Abstract
Background:
Diagnosis of eosinophilic esophagitis (EoE) requires manual quantification of tissue eosinophils. Eosinophil peroxidase (EPX) is an eosinophil-specific, cytoplasmic granule protein released during degranulation.
Aims:
The objective of this study was to evaluate image analysis of EPX immunohistochemistry as an automated method for histologic diagnosis of EoE.
Methods:
We performed a secondary analysis of prospectively collected esophageal biopsies obtained from adult subjects with EoE and controls. Tissue sections were stained with hematoxylin and eosin (H&E) and evaluated for peak eosinophils per high power field (eos/hpf). The same slides were de-stained and re-stained to detect EPX for direct comparison. Slides were digitized and EPX staining area/mm2 was quantified using image analysis. Paired samples were compared for changes in EPX staining in treatment responders and non-responders.
Results:
Thirty-eight EoE cases and 49 controls were analyzed. Among EoE subjects, matched post-treatment biopsies were available for 21 responders and 10 non-responders. Baseline EPX/mm2 was significantly increased in EoE subjects and decreased in treatment responders. EPX quantification correlated strongly with eos/hpf (r = 0.84, p < 0.0001) and identified EoE subjects with high diagnostic accuracy (AUC 0.95, p < 0.0001). The optimal diagnostic EPX-positive pixel/area threshold was 17,379 EPX/mm2. Several controls (5/49) with < 15 eos/hpf on H&E staining exceeded this cutoff.
Conclusions:
EPX/mm2 correlates strongly with eos/hpf, accurately identifies subjects with EoE, and decreases in treatment responders. Automated quantification of intact eosinophils and their degranulation products may enhance pathologic assessment. Future studies are needed to correlate EPX/mm2 with symptoms, endoscopic findings, and esophageal distensibility.
Keywords: Eosinophilic esophagitis, eosinophil peroxidase, image analysis, degranulation, biomarker
INTRODUCTION
Eosinophilic esophagitis (EoE) is a clinicopathologic diagnosis characterized by symptoms of esophageal dysfunction and eosinophilic inflammation [1]. Although eosinophils are resident in the stomach and intestines, any eosinophil infiltration of the esophagus may be indicative of pathology. The diagnosis of EoE requires a pathologic esophageal eosinophilic infiltrate in the appropriate clinical context with exclusion of other causes of esophageal eosinophilia. The histologic threshold for diagnosis of EoE is a peak esophageal eosinophil count of ≥ 15 eosinophils per high power field (eos/hpf).
Reliance on manual assessment of intact eosinophils within a high power field is problematic for several reasons:
Counting eosinophils on a hematoxylin and eosin (H&E) stained biopsy section is time consuming and labor intensive;
Due to the difficulty inherent to identifying eosinophils with H&E staining, eosinophilic inflammation may not be readily detected by pathologists [2];
The area of a high power field (ocular 10x and objective 40x = 400x magnification) may vary depending the biopsy section as well as the microscope and objective used [3]. This can be overcome by converting to a unit area (e.g. eos/mm2); however, this is not done routinely on a clinical basis;
Eosinophilic inflammation in EoE is patchy [4] and requires assessment of multiple biopsies in order to achieve adequate diagnostic sensitivity [5];
Exact peak eos/hpf are more difficult to report when eosinophil counts are extremetly elevated;
Up to 80% of tissue eosinophils undergo cytolytic degranulation [6]; therefore, measuring the density of intact eosinophils may underestimate the extent of eosinophilic inflammation and, potentially, misclassify individuals with active disease manifested primarily by extensive eosinophil tissue degranulation [7].
Eosinophil peroxidase (EPX) is an eosinophil secondary granule protein (ESGP) stored in cytoplasmic granules and released during degranulation. Unlike other ESGPs, which are present in other cell types, EPX is exclusively produced by eosinophils [8]. We have demonstrated the utility of EPX immunohistochemistry (IHC) as a marker of both intact eosinophils and their degranulation products in pediatric and adult studies of EoE [9–12]. In addition to EoE, we have shown that image analysis of EPX IHC may also have utility in other disease states such as food allergy [13] and chronic rhinosinusitis [14]. Importantly, eosinophil degranulation assessed by EPX IHC correlates better with dysphagia in EoE than eos/hpf [10]. Our previous efforts to quantify EPX immunostaining in tissue utilized a novel scoring system that assessed multiple elements [9]. While this method comprehensively evaluates EPX involvement, it is also time consuming, subjective, and more labor intensive than manual eosinophil counts of H&E stained tissue sections. We have developed an automated technique using image analysis of digitized slides that allows for rapid and thorough quantification of EPX staining in tissue [13,14]. The objective of this study was to evaluate image analysis of EPX IHC as an automated method for histologic diagnosis of EoE. We hypothesized that EPX staining would correlate with manual eosinophil counts and identify subjects with EoE.
