FOXA1 functions as a pioneer transcription factor by facilitating the access to chromatin for steroid hormone receptors, such as androgen receptor (AR) and estrogen receptor (ER) 1–4, but mechanisms regulating its binding to chromatin remain elusive. LSD1 (KDM1A) acts as a transcriptional repressor by demethylating mono/di-methylated histone H3 lysine 4 (H3K4me1/2) 5,6, but also acts as a steroid hormone receptor coactivator through unclear mechanisms. Here we show in prostate cancer (PCa) cells that LSD1 associates with FOXA1 and active enhancer markers, and that LSD1 inhibition globally disrupts FOXA1 chromatin binding. Mechanistically, we demonstrate that LSD1 positively regulates FOXA1 binding by demethylating lysine 270, adjacent to the wing2 region of FOXA1 DNA binding domain. Acting through FOXA1, LSD1 inhibition broadly disrupted AR binding and its transcriptional output, and dramatically decreased prostate cancer growth alone and in synergy with AR antagonist treatment in vivo. These mechanistic insights suggest new therapeutic strategies in steroid-driven cancers.
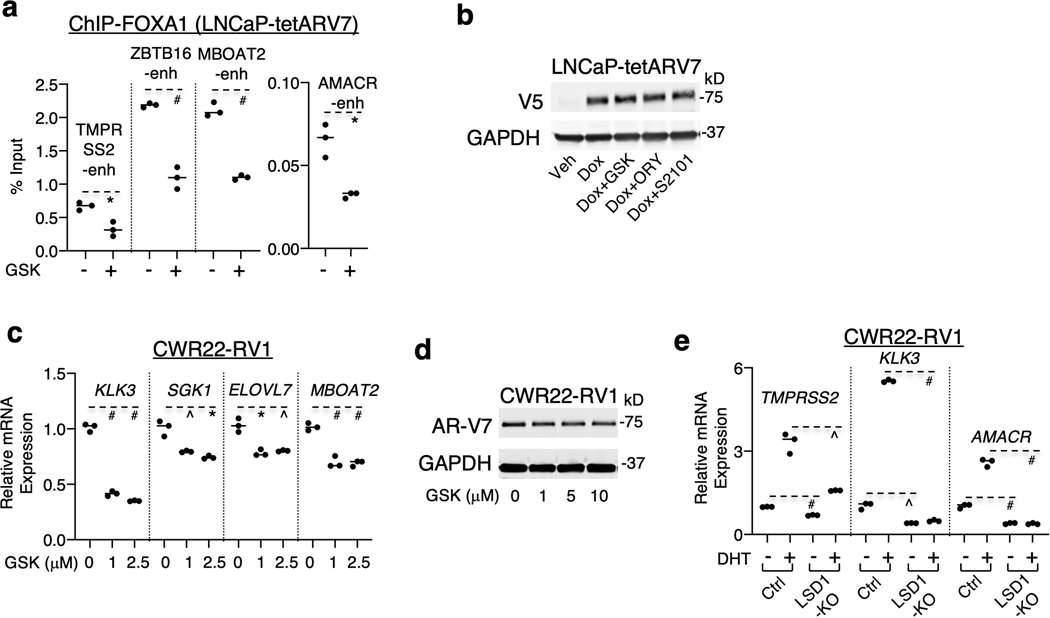
The AR coactivator function of LSD1 has been attributed to phosphorylation of histone H3 on threonine 6 (H3T6ph) and threonine 11, which may switch LSD1 substrate specificity from H3K4me1/2 to H3K9me1/2 7–10. However, we reported that the H3K4 demethylase activity of LSD1 persists at AR-regulated enhancers, including sites marked by H3T6ph 11, arguing against this model. As FOXA1 binds to AR-regulated enhancers marked with H3K4me1/2 prior to AR binding 12, and LSD1 chromatin binding substantially overlaps with FOXA1 binding sites 11, we hypothesized that LSD1 may regulate the accessibility of AR-mediated enhancers through interactions with FOXA1. Comprehensive ChIP-seq studies were carried out in LNCaP PCa cells grown in steroid-depleted medium to minimize the feedback effect of AR on FOXA1 chromatin binding 13. As shown in Figure 1a, LSD1 binding sites were associated with high levels of FOXA1 and active enhancer marks. Although we previously showed that FOXA1 binding is decreased by LSD1 silencing 11, it was unclear whether the catalytic activity of LSD1 is required (LSD1 can also function as a scaffold protein 14,15). As shown in Figure 1b–d and Extended Data Figure 1a–d, LSD1 inhibitor treatments (GSK2879552, S2101 16,17) led to a rapid and dramatic decrease of global FOXA1 binding. This finding was subsequently validated in LNCaP and CWR22RV1 PCa cells, and FOXA1 or AR protein expression was not affected by the treatments (Fig. 1e–g and Extended Data Fig. 1e–k). Moreover, overexpressing wildtype (WT) LSD1 but not the demethylase-deficient K661A mutant increased FOXA1 binding (Extended Data Fig. 1l–m).
Figure 1. LSD1 inhibition disrupts global chromatin binding of FOXA1.
a, Heatmap view of FOXA1, LSD1, H3K4me2, H3K27ac, and DHS (DNase hypersensitivity) ChIP-seq signal intensity at FOXA1 binding sites in LNCaP cells (FOXA1/AR positive, AR-V7 negative) grown in 5% CSS (hormone-depleted FBS). b, c, d, FOXA1 ChIP-seq (using antibody #1) was performed in LNCaP cells treated with 50 μM GSK2879552 at 4 h or 48 h. (b) Overlap of FOXA1 binding sites between vehicle and LSD1 inhibitors treated cells, (c) heatmap view for FOXA1 ChIP-seq signal intensity, and (d) the mean of FOXA1 ChIP-seq signals at all FOXA1 binding sites were shown (Veh vs. GSK-4h: P = 2.5 × 10−5; Veh vs. GSK-48h: P = 0.056). e, f, ChIP-qPCR for FOXA1 binding at KLK3/NKX3.1 enhancer sites in (e) LNCaP or (f) CWR22-RV1 cells (FOXA1/AR positive, AR-V7 positive) treated with GSK2879552 for the indicated time. g, LSD1-KO cell line was established in CWR22-RV1 cells using CRISPR/Cas9 approach (by sgRNA targeting LSD1). LSD1 expression was immunoblotted in the LSD1-KO line versus the control line. h, ATAC-seq was performed in LNCaP cells treated with vehicle or GSK2879552 at 4 h. Heatmap view for ATAC-seq signal intensity at FOXA1-binding sites was shown. i, The mean of ATAC-seq signals at FOXA1-binding sites (Veh vs. GSK-4h: P = 6.2 × 10−20). j, The mean of H3K4me2 ChIP-seq signals at LSD1 binding sites (Veh vs. GSK-4h: P = 0.86; Veh vs. GSK-48h: P = 6.1 × 10−14) or LSD1/FOXA1 overlapping sites (Veh vs. GSK-4h: P = 7.8 × 10−5; Veh vs. GSK-48h: P = 1.9 × 10−22).
