presence of accumulated Aβ and in APP Swe/Lon mice, confirming results obtained with human fibroblasts isolated from AD patients (Hedskog et al., 2013). Moreover, Aβ exposure significantly increases Ca2+ shuttling from the ER to mitochondria and expression of MAM-associated proteins in neurons. One common protein regulating communication between the two organelles is Sig-1R, which is highly enriched at MAMs (Hayashi, 2019). This protein interacts with BiP/GRP78/HSPA5, an UPR mediator in its dormant state. Upon dissociation from BiP, Sig-1R becomes activated and exerts innate chaperoning activity. It stabilizes IP3-R3 at MAMs and thereby regulates Ca2+ entry into mitochondria and ATP synthesis. In addition, Ser-1R controls expression of the transcription factors Xbp-1 and NF-KB.
In addition to physical contact and regulation of Ca2+ flux, the synthesis of lipids, including phosphatidylserine, steroids, and sphingolipids, also relies on communication between the ER and mitochondria (Hayashi, 2019). Despite that mitochondrial dysfunction may contribute to photoreceptor and other retinal cell degeneration in varied retinal degenerative diseases, the crosstalk between these two organelles is not understood in depth. Our laboratory and others reveal that activated apoptosis in photoreceptors arises from two apoptotic pathways—one related to the mitochondrial apoptosis-inducing factor (AIF) and other to ER-associated caspase-12 (Bhootada et al., 2015). Both AIF and caspase-12 translocate to the nucleus, which depends upon intracellular Ca2+ homeostasis and calpain activity (Sanges et al., 2006). However, more detailed studies characterizing ER-mitochondrial tethers in degenerating photoreceptors are necessary to determine whether all retinal degenerative diseases affect the ER-mitochondrial axis in a similar manner and how ER-mitochondrial crosstalk is regulated.
6. UPR and diseases
6.1. UPR and neurodegeneration
An activated UPR in general, and ISR in particular, are hallmarks of many neurodegenerative diseases including AD (Hoozemans et al., 2009; Lee et al., 2010), amyotrophic lateral sclerosis (ALS) (Atkin et al., 2008), and PD (Hoozemans et al., 2007), which belong to a group of diseases termed protein misfolding diseases (Chaudhuri and Paul, 2006). AD is estimated to affect as many 25 million people and is characterized by dysfunctional synapses and accumulation of phosphorylated Tau protein or β-amyloid plaque in the brain (Gerakis and Hetz, 2018). Upregulation of IRE1 and PERK signaling are detected in postmortem AD brain tissue (Hoozemans et al., 2009). In animal models, PERK signaling is activated in response to Tau accumulation (Ho et al., 2012; Hoozemans et al., 2009) but appears to precede the formation of tangles (Ho et al., 2012). In addition, PERK activation precedes localized expression of ATF4 (Baleriola et al., 2014), indicating the onset of ISR signaling.
ALS culminates in the death of motor neurons and is characterized by aggregation of ubiquitinated inclusions (Jaronen et al., 2014). ER stress is detected in brain tissue from postmortem ALS patients and mouse models of ALS as evidenced by ER dilation, ER/Golgi fragmentation, and ribosomal detachment (Lautenschlaeger et al., 2012; Oyanagi et al., 2008). These changes to the rough ER and Golgi apparatus are involved in ALS pathogenesis in a mouse model, suggesting that ER stress could drive cell death (Stieber et al., 2000). Activation of all three arms of the UPR—PERK, IRE1, and ATF6—are reported in animal models of ALS (Jaronen et al., 2014). We also found UPR activation in animal models of PD involving α-synuclein-associated toxicity (Gorbatyuk et al., 2012b; Gully et al., 2016).
PD is characterized by the death of neurons in the substantia nigra pars compacta, which contains neurons responsible for producing dopamine, a neurotransmitter that plays a key role in regulating voluntary movement. In degenerating neurons in PD patients, misfolded α-synuclein forms inclusions known as Lewy bodies (Mercado et al., 2016). Neurons with Lewy bodies show elevated levels of phosphorylated PERK as well as ATF4 upregulation in nigral tissue from postmortem PD patients (Hoozemans et al., 2007). Activation of PERK signaling is also detected in rat (Gorbatyuk et al., 2012b) and mouse (Sun et al., 2013) models of PD. Intriguingly, induction of the pro-apoptotic protein CHOP appears to occur independently of ATF4 induction in PD (Sun et al., 2013), suggesting that ATF4 plays a pro-survival role in PD. Parkin, which is downstream of ATF4 activation, plays an important role in the survival of dopaminergic neurons. ATF4 is necessary for maintaining Parkin levels in cells after treatment with dopaminergic toxins (Sun et al., 2013). However, we show that sustained overexpression of ATF4 can also cause severe loss of dopaminergic nigral neurons (Gully et al., 2016). By contrast, IRE1-sXBP1 signaling was found to promote survival of dopaminergic neurons, as knocking out the gene encoding either of these proteins makes substantia nigra neurons more susceptible to ER stress-associated cell death (Valdes et al., 2014), indicating that the role of the IRE1-sXBP1 arm in regulating protein homeostasis is likely vital to substantia nigra neurons. This may be true of many neurons, as knocking out XBP1 in adult mice results in chronic ER stress and neurodegeneration (Valdes et al., 2014).
6.2. UPR and retinal degeneration
Interest in the UPR as a contributing factor to ocular disease progression is growing. In addition to IRD characterized by photoreceptor degeneration, other pathophysiological conditions such as glaucoma associated with deterioration of retinal ganglion cells (Marola et al., 2019); AMD driven by RPE damage (Periyasamy and Shinohara, 2017); diabetic retinopathy, a neuro-vascular diabetic complication (Zhang et al., 2015); and cataracts resulting from stressed lens epithelial cells (Berthoud et al., 2016; Ma et al., 2018; Zhou et al., 2016) have been associated with UPR activation. Activated UPR and ISR signaling is observed in several animal models of retinal degeneration including P23H RHO mice and rats (Athanasiou et al., 2018; Lin et al., 2007), T17M RHO mice (Choudhury et al., 2013), rd10 mice (Yu et al., 2018), Ter349Glu RHO mice (Rana et al., 2014), and Tubby mice expressing TULP1 missense mutations (Cai et al., 2015; Lobo et al., 2016). Rhodopsin misfolding due to mutations in RHO is thought to play a role in photoreceptor cell death in human retinitis pigmentosa. P23H RHO mutation causes rhodopsin to misfold and accumulate in the ER, cell body, and nerve terminals (Gorbatyuk et al., 2010; Goto et al., 1995; Olsson et al., 1992). Lin et al. were the first to demonstrate that the retinas of P23H RHO rats exhibit UPR activation as shown by an elevation in both Chop and BiP mRNA (Lin et al., 2007). One study describes three lines of P23H RHO transgenic rats expressing different copy numbers of the transgene (LaVail et al., 2018). Among these rats, the severity of retinal degeneration correlates with the ratio of transgene-to-endogenous opsin mRNA. Given that both P23H RHO lines 1 and 3 show activation of the UPR, it would be interesting to compare the degree of activated UPR markers in retinas with the extent of retinal degeneration and the ratio of transgene-to-wild-type opsin mRNA expression. Unfortunately, studies investigating the UPR in P23H Rho line 1 do not perform comparisons of UPR markers with wild-type retinas (Athanasiou et al., 2017a; Athanasiou et al., 2017c).
One study reported a broad list of RHO mutations trapped in the ER of COS-7-transfected cells (Behnen et al., 2018). However, additional experiments confirming their mislocalization in the retina should be conducted. Another study describes the definitive activation of PERK signaling in P23H RHO rats, as their treatment with a PERK inhibitor resulted in a decline in the level of phosphorylated eIF2α (Athanasiou et al., 2017b). In addition, we alleviated retinal degeneration in P23H RHO rats by overexpressing Grp78 via delivery of AAV vector to photoreceptors (Gorbatyuk et al., 2010), indicating that increasing the protein folding capacity of the ER is sufficient to promote survival of P23H RHO photoreceptors.
Several studies have sought to identify the role of PERK in degenerating retinas. One study reports that PERK inhibition exacerbates retinal degeneration in transgenic P23H RHO rats (Athanasiou et al., 2017b) and demonstrates that inhibiting the dephosphorylation of eIF2α via administration of salubrinal, a Gadd34/PP1 inhibitor, slightly ameliorates the disease. This signifies that in P23H-1 rats, PERK activation may promote photoreceptor survival. Another study confirmed these results, reporting that P23H RHO knock-in mice show UPR activation (Comitato et al., 2019) and that long-term stimulation of the PERK arm exerts a protective effect by phosphorylating NRF2 transcription, providing an antioxidant response. Our study indicates that PERK ablation in rd16 mouse photoreceptors does not change a- or b-wave scotopic ERG amplitudes (Starr and Gorbatyuk, 2019).
Moreover, photoreceptor-specific ablation of PERK in C57BL/6J mice does not affect ERG recordings at P60. As these studies utilize different animal models, the role of PERK during the course of IRD requires further clarity. In addition, the primary mode of PERK activation in these studies appears to engage different molecular players. Whereas sustained activation of phosphorylated eIF2α is proposed to be neuroprotective to P23H-1 rat retinas, the study with P23H RHO knock-in mice suggests that PERK exerts an anti-apoptotic effect by up-regulating NRF2. Therefore, we explored the role of phosphorylated eIF2α in retinal degeneration in rd16 mice (Starr and Gorbatyuk, 2019). Although the aim of our study was to delineate the role of phosphorylated eIF2α in protein synthesis, we demonstrate that specific and sustained up-regulation of phosphorylated eIF2α reduces apoptosis in rd16 mouse retinas.
Although the mechanisms of UPR activation in many neurodegenerative diseases have been characterized, they are likely much more complicated in models of retinal degeneration in which mutant proteins do not inherently misfold. One example is the Ter349Glu RHO transgenic mouse line, which shows prominent activation of the ATF6 arm and ISR (Rana et al., 2014). These mice express transgenic rhodopsin in which the stop codon is replaced by a codon coding for glutamine, resulting in the addition of dozens of amino acids to the end of the protein (Bessant et al., 1999). In addition, Mockel et al. show that PERK signaling is at least partially responsible for the induction of apoptosis in retinal explants lacking the gene encoding Bardet-Biedl syndrome 12 (Mockel et al., 2012). We also found that rd16 mouse retinas expressing truncated CEP290 show activation of the UPR and ISR (Starr et al., 2018), resulting in sustained elevation of phosphorylated eIF2α, ATF4, and CHOP at P15 and P20.
It is completely unknown how the UPR become activated in these animal models. One similarity shared between models is that rhodopsin accumulates in the cell body due to mistrafficking (Hollingsworth and Gross, 2013; Mockel et al., 2012), but how this results in ER stress remains elusive. One possibility is the existence of a signaling network between the primary cilium and the ER. For example, Nakao et al. proposed that mislocalized phototransduction proteins such as rhodopsin could cause photoreceptor cell death by activating the adenylyl cyclase and cAMP pathway that leads to apoptosis (Nakao et al., 2012). cAMP is elevated in retinas expressing truncated rhodopsin (S334ter-4), which is known for its abnormal cellular distribution in photoreceptors due to mis-sorting problems (Green et al., 2000; Traverso et al., 2002). cAMP could further trigger the ER stress response through the cAMP/PKA pathway (Song et al., 2017).
The severity of rhodopsin mis-localization could also correlate with the rate of cell death in degenerating retinas. That is, a study shows that ERG responses in cyclic dark-reared S334ter-4 rats are 116% and 87% (a- and b-waves, respectively) greater than those in light/dark-reared rats (Green et al., 2000). The researchers proposed three scenarios by which apoptosis could occur in photoreceptors. In the first scenario, production of mutant proteins overwhelms the normal vesicular machinery of the plasma membrane and interferes with the routing of cargo. In the second scenario, the physical presence of high levels of mutant opsin in the plasma membrane interfere with normal neurotransmitter release in the synaptic zone. For example, mutations in genes encoding PDE6 subunits (i.e., PDE6A, PDE6B, PDE6C, PDE6G, PDE6H) prevent cGMP hydrolysis and thus lead to excessive accumulation in photoreceptors (Power et al., 2020). In the third scenario, the degradation of large amounts of mis-sorted proteins produces damage resulting from the metabolic load placed on photoreceptors.
Consistently, we found that rd16 mouse retinas with ongoing UPR exhibit both mis-localization of rhodopsin and up-regulation of adenylyl cyclase (Figure 3). Another possible explanation for UPR activation in these retinas is that the accumulation of rhodopsin in the cell body results in the back-up of proteins in the ER. However, neither of these two possibilities has been investigated. Finally, accumulation of proteins could affect cellular processes that signal for or result in UPR activation. For example, in a model of AD, β-amyloid oligomers or fibrils directly interact with N-methyl-D-aspartate receptors and perturb cytoplasmic Ca2+ balance, resulting in ER stress-dependent apoptosis (Alberdi et al., 2013).
Our research shows that targeting the UPR can retard retinal degeneration. One therapeutic strategy is to diminish ATF4 expression in T7M RHO retinas, which delays the onset of IRD by up to 3 months (Bhootada et al., 2016). Although this is also true in rd16 mice (Figure 3), the precise mechanism of ATF4-mediated cytoprotection is unknown, and this would be challenging to determine as ATF4, together with CHOP, controls the expression of 575 genes (Han et al., 2013). For example, the delay of photoreceptor cell death in rd16 mice is associated with enhanced synthesis of proteins in general and rhodopsin in particular (Figure 3). Among those proteins with altered expression levels could be those with anti-apoptotic activity. It is also worth noting that an increased translational rate is associated with accumulation of RHO in rd16 ATF4+/− retinas. Therefore, future studies are needed to understand modifications in translational profiles and verify whether expression and activation of the translational repressor 4e-BP1 is altered in rd16 ATF4+/− retinas. Our study of rd16 mice deficient in this unique ATF4 downstream target (Han et al., 2013) demonstrates not only an enhanced translational rate but also preservation of photoreceptors (Starr et al., 2019).
Therefore, similarities in stress signaling among animal models of neurodegeneration of the brain and retina can provide researchers with potential targets that could impact many diseases.
7. Animal models for studying the UPR in the retina
In this section, we describe mouse models available for studying the UPR in the retina to improve our understanding of IRD.
7.1. PERK KO mice
In a pioneering study, Zhang et al. examined the effect of systemically knocking out Perk in mice, providing the field with clues as to the importance of this protein (Zhang et al., 2002). Most PERK KO mice die before or shortly after birth (Zhang et al., 2002). As PERK is required for proper function of the pancreas (Gao et al., 2012; Zhang et al., 2002) and skeletal system development (Xiong et al., 2017; Zhang et al., 2002), pancreatic atrophy is likely the primary cause of lethality in these mice. The fact that PERK KO mice are not viable makes studying the role of PERK in specific cells and tissues challenging. However, the invention of the CRE-lox system helped overcome this issue and enables further investigation of the role of PERK (Gao et al., 2012; Nguyen et al., 2018; Sidoli et al., 2016; Xiong et al., 2017; Yang et al., 2016b; Zhang et al., 2002). By crossing Perk-floxed mice with those expressing Cre recombinase, Perk can be knocked out at specific times or in specific tissues or cell types. Although compounds that inhibit PERK were discovered (Axten et al., 2012), it was quickly realized that these compounds are highly toxic and have targets other than PERK (Rojas-Rivera et al., 2017), again making research on the role of PERK in certain tissues difficult without introducing a Cre-flox system through breeding, which is time-consuming. Despite the complicated breeding needed to introduce Cre into an animal, this system is an extremely powerful tool for studying proteins of interest without affecting other tissues, making data analysis much more streamlined. A mouse model was developed to express Cre on the mouse opsin promoter, iCre75, which restricts Cre expression to rods (Li et al., 2005). Like rhodopsin, these mice start to express Cre around P7, making them particularly useful for knocking out genes that could be involved in retinal development but problematic for studying the development of photoreceptors. Our recent work with the rd16 mouse model of IRD highlights the role of PERK in photoreceptor homeostasis using a conditional PERK KO to avoid the issue of pancreatic atrophy. We demonstrate that knocking down PERK does not affect the loss of retinal function (Starr and Gorbatyuk, 2019). However, given that recombination occurs around loxP sites at P7 and persists as late as P18 (Li et al., 2005), we could not assess whether PERK is important for photoreceptor development. Moreover, we found that PERK deficits in unstressed C57BL6 mouse retinas do not cause any retinal abnormalities at P60. Two recent studies examining the role of PERK in mouse and rat models of IRD using PERK inhibitors (Athanasiou et al., 2017a; Comitato et al., 2019) show that PERK plays a protective role in the survival of P23H rhodopsin photoreceptors. In one study, inhibiting PERK with GSK2606414A led to inhibited eIF2α phosphorylation, reduced ERG function, and decreased photoreceptor survival. In the other study, long-term stimulation of the PERK arm of the ER stress response exerted a protective effect by phosphorylating NRF2 and thus bestowing an antioxidant response. In our previous study of rd16 mice with chronically activated UPR compared with wild-type mice, we found that ablation of PERK had no deleterious effects in C57BL6 mice and did not exacerbate IRD (Starr and Gorbatyuk, 2019). However, we must mention that the aim of this study was not to assess whether PERK ablation affects cell viability but to examine the role of up- or down-regulated phosphorylated eIF2α in promoting retinal degeneration, and we proposed that the role of PERK must be validated in other animal models of retinal degeneration. Although we emphasized in this study that deficient PERK does not kill photoreceptors in C57BL6 mice, this deficit must be studied under stress conditions in C57BL6 mice to understand the physiological meaning of PERK knockdown in healthy and diseased retinas.