METHODS
Study population
We performed a retrospective analysis of prospectively banked esophageal tissue biopsies collected from adults at the University of North Carolina (UNC) as previously described [15–18]. In brief, biopsies were previously obtained from adult subjects (ages 18 to 80) with symptoms of esophageal dysfunction (dysphagia, heartburn, reflux, etc.) referred for outpatient esophagogastroduodenoscopy (EGD). Subjects were excluded from prospective enrollment in the UNC EoE biobank for any of the following reasons: known diagnosis of EoE or another eosinophilic gastrointestinal disorder; gastrointestinal bleeding; active anticoagulation; known esophageal cancer; prior esophageal surgery; known esophageal varices; medical instability or multiple comorbidities precluding enrollment in the clinical opinion of the endoscopist; or inability to provide consent. Consent was obtained for banking of research specimens prior to endoscopy. Slides from esophageal biopsies were selected as a convenience sample. This study was approved by the UNC Institutional Review Board.
Case definitions, clinical data, and biospecimen collection
EoE was diagnosed based on previous consensus guidelines at the time the patients were recruited and samples were collected (symptoms of esophageal dysfunction and > 15 eos/hpf in esophageal biopsies following an 8-week trial of high dose proton pump inhibitor (PPI) therapy) [19]. Demographics and comorbid atopic disorders were assessed by a baseline questionnaire administered after the PPI trial at the time of the diagnostic endoscopy. Controls were subjects referred for EGD with symptoms of dysphagia or gastroesophageal reflux disease (GERD) and < 15 eos/hpf on esophageal biopsy. Because samples were collected prior to the recent change eliminating a PPI trial from the diagnostic criteria [1], subjects with PPI-responsive EoE were excluded from this study. Longitudinal samples collected from patients treated with either topical swallowed steroids or dietary elimination after at least 6 weeks, as per standard clinical protocol were also analyzed. A positive treatment response was defined as a peak eosinophil count of < 15 eos/hpf in the subsequent esophageal biopsy [20,21].
Standardized case report forms were used to collect clinical data and a prospectively administered questionnaire was used to assess demographics, medical history, symptoms, allergic conditions, indications for endoscopy, endoscopic findings, and final diagnoses.Food allergies were assessed by subject self-report. Five esophageal biopsies were obtained for research purposes from the proximal (2 biopsies), mid (1 biopsy) and distal (2 biopsies) esophagus during the time of EGD. Tissue biopsies were fixed in formalin and embedded in paraffin. Gastric and duodenal biopsies were also collected to exclude concomitant eosinophilic gastroenteritis. Esophageal eosinophil counts were determined using H&E stains by a gastrointestinal pathologist according to a previously validated protocol [22,23]. Study personnel were blinded as to case/control status and EPX measurements during sample analysis.
EPX immunohistochemistry (IHC)
EPX was assessed by IHC using a mouse monoclonal anti-eosinophil peroxidase antibody (EPX-mAb; clone 82.2.1). This antibody stains EPX deposited in tissue, in free cytoplasmic granules, and within intact eosinophils. H&E stains from the esophageal segment with peak eosinophil counts were de-stained and re-stained for EPX for direct comparison. Slides were immersed in xylene until coverslips were easily removed (1–5 days). The tissue was rehydrated by bathing slides in 50% xylene:50% ethanol, 100% ethanol, 95% ethanol, and 70% ethanol for two minutes each. Slides were soaked in 1% HCl in 70% ethanol for 1 minute and then washed with deionized water. Antigen retrieval and EPX staining was performed as previously described [9,11,12].
Analysis of EPX stains
Tissue EPX was quantified as previously described [13]. Briefly, tissue sections were digitized (Aperio AT Turbo, Leica Biosystems, Buffalo Grove, IL) and EPX staining in an area equivalent to a high power field (0.24mm2) was measured using an automated pixel count algorithm with Aperio ImageScope software (version 11.2.0.780, Aperio Technologies, Vista, CA). The high power field with the most EPX staining was selected manually. For slides with multiple areas of eosinophilic involvement, several high power fields were analyzed to ensure the peak level of EPX staining was recorded. The number of pixels staining positive for EPX was divided by the area analyzed to calculate the peak EPX staining density (EPX/mm2). The research technician was blinded to clinical information and eos/hpf based on H&E stains.
Statistical analysis
Descriptive statistics were used to summarize baseline characteristics. A Chi-square test was used to compare categorical variables. Comparisons of median eos/hpf and EPX/mm2 were made using a non-parametric Mann-Whitney test. Tissue EPX/mm2 pre-and post-treatment were compared using a Wilcoxon Signed-Rank test. Correlations between peak eos/hpf and EPX/mm2 were obtained using Spearman’s correlation analysis. A receiver operating characteristic (ROC) curve for EPX/mm2 was generated and Youden’s index was used to determine an EPX/mm2 cutoff for EoE diagnosis. In order to detect an AUC of 0.7 with 90% power (alpha = 0.05) we calculated we would need at least 33 EoE cases and 33 controls. Statistical comparisons and plots were made with GraphPad Prism version 8.0.2 for Windows (GraphPad software, San Diego, CA, USA).