Since FOXA1 decompacts chromosome at enhancers, we next determined whether LSD1 inhibition suppresses chromatin opening at the binding sites. Using ATAC-seq, we show that LSD1 inhibition markedly decreased the chromosome accessibility at FOXA1-occupied sites before androgen stimulation (Fig. 1h–i). LSD1 inhibition also led to rapid decreases of DNase hypersensitivity (DHS) and acetylated H3K27 (H3K27ac), confirming reduced opening of these regions before AR binding (Extended Data Fig. 2a–d). Importantly, the levels of H3K4me2 at LSD1-FOXA1 co-occupied sites were not strongly affected by LSD1 inhibition at 4 h but eventually increased at 24 h (Fig. 1j), indicating that the disruption of FOXA1 binding by LSD1 inhibition (maximum reduction at 4 h) is mediated by a mechanism that is independent of H3K4 demethylation. Moreover, FOXA1 binding at AR-independent regulatory sites in PCa cells or ER-mediated enhancer sites in breast cancer (BCa) cells were also decreased by LSD1 inhibition (Extended Data Fig. 3a–c). Together, these data suggest that LSD1 globally maintains FOXA1 chromatin binding and regulates the opening of FOXA1-dependent enhancers via a specific molecular mechanism.
Since AR chromatin binding is dependent on FOXA1, we next hypothesized that LSD1 inhibition may impair AR recruitment to enhancers. Significantly, LSD1 inhibitor treatment in the presence of androgens markedly decreased the global AR chromatin binding, the levels of AR-induced DHS 18, and the recruitments of an AR coactivator, p300 (Fig. 2a–e and Extended Data Fig. 4a–b). Interestingly, this observation is in contrast to studies using approaches to silence FOXA1 expression, which indicate major reprogramming of AR cistrome 19,20. A possible explanation is that LSD1 inhibition leads to the decreased FOXA1 binding, which may still be sufficient to promote weaker recruitment of AR, while FOXA1 silencing forces AR to occupy alternative sites pioneered by other factors. Nonetheless, decreased AR binding by LSD1 inhibition broadly impaired the expression of direct AR-activated genes (Extended Data Fig. 4c–f). Moreover, overexpression of LSD1-WT but not K661A mutant enhanced AR binding and increased androgen-induced gene expression (Extended Data Fig. 4g–h). The effects of LSD1 inhibition on AR activity and cell growth were dependent on the doses of inhibitors and the levels of androgens or LSD1 expression (Fig. 2f and Extended Data Fig. 4i). Interestingly, the prolonged treatment of LSD1 inhibitor (~2 weeks) at much lower doses (1 μM) can recapitulate the effect of higher doses of inhibitor (50 μM for 48 h) on FOXA1/AR chromatin binding, AR transcriptional activity, and PCa cell growth (Fig. 2g–l and Extended Data Fig. 4j), suggesting that the effect of LSD1 inhibitor treatment is sustainable and accumulative. Genes repressed or enhanced by the short-term or prolonged treatment were also similarly enriched for unfolded protein responses or immune responses, respectively. The latter functions were also reported in a recent study showing LSD1 ablation enhances anti-tumor immunity 21 (Extended Data Fig. 4k).
Figure 2. LSD1 inhibition broadly impairs AR recruitment and suppresses AR transcriptional activity.
a, b, c, AR ChIP-seq analyses were performed in LNCaP cells treated with vehicle, DHT (10 nM for 4 h), or DHT (4 h) with pretreated GSK2879552 (50 μM, 0.5 or 48 h). (a) overlap of AR peaks in treated cells, (b) heatmap view of AR and FOXA1 binding intensity at AR binding sites, and (c) the mean of AR binding intensity were shown (Veh vs. GSK-4h: P = 8.8 × 10−33; Veh vs. GSK-48h: P = 9.9 × 10−267). d, e, ChIP-qPCR for (d) AR or (e) p300 at AR-mediated enhancers in LNCaP cells treated with/out DHT (10 nM, 4 h) and pretreated with GSK2879552 (48 h). f, RT-PCR for KLK3 in LNCaP cells treated with 1–10 nM DHT (24 h) and pretreated with GSK2879552 (0–50 μM, 24 h). g, h, i, j, LNCaP cells were maintained in the medium containing vehicle or 1 μM GSK2879552 for ~2 weeks. The following experiments were performed: (g) qRT-PCR for KLK3; (h) immunoblotting for KLK3; (i) ChIP-qPCR for FOXA1; (j) ChIP-qPCR for AR (with 10 nM DHT). k, RNA-seq analyses were done in these long-term treated cells (10 nM DHT, 24 h) in comparison with parental LNCaP cells treated with/out DHT (24 h) and pretreated with 50 μM GSK2879552 (24 h). Androgen-upregulated genes were identified from parental LNCaP cells using 2-fold cut-off (DHT/Vehicle). The heatmap for the change in expression in response to GSK2879552 treatment was shown. l, Cell density was examined under the indicated conditions (mean±SD). m, LNCaP cells stably expressing doxycycline-inducible AR-V7 (V5 tagged) (LNCaP-tetARV7) were subjected to immunoblotting. n, ChIP-qPCR for V5 binding in LNCaP-tetARV7 cells treated with doxycycline versus doxycycline plus GSK2879552 (10 μM, 24 h). o, qRT-PCR for AR-V7-regulated genes in these cells. p, ChIP-qPCR for AR-V7 binding in CWR22-RV1 cells treated with GSK2879552 (2.5 μM, 24 h). q, ChIP-qPCR for AR-V7 binding in the LSD1-KO line versus the control line.
We next sought to determine whether LSD1 inhibition affects chromatin binding of the constitutively active form of AR splice variants lacking the ligand-binding domain (such as AR-V7, NM_001348061.1) 22–24. As shown in Figure 2m–q and Extended Data Figure 5a–e, LSD1 inhibition or silencing decreased ligand-independent AR-V7 chromatin binding and its transcriptional activity in either AR-V7 overexpressing LNCaP cells or in CWR22-RV1 cells (with high levels of endogenous AR-V7 25,26) without significantly affecting AR-V7 protein expression. Overall, these results demonstrate that AR signaling is suppressed by LSD1 inhibition through globally impairing the chromatin recruitment of AR and AR splice variants.