7.2. GADD34 KO mice
The primary function of GADD34—initiation of PP1-mediated dephosphorylation of eIF2α—was not adequately demonstrated until the gene encoding Gadd34 (Ppp1r15a) was knocked out in mice (Kojima et al., 2003). Systemically knocking out Gadd34 results in viable mice (Kojima et al., 2003) that are generally healthy with a few exceptions. Mice lacking GADD34 eventually become obese and are more likely to develop nonalcoholic fatty liver disease (Nishio and Isobe, 2015). In addition, they are more susceptible to developing hepatic carcinoma (Nishio et al., 2017; Nishio and Isobe, 2015) and insulin resistance (Nishio and Isobe, 2015). Furthermore, mice lacking GADD34 develop a blood disease resembling thalassemia syndrome (Patterson et al., 2006). Additionally, female Gadd34−/− mice exhibit systemic inflammation (Nishio et al., 2017). These mice likely exhibit other problems that are not yet characterized. Nevertheless, they are currently the best option for studying the role of GADD34 even though we cannot discount the possibility that these abnormalities could have systemic consequences. Future endeavors employing Gadd34-floxed mice may provide a better understanding of the role of this protein in various tissues, including the mechanism of translational control in rd16 mouse retinas (Starr and Gorbatyuk, 2019).
7.3. CHOP KO mice
CHOP (also known as GADD153), a bZIP transcription factor, is strongly induced by ER stress (Wang and Ron, 1996). It is upregulated by the PERK pathway, although other UPR pathways may also contribute to its expression. It is a pro-apoptotic factor that promotes the transcription of several stress-related genes in response to stress (Bartlett et al., 1992; Ron and Habener, 1992; Wang et al., 1996). CHOP-deficient mice exhibit no abnormalities in development and display reduced apoptosis due to a lack of repression of the anti-apoptotic protein BCL-2, which could contribute to CHOP-mediated apoptosis (McCullough et al., 2001). Despite this fact, apoptosis still occurs in Chop−/− animals with ongoing ER stress, suggesting that other CHOP-independent mechanisms of cell death are activated in response to ER stress.
7.4. ATF4 KO mice
ATF4 KO mice show abnormalities in perinatal and postnatal development (Masuoka and Townes, 2002). Around 70% of animals die between embryonic day 17.5 and P14, and the remaining 30% survive until adulthood but display growth retardation and severe micropthalmia. Therefore, we and other research groups have used ATF4 hemizygous animals to validate role of this UPR marker.
8. Translational apparatus and control of protein synthesis
8.1. 5’ cap and IRES translation
Eukaryotic protein synthesis occurs through two modes of translation that coexist in cells: canonical or 5’-cap-dependent translation and non-canonical translation requiring an internal ribosome entry site (IRES) sequence located on the 5′ untranslated region (UTR) of mRNA. The difference between the two modes relies on their utilization of the 5’ cap initiation complex. The first mode is responsible for 85–90% of all synthetized proteins. The second mode requires a reduced subset of initiation factors and accounts for up to 10–15% of the remaining proteins (Komar and Hatzoglou, 2015; Leppek et al., 2018). Translation is generally achieved in three steps: initiation, elongation, and termination. Due to our interest in the initiation step of translation in healthy and diseased retinas, we focus primarily on this step.
Initiation of 5’ cap-dependent translation occurs through multiple steps including mRNA activation, scanning, ATG recognition, and joining to the 60S ribosomal subunit for further elongation (Figure 2). The TC is formed when GTP-bound eIF2 binds the initiator Met-tRNA (Barrieux and Rosenfeld, 1977; Kaempfer et al., 1978; Lorsch and Dever, 2010; Walton and Gill, 1976). Upon activation, the TC interacts with the 40S ribosomal subunit and other translation initiation factors, including eIF1, eIF1A, and the eIF3 complex (13 subunits from a-m) (Phan et al., 2001; Valasek et al., 2001). Together, these components form the 43S pre-initiation complex (PIC). All factors in the PIC are important for start codon recognition (Lorsch and Dever, 2010). The 43S PIC is then recruited to the 5’ prime end of a 7-methyl guanosine (m7G) cap of an mRNA by the eIF4F complex (Lorsch and Dever, 2010). The eIF4F complex consists of the cap binding protein eIF4E (Sonenberg et al., 1978), the helicase eIF4A (Lorsch and Herschlag, 1998), and the scaffolding protein eIF4G (Tarun et al., 1997). eIF4G creates a loop in the entirety of the mRNA by binding the poly A-tail and m7G cap, which are located at the 3’ and 5’ regions of a typical mRNA, respectively (Le et al., 1997; Tarun and Sachs, 1996). Once recruited to the mRNA, the PIC scans for a start codon in an ATP-dependent manner (Walton and Gill, 1976). When the anti-codon for Met-tRNA is found and bound in the P, or peptidyl, site of the ribosome, the GTP associated with eIF2 is hydrolyzed, releasing a phosphate in the process (Marcus and Feeley, 1966; Merrick, 1979). This process is mediated by eIF5, the GTPase-activating protein for eIF2 (Asano et al., 1999). For the translation elongation step to begin, the 60S ribosomal subunit must join the complex, which requires the displacement of several factors including eIF2-GDP, eIF1A, eIF3, eIF1, and eIF5 (Kearse and Wilusz, 2017). Recruitment of the 60S ribosomal subunit to the complex utilizes GTP, a process mediated by eIF5B (Asano et al., 1999).
Most mechanisms that negatively regulate translation target components of the initiation complex, which benefits cells energetically. Several steps in translation utilize high-energy phosphates in the form of ATP or GTP. Stopping translation before these hydrolyzation events is critical for the survival of energy-starved cells (Kearse and Wilusz, 2017). The best studied mechanisms of protein synthesis inhibition involve the regulation of eIF2 and eIF4E (Gingras et al., 1999; Sonenberg and Hinnebusch, 2009; Taniuchi et al., 2016). Following a cellular stressor such as nutrient deprivation, viral infection, or ER stress, one or both of these factors can be targeted to reduce translation rates (Sonenberg and Hinnebusch, 2009). Control of protein synthesis through these two factors has been a focus of our resent research (Starr and Gorbatyuk, 2019; Starr et al., 2018).
IRES mediates translation initiation independently of the cap through binding eIF3. Similar to 5’ cap-dependent translation, eIF3 controls assembly of mRNA with a 40S ribosome and formation of an IRES RNA-40S-eIF3 TC. In addition to varied eIFs, IRES translation also requires IRES trans-acting factors, the role of which is still under investigation. Such RNA chaperones could remodel the IRES structure to prepare a landing platform for the ribosome. The IRES element is commonly located on the 5’ UTR of RNA. Recruitment of the internal ribosome by IRES-containing mRNAs may be promoted by environmental changes favoring bypassing the 5′ cap region to keep up with protein expression when cap-dependent translation is weakened (Leppek et al., 2018). Such conditions could be cellular stress, mitosis, or apoptosis, resulting in reduced global cap-dependent translation. Cells may also increase the translation of certain proteins during programmed cell death. Ten to 15% of mammalian mRNAs are predicted to contain an IRES element (Leppek et al., 2018). Over 100 proposed IRES-containing eukaryotic mRNAs have been identified (Mokrejs et al., 2010). Among them are varied transcription (ATF4, NF-KB, C-JUN) and translational (eIFs) factors, chaperones (Grp78, HSP70, HSP90), growth factors (FGF-1/2, PDGF, VEGF, IGF2), apoptosis inhibitors (XIAP, Bcl-xL, Bcl2), apoptosis activators (Apaf1), and other regulatory molecules. Although the evidence for IRES-regulated mRNA translation was found more than a decade ago, the importance of eukaryotic RNA structure for IRES activity has only been discovered for some RNA structures and cellular IRESs.
Several well-studied mechanisms of non-canonical translation provide clues as to how ATF4 mRNA is regulated. Normally, genes with multiple uORFs with AUG start codons allow translation to begin, but downstream sequences have certain properties that make the ribosome stall, preventing the translation of downstream reading frames. The reason for the stall is often due to the existence of regions of RNA that form stable hairpins shortly after the start codon(Kearse and Wilusz, 2017). Although the mechanism that results in re-initiation at the downstream ORF is also a mystery, it likely involves a well-studied set of factors. Canonical translation requires Met-tRNA to be brought to the 43S PIC by eIF2. The anti-codon on Met-tRNA binds the triplet AUG. Therefore, when stress results in the phosphorylation of eIF2α, the PIC must find another way to initiate translation. A mechanism known as non-AUG translation is a process that occurs in all cells at a lesser frequency than canonical translation. One mechanism of non-AUG translation involves eIF2α (Komar et al., 2005) or eIF2D (Dmitriev et al., 2010) interacting with the PIC and binding non-AUG start sites. Interestingly, these factors do not require GTP to initiate translation, which differs from eIF2-mediated initiation or translation elongation (Dmitriev et al., 2010; Kearse and Wilusz, 2017; Komar et al., 2005). In addition, both of these factors are upregulated following cellular stress, consistent with a potential role of ATF4 activation. Interestingly, translation of BiP, another protein whose mRNA has uORFs, depends on eIF2A (Starck et al., 2016), further suggesting a role for eIF2A and non-AUG initiation in ER stress.
8.2. Two modes of regulation of 5’ cap-dependent translation
Protein synthesis requires cells to expend a great amount of energy, and the translation process is tightly regulated (Hinnebusch, 2017; Hinnebusch and Lorsch, 2012; Ryoo and Vasudevan, 2017; Sonenberg and Hinnebusch, 2009). Two major modes of translational control are currently known (Figure 4). In one approach, PERK phosphorylates and activates eIF2α during ISR. In the other approach, the phosphorylation state of translational repressors, eIF4E-binding proteins (4E-BP1, 2, and 3), is altered (Gingras et al., 1999; Gingras et al., 1998). Interestingly, ATF4 elevation during ER stress could connect both pathways via back loop-mediated control of phosphorylated eIF2α and the direct transcription of 4E-BPs as it has been shown in tissue culture (Han et al., 2013) and Drosophila (Ryoo and Vasudevan, 2017) systems. Recent research demonstrates that both mechanisms are dysfunctional in mice with IRD (Kunte et al., 2012; Lin et al., 2018; Punzo et al., 2009; Starr et al., 2018). Phosphorylated eIF2α prevents the eIF2 complex from directing Met-tRNA to the pre-initiation complex, resulting in translational inhibition. We and other investigators observe elevated phosphorylated eIF2α in the retinas of mice with IRD (Athanasiou et al., 2017b; Kunte et al., 2012; Starr et al., 2018). Despite this fact, the role of eIF2α in retinal pathogenesis has not been addressed until recently.
Figure 4.
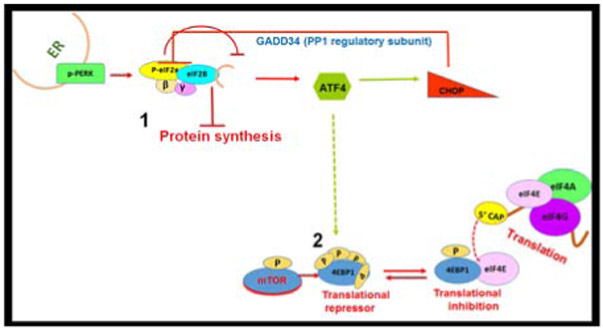
Two modes of translational control in photoreceptors. The first mode operates through eIF2α. Phosphorylation of eIF2α leads to translational attenuation during the ER stress response (1). The second mode regulates protein synthesis through eIF4-BP, a translational repressor that binds to eIF4 in its hypo-phosphorylated form and has a reduced binding affinity when is hyper-phosphorylated (2). ATF4 elevation during ER stress could connect both pathways via back loop-mediated control of phosphorylated eIF2α and the direct transcription of 4E-BPs.
8.2.1. Regulation of protein synthesis through phosphorylated eIF2α
Phosphorylated eIF2α can stop the global production of protein synthesis to allow cells to recover from accumulation of misfolded proteins during acute stress (Figure 4). Interestingly, varied animal models of retinal degenerative diseases and neurodegenerative disorders, including PD and AD, exhibit chronic activation of phosphorylated eIF2α along with a reduced translational rate. Therefore, we became interested in the role of phosphorylated eIF2α in controlling the translational rate during chronic stress. Our previous study indicates that the primary point of translational control in IRD may not occur through eIF2α (Starr et al., 2018). That is, we found that although PERK is responsible for phosphorylating eIF2α, it does not play a major role in regulating protein synthesis in degenerating retinas with chronic UPR activation. Furthermore, using two genetic approaches—systemic GADD34 KO and photoreceptor-specific conditional PERK KO—we demonstrate that altering phosphorylated eIF2α levels in rd16 mice is insufficient to delay retinal degeneration (Starr and Gorbatyuk, 2019). Nevertheless, multiple research groups report that PERK-mediated modulation of eIF2α and downstream signaling may be a viable strategy for treating neurodegenerative diseases (Athanasiou et al., 2017a; Comitato et al., 2019).
8.2.2. Regulation of protein synthesis through eIF4E
Global rates of protein synthesis in the retina have not been assessed until recently. Our group was the first to demonstrate chronic inhibition of translation in various models of IRD (Starr et al., 2018) (Figure 5). This study fueled our interest in further investigating the molecular control of protein synthesis in degenerating retinas (Starr and Gorbatyuk, 2019; Starr et al., 2019; Starr et al., 2018).
Figure 5.
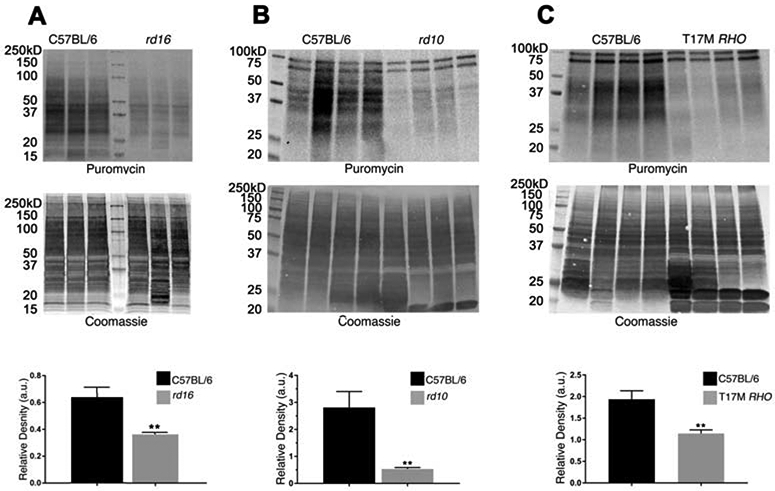
Translational attenuation in retinas of various models of IRD. Measurement of newly synthesized polypeptides using the SUnSET method shows translational attenuation in the retinas of rd16 mice at P15 (A and B), T17M RHO mice at P15 (C and D), and rd10 mice at P20 (E and F). Published with copyright permission from Cell Death and Disease (2018, May 1;9(5):484).