RESULTS
Table 1 details the baseline clinical characteristics of the study population. We evaluated esophageal biopsies from 38 EoE subjects and 49 non-EoE controls. Thirty-one EoE subjects had post-treatment biopsies available and 21 were categorized as treatment responders. Among the EoE subjects with longitudinal samples, 30 were treated with swallowed steroids and one was treated with diet elimination. The participants analyzed in this study are summarized in a CONSORT diagram (Supplemental Figure 1A). EoE subjects were more likely to be male and have comorbid food allergies when compared to controls. Rates of asthma, allergic rhinitis and atopic dermatitis were similar in cases (n = 38) and controls (n = 49). Dysphagia was the most common indication for endoscopy in both groups. The most common diagnosis among control subjects was GERD.
Table 1 –
Characteristics of the study population
| EoE cases (n = 38) | Controls (n = 49) | P* | |
|---|---|---|---|
| Mean age (± SD) | 36.9 ± 11.8 | 57.5 ± 12.1 | < 0.001 |
| Males (n, %) | 20 (52) | 14 (29) | 0.02 |
| White (n, %) | 36 (95) | 41 (84) | 0.11 |
| Symptoms (n, %) | |||
| Dysphagia | 38 (100) | 37 (76) | 0.001 |
| Heartburn | 2 (5) | 6 (12) | 0.26 |
| Atopic conditions (n, %) | |||
| Allergic rhinitis | 26 (68) | 23 (52) | 0.52 |
| Asthma | 16 (42) | 17 (39) | 0.75 |
| Atopic dermatitis | 4 (11) | 5 (11) | 0.9 |
| Food allergies | 16 (42) | 5 (11) | 0.002 |
| Any atopic condition | 30 (79) | 30 (68) | 0.27 |
| Endoscopic findings (n, %) | |||
| Exudates | 21 (55) | 2 (4) | < 0.001 |
| Rings | 37 (97) | 9 (18) | < 0.001 |
| Edema | 18 (47) | 1 (2) | < 0.001 |
| Furrows | 37 (97) | 2 (4) | < 0.001 |
| Stricture | 13 (34) | 6 (12) | 0.01 |
| Narrowing | 16 (42) | 2 (4) | < 0.001 |
| Erosive esophagitis | 9 (18) | 0 (0) | 0.005 |
| Schatzki’s ring | 5 (10) | 1 (3) | 0.17 |
| Hiatal hernia | 7 (18) | 18 (37) | 0.06 |
| Dilation performed | 11 (29) | 15 (31) | 0.87 |
| Diagnoses (n, %) | |||
| EoE | 38 (100) | 0 (0) | -- |
| GERD | -- | 30 (61) | |
| Esophageal dysmotility | -- | 12 (25) | |
| Functional | -- | 1 (2) | |
| Schatzki’s ring | -- | 4 (8) | |
| Normal | -- | 2 (4) |
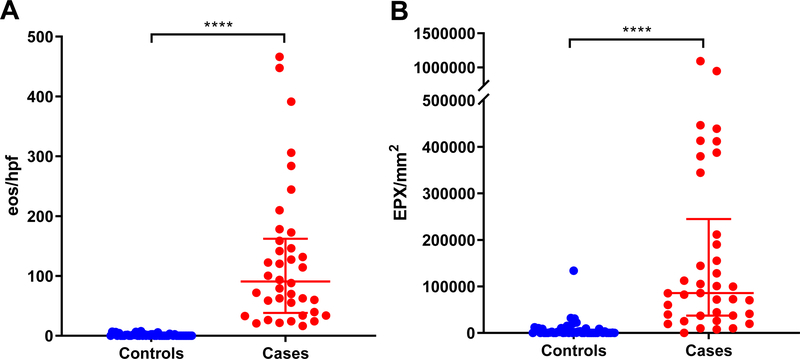
EPX staining identified tissue eosinophil infiltration and extracelluar deposition of the protein in subjects with active EoE (Figure 1). Subjects with EoE had marked eosinophilic inflammation assessed by both peak eosinophil counts from an H&E stain and EPX/mm2 (Figure 2). EoE subjects had a median of 90.66 (IQR 38.22–162.2) vs 0 (IQR 0–2.2) eos/hpf in controls (p < 0.0001) (Figure 2A). Similarly, EPX/mm2 was increased in EoE subjects vs. controls (median EPX/mm2 = 85,667 (IQR 37,237–244,825) vs. 412.4 (IQR 64.63–6,913) (p<0.0001)) (Figure 2B). Importantly, some subjects in the control group had increased EPX/mm2 (Figures 2 and 3), accounting for the greater degree of overlap when compared to eos/hpf. Two patterns of EPX staining were observed; a dense, punctate staining pattern and a lighter diffuse deposition (Figure 3). The latter was more commonly seen in the peripapillary squamous epithelium of subjects with elevated EPX/mm2, but without significant tissue eosinophilia based on H&E stains. Interestingly, these areas also had dilated intercellular spaces.