In addition to its classic function of demethylating H3K4, LSD1 can also demethylate non-histone proteins 27–29. Since LSD1 physically interacts with FOXA1 11,30, we hypothesized that LSD1 may regulate FOXA1 binding through the demethylation of FOXA1. As seen in Figure 3a and Extended Data Figure 6a–b, the levels of lysine-methylated FOXA1 were increased by LSD1 inhibitors and decreased by overexpression of LSD1-WT but not K661A mutant. A mass spectrometry analysis was then performed on the immunopurified FOXA1, leading to the identification of methylated lysine 270 (K270me) as a potential substrate of LSD1 (Extended Data Fig. 6c). This residue resides at the carboxyl-end of the wing2 region (aa 247–269) of Forkhead DNA binding domain (FKHD), a hot spot region for recurrent FOXA1 mutations in PCa 31–36 (Fig. 3b), and was recently reported as a critical residue for FOXA1 interaction with the nucleosome core 37. The demethylase activity of LSD1 on FOXA1 peptide containing K270me was subsequently determined and validated (Fig. 3c–d). Moreover, the level of lysine-methylated FOXA1 was significantly reduced in cells expressing methylation-deficient mutant K270R, suggesting K270 is a major methylation site (Fig. 3e). We then generated a FOXA1-K270me-specific antibody to confirm LSD1-mediated demethylation in PCa cells. As seen in Figure 3f, K270me was only detected in immunoprecipitated FOXA1-WT but not K270R mutant and its level was significantly increased by LSD1 inhibition, confirming that K270me is demethylated by LSD1 in vivo. In contrast to the suppression effect on FOXA1-WT, LSD1 inhibition or depletion did not decrease the chromatin binding of FOXA1-K270R at AR-regulated enhancers (Fig. 3g and Extended Data Fig. 6d–g).
Figure 3. LSD1 regulates FOXA1 chromatin binding through directly demethylating its lysine 270.
a, FOXA1 was immunoprecipitated from CWR22-RV1 cells treated with GSK2879552 (0–100 μM), followed by immunoblotting for methyl-lysine. b, Mass spectrometry analyses for immunoprecipitated FOXA1 from LNCaP cells stably overexpressing V5-FOXA1 identified methylated K270. c, The in vitro demethylation assay (measuring formaldehyde production) using synthetic H3K4me2 peptide (1–21 aa) or K270-methylated FOXA1 peptide (263–281 aa) as substrates incubated with recombinant LSD1 protein. d, The K270-methylated FOXA1 peptide incubated with/out 75 nM LSD1 (1 h) was analyzed by mass spectrometry. ‘me’ indicates a shift in mass equivalent to one methyl group. e, CWR22-RV1 cells stably expressing doxycycline-inducible V5-tagged FOXA1-WT or -K270R (CWR22RV1-tetFOXA1WT or CWR22RV1-tetFOXA1K270R cells) were generated for the following experiments. Immunoblotting for methyl-lysine on immunopurified V5-FOXA1 was shown (images cropped from the same blot). f, Immunoblotting for K270-methylated FOXA1 on immunopurified V5-FOXA1 protein. g, ChIP-qPCR for FOXA1-WT or K270R binding at AR-regulated enhancers in these stable cells treated with GSK2879552 (24 h). h, i, V5 ChIP-seq analyses were performed in the stable cells treated with/out GSK2879552 (50 μM, 4 h). (h) The venn-diagram for V5 binding peaks and (i) the heatmap view for V5-FOXA1 binding intensity (vehicle treated) were shown. j, AR ChIP-seq analyses were performed in these stable cells treated with DHT (10 nM, 4 h), and the heatmap view for AR binding intensity was shown. k, ChIP-qPCR for AR binding in these stable cells treated with DHT (10 nM) versus DHT plus enzalutamide (10 μM). Note: cells in g-k were all grown in the doxycycline-supplemented hormone-depleted medium. l, m, SCID male mice bearing xenograft tumors established from these CWR22-RV1 stable cell lines were fed with doxycycline supplemented diet and received daily enzalutamide treatment (10 mg/kg) via intraperitoneal injection (n = 4 independent tumors). The tumor volume (mean ± SD) was measured at the indicated time.
We next sought to determine the role of K270me on FOXA1 chromatin binding and the subsequent recruitment of AR. Using biochemical fractionation assays, we show that the K270R mutant was more tightly associated with chromatin (Extended Data Fig. 7a–b). Moreover, K270R-expressing cells contained significantly more FOXA1 binding sites and higher binding intensity than WT-expressing cells and were less affected by LSD1 inhibition (~30% loss versus ~51% loss with WT) (Fig. 3h–i), indicating that K270me may disrupt FOXA1 binding to chromatin. Significantly, overexpressing K270R mutant also resulted in a global increase of AR recruitments to chromosome (Fig. 3j and Extended Data Fig. 7c). Moreover, AR recruitment and activity in FOXA1-K270R cells were more resistant to a potent AR antagonist, enzalutamide 38, and overexpressing K270R mutant resulted in enzalutamide resistance in AR-mediated PCa tumor growth (Fig. 3k–m and Extended Data Fig. 7d–e), indicating that K270me may impair AR recruitment and increase its sensitivity to the antagonist treatment. Overall, these results indicate that LSD1-mediated K270-demethylation stabilizes FOXA1 binding and enhances the chromatin recruitment of AR.
The FKHD domain is conserved within members of FOXA proteins. As seen in Extended Data Figure 8a, FOXA2 shares the same sequence of wing2 loop and the lysine adjacent to it (K265). Therefore, we hypothesized that FOXA2 chromatin binding may also be regulated by LSD1 mediated demethylation. In PCa, FOXA2 activity is linked to the small cell neuroendocrine subtype 39 and it is expressed in AR-negative PC-3 cells (Extended Data Fig. 8b). Significantly, FOXA2 chromatin binding in PC-3 cells was suppressed by LSD1 inhibition while the chromatin binding of HOXB13, another important pioneer factor of AR 40, was not suppressed by LSD1 inhibition in LNCaP cells (Extended Data Fig. 8c–f), suggesting LSD1-mediated demethylation is a specific mechanism regulating FOXA1/2.