In 1978, Sonenberg et al. discovered that eIF4E is the m7G cap binding protein of the heterotrimeric eIF4F complex (Sonenberg, 2008; Sonenberg et al., 1978) and the canonical rate-limiting initiation factor. Thus, by negatively regulating eIF4E, cells can shut off synthesis of the vast majority of proteins (Gingras et al., 1998; Sonenberg, 2008; Sonenberg and Hinnebusch, 2009) (Figure 4). Depending on their hyper- or hypo-phosphorylated status, 4E-BPs can interact with eIF4E and prevent it from joining the cap-binding complex, reducing protein synthesis (Gingras et al., 1999; Gingras et al., 1998). The more Ser/Thr sites become phosphorylated, the weaker the affinity of 4E-BPs for eIF4E (Gingras et al., 1998). Also, 4E-BP1 phosphorylation is downregulated in both rd16 and rd10 mouse retinas (Starr et al., 2018). Despite advances in our knowledge of translation inhibition in neurodegeneration (Jan et al., 2018; Ma et al., 2013; Mercado et al., 2018; Moreno et al., 2012; Radford et al., 2015; Sharma et al., 2018), it is unclear whether a decline in protein synthesis is protective or whether increasing translation rates could be a viable neuroprotective strategy in IRD. Therefore, we conducted a study to decipher the role of translational attenuation in degenerating retinas during the course of IRD (Starr et al., 2019) using a comprehensive approach including applying pharmacological compounds, inhibiting protein synthesis, overexpressing constitutively active (i.e., less phosphorylated) 4E-BP1 in wild-type retinas via AAV gene delivery, and genetically ablating 4e-BP1/2 in the retinas of rd16 mice. For the first time, we reported that restoring protein synthesis rates delays retinal degeneration in mice (Starr et al., 2019).
The activity of 4E-BPs is regulated by mTOR phosphorylation. The mechanistic target of mTOR is a kinase that is coined the master regulator of protein synthesis and cellular metabolism (Chen, 2004; Sandsmark et al., 2007). mTOR is a kinase that forms the core of two complexes known as mTOR complex 1 (Kim et al., 2002; Nojima et al., 2003) and 2 (Jacinto et al., 2004; Sarbassov et al., 2004) (mTORC1 and mTORC2), which are involved in the regulation of protein synthesis and autophagy, respectively. mTOR forms a complex with multiple proteins including regulatory-associated protein of mTOR (Raptor), DEP domain-containing mTOR-interacting protein (DEPTOR), proline-rich AKT1 substrate 1 (PRAS40), and mammalian lethal with SEC13 protein 8 to form mTORC1 (Figure 6). Each of these proteins regulates the activity of mTOR in some way. Raptor promotes the phosphorylation of mTORC1 targets (Hara et al., 2002; Kim et al., 2002), whereas DEPTOR inhibits mTOR (Liu et al., 2010; Peterson et al., 2009). PRAS40 is a target of mTOR and a positive intermediate of mTORC1 signaling (Fonseca et al., 2007). Various external stimuli signal for the activation of mTOR through phosphorylation at multiple sites. Interestingly, we and other investigators observe downregulation of mTOR in degenerating retinas (Lin et al., 2018; Punzo et al., 2009; Starr et al., 2018). Moreover, the stimulation of the insulin/mTOR pathway delays cone death in a mouse model of RP (Punzo et al., 2009).
Figure 6.
mTORC1 regulation of protein synthesis. mTOR is a heavily regulated kinase that promotes cell growth and division following cell receipt of nutrients or growth factors. This schematic depicts a growth factor resulting in AKT and PDK1 activation by PI3K. AKT inhibits TSC1/2, enabling mTOR activation. mTOR regulates protein synthesis through S6K and 4E-BPs.
One well-studied activator of mTORC1 is protein kinase B, or AKT, a serine/threonine protein kinase (Inoki et al., 2002). Many external stimuli, which are often growth factors, initiate signaling cascades that converge to activate AKT (Burgering and Coffer, 1995), making it a central mediator of metabolic signaling. The general model of AKT activation involves a growth factor or hormone, such as insulin, that activates a receptor tyrosine kinase (Burgering and Coffer, 1995). Full activation of AKT is not achieved until it is phosphorylated at Ser473 by PDK2 or mTOR (Alessi et al., 1997; Partovian and Simons, 2004; Rane et al., 2001; Zhang et al., 2012). When AKT is active, it phosphorylates and alters the activity of several key players in many signaling cascades. For example, AKT activates mTORC1 by phosphorylating and inhibiting tuberous sclerosis 1 and 2 (TSC1/2), key negative regulators of mTORC1 (Gao and Pan, 2001; Inoki et al., 2002; Zhang et al., 2003a). When no longer inhibited by TSC1/2, mTORC1 promotes cell growth and division. To regulate translation, mTOR directly phosphorylates regulators of protein synthesis, including 4E-BPs (Gingras et al., 1999; Gingras et al., 1998) and S6 kinase (Garami et al., 2003). We demonstrated that the pAKT axis is downregulated in degenerating retinas of mouse models of IRD (Starr et al., 2018), and others validated the AKT gene transfer approach to rescue degenerating photoreceptors (McDougald et al., 2019).
9. Current therapeutic strategies for neuroprotection from degenerating photoreceptors
Currently, there are no therapies that effectively cure IRD. Thus, supportive care is the main approach to managing these diseases. Some evidence indicates that antioxidant vitamins slow IRD progression (Fahim, 2018). Recent studies show that 20 mg β-carotene, a less toxic form of vitamin A, increases scotopic and photopic b-wave ERG amplitudes (Rotenstreich et al., 2013). Also, the concentration of docosahexaenoic acid in red blood cells is associated with the rate of RP progression (Berson et al., 2010; Berson et al., 2004). As smoking and UV light are believed to be causative factors for retinal degeneration, precautions such as sunglasses are recommended for RP patients. Retinal prostheses have recently been designed to manage end-stage patients and hold promise for improving their quality of life. While these approaches may help manage RP, gene and stem cell therapy are the only currently available options for slowing down the rate of IRD progression. Since the first FDA-approved clinical trial for RPE65-associated LCA, several other clinical trials with IRD patients have begun (Vazquez-Dominguez et al., 2019), including those testing the use of AAV gene delivery and pharmacological compounds targeting ABCA4, CHM, CNGA3, CNGB3, MERTK, PDE6B, RPE65, RLBP1, RPGR, RS1, CEP290, and non-syndromic RP. Due to our interest in AAV-mediated gene transfer, we generated a double-mutant R585, 588A AAV vector that shows better transduction in the eye compared with other wild-type and mutant viruses (Gorbatyuk et al., 2019). Our laboratory is also investigating a new generation of viruses (Gorbatyuk et al., 2019) that could target the UPR-associated markers ATF4 and 4e-BP1 in mice with IRD and ongoing UPR activation.
9.1. UPR as a target in retinal degeneration
Suppression of chronic UPR activation as a therapeutic strategy was validated in different preclinical animal and cellular models of retinal degeneration. We demonstrated that AAV-mediated delivery of Grp78/BiP protein, a UPR marker, protects P23H RHO transgenic rats from photoreceptor deterioration and vision decline (Gorbatyuk et al., 2010). Later, another group found that BiP prevents rod opsin aggregation in vitro (Athanasiou et al., 2012). A more recent study proposes that pharmacological interventions against pathogenic ER stress should promote a favorable ratio of BiP to its disease-causing client protein (Vitale et al., 2019). Therefore, future experiments should determine the BiP-to-mutant rhodopsin ratio that provides therapeutic effects in degenerating retinas.
The FDA-approved 4-phenylbutyrate (PBA) is another chaperone validated for mutant rhodopsin folding in vitro (Mendes and Cheetham, 2008). P23H RHO folding is improved in SK-N-SH cells expressing the P23H-GFP opsin treated with 4-PBA. Additionally, this chaperone, together with Hsp90 inhibitor and 17-AAG compound, was recently tested in vitro and in vivo (Aguila et al., 2014); rats expressing the R135L rod opsin treated with HSP90 inhibitor in combination with 17-AAG show an improvement in P135L RHO mislocalization compared with wild-type rats. In 661W cells, 4-PBA reduces thapsigargin-induced elevations in CXCL10 and CCL2 expression and diminishes ER stress (Zhu et al., 2017). Recently, the ability of 4-PBA to improve P23H RHO folding and reduce ER stress was directly tested in mice (Qiu et al., 2019); treatment with this molecule significantly reduces the number of apoptotic cells and normalizes the autophagy flux-to-proteasome activity ratio, resulting in photoreceptor survival. Treatment with curcumin also improves the folding and proper localization of newly synthetized mutant rhodopsin in P23H RHO rats as well as retinal morphology, physiology, and gene expression (Vasireddy et al., 2011).
Over the course of IRD, other retinal cells, such as RPE and retinal ganglion cells, could also degenerate through activation of the ER stress response, suggesting that an AAV-mediated approach targeting UPR reprogramming could also be applied to these cell types. For example, overexpression of p58IPK (McLaughlin et al., 2018), GRP78 (Ha et al., 2018), or X-box binding protein 1 (Yang et al., 2016a) promotes retinal ganglion cell survival, whereas overexpression of GRP78/BIP increases RPE viability under ER stress (Ghaderi et al., 2018). In human RPE cells expressing mutant L22P RPE65 protein, application of 4-BPA chaperone in combination with a low temperature rescues enzymatic activity and may be a promising “protein repair therapy” that could enhance the efficacy of gene therapy (Jin et al., 2016).
Small molecules such as integrated stress response inhibitor (ISRIB) have been validated in patient keratoconic fibroblasts (Soiberman et al., 2019); inhibition of the ISR relieves many hallmarks of the keratoconic phenotype in affected cells, suggesting that targeting the ISR through ISRIB could have therapeutic potential. This molecule, however, has not been extensively tested in animal models of IRD. Our unpublished data from T17M RHO transgenic mice show that application of ISRIB does not improve rod function. However, this treatment slows down cone functional loss, suggesting that future experiments are required to carefully investigate the therapeutic potential of ISRIB.
Finally, a mini-peptide targeting ATF4, H2-MRAKWRKKRMRRLKRKRRKMRQ RSK-OH, was validated in an animal model of oxygen-induced retinopathy (Geng et al., 2018). Although different from IRD, this model is associated with ATF4 elevation (Wang et al., 2013). Thus, it would be interesting to test this peptide in preclinical animal models of IRD known to show ATF4 upregulation. This approach could be particularly valuable when large cDNA insertions, such as those coding for CEP290 or RPGR (290 and 113 kD, respectively), would render AAV gene delivery problematic due to instability and difficulties in maintaining a recombinant vector.
Conclusion:
Pharmacologically based therapies targeting a chronic ER stress response alone or in a combination with gene therapy could be valuable neuroprotective strategies for degenerating retinas.
Acknowledgements:
This work was supported by the National Eye Institute, grants RO1 EY027763 and P30 EY003039.
List of abbreviations
- PERK
Pancreatic endoplasmic reticulum kinase-like endoplasmic reticulum kinase
- IRE1
Inositol-requiring kinase/endoRNase 1
- ATF6
Activating transcription factor 6
- GRP78
Glucose-regulated protein 78
- BiP
Binding immunoglobulin protein
- eIF2α
Eukaryotic transcription factor 2α
- ATF4
Activating transcription factor 4
- CHOP
C/EBP homologous protein
- GADD153
Growth arrest and DNA damage inducible gene 153
- GADD34
Growth arrest and DNA damage inducible gene 34
- PP1
Protein phosphatase 1
- XBP1
X box-binding protein 1, transcription factor
- 4eIF-BPs
Eukaryotic translation initiation factor 4E-binding proteins
- IRD
Inherited retinal degeneration
- RP
Retinitis pigmentosa
- LCA
Leber’s congenital amaurosis
- TRB3
Tribbles homolog 3 protein
- ERAD
Endoplasmic reticulum-associated degradation
- eIF4E
Eukaryotic translation initiation factor 4E
- mTOR
Mammalian target of rapamycin
- AKT
Protein kinase B
- UPR
Unfolded protein response
- ISR
Integrated stress response
- TM
Tunicamycin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: The authors declare no competing or financial interests.
References
- A L, Zou T, He J, Chen X, Sun D, Fan X, Xu H, 2019. Rescue of Retinal Degeneration in rd1 Mice by Intravitreally Injected Metformin. Front Mol Neurosci 12, 102. 10.3389/fnmol.2019.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abisambra JF, Jinwal UK, Blair LJ, O'Leary JC 3rd, Li Q, Brady S, Wang L, Guidi CE, Zhang B, Nordhues BA, Cockman M, Suntharalingham A, Li P, Jin Y, Atkins CA, Dickey CA, 2013. Tau accumulation activates the unfolded protein response by impairing endoplasmic reticulum-associated degradation. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 9498–9507. 10.1523/jneurosci.5397-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Safieh L, Alrashed M, Anazi S, Alkuraya H, Khan AO, Al-Owain M, Al-Zahrani J, Al-Abdi L, Hashem M, Al-Tarimi S, Sebai MA, Shamia A, Ray-Zack MD, Nassan M, Al-Hassnan ZN, Rahbeeni Z, Waheeb S, Alkharashi A, Abboud E, Al-Hazzaa SA, Alkuraya FS, 2013. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res 23, 236–247. 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adekeye A, Haeri M, Solessio E, Knox BE, 2014. Ablation of the proapoptotic genes CHOP or Ask1 does not prevent or delay loss of visual function in a P23H transgenic mouse model of retinitis pigmentosa. PLoS One 9, e83871. 10.1371/journal.pone.0083871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguila M, Bevilacqua D, McCulley C, Schwarz N, Athanasiou D, Kanuga N, Novoselov SS, Lange CA, Ali RR, Bainbridge JW, Gias C, Coffey PJ, Garriga P, Cheetham ME, 2014. Hsp90 inhibition protects against inherited retinal degeneration. Human molecular genetics 23, 2164–2175. 10.1093/hmg/ddt613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberdi E, Wyssenbach A, Alberdi M, Sanchez-Gomez MV, Cavaliere F, Rodriguez JJ, Verkhratsky A, Matute C, 2013. Ca(2+) -dependent endoplasmic reticulum stress correlates with astrogliosis in oligomeric amyloid beta-treated astrocytes and in a model of Alzheimer's disease. Aging cell 12, 292–302. 10.1111/acel.12054. [DOI] [PubMed] [Google Scholar]
- Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P, 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Current biology : CB 7, 261–269. [DOI] [PubMed] [Google Scholar]
- Ali MM, Bagratuni T, Davenport EL, Nowak PR, Silva-Santisteban MC, Hardcastle A, McAndrews C, Rowlands MG, Morgan GJ, Aherne W, Collins I, Davies FE, Pearl LH, 2011. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. The EMBO Journal 30, 894–905. 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alone PV, Dever TE, 2006. Direct binding of translation initiation factor eIF2gamma-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B epsilon. The Journal of biological chemistry 281, 12636–12644. 10.1074/jbc.M511700200. [DOI] [PubMed] [Google Scholar]
- Amin-Wetzel N, Neidhardt L, Yan Y, Mayer MP, Ron D, 2019. Unstructured regions in IRE1alpha specify BiP-mediated destabilisation of the luminal domain dimer and repression of the UPR. Elife 8 10.7554/eLife.50793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum T, Santana E, Aguirre GD, 2017. Strong upregulation of inflammatory genes accompanies photoreceptor demise in canine models of retinal degeneration. PLoS One 12, e0177224. 10.1371/journal.pone.0177224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Simmen T, 2016. ER-luminal thiol/selenol-mediated regulation of Ca2+ signalling. Biochemical Society transactions 44, 452–459. 10.1042/bst20150233. [DOI] [PubMed] [Google Scholar]
- Arango-Gonzalez B, Trifunovic D, Sahaboglu A, Kranz K, Michalakis S, Farinelli P, Koch S, Koch F, Cottet S, Janssen-Bienhold U, Dedek K, Biel M, Zrenner E, Euler T, Ekstrom P, Ueffing M, Paquet-Durand F, 2014. Identification of a common non-apoptotic cell death mechanism in hereditary retinal degeneration. PLoS One 9, e112142. 10.1371/journal.pone.0112142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshavsky VY, Burns ME, 2014. Current understanding of signal amplification in phototransduction. Cellular logistics 4, e29390. 10.4161/cl.29390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG, 1999. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. The EMBO Journal 18, 1673–1688. 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Bellingham J, Kanuga N, Adamson P, Cheetham ME, 2017a. The role of the ER stress-response protein PERK in rhodopsin retinitis pigmentosa. Hum Mol Genet 26, 4896–4905. 10.1093/hmg/ddx370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Bellingham J, Kanuga N, Adamson P, Cheetham ME, 2017b. The role of the ER stress response protein PERK in rhodopsin retinitis pigmentosa. Hum Mol Genet. 10.1093/hmg/ddx370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Bellingham J, Li W, McCulley C, Reeves PJ, Cheetham ME, 2018. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog Retin Eye Res 62, 1–23. 10.1016/j.preteyeres.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Aguila M, Opefi CA, South K, Bellingham J, Bevilacqua D, Munro PM, Kanuga N, Mackenzie FE, Dubis AM, Georgiadis A, Graca AB, Pearson RA, Ali RR, Sakami S, Palczewski K, Sherman MY, Reeves PJ, Cheetham ME, 2017c. Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration. Human molecular genetics 26, 305–319. 10.1093/hmg/ddw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou D, Kosmaoglou M, Kanuga N, Novoselov SS, Paton AW, Paton JC, Chapple JP, Cheetham ME, 2012. BiP prevents rod opsin aggregation. Molecular biology of the cell 23, 3522–3531. 10.1091/mbc.E12-02-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK, 2008. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiology of disease 30, 400–407. 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Aviram N, Schuldiner M, 2017. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J Cell Sci 130, 4079–4085. 10.1242/jcs.204396. [DOI] [PubMed] [Google Scholar]
- Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, Atkins C, Liu Q, Rabindran S, Kumar R, Hong X, Goetz A, Stanley T, Taylor JD, Sigethy SD, Tomberlin GH, Hassell AM, Kahler KM, Shewchuk LM, Gampe RT, 2012. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). Journal of medicinal chemistry 55, 7193–7207. 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A, 2013. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res 41, 7683–7699. 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CC, Sparrow JM, 2001. Visual symptomatology in patients with sight-threatening diabetic retinopathy. Diabet Med 18, 883–888. [DOI] [PubMed] [Google Scholar]
- Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, Hengst U, 2014. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell 158, 1159–1172. 10.1016/j.cell.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrieux A, Rosenfeld MG, 1977. Characterization of GTP-dependent Met-tRNAf binding protein. The Journal of biological chemistry 252, 3843–3847. [PubMed] [Google Scholar]
- Barrieux A, Rosenfeld MG, 1978. mRNA-induced dissociation of initiation factor 2. The Journal of biological chemistry 253, 6311–6314. [PubMed] [Google Scholar]
- Bartlett JD, Luethy JD, Carlson SG, Sollott SJ, Holbrook NJ, 1992. Calcium ionophore A23187 induces expression of the growth arrest and DNA damage inducible CCAAT/enhancerbinding protein (C/EBP)-related gene, gadd153. Ca2+ increases transcriptional activity and mRNA stability. The Journal of biological chemistry 267, 20465–20470. [PubMed] [Google Scholar]
- Bause E, Lehle L, 1979. Enzymatic N-glycosylation and O-glycosylation of synthetic peptide acceptors by dolichol-linked sugar derivatives in yeast. European journal of biochemistry 101, 531–540. [DOI] [PubMed] [Google Scholar]
- Baylor DA, Lamb TD, Yau KW, 1979. Responses of retinal rods to single photons. J Physiol 288, 613–634. [PMC free article] [PubMed] [Google Scholar]
- Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA, 1990. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A 87, 1620–1624. 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnen P, Felline A, Comitato A, Di Salvo MT, Raimondi F, Gulati S, Kahremany S, Palczewski K, Marigo V, Fanelli F, 2018. A Small Chaperone Improves Folding and Routing of Rhodopsin Mutants Linked to Inherited Blindness. iScience 4, 1–19. 10.1016/j.isci.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C, Ziegler S, Martrus Zapater G, Chapel A, Griesbeck M, Hildebrandt H, Lohse AW, Altfeld M, 2017. TLR7-mediated activation of XBP1 correlates with the IFNalpha production in humans. Cytokine 94, 55–58. 10.1016/j.cyto.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Brockhurst RJ, Hayes KC, Johnson EJ, Anderson EJ, Johnson CA, Gaudio AR, Willett WC, Schaefer EJ, 2010. Clinical trial of lutein in patients with retinitis pigmentosa receiving vitamin A. Arch Ophthalmol 128, 403–411. 10.1001/archophthalmol.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson EL, Rosner B, Sandberg MA, Weigel-DiFranco C, Moser A, Brockhurst RJ, Hayes KC, Johnson CA, Anderson EJ, Gaudio AR, Willett WC, Schaefer EJ, 2004. Further evaluation of docosahexaenoic acid in patients with retinitis pigmentosa receiving vitamin A treatment: subgroup analyses. Arch Ophthalmol 122, 1306–1314. 10.1001/archopht.122.9.1306. [DOI] [PubMed] [Google Scholar]
- Berthoud VM, Minogue PJ, Lambert PA, Snabb JI, Beyer EC, 2016. The Cataract-linked Mutant Connexin50D47A Causes Endoplasmic Reticulum Stress in Mouse Lenses. J Biol Chem 291, 17569–17578. 10.1074/jbc.M115.707950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D, 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nature cell biology 2, 326–332. 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Bessant DA, Khaliq S, Hameed A, Anwar K, Payne AM, Mehdi SQ, Bhattacharya SS, 1999. Severe autosomal dominant retinitis pigmentosa caused by a novel rhodopsin mutation (Ter349Glu). Mutations in brief no. 208. Online. Human mutation 13, 83.. [DOI] [PubMed] [Google Scholar]
- Bhootada Y, Choudhury S, Gully C, Gorbatyuk M, 2015. Targeting Caspase-12 to Preserve Vision in Mice With Inherited Retinal Degeneration. Invest Ophthalmol Vis Sci 56, 4725–4733. 10.1167/iovs.15-16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhootada Y, Kotla P, Zolotukhin S, Gorbatyuk O, Bebok Z, Athar M, Gorbatyuk M, 2016. Limited ATF4 Expression in Degenerating Retinas with Ongoing ER Stress Promotes Photoreceptor Survival in a Mouse Model of Autosomal Dominant Retinitis Pigmentosa. PLoS One 11, e0154779. 10.1371/journal.pone.0154779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtel J, Gliem M, Mangold E, Muller PL, Holz FG, Neuhaus C, Lenzner S, Zahnleiter D, Betz C, Eisenberger T, Bolz HJ, Charbel Issa P, 2018. Next-generation sequencing identifies unexpected genotype-phenotype correlations in patients with retinitis pigmentosa. PloS one 13, e0207958. 10.1371/journal.pone.0207958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield SA, Dacheux RF, 2001. Rod vision: pathways and processing in the mammalian retina. Progress in retinal and eye research 20, 351–384. [DOI] [PubMed] [Google Scholar]
- Bogorad AM, Lin KY, Marintchev A, 2017. Novel mechanisms of eIF2B action and regulation by eIF2alpha phosphorylation. Nucleic acids research 45, 11962–11979. 10.1093/nar/gkx845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad AM, Lin KY, Marintchev A, 2018. eIF2B Mechanisms of Action and Regulation: A Thermodynamic View. Biochemistry 57, 1426–1435. 10.1021/acs.biochem.7b00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollimuntha S, Pani B, Singh BB, 2017. Neurological and Motor Disorders: Neuronal Store-Operated Ca(2+) Signaling: An Overview and Its Function. Advances in experimental medicine and biology 993, 535–556. 10.1007/978-3-319-57732-6_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Esteban-Martinez L, Serrano-Puebla A, Gomez-Sintes R, Villarejo-Zori B, 2016. Autophagy in the eye: Development, degeneration, and aging. Prog Retin Eye Res 55, 206–245. 10.1016/j.preteyeres.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Brecelj J, Stirn-Kranjc B, 1999. ERG and VEP follow-up study in children with Leber's congenital amaurosis. Eye (London, England) 13 ( Pt 1), 47–54. 10.1038/eye.1999.10. [DOI] [PubMed] [Google Scholar]
- Bryda EC, LaVail MM, 2019. Letter to the editor announcing the availability of RCS and transgenic rats with P23H and S334ter rhodopsin mutations with inherited retinal degenerations. Exp Eye Res 178, 176. 10.1016/j.exer.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Bunel M, Chaudieu G, Hamel C, Lagoutte L, Manes G, Botherel N, Brabet P, Pilorge P, Andre C, Quignon P, 2019. Natural models for retinitis pigmentosa: progressive retinal atrophy in dog breeds. Hum Genet 138, 441–453. 10.1007/s00439-019-01999-6. [DOI] [PubMed] [Google Scholar]
- Burchell A, Allan BB, Hume R, 1994. Glucose-6-phosphatase proteins of the endoplasmic reticulum. Mol Membr Biol 11, 217–227. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Coffer PJ, 1995. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376, 599–602. 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- Burgoyne T, Meschede IP, Burden JJ, Bailly M, Seabra MC, Futter CE, 2015. Rod disc renewal occurs by evagination of the ciliary plasma membrane that makes cadherin-based contacts with the inner segment. Proc Natl Acad Sci U S A 112, 15922–15927. 10.1073/pnas.1509285113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MR, Ma H, Yang F, Belcher J, Le YZ, Mikoshiba K, Biel M, Michalakis S, Iuso A, Krizaj D, Ding XQ, 2017. Endoplasmic reticulum (ER) Ca(2+)-channel activity contributes to ER stress and cone death in cyclic nucleotide-gated channel deficiency. J Biol Chem 292, 11189–11205. 10.1074/jbc.M117.782326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bygrave FL, Benedetti A, 1996. What is the concentration of calcium ions in the endoplasmic reticulum? Cell calcium 19, 547–551. [DOI] [PubMed] [Google Scholar]
- Cai X, Chen L, McGinnis JF, 2015. Correlation of ER stress and retinal degeneration in tubby mice. Exp Eye Res 140, 130–138. 10.1016/j.exer.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campochiaro PA, Mir TA, 2018. The mechanism of cone cell death in Retinitis Pigmentosa. Prog Retin Eye Res 62, 24–37. 10.1016/j.preteyeres.2017.08.004. [DOI] [PubMed] [Google Scholar]
- Campochiaro PA, Strauss RW, Lu L, Hafiz G, Wolfson Y, Shah SM, Sophie R, Mir TA, Scholl HP, 2015. Is There Excess Oxidative Stress and Damage in Eyes of Patients with Retinitis Pigmentosa? Antioxid Redox Signal 23, 643–648. 10.1089/ars.2015.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Ali MM, 2015. Crystal structures reveal transient PERK luminal domain tetramerization in endoplasmic reticulum stress signaling. The EMBO Journal 34, 1589–1600. 10.15252/embj.201489183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM, Sidman RL, 1978. Differential effect of the rd mutation on rods and cones in the mouse retina. Invest Ophthalmol Vis Sci 17, 489–498. [PubMed] [Google Scholar]
- Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, Sayer JA, Otto EA, Attanasio M, O’Toole JF, Jin G, Shou C, Hildebrandt F, Williams DS, Heckenlively JR, Swaroop A, 2006. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Human molecular genetics 15, 1847–1857. 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GQ, Hao Y, Wong F, 1993. Apoptosis: final common pathway of photoreceptor death in rd, rds, and rhodopsin mutant mice. Neuron 11, 595–605. 10.1016/0896-6273(93)90072-y. [DOI] [PubMed] [Google Scholar]
- Chaudhuri TK, Paul S, 2006. Protein-misfolding diseases and chaperone-based therapeutic approaches. The FEBS journal 273, 1331–1349. 10.1111/j.1742-4658.2006.05181.x. [DOI] [PubMed] [Google Scholar]
- Chen CK, Zhang K, Church-Kopish J, Huang W, Zhang H, Chen YJ, Frederick JM, Baehr W, 2001. Characterization of human GRK7 as a potential cone opsin kinase. Mol Vis 7, 305–313. [PubMed] [Google Scholar]
- Chen J, 2004. Novel regulatory mechanisms of mTOR signaling. Current topics in microbiology and immunology 279, 245–257. [DOI] [PubMed] [Google Scholar]
- Cheung KH, Shineman D, Muller M, Cardenas C, Mei L, Yang J, Tomita T, Iwatsubo T, Lee VM, Foskett JK, 2008. Mechanism of Ca2+ disruption in Alzheimer's disease by presenilin regulation of InsP3 receptor channel gating. Neuron 58, 871–883. 10.1016/j.neuron.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Chan P, Wissinger B, Vincent A, Skorczyk-Werner A, Krawczynski MR, Kaufman RJ, Tsang SH, Heon E, Kohl S, Lin JH, 2017. Achromatopsia mutations target sequential steps of ATF6 activation. Proc Natl Acad Sci U S A 114, 400–405. 10.1073/pnas.1606387114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Joseph V, Yasumura D, Matthes MT, Lewin AS, Gorbatyuk MS, Ahern K, LaVail MM, Lin JH, 2016. Ablation of Chop Transiently Enhances Photoreceptor Survival but Does Not Prevent Retinal Degeneration in Transgenic Mice Expressing Human P23H Rhodopsin. Adv Exp Med Biol 854, 185–191. 10.1007/978-3-319-17121-0_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Kroeger H, Sakami S, Messah C, Yasumura D, Matthes MT, Coppinger JA, Palczewski K, LaVail MM, Lin JH, 2015. Robust Endoplasmic Reticulum-Associated Degradation of Rhodopsin Precedes Retinal Degeneration. Mol Neurobiol 52, 679–695. 10.1007/s12035-014-8881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Bhootada Y, Gorbatyuk O, Gorbatyuk M, 2013. Caspase-7 ablation modulates UPR, reprograms TRAF2-JNK apoptosis and protects T17M rhodopsin mice from severe retinal degeneration. Cell Death Dis 4, e528. 10.1038/cddis.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Nashine S, Bhootada Y, Kunte MM, Gorbatyuk O, Lewin AS, Gorbatyuk M, 2014. Modulation of the rate of retinal degeneration in T17M RHO mice by reprogramming the unfolded protein response. Adv Exp Med Biol 801, 455–462. 10.1007/978-1-4614-3209-8_58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR, 2011. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol 14, 93–105. 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comitato A, Schiroli D, Montanari M, Marigo V, 2019. Calpain Activation Is the Major Cause of Cell Death in Photoreceptors Expressing a Rhodopsin Misfolding Mutation. Mol Neurobiol. 10.1007/s12035-019-01723-5. [DOI] [PubMed] [Google Scholar]
- Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB, 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. The Journal of cell biology 190, 927–940. 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P, 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America 102, 18773–18784. 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Li J, Ron D, Sha B, 2011. The structure of the PERK kinase domain suggests the mechanism for its activation. Acta crystallographica. Section D, Biological crystallography 67, 423–428. 10.1107/s0907444911006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio CA, Sloan KR Jr., Packer O, Hendrickson AE, Kalina RE, 1987. Distribution of cones in human and monkey retina: individual variability and radial asymmetry. Science (New York, N.Y.) 236, 579–582. [DOI] [PubMed] [Google Scholar]
- Daiger SP, Sullivan LS, Bowne SJ, 2013. Genes and mutations causing retinitis pigmentosa. Clin Genet 84, 132–141. 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Aydin Y, Wu T, 2019. Integrated stress response in hepatitis C promotes Nrf2-related chaperone-mediated autophagy: A novel mechanism for host-microbe survival and HCC development in liver cirrhosis. Semin Cell Dev Biol. 10.1016/j.semcdb.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson WW, Hazariwala K, Karges S, 2000. Human photopic response to circulating glucose. Doc Ophthalmol 101, 155–163. [DOI] [PubMed] [Google Scholar]
- Dedigama-Arachchige PM, Acharige NPN, Pflum MKH, 2018. Identification of PP1-Gadd34 substrates involved in the unfolded protein response using K-BIPS, a method for phosphatase substrate identification. Molecular omics 14, 121–133. 10.1039/c7mo00064b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan S, Saveljeva S, Gorman AM, Samali A, 2013. Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress. Cell Mol Life Sci 70, 2425–2441. 10.1007/s00018-012-1173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca AP, Whitmore SS, Barnes J, Sharma TP, Westfall TA, Scott CA, Weed MC, Wiley JS, Wiley LA, Johnston RM, Schnieders MJ, Lentz SR, Tucker BA, Mullins RF, Scheetz TE, Stone EM, Slusarski DC, 2016. Hypomorphic mutations in TRNT1 cause retinitis pigmentosa with erythrocytic microcytosis. Hum Mol Genet 25, 44–56. 10.1093/hmg/ddv446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries SH, 2000. Bipolar cells use kainate and AMPA receptors to filter visual information into separate channels. Neuron 28, 847–856. [DOI] [PubMed] [Google Scholar]
- Dickhout JG, Carlisle RE, Jerome DE, Mohammed-Ali Z, Jiang H, Yang G, Mani S, Garg SK, Banerjee R, Kaufman RJ, Maclean KN, Wang R, Austin RC, 2012. Integrated stress response modulates cellular redox state via induction of cystathionine gamma-lyase: cross-talk between integrated stress response and thiol metabolism. J Biol Chem 287, 7603–7614. 10.1074/jbc.M111.304576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JD, Salinas RY, Arshavsky VY, 2015. Discs of mammalian rod photoreceptors form through the membrane evagination mechanism. The Journal of cell biology 211, 495–502. 10.1083/jcb.201508093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN, 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. The Journal of biological chemistry 285, 26779–26787. 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerrler WT, Lehrman MA, 1999. Regulation of the dolichol pathway in human fibroblasts by the endoplasmic reticulum unfolded protein response. Proceedings of the National Academy of Sciences of the United States of America 96, 13050–13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Cai X, Wu Y, Liu Y, Deng L, Chen H, 2017. Insights from Genetic Model Systems of Retinal Degeneration: Role of Epsins in Retinal Angiogenesis and VEGFR2 Signaling. J Nat Sci 3. [PMC free article] [PubMed] [Google Scholar]