Figure 1. EPX immunohistochemistry (IHC) highlights eosinophil tissue infiltration and degranulation in active EoE.
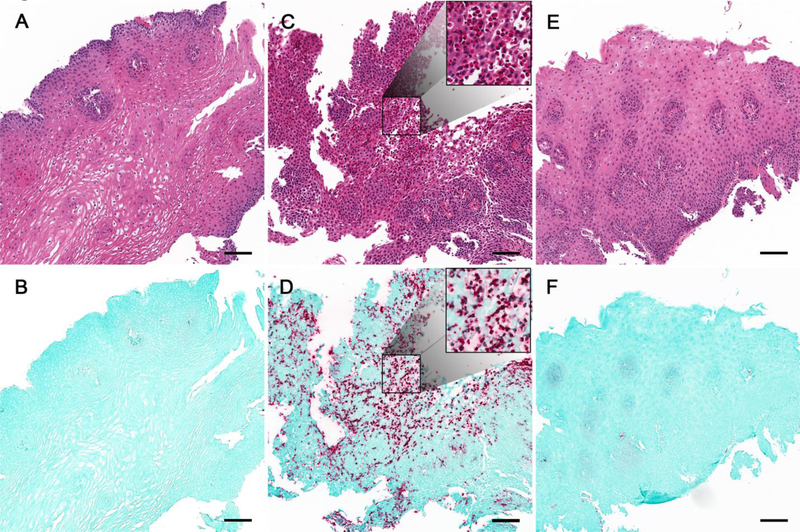
H&E (A, C, E) and EPX IHC (B, D, F) performed on the same esophageal tissue section. Histopathology shown for a representative control subject (A, B) and an EoE subject pre-(C, D) and post-treatment (E, F) with swallowed budesonide. EPX staining revealed marked eosinophilic inflammation in active EoE subject with complete resolution following treatment.
Figure 2. Esophageal eosinophilia and EPX staining are markedly increased in EoE subjects.
Manual peak eosinophil counts (A) and automated EPX assessments (EPX/mm2, B) in EoE cases and controls. Note that some subjects with < 15 eos/hpf have evidence for eosinophilic inflammation by EPX staining. Median eos/hpf and EPX/mm2 compared using a Mann-Whitney test. ****p < 0.0001
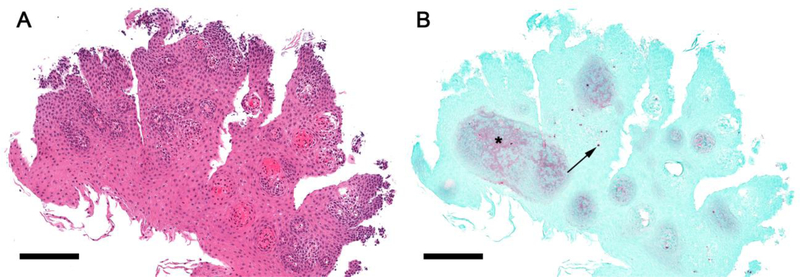
Figure 3. EPX staining reveals eosinophilic inflammation in a control subject.
H&E (A) and EPX (B) stains of a section from an esophageal biopsy from a subject with dysphagia initially classified as a non-EoE control. EPX immunohistochemistry reveals dense punctate deposition (B, arrow) and diffuse staining (B, asterisk). Diffuse EPX staining concentrates in the peripapillary squamous epithelium and co-localizes with areas where there are also dilated intercellular spaces.
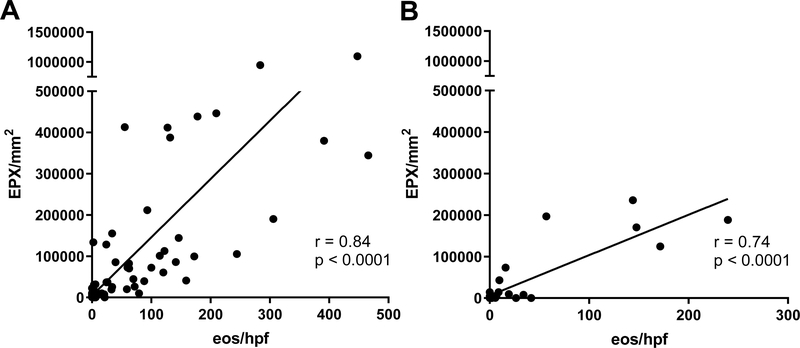
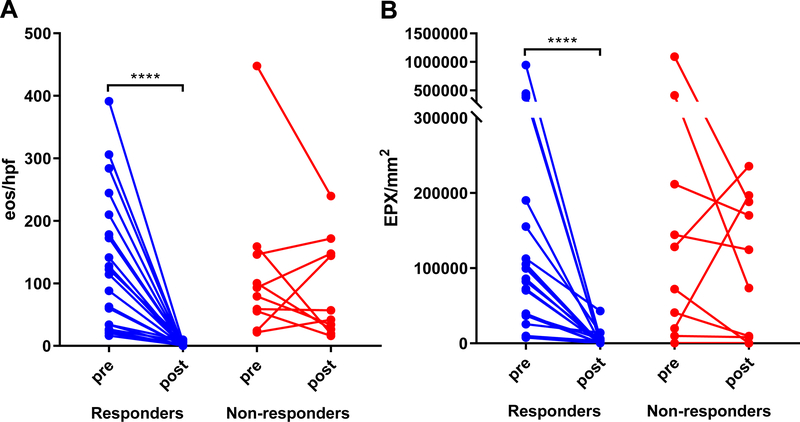
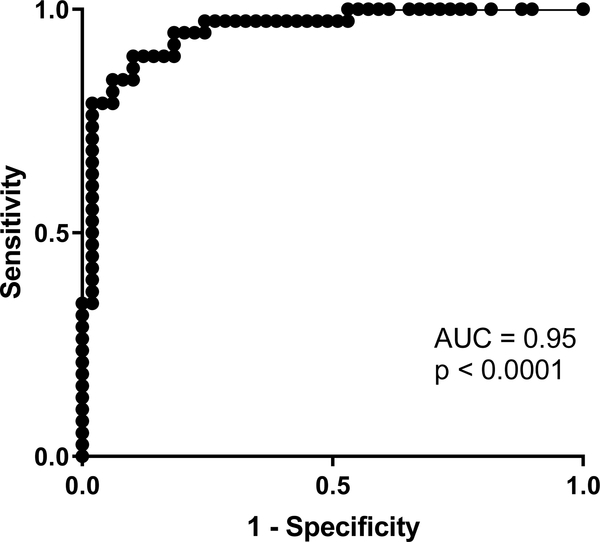
EPX/mm2 correlated strongly with peak eos/hpf (r = 0.84, p < 0.0001) in pre-(Figure 4A) and post-treatment biopsies (Figure 4B). Longitudinal analysis demonstrated that both peak eos/hpf and EPX/mm2 significantly decreased in response to therapy (Figure 5). Significant post-treatment differences were not observed among non-responders (Figure 5). EPX/mm2 identified subjects with high diagnostic accuracy, as indicated by an AUC of 0.95 (Figure 6). Based on a Youden’s index value of 0.793, the EPX cutoff that maximized diagnostic sensitivity (0.89) and specificity (0.90) was 17,379 EPX-positive pixels/mm2. Five of the 49 controls had an EPX/mm2 measurement above this threshold with one control EPX/mm2 = 133,668.9 (Figure 2B). The range for peak eosinophil counts in these subjects was 0–6 eos/hpf. Of the five control subjects, two had rings and one mild furrows. Four of five were diagnosed with reflux. The control subject with the highest EPX/mm2 had achalasia.