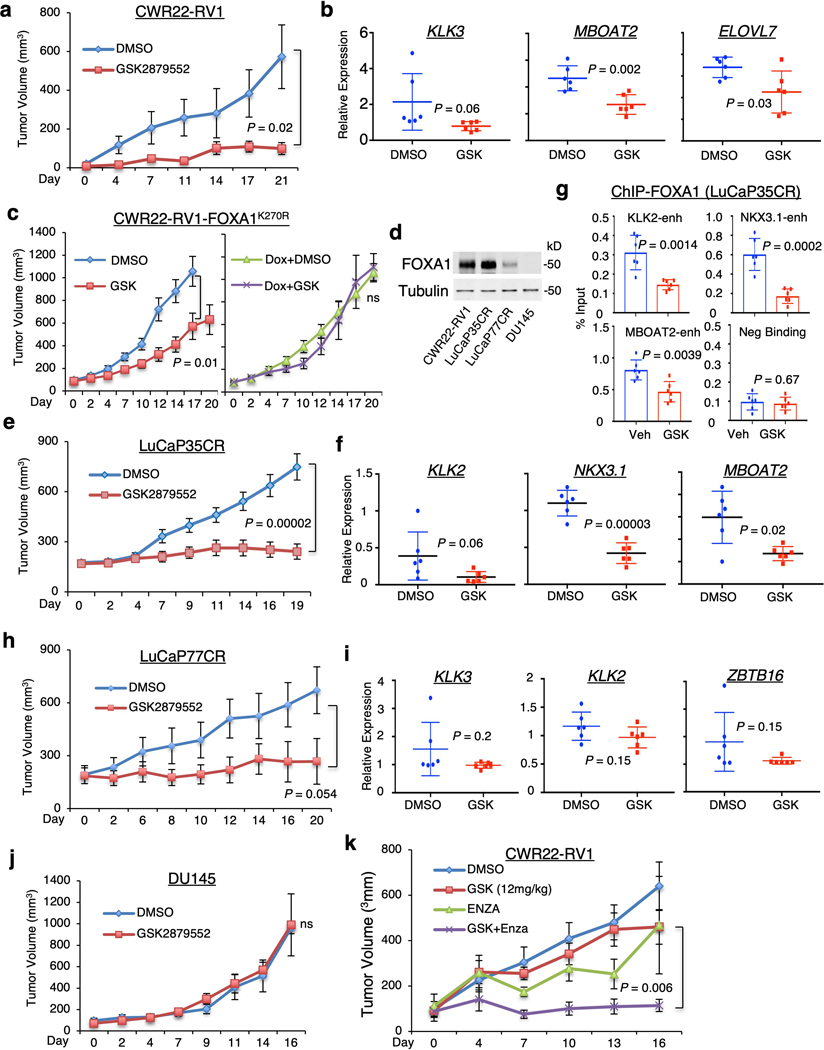
FOXA1 is highly expressed in PCa, BCa, and small cell lung cancer (SCLC), and its expression is correlated with LSD1 (Extended Data Fig. 9a–c). Several LSD1 inhibitors (GSK2879552, ORY-1001, INCB059872, SP2577) are being tested in clinical trials for cancer treatment 15,17,41,42. To examine how FOXA1-positive castration-resistant PCa (CRPC) respond to LSD1 inhibition, we generated CWR22-RV1 xenograft tumors in castrated mice and treated them with GSK2879552. As shown in Figure 4a, CWR22-RV1 tumor growth was markedly repressed by LSD1 inhibition, which led to an increase of H3K4me2 and a decrease of AR-FL/V7-regulated gene expression (Fig. 4b and Extended Data Fig. 9d–f). In contrast, LSD1 inhibition did not affect the growth of CWR22-RV1 tumors expressing K270R mutant, indicating that the effect of LSD1 inhibitor treatment is mediated via blocking FOXA1 K270-demethylation (Fig. 4c).
Figure 4. LSD1 inhibitor treatment suppresses tumor growth alone or in synergy with enzalutamide in FOXA1-high CRPC models.
a, b, Castrated SCID male mice bearing CWR22-RV1 xenograft tumors received daily DMSO or GSK2879552 (33 mg/kg) via intraperitoneal injection (n = 6 independent tumors). (a) The tumor growth was recorded. (b) After the mice were sacrificed, tumor samples were subjected to qRT-PCR for indicated AR-FL/V7 regulated genes. c, Castrated SCID male mice bearing CWR22RV1-tetFOXA1K270R xenograft tumors fed with regular or doxycycline-supplemented diet were treated with daily DMSO or GSK2879552 (33 mg/kg) via intraperitoneal injection (n = 8 independent tumors). The tumor growth was recorded. d, The protein expression of FOXA1 was examined in tumor samples from vehicle-treated xenograft tumors of four CRPC models. e, f, g, Castrated SCID male mice bearing LuCaP35CR xenograft tumors received daily DMSO or GSK2879552 (33 mg/kg) via intraperitoneal injection (n = 12 independent tumors). (e) The tumor growth was recorded, and the tumor samples were subjected to (f) qRT-PCR for indicated AR-FL/V7 regulated genes and (g) ChIP-qPCR for FOXA1 binding at indicated sites. h, i, Castrated SCID male mice bearing LuCaP77CR xenograft tumors received daily DMSO or GSK2879552 (33 mg/kg) via intraperitoneal injection (n = 7 independent tumors). (h) The tumor growth was recorded and (i) tumor samples were subjected to qRT-PCR for indicated AR-FL/V7 regulated genes. j, Castrated SCID male mice bearing DU145 xenograft tumors received daily DMSO or GSK2879552 (33 mg/kg) via intraperitoneal injection (n = 6 independent tumors). k, SCID mice bearing CWR22-RV1 xenograft tumors received daily DMSO, GSK2879552 (12 mg/kg), enzalutamide (10 mg/kg), or the combination via intraperitoneal injection (n = 6 independent tumors). Note: the statistical difference for the tumor growth (mean ± SD) at the final time point was determined by a two-tailed Student’s t test.