- Drivas TG, Holzbaur EL, Bennett J, 2013. Disruption of CEP290 microtubule/membrane-binding domains causes retinal degeneration. The Journal of clinical investigation 123, 4525–4539. 10.1172/jci69448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupzyk A, Tsai B, 2016. How Polyomaviruses Exploit the ERAD Machinery to Cause Infection. Viruses 8 10.3390/v8090242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahim A, 2018. Retinitis pigmentosa: recent advances and future directions in diagnosis and management. Curr Opin Pediatr 30, 725–733. 10.1097/MOP.0000000000000690. [DOI] [PubMed] [Google Scholar]
- Fain GL, Dowling JE, 1973. Intracellular recordings from single rods and cones in the mudpuppy retina. Science 180, 1178–1181. 10.1126/science.180.4091.1178. [DOI] [PubMed] [Google Scholar]
- Farook JM, Shields J, Tawfik A, Markand S, Sen T, Smith SB, Brann D, Dhandapani KM, Sen N, 2013. GADD34 induces cell death through inactivation of Akt following traumatic brain injury. Cell death & disease 4, e754. 10.1038/cddis.2013.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG, 2007. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. The Journal of biological chemistry 282, 24514–24524. 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- Fusakio ME, Willy JA, Wang Y, Mirek ET, Al Baghdadi RJ, Adams CM, Anthony TG, Wek RC, 2016. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Molecular biology of the cell 27, 1536–1551. 10.1091/mbc.E16-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Pan D, 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes & development 15, 1383–1392. 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sartori DJ, Li C, Yu QC, Kushner JA, Simon MC, Diehl JA, 2012. PERK is required in the adult pancreas and is essential for maintenance of glucose homeostasis. Molecular and cellular biology 32, 5129–5139. 10.1128/mcb.01009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G, 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Molecular cell 11, 1457–1466. [DOI] [PubMed] [Google Scholar]
- Garcia-Huerta P, Bargsted L, Rivas A, Matus S, Vidal RL, 2016. ER chaperones in neurodegenerative disease: Folding and beyond. Brain research 1648, 580–587. 10.1016/j.brainres.2016.04.070. [DOI] [PubMed] [Google Scholar]
- Geng W, Qin F, Ren J, Xiao S, Wang A, 2018. Mini-peptide RPL41 attenuated retinal neovascularization by inducing degradation of ATF4 in oxygen-induced retinopathy mice. Experimental cell research 369, 243–250. 10.1016/j.yexcr.2018.05.027. [DOI] [PubMed] [Google Scholar]
- Gerakis Y, Hetz C, 2018. Emerging roles of ER stress in the etiology and pathogenesis of Alzheimer's disease. The FEBS journal 285, 995–1011. 10.1111/febs.14332. [DOI] [PubMed] [Google Scholar]
- Ghaderi S, Ahmadian S, Soheili ZS, Ahmadieh H, Samiei S, Kheitan S, Pirmardan ER, 2018. AAV delivery of GRP78/BiP promotes adaptation of human RPE cell to ER stress. Journal of cellular biochemistry 119, 1355–1367. 10.1002/jcb.26296. [DOI] [PubMed] [Google Scholar]
- Ghodrati M, Khaligh-Razavi SM, Lehky SR, 2017. Towards building a more complex view of the lateral geniculate nucleus: Recent advances in understanding its role. Progress in neurobiology 156, 214–255. 10.1016/j.pneurobio.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N, 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N, 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes & development 12, 502–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Arnaud Sampaio VF, Lameu C, Ulrich H, 2018. Calcium signalling: A common target in neurological disorders and neurogenesis. Seminars in cell & developmental biology. 10.1016/j.semcdb.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk M, Gorbatyuk O, 2013. Review: retinal degeneration: focus on the unfolded protein response. Mol Vis 19, 1985–1998. [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Gorbatyuk OS, LaVail MM, Lin JH, Hauswirth WW, Lewin AS, 2012a. Functional rescue of P23H rhodopsin photoreceptors by gene delivery. Adv Exp Med Biol 723, 191–197. 10.1007/978-1-4614-0631-0_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Knox T, LaVail MM, Gorbatyuk OS, Noorwez SM, Hauswirth WW, Lin JH, Muzyczka N, Lewin AS, 2010. Restoration of visual function in P23H rhodopsin transgenic rats by gene delivery of BiP/Grp78. Proc Natl Acad Sci U S A 107, 5961–5966. 10.1073/pnas.0911991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk MS, Shabashvili A, Chen W, Meyers C, Sullivan LF, Salganik M, Lin JH, Lewin AS, Muzyczka N, Gorbatyuk OS, 2012b. Glucose regulated protein 78 diminishes alphasynuclein neurotoxicity in a rat model of Parkinson disease. Mol Ther 20, 1327–1337. 10.1038/mt.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk OS, Warrington KH Jr., Gorbatyuk MS, Zolotukhin I, Lewin AS, Muzyczka N, 2019. Biodistribution of adeno-associated virus type 2 with mutations in the capsid that contribute to heparan sulfate proteoglycan binding. Virus Res 274, 197771. 10.1016/j.virusres.2019.197771. [DOI] [PubMed] [Google Scholar]
- Goto Y, Peachey NS, Ripps H, Naash MI, 1995. Functional abnormalities in transgenic mice expressing a mutant rhodopsin gene. Investigative ophthalmology & visual science 36, 62–71. [PubMed] [Google Scholar]
- Green ES, Menz MD, LaVail MM, Flannery JG, 2000. Characterization of rhodopsin mis-sorting and constitutive activation in a transgenic rat model of retinitis pigmentosa. Invest Ophthalmol Vis Sci 41, 1546–1553. [PubMed] [Google Scholar]
- Greene CM, McElvaney NG, 2010. Protein misfolding and obstructive lung disease. Proc Am Thorac Soc 7, 346–355. 10.1513/pats.201002-019AW. [DOI] [PubMed] [Google Scholar]
- Gregory CY, Bird AC, 1995. Cell loss in retinal dystrophies by apoptosis--death by informed consent! Br J Ophthalmol 79, 186–190. 10.1136/bjo.79.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenell A, Wang Y, Yam M, Swarup A, Dilan TL, Hauer A, Linton JD, Philp NJ, Gregor E, Zhu S, Shi Q, Murphy J, Guan T, Lohner D, Kolandaivelu S, Ramamurthy V, Goldberg AFX, Hurley JB, Du J, 2019. Loss of MPC1 reprograms retinal metabolism to impair visual function. Proc Natl Acad Sci U S A 116, 3530–3535. 10.1073/pnas.1812941116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Ouyang C, Lin W, Zhang T, Cao X, Xia Z, Wang X, 2014. Phosphatase holoenzyme PP1/GADD34 negatively regulates TLR response by inhibiting TAK1 serine 412 phosphorylation. Journal of immunology (Baltimore, Md. : 1950) 192, 2846–2856. 10.4049/jimmunol.1302537. [DOI] [PubMed] [Google Scholar]
- Gully JC, Sergeyev VG, Bhootada Y, Mendez-Gomez H, Meyers CA, Zolotukhin S, Gorbatyuk MS, Gorbatyuk OS, 2016. Up-regulation of activating transcription factor 4 induces severe loss of dopamine nigral neurons in a rat model of Parkinson's disease. Neurosci Lett 627, 36–41. 10.1016/j.neulet.2016.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie LN, Abiraman K, Plyler ES, Sprenkle NT, Gibson SA, McFarland BC, Rajbhandari R, Rowse AL, Benveniste EN, Meares GP, 2016. Attenuation of PKR-like ER Kinase (PERK) Signaling Selectively Controls Endoplasmic Reticulum Stress-induced Inflammation Without Compromising Immunological Responses. J Biol Chem 291, 15830–15840. 10.1074/jbc.M116.738021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Y, Liu W, Liu H, Zhu S, Xia F, Gerson JE, Azhar NA, Tilton RG, Motamedi M, Kayed R, Zhang W, 2018. AAV2-mediated GRP78 Transfer Alleviates Retinal Neuronal Injury by Downregulating ER Stress and Tau Oligomer Formation. Invest Ophthalmol Vis Sci 59, 4670–4682. 10.1167/iovs.18-24427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, Kilberg MS, Sartor MA, Kaufman RJ, 2013. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 15, 481–490. 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneda M, Xiao H, Hasegawa T, Kimura Y, Nakashima I, Isobe K, 2004. Regulation of mouse GADD34 gene transcription after DNA damaging agent methylmethane sulfonate. Gene 336, 139–146. 10.1016/j.gene.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K, 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Scheuner D, Chen JJ, Kaufman RJ, Ron D, 2009. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2alpha) dephosphorylation in mammalian development. Proceedings of the National Academy of Sciences of the United States of America 106, 1832–1837. 10.1073/pnas.0809632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D, 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11, 619–633. [DOI] [PubMed] [Google Scholar]
- Harosi FI, 1975. Absorption spectra and linear dichroism of some amphibian photoreceptors. J Gen Physiol 66, 357–382. 10.1085/jgp.66.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartong DT, Berson EL, Dryja TP, 2006. Retinitis pigmentosa. Lancet 368, 1795–1809. 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- Hayashi T, 2019. The Sigma-1 Receptor in Cellular Stress Signaling. Frontiers in neuroscience 13, 733. 10.3389/fnins.2019.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K, 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Molecular biology of the cell 10, 3787–3799. 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedskog L, Pinho CM, Filadi R, Ronnback A, Hertwig L, Wiehager B, Larssen P, Gellhaar S, Sandebring A, Westerlund M, Graff C, Winblad B, Galter D, Behbahani H, Pizzo P, Glaser E, Ankarcrona M, 2013. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer's disease and related models. Proc Natl Acad Sci U S A 110, 7916–7921. 10.1073/pnas.1300677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, 2017. Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends in biochemical sciences 42, 589–611. 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR, 2012. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor perspectives in biology 4 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YS, Yang X, Lau JC, Hung CH, Wuwongse S, Zhang Q, Wang J, Baum L, So KF, Chang RC, 2012. Endoplasmic reticulum stress induces tau pathology and forms a vicious cycle: implication in Alzheimer's disease pathogenesis. Journal of Alzheimer's disease : JAD 28, 839–854. 10.3233/jad-2011-111037. [DOI] [PubMed] [Google Scholar]
- Hojmann Larsen A, Frandsen A, Treiman M, 2001. Upregulation of the SERCA-type Ca2+ pump activity in response to endoplasmic reticulum stress in PC12 cells. BMC biochemistry 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth TJ, Gross AK, 2013. The severe autosomal dominant retinitis pigmentosa rhodopsin mutant Ter349Glu mislocalizes and induces rapid rod cell death. J Biol Chem 288, 29047–29055. 10.1074/jbc.M113.495184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjo Y, Ayaki T, Tomiyama T, Horibe T, Ito H, Mori H, Takahashi R, Kawakami K, 2015. Increased GADD34 in oligodendrocytes in Alzheimer's disease. Neuroscience letters 602, 50–55. 10.1016/j.neulet.2015.06.052. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemuller JM, Scheper W, 2007. Activation of the unfolded protein response in Parkinson's disease. Biochemical and biophysical research communications 354, 707–711. 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelenboom P, Scheper W, 2009. The unfolded protein response is activated in pretangle neurons in Alzheimer's disease hippocampus. The American journal of pathology 174, 1241–1251. 10.2353/ajpath.2009.080814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins CJ, Mayekar MK, Martin N, Saylor KL, Gonit M, Jailwala P, Kasoji M, Haines DC, Quinones OA, Johnson PF, 2015. C/EBPgamma Is a Critical Regulator of Cellular Stress Response Networks through Heterodimerization with ATF4. Mol Cell Biol 36, 693–713. 10.1128/MCB.00911-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D, Mallucci GR, 2019. The unfolded protein response in neurodegenerative disorders - therapeutic modulation of the PERK pathway. FEBS J 286, 342–355. 10.1111/febs.14422. [DOI] [PubMed] [Google Scholar]
- Ingram NT, Sampath AP, Fain GL, 2016. Why are rods more sensitive than cones? J Physiol 594, 5415–5426. 10.1113/JP272556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL, 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature cell biology 4, 648–657. 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN, 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nature cell biology 6, 1122–1128. 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jan A, Jansonius B, Delaidelli A, Bhanshali F, An YA, Ferreira N, Smits LM, Negri GL, Schwamborn JC, Jensen PH, Mackenzie IR, Taubert S, Sorensen PH, 2018. Activity of translation regulator eukaryotic elongation factor-2 kinase is increased in Parkinson disease brain and its inhibition reduces alpha synuclein toxicity. Acta neuropathologica communications 6, 54. 10.1186/s40478-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaronen M, Goldsteins G, Koistinaho J, 2014. ER stress and unfolded protein response in amyotrophic lateral sclerosis-a controversial role of protein disulphide isomerase. Frontiers in cellular neuroscience 8, 402. 10.3389/fncel.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Li S, Hu J, Jin HH, Jacobson SG, Bok D, 2016. Functional Rescue of Retinal Degeneration-Associated Mutant RPE65 Proteins. Adv Exp Med Biol 854, 525–532. 10.1007/978-3-319-17121-0_70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LE, van Veen T, Ekstrom PA, 2005. Differential Akt activation in the photoreceptors of normal and rd1 mice. Cell Tissue Res 320, 213–222. 10.1007/s00441-004-1046-8. [DOI] [PubMed] [Google Scholar]
- Jones BW, Marc RE, 2005. Retinal remodeling during retinal degeneration. Exp Eye Res 81, 123–137. 10.1016/j.exer.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Joshi AU, Kornfeld OS, Mochly-Rosen D, 2016. The entangled ER-mitochondrial axis as a potential therapeutic strategy in neurodegeneration: A tangled duo unchained. Cell Calcium 60, 218–234. 10.1016/j.ceca.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousse C, Deval C, Maurin AC, Parry L, Cherasse Y, Chaveroux C, Lefloch R, Lenormand P, Bruhat A, Fafournoux P, 2007. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J Biol Chem 282, 15851–15861. 10.1074/jbc.M611723200. [DOI] [PubMed] [Google Scholar]
- Kaempfer R, Rosen H, Israeli R, 1978. Translational control: recognition of the methylated 5' end and an internal sequence in eukaryotic mRNA by the initiation factor that binds methionyl-tRNAfMet. Proceedings of the National Academy of Sciences of the United States of America 75, 650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Seol HS, Lee SE, Suh YA, Kim J, Jang SJ, Yu E, 2019. Guanabenz Acetate Induces Endoplasmic Reticulum Stress-Related Cell Death in Hepatocellular Carcinoma Cells. Journal of pathology and translational medicine. 10.4132/jptm.2019.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Ryoo HD, 2009. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc Natl Acad Sci U S A 106, 17043–17048. 10.1073/pnas.0905566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2012. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends in cell biology 22, 407–417. 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE, 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes & development 31, 1717–1731. 10.1101/gad.305250.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefalov VJ, 2012. Rod and cone visual pigments and phototransduction through pharmacological, genetic, and physiological approaches. J Biol Chem 287, 1635–1641. 10.1074/jbc.R111.303008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E, 2004. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem 279, 3525–3534. 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM, 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175. [DOI] [PubMed] [Google Scholar]
- Koenekoop RK, Fishman GA, Iannaccone A, Ezzeldin H, Ciccarelli ML, Baldi A, Sunness JS, Lotery AJ, Jablonski MM, Pittler SJ, Maumenee I, 2002. Electroretinographic abnormalities in parents of patients with Leber congenital amaurosis who have heterozygous GUCY2D mutations. Archives of ophthalmology (Chicago, Ill. : 1960) 120, 1325–1330. [DOI] [PubMed] [Google Scholar]
- Kohno K, Normington K, Sambrook J, Gething MJ, Mori K, 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Molecular and cellular biology 13, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima E, Takeuchi A, Haneda M, Yagi A, Hasegawa T, Yamaki K, Takeda K, Akira S, Shimokata K, Isobe K, 2003. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 17, 1573–1575. 10.1096/fj.02-1184fje. [DOI] [PubMed] [Google Scholar]
- Komar AA, Gross SR, Barth-Baus D, Strachan R, Hensold JO, Goss Kinzy T, Merrick WC, 2005. Novel characteristics of the biological properties of the yeast Saccharomyces cerevisiae eukaryotic initiation factor 2A. The Journal of biological chemistry 280, 15601–15611. 10.1074/jbc.M413728200. [DOI] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M, 2015. Exploring Internal Ribosome Entry Sites as Therapeutic Targets. Front Oncol 5, 233. 10.3389/fonc.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Safer B, 1983. Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. The Journal of biological chemistry 258, 3402–3408. [PubMed] [Google Scholar]