Figure 4. EPX/mm2 correlates strongly with peak eosinophil counts.
Correlation plot of EPX/mm2 (y-axis) vs. eos/hpf (x-axis) from EoE subjects pre-(A) and post-(B) treatment. Spearman’s rho value reported.
Figure 5. Peak eosinophil counts and EPX/mm2 decline in response to treatment in EoE subjects.
Paired comparisons (Wilcoxon signed-rank test) of median peak eos/hpf (A) and EPX/mm2 (B) in EoE subjects (n = 31) before and after therapy. Significant decreases were seen in treatment responders (n = 21), but not in non-responders (n = 10). **** p < 0.0001
Figure 6. EPX/mm2 is an accurate diagnostic marker of EoE.
The ROC curve for EPX/mm2 suggests automated analysis of EPX is a highly accurate diagnostic strategy.
DISCUSSION
Histologic diagnosis of EoE requires manual quantification of peak eos/hpf. Though reliability with careful counting using standard protocols can be sufficient for EoE diagnosis [23], counting eosinophils is time-consuming, labor-intensive, and subjective [2]; thus, there is a critical need to develop standardized methods that can rapidly and reliably assess eosinophilic tissue inflammation. Our laboratory has developed novel reagents for detection of eosinophil secondary granule proteins and methods for digital quantification of EPX staining [13,14]. In this study, we evaluated the utility of automated EPX analysis as a diagnostic tool for EoE. We found that EPX/mm2 was markedly increased in EoE subjects and correlated strongly with the current gold standard for histologic diagnosis (peak eos/hpf). We also demonstrated that EPX/mm2 decreases in response to treatment and is highly accurate as a diagnostic marker assessing EoE activity. Of particular interest, we found a subset of patients with very high EPX staining and eosinophil counts of < 15 eos/hpf. This raises the question, which would require future study, of whether EPX may enhance current diagnostic approaches.