To further determine whether FOXA1 expression affects the efficacy of LSD1 inhibitors in vivo, we established three additional CRPC xenograft models with various expression levels of FOXA1, including LuCaP35CR, LuCaP77CR 43,44, and DU145. LuCaP35CR tumors express high levels of FOXA1 whereas LuCaP77CR and DU145 tumors have a much weaker or undetectable expression of FOXA1, respectively (Fig. 4d). Similar to the CWR22-RV1 model, LuCaP35CR tumors were very sensitive to GSK2879552 and ORY-1001, and these treatments suppressed FOXA1 chromatin binding and the expression of AR-FL/V7-regulated genes (Fig. 4e–g and Extended Data Fig. 10a–f). However, the tumor responses to the LSD1 inhibitor in LuCaP77CR model were much weaker and the suppression of AR-FL/AR-V7 signaling was not significant, although this treatment can similarly increase H3K4me2 (Fig. 4h–i and Extended Data Fig. 10g). In sharp contrast to CWR22RV1 and LuCaP35CR, LSD1 inhibition did not affect the growth of FOXA1-negative DU145 tumors (Fig. 4j and Extended Data Fig. 10h). Taken together, these in vivo animal studies suggest that the efficacy of LSD1 inhibitor treatment in CRPC may be correlated with the expression levels of FOXA1.
Finally, we examined whether LSD1 inhibitor treatment can therapeutically synergize with enzalutamide. As seen in Extended Data Figure 10i, the additive effect of LSD1 inhibitor with enzalutamide on suppressing the growth of CWR22-RV1 cells was observed in cell culture. More importantly, treating the mice bearing CWR22-RV1 xenografts with GSK2879552 (at a lower dose) and enzalutamide significantly increased the efficacy of the treatment by enzalutamide alone (Fig. 4k), indicating a therapeutic potential for combining LSD1 inhibitor with the intense androgen deprivation therapies to treat PCa.
In summary, these findings indicate that FOXA1 chromatin binding is regulated by LSD1-mediated demethylation at K270 and by this mechanism, LSD1 maintains the enhancer accessibility to AR (and other steroid hormone receptors) and thus regulates its chromatin binding and transcriptional output. It is now well accepted that AR remains as a major driver of CRPC that relapses after castration therapies and treatments with abiraterone or enzalutamide 38,45. Importantly, recent studies revealed a significant portion of CRPC patient samples (~25%) contain chromatin rearrangements within a FOXA1 enhancer, which drive the overexpression of FOXA1 and the restoration of AR signaling 35. Therefore, there is a critical need for developing approaches and agents to target FOXA1 through novel mechanisms and LSD1 inhibition is an attractive approach to inhibit the FOXA1-AR axis in CRPC. Our findings also suggest that LSD1 inhibition is a promising therapeutic strategy to suppress the activity of the constitutively active AR splice variants as it directly targets the chromatin binding of AR-Vs. With more active and selective LSD1 inhibitors being developed and tested in the clinic, these preclinical findings may pave the way to improve treatment of PCa patients with FOXA1 overexpression.
METHODS
Cell culture and viral transduction
LNCaP, CWR22-RV1, DU145, and PC-3 cells were purchased from ATCC and cultured in RPMI-1640 with 10% FBS (fetal bovine serum). All lines were authenticated using short tandem repeat (STR) profiling and tested for mycoplasma contamination (MycoAlert kit, Lonza) every six months. MCF-7 cells were kindly gifted from Dr. X. Yuan at Beth Israel Deaconess Medical Center and were cultured in DMEM with 10% FBS. For androgen stimulation assays, cells were grown to 50–60% confluence in medium containing 5% CSS for 2–3 days and then treated with DHT for 0–48 hours. siRNAs against FOXA1 or LSD1 were obtained from Dharmacon RNAi Technologies (ON-TARGETplus).
Entry vectors of FOXA1 and LSD1 were purchased from the PlasmID Repository of Harvard Medical School. pLIX_403 lentiviral vectors expressing FOXA1-WT, FOXA1-K270R, LSD1-WT, or LSD1-K661A were generated using the Gateway Technology with Clonase II (Invitrogen #12535–029). FOXA1 K270R point mutation (A->G) and LSD1 K661A (A->G, A->C) were generated using QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) per the manufacturer’s protocol. The primers for generating FOXA1-K270R and LSD1-K661A were listed in Supplementary Table 1. Lentiviral particles were assembled in HEK293T cells using the 2nd generation system. In brief, 3 days after transfection in HEK293T cells, viral supernatant was centrifuged for 10 minutes at 3,000 RPM to remove any cell debris and passed through a 0.45μm filter. The collected lentiviral particles were used to infect the cells in the presence of polybrene, followed by selection with puromycin (Gibco).
Generation of CRISPR knockout clones
Lentivirus expressing Cas9 (lentiCas9-Blast; Addgene #52962) were assembled as mentioned above. CWR22RV1 cells stably expressing Cas9 were firstly generated by infection of lentiCas9 through selection of Blasticidin. Cas9 expression was confirmed by immunoblotting (Abcam #210752). CWR22-RV1-Cas9 cells were then transiently transfected using Lipofectamine2000 (Thermo Fisher) with lentiCRISPR-V2 (Addgene #52961) expressing sgLSD1 (5’-GGGGCCTGGCGGAACCGCCG-3’), which was described in a previous study 21. Cells were then selected by puromycin until single-cell clones emerged. Individual clones were further expanded and the loss of LSD1 expression was confirmed by immunoblotting.
In vitro demethylation assay
Formaldehyde production was measured using the Histone Demethylase Assay kit (Active Motif) following the manufacturer’s protocol. Synthetic FOXA1 peptide (263–281 aa) (GenScript, with >98% purity) or H3K4me2 (121 aa) peptide (Active Motif) were incubated with 0–150 nM recombinant LSD1 (Active Motif) in demethylation buffer for 1 hour at 37°C, and then detection buffer for 1 hour at 37°C, followed by fluorescence detection with excitation wavelength at 410 nm and emission wavelength at 480 nm. The reaction mixture was further analyzed by Matrix-Assisted Laser Desorption/Ionization (MALDI) mass spectrometry at the Molecular Biology Core of Dana-Farber Cancer Institute.
Immunoprecipitation and mass spectrometry
For endogenous FOXA1 immunoprecipitation, cells were lysed using RIPA lysis buffer (150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris pH 8.0) supplemented with protease inhibitor (Thermo Fisher), followed by pre-clearing using IgG-conjugated beads (Sigma) for 1 hour at 4°C. Equal amounts of protein (1 to 5 mg) were mixed with 25 μl FOXA1-conjugated beads (Santa Cruz) for overnight. Immunocomplexes were eluted with the sample buffer (Bio-Rad). For V5 pull-down, cells stably expressing doxycycline-induced V5-tagged FOXA1 were pretreated with doxycycline for 2 days, followed by GSK2879552 (50 μM) treatment for 16 hours. Extracts were incubated with 15 μl V5-conjugated beads overnight. For mass spectrometry, at least 6 × 108 cells were used for mapping the post-translational modification sites through Thermo Orbitrap Elite & Thermo QExactive HF Orbitrap microcapillary LC-MS/MS at the Mass Spectrometry Core of Beth Israel Deaconess Medical Center.