- Kopp MC, Larburu N, Durairaj V, Adams CJ, Ali MMU, 2019. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nature structural & molecular biology 26, 1053–1062. 10.1038/s41594-019-0324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MP, White DA, Kaushal S, 2009. Biphasic photoreceptor degeneration induced by light in a T17M rhodopsin mouse model of cone bystander damage. Invest Ophthalmol Vis Sci 50, 2956–2965. 10.1167/iovs.08-3116. [DOI] [PubMed] [Google Scholar]
- Krokowski D, Guan BJ, Wu J, Zheng Y, Pattabiraman PP, Jobava R, Gao XH, Di XJ, Snider MD, Mu TW, Liu S, Storrie B, Pearlman E, Blumental-Perry A, Hatzoglou M, 2017. GADD34 Function in Protein Trafficking Promotes Adaptation to Hyperosmotic Stress in Human Corneal Cells. Cell reports 21, 2895–2910. 10.1016/j.celrep.2017.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran N, Moore AT, Weleber RG, Michaelides M, 2017. Leber congenital amaurosis/early-onset severe retinal dystrophy: clinical features, molecular genetics and therapeutic interventions. The British journal of ophthalmology 101, 1147–1154. 10.1136/bjophthalmol-2016-309975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunchithapautham K, Rohrer B, 2007a. Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy 3, 433–441. 10.4161/auto.4294. [DOI] [PubMed] [Google Scholar]
- Kunchithapautham K, Rohrer B, 2007b. Autophagy is one of the multiple mechanisms active in photoreceptor degeneration. Autophagy 3, 65–66. 10.4161/auto.3431. [DOI] [PubMed] [Google Scholar]
- Kunte MM, Choudhury S, Manheim JF, Shinde VM, Miura M, Chiodo VA, Hauswirth WW, Gorbatyuk OS, Gorbatyuk MS, 2012. ER stress is involved in T17M rhodopsin-induced retinal degeneration. Investigative ophthalmology & visual science 53, 3792–3800. 10.1167/iovs.11-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labzin LI, Heneka MT, Latz E, 2018. Innate Immunity and Neurodegeneration. Annu Rev Med 69, 437–449. 10.1146/annurev-med-050715-104343. [DOI] [PubMed] [Google Scholar]
- Lautenschlaeger J, Prell T, Grosskreutz J, 2012. Endoplasmic reticulum stress and the ER mitochondrial calcium cycle in amyotrophic lateral sclerosis. Amyotrophic lateral sclerosis : official publication of the World Federation of Neurology Research Group on Motor Neuron Diseases 13, 166–177. 10.3109/17482968.2011.641569. [DOI] [PubMed] [Google Scholar]
- LaVail MM, Nishikawa S, Steinberg RH, Naash MI, Duncan JL, Trautmann N, Matthes MT, Yasumura D, Lau-Villacorta C, Chen J, Peterson WM, Yang H, Flannery JG, 2018. Phenotypic characterization of P23H and S334ter rhodopsin transgenic rat models of inherited retinal degeneration. Exp Eye Res 167, 56–90. 10.1016/j.exer.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVail MM, Yasumura D, Matthes MT, Drenser KA, Flannery JG, Lewin AS, Hauswirth WW, 2000. Ribozyme rescue of photoreceptor cells in P23H transgenic rats: long-term survival and late-stage therapy. Proc Natl Acad Sci U S A 97, 11488–11493. 10.1073/pnas.210319397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR, 1997. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. The Journal of biological chemistry 272, 16247–16255. [DOI] [PubMed] [Google Scholar]
- Lee JH, Won SM, Suh J, Son SJ, Moon GJ, Park UJ, Gwag BJ, 2010. Induction of the unfolded protein response and cell death pathway in Alzheimer's disease, but not in aged Tg2576 mice. Experimental & molecular medicine 42, 386–394. 10.3858/emm.2010.42.5.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, Barna M, 2018. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19, 158–174. 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, Shen S, Nguyen V, Backes BJ, Heiman M, Heintz N, Greengard P, Hui S, Tang Q, Trusina A, Oakes SA, Papa FR, 2012. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab 16, 250–264. 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveillard T, 2015. Cancer metabolism of cone photoreceptors. Oncotarget 6, 32285–32286. 10.18632/oncotarget.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen D, Sauve Y, McCandless J, Chen YJ, Chen CK, 2005. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis 41, 73–80. 10.1002/gene.20097. [DOI] [PubMed] [Google Scholar]
- Li WW, Hsiung Y, Zhou Y, Roy B, Lee AS, 1997. Induction of the mammalian GRP78/BiP gene by Ca2+ depletion and formation of aberrant proteins: activation of the conserved stress-inducible grp core promoter element by the human nuclear factor YY1. Molecular and cellular biology 17, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Xiong G, Yang W, 2018. Ribosomal protein S6 kinase 1 promotes the survival of photoreceptors in retinitis pigmentosa. Cell Death Dis 9, 1141. 10.1038/s41419-018-1198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P, 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science (New York, N.Y.) 318, 944–949. 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ, 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. The Journal of biological chemistry 275, 24881–24885. 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- Liu M, Weiss MA, Arunagiri A, Yong J, Rege N, Sun J, Haataja L, Kaufman RJ, Arvan P, 2018a. Biosynthesis, structure, and folding of the insulin precursor protein. Diabetes, obesity & metabolism 20 Suppl 2, 28–50. 10.1111/dom.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wilk SA, Wang A, Zhou L, Wang RH, Ogawa W, Deng C, Dong LQ, Liu F, 2010. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. The Journal of biological chemistry 285, 36387–36394. 10.1074/jbc.M110.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen Y, Chen Z, Xing Y, Shen Y, 2018b. Immunohistochemical profile of long-standing traumatic retinal detachment in atrophic globe in a young patient. Exp Ther Med 16, 2387–2391. 10.3892/etm.2018.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo GP, Ebke LA, Au A, Hagstrom SA, 2016. TULP1 Missense Mutations Induces the Endoplasmic Reticulum Unfolded Protein Response Stress Complex (ER-UPR). Adv Exp Med Biol 854, 223–230. 10.1007/978-3-319-17121-0_30. [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE, 2010. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. The Journal of biological chemistry 285, 21203–21207. 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D, 1998. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNAdependent conformational changes. Biochemistry 37, 2194–2206. 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- Luhr M, Torgersen ML, Szalai P, Hashim A, Brech A, Staerk J, Engedal N, 2019. The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress. J Biol Chem 294, 8197–8217. 10.1074/jbc.RA118.002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre P, Cavener DR, Klann E, 2013. Suppression of eIF2alpha kinases alleviates Alzheimer's disease-related plasticity and memory deficits. Nature neuroscience 16, 1299–1305. 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma TJ, Lan DH, He SZ, Ye Z, Li P, Zhai W, Chen WQ, Huang Y, Fu Y, Sun A, Wang YB, Ye Z, Li JL, Gao Y, Yan XL, Li ZH, 2018. Nrf2 protects human lens epithelial cells against H2O2-induced oxidative and ER stress: The ATF4 may be involved. Exp Eye Res 169, 28–37. 10.1016/j.exer.2018.01.018. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM, 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. The Journal of biological chemistry 278, 34864–34873. 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- Ma Y, Kawasaki R, Dobson LP, Ruddle JB, Kearns LS, Wong TY, Mackey DA, 2012. Quantitative analysis of retinal vessel attenuation in eyes with retinitis pigmentosa. Invest Ophthalmol Vis Sci 53, 4306–4314. 10.1167/iovs.11-8596. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Stefanidakis M, Wilson CJ, Baral R, Barrera LA, Bounoutas GS, Bumcrot D, Chao H, Ciulla DM, DaSilva JA, Dass A, Dhanapal V, Fennell TJ, Friedland AE, Giannoukos G, Gloskowski SW, Glucksmann A, Gotta GM, Jayaram H, Haskett SJ, Hopkins B, Horng JE, Joshi S, Marco E, Mepani R, Reyon D, Ta T, Tabbaa DG, Samuelsson SJ, Shen S, Skor MN, Stetkiewicz P, Wang T, Yudkoff C, Myer VE, Albright CF, Jiang H, 2019. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nature medicine 25, 229–233. 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ, 2007. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal 9, 2277–2293. 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- Mao H, Gorbatyuk MS, Rossmiller B, Hauswirth WW, Lewin AS, 2012. Long-term rescue of retinal structure and function by rhodopsin RNA replacement with a single adeno-associated viral vector in P23H RHO transgenic mice. Hum Gene Ther 23, 356–366. 10.1089/hum.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus A, Feeley J, 1966. Ribosome activation and polysome formation in vitro: requirement for ATP. Proceedings of the National Academy of Sciences of the United States of America 56, 1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marola OJ, Syc-Mazurek SB, Libby RT, 2019. DDIT3 (CHOP) contributes to retinal ganglion cell somal loss but not axonal degeneration in DBA/2J mice. Cell death discovery 5, 140. 10.1038/s41420-019-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Chen X, Lee AH, Glimcher LH, 2010. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol 11, 411–418. 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH, 2012. The tasks of amacrine cells. Visual neuroscience 29, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuoka HC, Townes TM, 2002. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99, 736–745. 10.1182/blood.v99.3.736. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Sugio S, Seghers F, Krizaj D, Akiyama H, Ishizaki Y, Gailly P, Shibasaki K, 2018. Retinal Detachment-Induced Muller Glial Cell Swelling Activates TRPV4 Ion Channels and Triggers Photoreceptor Death at Body Temperature. J Neurosci 38, 8745–8758. 10.1523/JNEUROSCI.0897-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL, 1996. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. The Journal of cell biology 132, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ, 2001. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol 21, 1249–1259. 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougald DS, Papp TE, Zezulin AU, Zhou S, Bennett J, 2019. AKT3 Gene Transfer Promotes Anabolic Reprogramming and Photoreceptor Neuroprotection in a Pre-clinical Model of Retinitis Pigmentosa. Mol Ther 27, 1313–1326. 10.1016/j.ymthe.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin T, Dhimal N, Li J, Wang JJ, Zhang SX, 2018. p58(IPK) Is an Endogenous Neuroprotectant for Retinal Ganglion Cells. Frontiers in aging neuroscience 10, 267. 10.3389/fnagi.2018.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meares GP, Liu Y, Rajbhandari R, Qin H, Nozell SE, Mobley JA, Corbett JA, Benveniste EN, 2014. PERK-dependent activation of JAK1 and STAT3 contributes to endoplasmic reticulum stress-induced inflammation. Mol Cell Biol 34, 3911–3925. 10.1128/MCB.00980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes HF, Cheetham ME, 2008. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Human molecular genetics 17, 3043–3054. 10.1093/hmg/ddn202. [DOI] [PubMed] [Google Scholar]
- Mercado G, Castillo V, Soto P, Lopez N, Axten JM, Sardi SP, Hoozemans JJM, Hetz C, 2018. Targeting PERK signaling with the small molecule GSK2606414 prevents neurodegeneration in a model of Parkinson's disease. Neurobiology of disease 112, 136–148. 10.1016/j.nbd.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Mercado G, Castillo V, Soto P, Sidhu A, 2016. ER stress and Parkinson's disease: Pathological inputs that converge into the secretory pathway. Brain research 1648, 626–632. 10.1016/j.brainres.2016.04.042. [DOI] [PubMed] [Google Scholar]
- Merrick WC, 1979. Evidence that a single GTP is used in the formation of 80 S initiation complexes. The Journal of biological chemistry 254, 3708–3711. [PubMed] [Google Scholar]
- Miraldi Utz V, Coussa RG, Antaki F, Traboulsi EI, 2018. Gene therapy for RPE65-related retinal disease. Ophthalmic genetics 39, 671–677. 10.1080/13816810.2018.1533027. [DOI] [PubMed] [Google Scholar]
- Mizobuchi K, Katagiri S, Hayashi T, Yoshitake K, Fujinami K, Kuniyoshi K, Mishima R, Tsunoda K, Iwata T, Nakano T, 2019. Clinical findings of end-stage retinitis pigmentosa with a homozygous PDE6A variant (p.R653X). American journal of ophthalmology case reports 13, 110–115. 10.1016/j.ajoc.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockel A, Obringer C, Hakvoort TBM, Seeliger M, Lamers WH, Stoetzel C, Dollfus H, Marion V, 2012. Pharmacological Modulation of the Retinal Unfolded Protein Response in Bardet-Biedl Syndrome Reduces Apoptosis and Preserves Light Detection Ability. Journal of Biological Chemistry 287, 37483–37494. 10.1074/jbc.M112.386821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrejs M, Masek T, Vopalensky V, Hlubucek P, Delbos P, Pospisek M, 2010. IRESite--a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res 38, D131–136. 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookherjee S, Chen HY, Isgrig K, Yu W, Hiriyanna S, Levron R, Li T, Colosi P, Chien W, Swaroop A, Wu Z, 2018. A CEP290 C-Terminal Domain Complements the Mutant CEP290 of Rd16 Mice In Trans and Rescues Retinal Degeneration. Cell reports 25, 611–623.e616. 10.1016/j.celrep.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, Barrett DA, Tsaytler P, Bertolotti A, Willis AE, Bushell M, Mallucci GR, 2012. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature 485, 507–511. 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshedian A, Fain GL, 2015. Single-photon sensitivity of lamprey rods with cone-like outer segments. Curr Biol 25, 484–487. 10.1016/j.cub.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Matsumoto H, Roh M, Suzuki J, Hisatomi T, Ikeda Y, Miller JW, Vavvas DG, 2012. Receptor interacting protein kinase mediates necrotic cone but not rod cell death in a mouse model of inherited degeneration. Proc Natl Acad Sci U S A 109, 14598–14603. 10.1073/pnas.1206937109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga-Zamalloa CA, Ghosh AK, Patil SB, Reed NA, Chan LS, Davuluri S, Peranen J, Hurd TW, Rachel RA, Khanna H, 2011. Accumulation of the Raf-1 kinase inhibitory protein (Rkip) is associated with Cep290-mediated photoreceptor degeneration in ciliopathies. The Journal of biological chemistry 286, 28276–28286. 10.1074/jbc.M111.237560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AR, Vuong L, Brobst D, Fliesler SJ, Peachey NS, Gorbatyuk MS, Naash MI, Al-Ubaidi MR, 2015. Glycosylation of rhodopsin is necessary for its stability and incorporation into photoreceptor outer segment discs. Hum Mol Genet 24, 2709–2723. 10.1093/hmg/ddv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarella MA, Macdonald IM, 2011. Current concepts in the treatment of retinitis pigmentosa. J Ophthalmol 2011, 753547. 10.1155/2011/753547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Shimazawa M, Sugitani S, Kudo T, Imai S, Inokuchi Y, Tsuruma K, Hara H, 2013. Role of endoplasmic reticulum stress in light-induced photoreceptor degeneration in mice. J Neurochem 125, 111–124. 10.1111/jnc.12116. [DOI] [PubMed] [Google Scholar]
- Nakao T, Tsujikawa M, Notomi S, Ikeda Y, Nishida K, 2012. The role of mislocalized phototransduction in photoreceptor cell death of retinitis pigmentosa. PLoS One 7, e32472. 10.1371/journal.pone.0032472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashine S, Bhootada Y, Lewin AS, Gorbatyuk M, 2013. Ablation of C/EBP homologous protein does not protect T17M RHO mice from retinal degeneration. PLoS One 8, e63205. 10.1371/journal.pone.0063205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HG, Conn CS, Kye Y, Xue L, Forester CM, Cowan JE, Hsieh AC, Cunningham JT, Truillet C, Tameire F, Evans MJ, Evans CP, Yang JC, Hann B, Koumenis C, Walter P, Carroll PR, Ruggero D, 2018. Development of a stress response therapy targeting aggressive prostate cancer. Science translational medicine 10.10.1126/scitranslmed.aar2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio N, Hasegawa T, Tatsuno I, Isaka M, Isobe KI, 2017. Female GADD34 mice develop age-related inflammation and hepatocellular carcinoma. Geriatrics & gerontology international 17, 2593–2601. 10.1111/ggi.13080. [DOI] [PubMed] [Google Scholar]
- Nishio N, Isobe K, 2015. GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance. Scientific reports 5, 13519. 10.1038/srep13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noell WK, 1951. The effect of iodoacetate on the vertebrate retina. J Cell Comp Physiol 37, 283–307. [DOI] [PubMed] [Google Scholar]
- Noell WK, 1952a. Electrophysiologic study of the retina during metabolic impairment. Am J Ophthalmol 35, 126–133. 10.1016/0002-9394(52)90265-1. [DOI] [PubMed] [Google Scholar]
- Noell WK, 1952b. The impairment of visual cell structure by iodoacetate. J Cell Comp Physiol 40, 25–55. 10.1002/jcp.1030400104. [DOI] [PubMed] [Google Scholar]
- Noiva R, Kimura H, Roos J, Lennarz WJ, 1991. Peptide binding by protein disulfide isomerase, a resident protein of the endoplasmic reticulum lumen. The Journal of biological chemistry 266, 19645–19649. [PubMed] [Google Scholar]
- Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K, 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. The Journal of biological chemistry 278, 15461–15464. 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- Nomura A, Shigemoto R, Nakamura Y, Okamoto N, Mizuno N, Nakanishi S, 1994. Developmentally regulated postsynaptic localization of a metabotropic glutamate receptor in rat rod bipolar cells. Cell 77, 361–369. [DOI] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding HP, Ron D, 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. The Journal of cell biology 153, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H, 2005. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 24, 1243–1255. 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T, 2009. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Experimental cell research 315, 2496–2504. 10.1016/j.yexcr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP, 1992. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron 9, 815–830. [DOI] [PubMed] [Google Scholar]
- Onate M, Catenaccio A, Martinez G, Armentano D, Parsons G, Kerr B, Hetz C, Court FA, 2016. Activation of the unfolded protein response promotes axonal regeneration after peripheral nerve injury. Sci Rep 6, 21709. 10.1038/srep21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyanagi K, Yamazaki M, Takahashi H, Watabe K, Wada M, Komori T, Morita T, Mizutani T, 2008. Spinal anterior horn cells in sporadic amyotrophic lateral sclerosis show ribosomal detachment from, and cisternal distention of the rough endoplasmic reticulum. Neuropathology and applied neurobiology 34, 650–658. 10.1111/j.1365-2990.2008.00941.x. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M, 2000. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745. 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Paquet-Durand F, Azadi S, Hauck SM, Ueffing M, van Veen T, Ekstrom P, 2006. Calpain is activated in degenerating photoreceptors in the rd1 mouse. J Neurochem 96, 802–814. 10.1111/j.1471-4159.2005.03628.x. [DOI] [PubMed] [Google Scholar]
- Partaledis JA, Berlin V, 1993. The FKB2 gene of Saccharomyces cerevisiae, encoding the immunosuppressant-binding protein FKBP-13, is regulated in response to accumulation of unfolded proteins in the endoplasmic reticulum. Proceedings of the National Academy of Sciences of the United States of America 90, 5450–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partovian C, Simons M, 2004. Regulation of protein kinase B/Akt activity and Ser473 phosphorylation by protein kinase Calpha in endothelial cells. Cellular signalling 16, 951–957. 10.1016/j.cellsig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Patterson AD, Hollander MC, Miller GF, Fornace AJ Jr., 2006. Gadd34 requirement for normal hemoglobin synthesis. Molecular and cellular biology 26, 1644–1653. 10.1128/mcb.26.5.1644-1653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng B, Xiao J, Wang K, So KF, Tipoe GL, Lin B, 2014. Suppression of microglial activation is neuroprotective in a mouse model of human retinitis pigmentosa. J Neurosci 34, 8139–8150. 10.1523/JNEUROSCI.5200-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyasamy P, Shinohara T, 2017. Age-related cataracts: Role of unfolded protein response, Ca(2+) mobilization, epigenetic DNA modifications, and loss of Nrf2/Keap1 dependent cytoprotection. Prog Retin Eye Res 60, 1–19. 10.1016/j.preteyeres.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins GA, Ellisman MH, Fox DA, 2003. Three-dimensional analysis of mouse rod and cone mitochondrial cristae architecture: bioenergetic and functional implications. Mol Vis 9, 60–73. [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Gerber S, Ghazi I, Leowski C, Ducroq D, Souied E, Dufier JL, Munnich A, Kaplan J, 1999. Leber congenital amaurosis. Molecular genetics and metabolism 68, 200–208. 10.1006/mgme.1999.2906. [DOI] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM, 2009. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886. 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer RL, Marc RE, Jones BW, 2019. Persistent remodeling and neurodegeneration in late-stage retinal degeneration. Prog Retin Eye Res. 10.1016/j.preteyeres.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Schoenfeld LW, Valasek L, Nielsen KH, Hinnebusch AG, 2001. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNA(i)Met. The EMBO Journal 20, 2954–2965. 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitale PM, Gorbatyuk O, Gorbatyuk M, 2017. Neurodegeneration: Keeping ATF4 on a Tight Leash. Front Cell Neurosci 11, 410. 10.3389/fncel.2017.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobre KFR, Poet GJ, Hendershot LM, 2019. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. The Journal of biological chemistry 294, 2098–2108. 10.1074/jbc.REV118.002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS, 1998. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell 1, 171–182. 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- Power M, Das S, Schutze K, Marigo V, Ekstrom P, Paquet-Durand F, 2020. Cellular mechanisms of hereditary photoreceptor degeneration - Focus on cGMP. Prog Retin Eye Res 74, 100772. 10.1016/j.preteyeres.2019.07.005. [DOI] [PubMed] [Google Scholar]
- Punzo C, Kornacker K, Cepko CL, 2009. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci 12, 44–52. 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Yao J, Jia L, Thompson DA, Zacks DN, 2019. Shifting the balance of autophagy and proteasome activation reduces proteotoxic cell death: a novel therapeutic approach for restoring photoreceptor homeostasis. Cell Death Dis 10, 547. 10.1038/s41419-019-1780-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachel RA, Yamamoto EA, Dewanjee MK, May-Simera HL, Sergeev YV, Hackett AN, Pohida K, Munasinghe J, Gotoh N, Wickstead B, Fariss RN, Dong L, Li T, Swaroop A, 2015. CEP290 alleles in mice disrupt tissue-specific cilia biogenesis and recapitulate features of syndromic ciliopathies. Hum Mol Genet 24, 3775–3791. 10.1093/hmg/ddv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford H, Moreno JA, Verity N, Halliday M, Mallucci GR, 2015. PERK inhibition prevents taumediated neurodegeneration in a mouse model of frontotemporal dementia. Acta Neuropathologica 130, 633–642. 10.1007/s00401-015-1487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahate K, Bhatt LK, Prabhavalkar KS, 2019. SERCA stimulation: a potential approach in therapeutics. Chem Biol Drug Des. 10.1111/cbdd.13620. [DOI] [PubMed] [Google Scholar]
- Rana T, Kotla P, Fullard R, Gorbatyuk M, 2017. TNFa knockdown in the retina promotes cone survival in a mouse model of autosomal dominant retinitis pigmentosa. Biochim Biophys Acta Mol Basis Dis 1863, 92–102. 10.1016/j.bbadis.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana T, Shinde VM, Starr CR, Kruglov AA, Boitet ER, Kotla P, Zolotukhin S, Gross AK, Gorbatyuk MS, 2014. An activated unfolded protein response promotes retinal degeneration and triggers an inflammatory response in the mouse retina. Cell Death Dis 5, e1578. 10.1038/cddis.2014.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane MJ, Coxon PY, Powell DW, Webster R, Klein JB, Pierce W, Ping P, McLeish KR, 2001. p38 Kinase-dependent MAPKAPK-2 activation functions as 3-phosphoinositide-dependent kinase-2 for Akt in human neutrophils. The Journal of biological chemistry 276, 3517–3523. 10.1074/jbc.M005953200. [DOI] [PubMed] [Google Scholar]
- Reme CE, Grimm C, Hafezi F, Marti A, Wenzel A, 1998. Apoptotic cell death in retinal degenerations. Prog Retin Eye Res 17, 443–464. [DOI] [PubMed] [Google Scholar]
- Rieke F, Baylor DA, 1998. Origin of reproducibility in the responses of retinal rods to single photons. Biophys J 75, 1836–1857. 10.1016/S0006-3495(98)77625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripps H, 2002. Cell death in retinitis pigmentosa: gap junctions and the 'bystander' effect. Exp Eye Res 74, 327–336. 10.1006/exer.2002.1155. [DOI] [PubMed] [Google Scholar]
- Robinson A, He M, Westwood OM, Austen BM, 1993. Changes in levels of pancreatic endoplasmic reticulum proteins that function in translocation and maturation of secretory proteins in response to cholecystokinin. Cytotechnology 11, 197–204. [DOI] [PubMed] [Google Scholar]
- Rojas-Rivera D, Delvaeye T, Roelandt R, Nerinckx W, Augustyns K, Vandenabeele P, Bertrand MJM, 2017. When PERK inhibitors turn out to be new potent RIPK1 inhibitors: critical issues on the specificity and use of GSK2606414 and GSK2656157. Cell death and differentiation 24, 1100–1110. 10.1038/cdd.2017.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Habener JF, 1992. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes & development 6, 439–453. [DOI] [PubMed] [Google Scholar]
- Ross JA, Bressler KR, Thakor N, 2018. Eukaryotic Initiation Factor 5B (eIF5B) Cooperates with eIF1A and eIF5 to Facilitate uORF2-Mediated Repression of ATF4 Translation. International journal of molecular sciences 19 10.3390/ijms19124032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenstreich Y, Belkin M, Sadetzki S, Chetrit A, Ferman-Attar G, Sher I, Harari A, Shaish A, Harats D, 2013. Treatment with 9-cis beta-carotene-rich powder in patients with retinitis pigmentosa: a randomized crossover trial. JAMA Ophthalmol 131, 985–992. 10.1001/jamaophthalmol.2013.147. [DOI] [PubMed] [Google Scholar]
- Rueda EM, Johnson JE Jr., Giddabasappa A, Swaroop A, Brooks MJ, Sigel I, Chaney SY, Fox DA, 2016. The cellular and compartmental profile of mouse retinal glycolysis, tricarboxylic acid cycle, oxidative phosphorylation, and ~P transferring kinases. Mol Vis 22, 847–885. [PMC free article] [PubMed] [Google Scholar]
- Rushton WA, 1965. Visual Adaptation. Proc R Soc Lond B Biol Sci 162, 20–46. 10.1098/rspb.1965.0024. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Vasudevan D, 2017. Two distinct nodes of translational inhibition in the Integrated Stress Response. BMB Rep 50, 539–545. 10.5483/bmbrep.2017.50.11.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L, 2011. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 20, 131–139. 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakami S, Kolesnikov AV, Kefalov VJ, Palczewski K, 2014. P23H opsin knock-in mice reveal a novel step in retinal rod disc morphogenesis. Hum Mol Genet 23, 1723–1741. 10.1093/hmg/ddt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, Roman AJ, Cideciyan AV, Jacobson SG, Palczewski K, 2011. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem 286, 10551–10567. 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samtleben S, Jaepel J, Fecher C, Andreska T, Rehberg M, Blum R, 2013. Direct imaging of ER calcium with targeted-esterase induced dye loading (TED). Journal of visualized experiments : JoVE, e50317. 10.3791/50317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandsmark DK, Pelletier C, Weber JD, Gutmann DH, 2007. Mammalian target of rapamycin: master regulator of cell growth in the nervous system. Histology and histopathology 22, 895–903. 10.14670/hh-22.895. [DOI] [PubMed] [Google Scholar]
- Sanges D, Comitato A, Tammaro R, Marigo V, 2006. Apoptosis in retinal degeneration involves cross-talk between apoptosis-inducing factor (AIF) and caspase-12 and is blocked by calpain inhibitors. Proc Natl Acad Sci U S A 103, 17366–17371. 10.1073/pnas.0606276103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM, 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Current biology : CB 14, 1296–1302. 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sato K, Li S, Gordon WC, He J, Liou GI, Hill JM, Travis GH, Bazan NG, Jin M, 2013. Receptor interacting protein kinase-mediated necrosis contributes to cone and rod photoreceptor degeneration in the retina lacking interphotoreceptor retinoid-binding protein. J Neurosci 33, 17458–17468. 10.1523/JNEUROSCI.1380-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F, 2006. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nature genetics 38, 674–681. 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- Scarpelli DG, Craig EL, 1963. The fine localization of nucleoside triphosphatase activity in the retina of the frog. J Cell Biol 17, 279–288. 10.1083/jcb.17.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapf JL, Kraft TW, Nunn BJ, Baylor DA, 1988. Spectral sensitivity of primate photoreceptors. Visual neuroscience 1, 255–261. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Blower MD, 2016. The endoplasmic reticulum: structure, function and response to cellular signaling. Cellular and molecular life sciences : CMLS 73, 79–94. 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D, Ronai ZA, 2015. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci 40, 141–148. 10.1016/j.tibs.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Ounallah-Saad H, Chakraborty D, Hleihil M, Sood R, Barrera I, Edry E, Kolatt Chandran S, Ben Tabou de Leon S, Kaphzan H, Rosenblum K, 2018. Local Inhibition of PERK Enhances Memory and Reverses Age-Related Deterioration of Cognitive and Neuronal Properties. The Journal of neuroscience : the official journal of the Society for Neuroscience 38, 648–658. 10.1523/jneurosci.0628-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R, 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Developmental cell 3, 99–111. [DOI] [PubMed] [Google Scholar]
- Shen J, Snapp EL, Lippincott-Schwartz J, Prywes R, 2005. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Molecular and cellular biology 25, 921–932. 10.1128/mcb.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X, 2004. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. The Journal of cell biology 164, 291–300. 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC, 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Molecular and cellular biology 18, 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V, Kotla P, Strang C, Gorbatyuk M, 2016. Unfolded protein response-induced dysregulation of calcium homeostasis promotes retinal degeneration in rat models of autosomal dominant retinitis pigmentosa. Cell Death Dis 7, e2085. 10.1038/cddis.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S, Cherepanova NA, Gilmore R, 2015. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Seminars in cell & developmental biology 41, 71–78. 10.1016/j.semcdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli M, Musner N, Silvestri N, Ungaro D, D'Antonio M, Cavener DR, Feltri ML, Wrabetz L, 2016. Ablation of Perk in Schwann Cells Improves Myelination in the S63del Charcot-Marie-Tooth 1B Mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 11350–11361. 10.1523/jneurosci.1637-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova OS, Shinde VM, Lenox AR, Gorbatyuk MS, 2014. Modulation of cellular signaling pathways in P23H rhodopsin photoreceptors. Cell Signal 26, 665–672. 10.1016/j.cellsig.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RG, Freed MA, Sterling P, 1986. Microcircuitry of the dark-adapted cat retina: functional architecture of the rod-cone network. J Neurosci 6, 3505–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soiberman US, Shehata AEM, Lu MX, Young T, Daoud YJ, Chakravarti S, Jun AS, Foster JW, 2019. Small Molecule Modulation of the Integrated Stress Response Governs the Keratoconic Phenotype In Vitro. Invest Ophthalmol Vis Sci 60, 3422–3431. 10.1167/iovs.19-27151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, 2008. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochemistry and cell biology = Biochimie et biologie cellulaire 86, 178–183. 10.1139/o08-034. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG, 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ, 1978. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5'-terminal cap in mRNA. Proceedings of the National Academy of Sciences of the United States of America 75, 4843–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YF, Hogstrand C, Wei CC, Wu K, Pan YX, Luo Z, 2017. Endoplasmic reticulum (ER) stress and cAMP/PKA pathway mediated Zn-induced hepatic lipolysis. Environ Pollut 228, 256–264. 10.1016/j.envpol.2017.05.046. [DOI] [PubMed] [Google Scholar]
- Sorensen NB, Christiansen AT, Kjaer TW, Klemp K, Cour M, Heegaard S, Warfvinge K, Kiilgaard JF, 2019. Loss of retinal tension and permanent decrease in retinal function: a new porcine model of rhegmatogenous retinal detachment. Acta Ophthalmol. 10.1111/aos.14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P, 2016. Translation from the 5' untranslated region shapes the integrated stress response. Science (New York, N.Y.) 351, aad3867. 10.1126/science.aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr CR, Gorbatyuk MS, 2019. Delineating the role of eIF2alpha in retinal degeneration. Cell Death Dis 10, 409. 10.1038/s41419-019-1641-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr CR, Nyankerh CNA, Qi X, Hu Y, Gorbatyuk OS, Sonenberg N, Boulton ME, Gorbatyuk MS, 2019. Role of Translational Attenuation in Inherited Retinal Degeneration. Invest Ophthalmol Vis Sci 60, 4849–4857. 10.1167/iovs.19-27512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr CR, Pitale PM, Gorbatyuk M, 2018. Translational attenuation and retinal degeneration in mice with an active integrated stress response. Cell Death Dis 9, 484. 10.1038/s41419-018-0513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieber A, Gonatas JO, Collard J, Meier J, Julien J, Schweitzer P, Gonatas NK, 2000. The neuronal Golgi apparatus is fragmented in transgenic mice expressing a mutant human SOD1, but not in mice expressing the human NF-H gene. Journal of the neurological sciences 173, 63–72. [DOI] [PubMed] [Google Scholar]