Several studies have highlighted the importance of evaluating eosinophil degranulation products in eosinophilic disorders [7,9,24,25]. Given the propensity for eosinophils to undergo cytolytic degranulation in tissue [6], occasionally patients with extensive eosinophilic inflammation may not cross established histologic thresholds for intact eosinophil counts (i.e. ≥ 15 eos/hpf) in EoE [7,24]. All of the subjects in this study had symptoms of esophageal dysfunction and some control subjects showed eosinophilic inflammation not appreciable by conventional H&E staining. Interestingly, several subjects without increased eosinophil counts had diffuse EPX staining concentrated in the peripapillary squamous epithelium. We speculate that this staining may represent subepithelial eosinophil activity contributing to clinical symptoms [26]. Interestingly, eosinophilic infiltration of the esophageal muscular layer has been reported in achalasia [27] which may be consistent with diffuse peripapillary staining pattern seen in one of our controls subjects with markedly elevated EPX/mm2. Conversely, we noted a few EoE cases where eosinophil counts were increased and EPX/mm2 seemed disproportionately low. These subjects were still detected by EPX staining; however, they did not have marked degranulation. This observation may have implications for eosinophil activities and correlation with clinical symptoms.
We and others have examined staining for EDN [25,28], MBP[28–31], ECP [32] and EPX [9,11,12] as tissue biomarkers in EoE. Previous studies have primarily used manual scoring systems to quantify eosinophil degranulation. Strasser et al recently used a method similar to ours and found that automated quantification of ECP in tissue also correlates strongly with manual eosinophil counts [32]. While the eosinophils are the primary source of tissue ECP in EoE, ECP is not eosinophil-specific [8] and may be released by other cell types including neutrophils [33]. Ours is the first study to evaluate automated image analysis of EPX staining in a controlled study using prospectively banked samples from EoE subjects. There has also been recognition, as evidenced by the development of the EoE Histologic Scoring System (EoEHSS) [34] that eosinophil counts are not the sine qua non for assessing histologic disease activity [35]. While the EoEHSS is both reliable and responsive to treatment [36,37], it quantifies 8 different mucosal parameters which increases time over eosinophil count alone and must also be done manually.
We acknowledge that our approach still requires some subjective assessment but serves as a starting point for application of digital pathology methods in EoE. Digital pathology techniques are increasingly being used by many centers to increase efficiency and diagnostic accuracy [22]. Automation reduces subjectivity and, in this case, may allow for a more granular assessment of pathologic processes. Widespread application of the methods described outside of our center requires optimization of both the staining and digital quantification methods. Algorithms for quantifying positive pixel staining should be adjusted depending on the chromagen used for IHC (e.g. DAB or permanent red) and standardized in order to define diagnostic thresholds. Pixel counts may also vary depending on the quality and consistency of the IHC stain. Finally, the costs associated with optimization, automation and performance of IHC must be balanced with the costs of a pathologist’s time.
This study has several strengths including analysis of a large number of well-characterized, prospective EoE samples. In addition to comparing baseline esophageal biopsies with controls we also assessed longitudinal changes in EPX IHC following treatment in both responders and non-responders. By de-staining existing H&E slides and re-staining them for EPX, were able to make direct comparisons of EPX density with manual eosinophil counts. We acknowledge certain limitations. This is a single center study of adult subjects which may affect generalizability. In addition, EPX staining was performed by hand in the laboratory which may have introduced some staining variability; future studies using automated EPX staining methods would be needed for more widespread implementation of this technique. Samples were batched and stained by the same lab personnel to minimize differences related to staining technique. We noted minor tissue artifact associated with destaining the re-staining identical sections; notwithstanding, this approach allowed for direct comparison of tissue sections. Given a very strong positive correlation between EPX/mm2 and tissue eosinophilia, we do not suspect that trauma due to handling significantly influenced the results. Due to recent changes in the diagnostic criteria, we also excluded subjects with PPI-responsive EoE. We do not suspect exclusion of these subjects would have altered the results, but this would also merit future study.
In summary, automated EPX analysis is a powerful tool for quantifying eosinophilic inflammation in EoE that may increase efficiency and reproducibility. Future studies employing this methodology in EoE will compare EPX staining in samples coupled with clinical symptoms [10,38], endoscopic reference scores [39], and functional assessments of esophageal distensibility [40] to determine whether it enhances diagnostic accuracy.
Supplementary Material
Acknowledgements:
We thank William E. LeSuer, B.S. and Sergei Ochkur, Ph.D. for their assistance with scanning slides and figure preparation.
Financial support: NIH K23 DK090073 (E.S.D.), NIH R01 DK101856, ADHS18–198880 New Investigator Award (B.L.W.), Mayo Clinic Arizona (B.L.W.), Donald Levin Family Foundation, Berger and Quayle Families. This work was independent of all funding sources.
Abbreviations used:
- AEC
absolute eosinophil count
- EoE
eosinophilic esophagitis
- EPX
eosinophil peroxidase
- EPX-mAb
monoclonal anti-eosinophil peroxidase antibody
- ESGPs
eosinophil secondary granule proteins
- eos/hpf
eosinophils per high power field
- GERD
gastroesophageal reflux disease
- IHC
immunohistochemistry
- PPI
proton pump inhibitor
- ROC
receiver operating characteristic
Footnotes
Disclosure of Potential Conflicts of Interest: The authors of this study have no relevant conflicts of interest.