Chromatin immunoprecipitation (ChIP)
Cells were treated with S2101 (Calbiochem), OG-L002 (Selleckchem), C12 (XcessBio), GSK2879552 (Selleck), and 10 nM of DHT. ChIP assay was performed based on the previously described protocol 46. Cells were crosslinked with 1% formaldehyde for 10 minutes and quenched by 125 mM glycine for 5 minutes at room temperature with gentle shaking. Cells were quickly rinsed in cold PBS twice and collected in PBS supplemented with protease inhibitors. Cells were then centrifuged and lysed in ice-cold lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris-HCl pH 8.1) supplemented with protease inhibitor for 10 minutes. The cell lysate was sonicated using Bioruptor Sonicator (Diagenode) to break DNA into ~300-bp fragments (~500 bp for ChIP-qPCR). Soluble chromatin was diluted in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8.1), and 4 μg ChIP-grade antibody was added and incubated at 4°C overnight with gentle shaking. 50 μl protein A or G beads flurry with yeast RNA (final concentration 100 μg/ml) (Thermo Fisher) was added and incubated for one hour at 4°C. The beads were then washed in following buffers for 10 minutes each at 4°C: TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8.1), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl pH 8.1), Buffer III (0.25 mM LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.1), and TE buffer (2 times). To elude DNA, beads were incubated in elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature with aggressive shaking for 15 minutes. The supernatant was then collected and incubated at 65°C for overnight to reverse-crosslink the DNA. Qiagen QIAquick Purification Kit was used for purifying the DNA for the subsequent sequencing or PCR approaches. For ChIP assay using tumor tissues, ~20 mg of frozen tissue sample was cut into small pieces and homogenized by handheld TissueRuptor II (Qiagen). Cells were washed in cold PBS two times and centrifuged. The cell pellet was resuspended in PBS supplemented with protease inhibitor and formaldehyde at a final concentration of 1%, and then incubated at room temperature for 10 minutes. The rest of the steps are the same as the ChIP assay using culture cells. The following antibodies were used in ChIP: anti-FOXA1 antibody #1 (Abcam, ab23738), anti-FOXA1 antibody #2 (Abcam, ab5089), anti-FOXA2 antibody (Millipore, 17–10258), anti-HOXB13 antibody (Cell Signaling, 90944), anti-H3K4me2 antibody (Millipore, 07–030), anti-H3K27ac antibody (Abcam, ab4729), anti-AR antibody (Santa Cruz, sc-815), anti-V5 (Thermofisher, R960–25), anti-AR-V7 (Precision Antibody, AG10008), anti-p300 (Abcam, ab10485) and anti-IgG (Millipore). The qPCR analysis was carried out using the SYBR Green method on the QuantStudio 3 Real-time PCR system (Thermo Fisher Scientific). ChIP-qPCR experiments were done in triplicates from independent tissue culture and the results were normalized to the input DNA.
For ChIP-seq analysis, library construction was prepared using ThruPLEX DNA-seq 48D Kit (Rubicon Genomics) based on the manufacturer’s protocol, and the sequencing was performed on HiSeq 2500 Illumina Genome Analyzer. Individual ChIP-seq was performed in technical duplicates and the raw data were merged for further analysis. All the detailed descriptions for ChIP-seq and other experiments can be found in Life Sciences Reporting Summary.
ATAC-seq
Omni ATAC-seq was performed following protocol as previously described 47. About 50,000 viable LNCaP cells (growing in 5% CSS) after GSK2879552 treatment were centrifuged at 500 RCF at 4°C. The pellet was lysed in 50 μl cold resuspension buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2 0.1% NP40, 0.1% Tween-20, and 0.01% Digitonin). The lysis solution was then diluted with 1 ml cold buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20). Nuclei were collected by centrifuged at 500 RCF at 4°C for 10 minutes. Pellet was resuspended in 50 μl of transposition mixture (25 μl 2× TD buffer, 2.5 μl transposase, 16.5 μl PBS, 0.5 μl 1% digitonin, 0.5 μl 10% Tween-20, 5 μl H2O) using Illumina Tagment DNA TDE1 Enzyme and Buffer Kit, and incubated at 37°C for 30 minutes in a thermomixer with 1,000 RPM mixing. DNA samples were cleaned immediately by Qiagen QIAquick Purification Kit and PCR Pre-amplified by NEBNext High-Fidelity 2× PCR Master Mix. qPCR amplification was used to determine the additional cycles to prevent over-amplification. The final PCR product was purified by Qiagen QIAquick Purification Kit and run on Agilent High Sensitivity Screen Tape for quality control. The libraries were sequenced on the HiSeq 2500 Illumina Genome Analyzer.
RT-PCR and RNA-seq
RNA was extracted with TRIzol Reagent (Invitrogen) based on the manufacturer’s protocol. For tissue RNA isolation, the RNeasy Mini kit (Qiagen) was used following the manufacturer’s protocol. Same amount of tissue samples were bead (5 mm) milled using TissueLyser LT (Qiagen). Gene expression was measured using realtime RT-PCR analyses with TaqMan one-step RT-PCR reagents on the QuantStudio 3 Real-time PCR system and results were normalized to co-amplified GAPDH. For RNA-seq analysis, RNA was purified using RNeasy Mini Kit (Qiagen). TruSeq® Strnd Total RNA LT (Illumina) was used for library construction based on manufacturer’s protocol, and sequencing was performed on HiSeq 2500 Illumina Genome Analyzer.
Subcellular Fractionation Assay
Subcellular Protein Fractionation Kit (Thermofisher) was used to extract the soluble nuclear fraction and chromatin-bound fraction. 2 × 106 cells were used for the fractionation. After harvesting cell with trypsin, the following fractionations were isolated per manufacturer’s protocol: the cytoplasmic extract was isolated using CEB buffer, the membrane extract was isolated using MEB buffer, the soluble nuclear extract was isolated using NEB buffer, the chromatin-bound extract was isolated using NEB buffer plus Micrococcal Nuclease, and the cytoskeletal extract was isolated using PEB buffer. The extracts were boiled with 4× Laemmli buffer at 95°C for 10 minutes before loading for immunoblotting.