- Su Q, Wang S, Gao HQ, Kazemi S, Harding HP, Ron D, Koromilas AE, 2008. Modulation of the eukaryotic initiation factor 2 alpha-subunit kinase PERK by tyrosine phosphorylation. The Journal of biological chemistry 283, 469–475. 10.1074/jbc.M704612200. [DOI] [PubMed] [Google Scholar]
- Subramanian B, Anand M, Khan NW, Khanna H, 2014. Loss of Raf-1 kinase inhibitory protein delays early-onset severe retinal ciliopathy in Cep290rd16 mouse. Investigative ophthalmology & visual science 55, 5788–5794. 10.1167/iovs.14-14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Kotler JLM, Liu S, Street TO, 2019. The ER chaperones BiP and Grp94 selectively associate when BiP is in the ADP conformation. The Journal of biological chemistry. 10.1074/jbc.RA118.007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Liu J, Crary JF, Malagelada C, Sulzer D, Greene LA, Levy OA, 2013. ATF4 protects against neuronal death in cellular Parkinson's disease models by maintaining levels of parkin. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 2398–2407. 10.1523/jneurosci.2292-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder AH, Wang RA, Kumar R, 2002. Expression and transactivating functions of the bZIP transcription factor GADD153 in mammary epithelial cells. Oncogene 21, 4289–4300. 10.1038/sj.onc.1205529. [DOI] [PubMed] [Google Scholar]
- Taniuchi S, Miyake M, Tsugawa K, Oyadomari M, Oyadomari S, 2016. Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Scientific reports 6, 32886. 10.1038/srep32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ Jr., Sachs AB, 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. The EMBO Journal 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ Jr., Wells SE, Deardorff JA, Sachs AB, 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proceedings of the National Academy of Sciences of the United States of America 94, 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayebi N, Akinrinade O, Khan MI, Hejazifar A, Dehghani A, Cremers FPM, Akhlaghi M, 2019. Targeted next generation sequencing reveals genetic defects underlying inherited retinal disease in Iranian families. Molecular vision 25, 106–117. [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ, 2000. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes & development 14, 2725–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo D, Ramon E, Aguila M, Cordomi A, Perez JJ, Mendes HF, Cheetham ME, Garriga P, 2011. Molecular mechanisms of disease for mutations at Gly-90 in rhodopsin. J Biol Chem 286, 39993–40001. 10.1074/jbc.M110.201517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso V, Bush RA, Sieving PA, Deretic D, 2002. Retinal cAMP levels during the progression of retinal degeneration in rhodopsin P23H and S334ter transgenic rats. Invest Ophthalmol Vis Sci 43, 1655–1661. [PubMed] [Google Scholar]
- Tsang SH, Sharma T, 2018. Leber Congenital Amaurosis. Advances in experimental medicine and biology 1085, 131–137. 10.1007/978-3-319-95046-4_26. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D, 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666. 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K, 2008. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321, 569–572. 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- Valasek L, Phan L, Schoenfeld LW, Valaskova V, Hinnebusch AG, 2001. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. The EMBO Journal 20, 891–904. 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes P, Mercado G, Vidal RL, Molina C, Parsons G, Court FA, Martinez A, Galleguillos D, Armentano D, Schneider BL, Hetz C, 2014. Control of dopaminergic neuron survival by the unfolded protein response transcription factor XBP1. Proceedings of the National Academy of Sciences of the United States of America 111, 6804–6809. 10.1073/pnas.1321845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela V, Collyer E, Armentano D, Parsons GB, Court FA, Hetz C, 2012. Activation of the unfolded protein response enhances motor recovery after spinal cord injury. Cell Death Dis 3, e272. 10.1038/cddis.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet AR, Giordano F, Gerlo S, Segura I, Van Eygen S, Molenberghs G, Rocha S, Houcine A, Derua R, Verfaillie T, Vangindertael J, De Keersmaecker H, Waelkens E, Tavernier J, Hofkens J, Annaert W, Carmeliet P, Samali A, Mizuno H, Agostinis P, 2017. The ER Stress Sensor PERK Coordinates ER-Plasma Membrane Contact Site Formation through Interaction with Filamin-A and F-Actin Remodeling. Molecular cell 65, 885–899.e886. 10.1016/j.molcel.2017.01.020. [DOI] [PubMed] [Google Scholar]
- Vasireddy V, Chavali VR, Joseph VT, Kadam R, Lin JH, Jamison JA, Kompella UB, Reddy GB, Ayyagari R, 2011. Rescue of photoreceptor degeneration by curcumin in transgenic rats with P23H rhodopsin mutation. PloS one 6, e21193. 10.1371/journal.pone.0021193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Dominguez I, Garanto A, Collin RWJ, 2019. Molecular Therapies for Inherited Retinal Diseases-Current Standing, Opportunities and Challenges. Genes (Basel) 10.10.3390/genes10090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenz L, Hartl FU, 2014. Sugarcoating ER Stress. Cell 156, 1125–1127. 10.1016/j.cell.2014.02.035. [DOI] [PubMed] [Google Scholar]
- Viringipurampeer IA, Gregory-Evans CY, Metcalfe AL, Bashar E, Moritz OL, Gregory-Evans K, 2019. Cell Death Pathways in Mutant Rhodopsin Rat Models Identifies Genotype-Specific Targets Controlling Retinal Degeneration. Mol Neurobiol 56, 1637–1652. 10.1007/s12035-018-1192-8. [DOI] [PubMed] [Google Scholar]
- Viringipurampeer IA, Metcalfe AL, Bashar AE, Sivak O, Yanai A, Mohammadi Z, Moritz OL, Gregory-Evans CY, Gregory-Evans K, 2016. NLRP3 inflammasome activation drives bystander cone photoreceptor cell death in a P23H rhodopsin model of retinal degeneration. Hum Mol Genet 25, 1501–1516. 10.1093/hmg/ddw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viringipurampeer IA, Shan X, Gregory-Evans K, Zhang JP, Mohammadi Z, Gregory-Evans CY, 2014. Rip3 knockdown rescues photoreceptor cell death in blind pde6c zebrafish. Cell Death Differ 21, 665–675. 10.1038/cdd.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale M, Bakunts A, Orsi A, Lari F, Tade L, Danieli A, Rato C, Valetti C, Sitia R, Raimondi A, Christianson JC, van Anken E, 2019. Inadequate BiP availability defines endoplasmic reticulum stress. Elife 8 10.7554/eLife.41168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volland S, Hughes LC, Kong C, Burgess BL, Linberg KA, Luna G, Zhou ZH, Fisher SK, Williams DS, 2015. Three-dimensional organization of nascent rod outer segment disk membranes. Proc Natl Acad Sci U S A 112, 14870–14875. 10.1073/pnas.1516309112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakai T, Fissore RA, 2013. Ca(2+) homeostasis and regulation of ER Ca(2+) in mammalian oocytes/eggs. Cell Calcium 53, 63–67. 10.1016/j.ceca.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D, 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science (New York, N.Y.) 334, 1081–1086. 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Walton GM, Gill GN, 1976. Regulation of ternary (Met-tRNAf - GTP - eukaryotic initiation factor 2) protein synthesis initiation complex formation by the adenylate energy charge. Biochimica et biophysica acta 418, 195–203. [DOI] [PubMed] [Google Scholar]
- Wang M, Kaufman RJ, 2016. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335. 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- Wang P, Li J, Tao J, Sha B, 2018. The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization. The Journal of biological chemistry 293, 4110–4121. 10.1074/jbc.RA117.001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Chen J, 2014. Induction of the unfolded protein response by constitutive G-protein signaling in rod photoreceptor cells. J Biol Chem 289, 29310–29321. 10.1074/jbc.M114.595207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Fernandez de Castro J, Vukmanic E, Zhou L, Emery D, Demarco PJ, Kaplan HJ, Dean DC, 2011. Selective rod degeneration and partial cone inactivation characterize an iodoacetic acid model of Swine retinal degeneration. Invest Ophthalmol Vis Sci 52, 7917–7923. 10.1167/iovs.11-7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang G, Kunte M, Shinde V, Gorbatyuk M, 2013. Modulation of angiogenesis by genetic manipulation of ATF4 in mouse model of oxygen-induced retinopathy [corrected]. Invest Ophthalmol Vis Sci 54, 5995–6002. 10.1167/iovs.13-12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Lawson B, Brewer JW, Zinszner H, Sanjay A, Mi LJ, Boorstein R, Kreibich G, Hendershot LM, Ron D, 1996. Signals from the stressed endoplasmic reticulum induce C/EBPhomologous protein (CHOP/GADD153). Mol Cell Biol 16, 4273–4280. 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Ron D, 1996. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP Kinase. Science 272, 1347–1349. 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- Wassle H, 2004. Parallel processing in the mammalian retina. Nature reviews. Neuroscience 5, 747–757. 10.1038/nrn1497. [DOI] [PubMed] [Google Scholar]
- Weisschuh N, Feldhaus B, Khan MI, Cremers FPM, Kohl S, Wissinger B, Zobor D, 2018. Molecular and clinical analysis of 27 German patients with Leber congenital amaurosis. PloS one 13, e0205380. 10.1371/journal.pone.0205380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werblin FS, Dowling JE, 1969. Organization of the retina of the mudpuppy, Necturus maculosus. II. Intracellular recording. Journal of neurophysiology 32, 339–355. 10.1152/jn.1969.32.3.339. [DOI] [PubMed] [Google Scholar]
- Wilson JH, Wensel TG, 2003. The nature of dominant mutations of rhodopsin and implications for gene therapy. Mol Neurobiol 28, 149–158. 10.1385/MN:28:2:149. [DOI] [PubMed] [Google Scholar]
- Win S, Than TA, Fernandez-Checa JC, Kaplowitz N, 2014. JNK interaction with Sab mediates ER stress induced inhibition of mitochondrial respiration and cell death. Cell Death Dis 5, e989. 10.1038/cddis.2013.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler BS, 1981. Glycolytic and oxidative metabolism in relation to retinal function. J Gen Physiol 77, 667–692. 10.1085/jgp.77.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Hou S, Orr BA, Kuo BR, Youn YH, Ong T, Roth F, Eberhart CG, Robinson GW, Solecki DJ, Taketo MM, Gilbertson RJ, Roussel MF, Han YG, 2017. mTORC1-Mediated Inhibition of 4EBP1 Is Essential for Hedgehog Signaling-Driven Translation and Medulloblastoma. Developmental cell 43, 673–688.e675. 10.1016/j.devcel.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Rapoport TA, 2018. Mechanistic insights into ER-associated protein degradation. Current opinion in cell biology 53, 22–28. 10.1016/j.ceb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zhong M, Li M, Huang H, Liao J, Lu A, Guo K, Ma N, Lin J, Duan J, Liu L, Xu F, Zhong Z, Chen J, 2018. Mutation Analysis of Pre-mRNA Splicing Genes PRPF31, PRPF8, and SNRNP200 in Chinese Families with Autosomal Dominant Retinitis Pigmentosa. Current molecular medicine 18, 287–294. 10.2174/1566524018666181024160452. [DOI] [PubMed] [Google Scholar]
- Xiong G, Hindi SM, Mann AK, Gallot YS, Bohnert KR, Cavener DR, Whittemore SR, Kumar A, 2017. The PERK arm of the unfolded protein response regulates satellite cell-mediated skeletal muscle regeneration. eLife 6 10.7554/eLife.22871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhang L, Yang Y, Li N, Lai W, Wang F, Zhu X, Wang T, 2019. ER complex proteins are required for rhodopsin biosynthesis and photoreceptor survival in Drosophila and mice. Cell Death Differ. 10.1038/s41418-019-0378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Xiao Z, Zou T, Huang Z, 2015. Induction of GADD34 Regulates the Neurotoxicity of Amyloid beta. American journal of Alzheimer's disease and other dementias 30, 313–319. 10.1177/1533317514545616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Kamiyama M, Nitta K, Yamada T, Hayasaka S, 1996. Selective reduction of the S cone electroretinogram in diabetes. Br J Ophthalmol 80, 973–975. 10.1136/bjo.80.11.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li S, Miao L, Huang H, Liang F, Teng X, Xu L, Wang Q, Xiao W, Ridder WH 3rd, Ferguson TA, Chen DF, Kaufman RJ, Hu Y, 2016a. Rescue of Glaucomatous Neurodegeneration by Differentially Modulating Neuronal Endoplasmic Reticulum Stress Molecules. J Neurosci 36, 5891–5903. 10.1523/JNEUROSCI.3709-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zhou X, Zimmermann HR, Cavener DR, Klann E, Ma T, 2016b. Repression of the eIF2alpha kinase PERK alleviates mGluR-LTD impairments in a mouse model of Alzheimer's disease. Neurobiology of aging 41, 19–24. 10.1016/j.neurobiolaging.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarfitz S, Hurley JB, 1994. Transduction mechanisms of vertebrate and invertebrate photoreceptors. The Journal of biological chemistry 269, 14329–14332. [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K, 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K, 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Molecular and cellular biology 20, 6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Ikeda Y, Notomi S, Ishikawa K, Murakami Y, Hisatomi T, Enaida H, Ishibashi T, 2013. Clinical evidence of sustained chronic inflammatory reaction in retinitis pigmentosa. Ophthalmology 120, 100–105. 10.1016/j.ophtha.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Yu M, Yan W, Beight C, 2018. Lutein and Zeaxanthin Isomers Reduce Photoreceptor Degeneration in the Pde6brd10 Mouse Model of Retinitis Pigmentosa. BioMed Research International 2018, 8. 10.1155/2018/4374087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Q, Lord KA, Alamo I Jr., Hollander MC, Carrier F, Ron D, Kohn KW, Hoffman B, Liebermann DA, Fornace AJ Jr., 1994. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Molecular and cellular biology 14, 2361–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Yang W, Jiang F, Zou P, Zeng Y, Ling X, Zhou Z, Cao J, Ao L, 2019. PERK regulates Nrf2/ARE antioxidant pathway against dibutyl phthalate-induced mitochondrial damage and apoptosis dependent of reactive oxygen species in mouse spermatocyte-derived cells. Toxicol Lett 308, 24–33. 10.1016/j.toxlet.2019.03.007. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ, 2003a. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. The Journal of clinical investigation 112, 1223–1233. 10.1172/jci17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR, 2002. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Molecular and cellular biology 22, 3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SX, Ma JH, Bhatta M, Fliesler SJ, Wang JJ, 2015. The unfolded protein response in retinal vascular diseases: implications and therapeutic potential beyond protein folding. Prog Retin Eye Res 45, 111–131. 10.1016/j.preteyeres.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li L, Su Q, Gao G, Khanna H, 2018. Gene Therapy Using a miniCEP290 Fragment Delays Photoreceptor Degeneration in a Mouse Model of Leber Congenital Amaurosis. Human gene therapy 29, 42–50. 10.1089/hum.2017.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wensel TG, Kraft TW, 2003b. GTPase regulators and photoresponses in cones of the eastern chipmunk. J Neurosci 23, 1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang G, Xu X, Su W, Yu B, 2012. mTOR-rictor is the Ser473 kinase for AKT1 in mouse one-cell stage embryos. Molecular and cellular biochemistry 361, 249–257. 10.1007/s11010-011-1110-0. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zabel MK, Wang X, Ma W, Shah P, Fariss RN, Qian H, Parkhurst CN, Gan WB, Wong WT, 2015a. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med 7, 1179–1197. 10.15252/emmm.201505298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhou J, Liu D, Dong F, Cheng H, Wang W, Pang Y, Wang Y, Mu X, Ni Y, Li Z, Xu H, Hao S, Wang X, Ma S, Wang QF, Xiao G, Yuan W, Liu B, Cheng T, 2015b. ATF4 plays a pivotal role in the development of functional hematopoietic stem cells in mouse fetal liver. Blood 126, 2383–2391. 10.1182/blood-2015-03-633354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan X, Zhang X, Hess ME, Bruning JC, Qian SB, 2018. N(6)-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response. Molecular cell 69, 636–647.e637. 10.1016/j.molcel.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Bennett TM, Shiels A, 2016. Lens ER-stress response during cataract development in Mip-mutant mice. Biochim Biophys Acta 1862, 1433–1442. 10.1016/j.bbadis.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Liu H, Sha H, Qi L, Gao DS, Zhang W, 2017. PERK and XBP1 differentially regulate CXCL10 and CCL2 production. Exp Eye Res 155, 1–14. 10.1016/j.exer.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]