REFERENCES:
- 1.Dellon ES, Liacouras CA, Molina-Infante J et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference Gastroenterology. 2018;155:1022–1033 e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stucke EM, Clarridge KE, Collins MH, Henderson CJ, Martin LJ, Rothenberg ME. Value of an Additional Review for Eosinophil Quantification in Esophageal Biopsies J Pediatr Gastroenterol Nutr. 2015;61:65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellon ES, Aderoju A, Woosley JT, Sandler RS, Shaheen NJ. Variability in diagnostic criteria for eosinophilic esophagitis: a systematic review Am J Gastroenterol. 2007;102:2300–2313. [DOI] [PubMed] [Google Scholar]
- 4.Saffari H, Peterson KA, Fang JC, Teman C, Gleich GJ, Pease LF 3rd. Patchy eosinophil distributions in an esophagectomy specimen from a patient with eosinophilic esophagitis: Implications for endoscopic biopsy J Allergy Clin Immunol. 2012;130:798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen JA, Lager DJ, Lewin M, Rendon G, Roberts CA. The optimal number of biopsy fragments to establish a morphologic diagnosis of eosinophilic esophagitis Am J Gastroenterol. 2014;109:515–520. [DOI] [PubMed] [Google Scholar]
- 6.Saffari H, Hoffman LH, Peterson KA et al. Electron microscopy elucidates eosinophil degranulation patterns in patients with eosinophilic esophagitis J Allergy Clin Immunol. 2014;133:1728–1734 e1721. [DOI] [PubMed] [Google Scholar]
- 7.Peterson KA, Cobell WJ, Clayton FC et al. Extracellular Eosinophil Granule Protein Deposition in Ringed Esophagus with Sparse Eosinophils Dig Dis Sci. 2015;60:2646–2653. [DOI] [PubMed] [Google Scholar]
- 8.Ochkur SI, Kim JD, Protheroe CA et al. A sensitive high throughput ELISA for human eosinophil peroxidase: a specific assay to quantify eosinophil degranulation from patient-derived sources J Immunol Methods. 2012;384:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Protheroe C, Woodruff SA, de Petris G et al. A novel histologic scoring system to evaluate mucosal biopsies from patients with eosinophilic esophagitis Clin Gastroenterol Hepatol. 2009;7:749–755 e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin LJ, Franciosi JP, Collins MH et al. Pediatric Eosinophilic Esophagitis Symptom Scores (PEESS v2.0) identify histologic and molecular correlates of the key clinical features of disease J Allergy Clin Immunol. 2015;135:1519–1528 e1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright BL, Ochkur SI, Olson NS et al. Normalized serum eosinophil peroxidase levels are inversely correlated with esophageal eosinophilia in eosinophilic esophagitis Dis Esophagus. 2018;31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen N, Baumgarten A, Wright BL et al. Histologic similarities in children with eosinophilic esophagitis and proton pump inhibitor-responsive esophageal eosinophilia J Allergy Clin Immunol. 2019;143:1237–1240 e1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright BL, Fernandez-Becker NQ, Kambham N et al. Baseline Gastrointestinal Eosinophilia Is Common in Oral Immunotherapy Subjects With IgE-Mediated Peanut Allergy Front Immunol. 2018;9:2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lal D, Wright BL, Shim KP et al. Eosinophil peroxidase, GATA3, and T-bet as tissue biomarkers in chronic rhinosinusitis J Allergy Clin Immunol. 2019;143:2284–2287 e2286. [DOI] [PubMed] [Google Scholar]
- 15.Wen T, Dellon ES, Moawad FJ, Furuta GT, Aceves SS, Rothenberg ME. Transcriptome analysis of proton pump inhibitor-responsive esophageal eosinophilia reveals proton pump inhibitor-reversible allergic inflammation J Allergy Clin Immunol. 2015;135:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dellon ES, Rusin S, Gebhart JH et al. Utility of a non-invasive serum biomarker panel for diagnosis and monitoring of EoE: A prospective study Am J Gastroenterol. 2015;110:821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dellon ES, Cotton CC, Gebhart JH et al. Accuracy of the Eosinophilic Esophagitis Endoscopic Reference Score in Diagnosis and Determining Response to Treatment Clin Gastroenterol Hepatol. 2016;14:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eluri S, Selitsky SR, Perjar I et al. Clinical and Molecular Factors Associated With Histologic Response to Topical Steroid Treatment in Patients With Eosinophilic Esophagitis Clin Gastroenterol Hepatol. 2019;17:1081–1088 e1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liacouras CA, Furuta GT, Hirano I et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults J Allergy Clin Immunol. 2011;128:3–20 e26; quiz 21–22. [DOI] [PubMed] [Google Scholar]