Immunoblotting
Cells were lysed with RIPA buffer with protease inhibitors. For tissue protein isolation, tissue samples were bead (5 mm) milled in RIPA buffer with protease inhibitors by TissueLyser LT (Qiagen). Protein concentration was measured using the Pierce BCA Protein Assay Kit. Samples with same volume and amount of protein were loaded in 4~15% Mini-PROTEAN TGX precast protein gels (Bio-Rad) and transferred to nitrocellulose membrane (Bio-Rad). Membranes were blocked in 5% non-fat milk and incubated with following antibodies at 4°C overnight: anti-AR (Millipore, 1:1,000), anti-histone H3 (1:5,000), anti-H3K4me2 (1:1,000), anti-methyl-lysine (1:200), anti-LSD1 (1:1,000), anti-FOXA1 (1:2,000), anti-β-Tubulin (1:5,000), anti-v5 (1:1,000), anti-GAPDH (1:5,000) (Abcam), anti-FOXA2 (1:1,000) (Millipore), and anti-KLK3 (1:1,000) (Meridian Life Science). The anti-FOXA1-K270me antibody was specially developed by Abcam. The company employed a phage library by using a “Rapid Liquid Screening” approach [target sequence for selection: Biotin-YLRRQKRFKCE(Kme1)QPGA]. The antibody tested in the experiment is from the first round of IgG conversion (Human/Rabbit chimeric) based on the scFv resulted from screening. The antibody concentration used in immunoblotting blotting is ~8 μg/ml. Membranes were then incubated with fluorescence-labeled secondary antibodies (LI-COR Biosciences) in 5% non-fat milk for 1 hour at room temperature. Gel images were taken by LI-COR Odyssey system under the wavelength of 680 nm or 800 nm. For detecting K270me-specific antibody (1:200), membranes were blocked in 5% BSA, and antibodies were also diluted in 5% BSA. For detecting FOXA1 in FOXA1-immunoprecipitation experiments, EasyBlot secondary antibody (GeneTex) was used to eliminate the overlapping band of IgG heavy chain. Blots shown are representative of at least 3 independent experiments. The full scan of Western blots can be found in Source Data.
Cell Counting
For testing prolonged inhibitor response, after treatments, cells were collected and fixed with 70% ethanol for 3 hours, followed by staining with Count & Viability Assay kit (proliferation), and cell counting by Muse Cell Analyzer (EMD Millipore). For testing cell viability in response to different drug doses, CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used. 20,000 cells were seeded in 96-well plates, and Glo Reagent was added to each well with a 1:1 ratio to the volume of the medium. After mixing on an orbital shaker for 5 minutes, the plate was incubated at room temperature for another 10 minutes, and the luminescence signal was recorded.
Xenograft tumors
CWR22-RV1 and DU145 xenograft tumors were established by subcutaneous injection of 1 × 106 cells in 6-week old castrated male SCID mice (Taconic). For passaging LuCaP35CR and LuCaP77CR tumors, fresh tumors were cut into small pieces (using sharp scissors) and were trypsinized for 30 minutes, followed by resuspending tumor cells in RPMI-1640 mixed with Matrigel (1:1) and subcutaneous injection in castrated male SCID mice. For inducing FOXA1 expression in xenograft tumors derived from CWR22-RV1 stable cells overexpressing WT or K270R FOXA1, mice were fed with mouse chow supplemented with doxycycline (Teklad Custom Diet). During the treatment period, tumor volume was measured by electronic manual caliper using formula L × W2/2. All animal experiments were approved by the UMass Boston Institutional Animal Care and Use Committee and were performed following institutional and national (USA) guidelines. The housing conditions were ambient temperatures of 65–75°F with 40–60% humidity and 12-hour light/12-hour dark cycle.
Analysis for ChIP-seq and ATAC-seq
Raw reads were aligned to hg19 using bwa (version 0.7.2) 48 with aln function followed by samse. For bwa aln, the first 32 subsequences were used as seed with trimming parameter setting as 5 (-l 32 -q 5). Default parameters were used for bwa samse. The resulted sam files are converted to bam with samtools (version 0.1.18) 49. MACS2 (version 2.1.2) 50 was used to call peak on the bam files with fix-bimodal turned on and extend size set at 100 (--bw 250 –mfold 10 30 –fix-bimodal –extsize 100). The q-value cutoff for peak significance is set as 0.05 (--qvalue 0.05). bedGraph files containing signal per million reads produced from MACS2 were converted to bigwig files with ucsctool kit (315). Briefly, bedGraph files were sorted with bedSort, clipped with bedClip, and finally converted using bedGraphToBigWig to bigwig.
For global chromatin binding and accessibility analyses, all ChIP-seq and ATAC-seq results were normalized to the same sequencing depths. A high correlation of above 0.9 was observed between duplicates for all ChIP-seq and ATAC-seq data. To measure differences between averaged profiles for ChIP-seq and ATAC-seq, the non-parametric Kolmogorov-Smirnov test (KS-test) was used to calculate one-sided P value using data from both replicates. The R package ChIPpeakAnno (version 3.10.1) 51 was used for analyzing peak intervals. The function findOverlapsOfPeaks was used to determine the overlapping among two or more peak sets. The deepTools suite (version 2.4.1) 52 was used to extract and visualize signals from bigwig files. The function computeMatrix was used with reference-point mode to calculate scores for each genomic region. Missing data were treated as zeros and regions with only zero values were skipped. The plotHeatmap and plotProfile functions were used to generate heatmap or a profile plot for scores over genomic regions.
RNA-seq analysis
Raw reads of RNA-seq were aligned to the hg19 reference genome using STAR (version 2.4.2a) 53 with gene counting function enabled (--quantMode GeneCounts). The infer_experiment.py function from RSeQC (version 2.6.1) 54 was used to determine how and whether the reads were stranded. Gene level count data were then pulled according to the strandness information from STAR output. Normalized counts per million were calculated before downstream analysis and a pseudo count of 1 is added when calculating fold change.
Target prediction
Binding and Expression Target Analysis (BETA, v1.0.7) 55 was used to predict targets from ChIP-seq and RNA-seq data. Peak interval files from MACS2 and differential analysis results from edgeR were used as input with the default parameter setting in running BETA.