- 20.Wolf WA, Cotton CC, Green DJ et al. Evaluation of histologic cutpoints for treatment response in eosinophilic esophagitis J Gastroenterol Hepatol Research. 2015;4:1780–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed CC, Wolf WA, Cotton CC et al. Optimal Histologic Cutpoints for Treatment Response in Patients With Eosinophilic Esophagitis: Analysis of Data From a Prospective Cohort Study Clin Gastroenterol Hepatol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dellon ES, Fritchie KJ, Rubinas TC, Woosley JT, Shaheen NJ. Inter-and intraobserver reliability and validation of a new method for determination of eosinophil counts in patients with esophageal eosinophilia Dig Dis Sci. 2010;55:1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusin S, Covey S, Perjar I et al. Determination of esophageal eosinophil counts and other histologic features of eosinophilic esophagitis by pathology trainees is highly accurate Hum Pathol. 2017;62:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson KA, Gleich GJ, Limaye NS et al. Eosinophil granule major basic protein 1 deposition in eosinophilic esophagitis correlates with symptoms independent of eosinophil counts Dis Esophagus. 2019. [DOI] [PubMed] [Google Scholar]
- 25.Kephart GM, Alexander JA, Arora AS et al. Marked deposition of eosinophil-derived neurotoxin in adult patients with eosinophilic esophagitis Am J Gastroenterol. 2010;105:298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoepfer AM, Simko A, Bussmann C et al. Eosinophilic Esophagitis: Relationship of Subepithelial Eosinophilic Inflammation With Epithelial Histology, Endoscopy, Blood Eosinophils, and Symptoms Am J Gastroenterol. 2018;113:348–357. [DOI] [PubMed] [Google Scholar]
- 27.Tottrup A, Fredens K, Funch-Jensen P, Aggestrup S, Dahl R. Eosinophil infiltration in primary esophageal achalasia. A possible pathogenic role Dig Dis Sci. 1989;34:1894–1899. [DOI] [PubMed] [Google Scholar]
- 28.Sridhara S, Ravi K, Smyrk TC et al. Increased numbers of eosinophils, rather than only etiology, predict histologic changes in patients with esophageal eosinophilia Clin Gastroenterol Hepatol. 2012;10:735–741. [DOI] [PubMed] [Google Scholar]
- 29.Dellon ES, Speck O, Woodward K et al. Markers of eosinophilic inflammation for diagnosis of eosinophilic esophagitis and proton pump inhibitor-responsive esophageal eosinophilia: a prospective study Clin Gastroenterol Hepatol. 2014;12:2015–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dellon ES, Chen X, Miller CR, Woosley JT, Shaheen NJ. Diagnostic utility of major basic protein, eotaxin-3, and leukotriene enzyme staining in eosinophilic esophagitis Am J Gastroenterol. 2012;107:1503–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller S, Aigner T, Neureiter D, Stolte M. Eosinophil infiltration and degranulation in oesophageal mucosa from adult patients with eosinophilic oesophagitis: a retrospective and comparative study on pathological biopsy J Clin Pathol. 2006;59:1175–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strasser DS, Seger S, Bussmann C et al. Eosinophilic oesophagitis: relevance of mast cell infiltration Histopathology. 2018;73:454–463. [DOI] [PubMed] [Google Scholar]
- 33.Sur S, Glitz DG, Kita H et al. Localization of eosinophil-derived neurotoxin and eosinophil cationic protein in neutrophilic leukocytes J Leukoc Biol. 1998;63:715–722. [DOI] [PubMed] [Google Scholar]
- 34.Collins MH, Martin LJ, Alexander ES et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring Dis Esophagus. 2017;30:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whelan KA, Godwin BC, Wilkins B et al. Persistent Basal Cell Hyperplasia Is Associated With Clinical and Endoscopic Findings in Patients With Histologically Inactive Eosinophilic Esophagitis Clin Gastroenterol Hepatol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warners MJ, Ambarus CA, Bredenoord AJ et al. Reliability of histologic assessment in patients with eosinophilic oesophagitis Aliment Pharmacol Ther. 2018;47:940–950. [DOI] [PubMed] [Google Scholar]
- 37.Hirano I, Collins MH, Assouline-Dayan Y et al. A randomized, double-blind, placebo-controlled trial of a novel recombinant, humanized, anti-interleukin-13 monoclonal antibody (RPC4046) in patients with active eosinophilic esophagitis: Results of the HEROES study United European Gastroenterol J. 2016;4 (Suppl):OP325. [Google Scholar]
- 38.Schoepfer AM, Straumann A, Panczak R et al. Development and validation of a symptom-based activity index for adults with eosinophilic esophagitis Gastroenterology. 2014;147:1255–1266 e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirano I, Moy N, Heckman MG, Thomas CS, Gonsalves N, Achem SR. Endoscopic assessment of the oesophageal features of eosinophilic oesophagitis: validation of a novel classification and grading system Gut. 2013;62:489–495. [DOI] [PubMed] [Google Scholar]
- 40.Lin Z, Kahrilas PJ, Xiao Y et al. Functional luminal imaging probe topography: an improved method for characterizing esophageal distensibility in eosinophilic esophagitis Therap Adv Gastroenterol. 2013;6:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.