Statistics and Reproducibility
All ChIP-qPCR and qRT-PCR data were presented as the dot-plot format from samples collected from 3 independent tissue cultures. Statistical analysis was performed using unpaired two-tailed Student’s t test by comparing treatment versus vehicle control or otherwise as indicated. We use ns (P > 0.05), ^ (0.01 < P < 0.05), * (0.001 < P < 0.01), and # (P < 0.001) to indicate the levels of P value. For ChIP-seq and ATAC-seq results, the non-parametric Kolmogorov-Smirnov test was used to calculate one-sided P value using data from both replicates. For animal studies, a two-tailed Student’s t test was performed to determine the statistical difference of tumor growth at the final time point. The results for immunoblotting are representative of at least three biologically independent experiments. All statistical analyses and visualization were performed by using GraphPad (Prism 7/8) or R (version 3.4.0) unless otherwise specified.
PCR primers
For ChIP-qPCR, primers for KLK3-enh, KLK2-enh, NKX3.1-enh, SGK1-enh, SGK3-enh, MBOAT2-enh, RAB11B-FBS, PDK4-FBS, and CDK1-FBS were listed in Supplementary Table 1. Primers for TMPRSS2-enh, ZBTB16-enh, and SH2B1-enh were described previously 11. Primers for TFF1-enh, NRIP1-enh, PGR-enh, and DSCAM-enh were described previously 56,57.
For RT-qPCR, primer and probe sets for KLK3 and ZBTB16 were listed in Supplementary Table 1. Primer and probe sets for MBOAT2 (Hs01027245_m1), ELOVL7 (Hs00405151_m1), FKBP5 (Hs01561006_m1), NKX3.1 (Hs00171834_m1), SGK1 (Hs00178612_m1), ACSL3 (Hs00244853_m1), ELOVL5 (Hs01094711_m1), NANS (Hs00219054_m1), and AR-V7 (AI6R0CI) were purchased from Thermo Fisher.
Extended Data
Extended Data Fig. 1|. LSD1 regulates FOXA1 chromatin binding in PCa cells.
a, b, FOXA1 ChIP-seq was performed using an anti-FOXA1 antibody (Ab#1) in LNCaP cells treated with vehicle or 50μM S2101 for 4 hours (h) or 48h. (a) Heatmap view for FOXA1 ChIP-seq peak intensity and (b) the mean of FOXA1 ChIP-seq signals at FOXA1 binding sites (Veh vs S2101–4h: p=0.09; Veh vs S2101–48h: p=2.5×10–5) were shown. c, d, FOXA1 ChIP-seq was performed using an anti-FOXA1 antibody (#2) in LNCaP cells treated with vehicle or GSK2879552 (50μM, 4h). (c) Heatmap view for FOXA1 ChIP-seq peak intensity and (d) the mean of FOXA1 ChIP-seq signals at FOXA1 binding sites (Veh vs GSK-4h: p=7.0×10–72) were shown. e, ChIP-qPCR for FOXA1 binding at KLK3/NKX3.1 enhancer site. f, ChIP-qPCR for FOXA1 binding in LNCaP cells treated with vehicle or LSD1 inhibitors for 4h (S2101, OG-L002 at 50μM and C12 at 5μM). g, Immunoblotting for H3K4me2 in LNCaP or CWR22-RV1 cells treated with GSK2879552 at indicated doses for 48h. h, Immunoblotting for FOXA1 or AR in LNCaP cells treated with 0–50μM GSK2879552 for 48h. i, Immunoblotting for FOXA1 or AR in LNCaP cells treated with GSK2879552 (50μM, 0–48h). j, Immunoblotting for FOXA1 or AR in CWR22-RV1 cells treated with GSK2879552 (0–5μM, 48h). k, Immunoblotting for FOXA1 or AR in CWR22-RV1 control cell line versus LSD1-KO cell line. l, m, LNCaP cells stably overexpressing doxycycline (dox)-regulated LSD1-WT or LSD1-K661A mutant treated with/out doxycycline were subjected to (l) immunoblotting or (m) ChIP-qPCR for FOXA1 at indicated sites. Note: Experiments described in this figure were all done under hormone-depleted conditions.
Extended Data Fig. 2|. LSD1 inhibition represses chromosome openness at FOXA1/AR mediated enhancers.
a, qPCR for DHS (DNA hypersensitivity) levels at indicated enhancer sites in LNCaP cells treated with S2101 for 4h. b, H3K27ac ChIP-seq was performed in LNCaP cells treated with vehicle or GSK2879552 (50μM, 4h). The mean values of peak intensities for FOXA1 ChIP-seq (Veh vs GSK-4h: p=0.03), H3K27ac ChIP-seq (Veh vs GSK-4h: p=3.9×10–12), and ATAC-seq at AR-binding sites (Veh vs GSK-4h: p=1.4×10–15) were shown. c, ChIP- qPCR for H3K27ac at indicated sites in LNCaP cells treated with vehicle or GSK2879552 (50μM, 4h). d, ChIP-qPCR for H3K27ac at indicated sites in CWR22-RV1 cells treated with vehicle or S2101 (50μM, 4h). Note: Experiments described in this figure were all done under hormone-depleted conditions.
Extended Data Fig. 3|. LSD1 promotes FOXA1 binding at AR-independent regulatory sites.
a, Heatmap view for FOXA1 ChIP-seq peaks at non-AR binding sites in LNCaP cells. b, ChIP-qPCR for FOXA1 binding at indicated non-AR binding sites (FBS: FOXA1-solo Binding Sites) in LNCaP cells treated with GSK2879552 or S2101 (50μM, 0–48h). c, ChIP-qPCR for FOXA1 binding at indicated ER-regulated enhancers in MCF-7 cells treated with GSK2879552 (50μM, 4h).
Extended Data Fig. 4|. LSD1 regulates AR chromatin binding and activity.
a, AR ChIP-seq analyses were performed in LNCaP cells treated with vehicle only, DHT (10nM, 4h), DHT (4h) with pretreatment of GSK2879552 (50μM, 0.5h), or DHT (4h) with pretreatment of GSK2879552 (50μM, 48h). Overlap of AR binding sites in vehicle and DHT treated cells was shown. b, qPCR for DHT-induced DHS (DNase HyperSensitivity) intensity at AR regulated enhancers in LNCaP cells pretreated with S2101 (50μM, 0.5h) and then treated with/out DHT (10nM, 4h). c, FOXA1, HOXB13, and AR ChIP-seq peaks as well as ATAC-seq peaks at KLK3 locus. d, Identification of the subset of directly AR-regulated genes by BETA (Binding and Expression Target Analysis) using AR ChIP-seq (DHT treated for 4h) and RNA-seq in LNCaP cells (vehicle versus DHT treated for 24h). e, RNA-seq analyses in LNCaP cells treated with/out DHT (10nM, 24h) and with/out pretreatment of GSK2879552 (50μM, 24h) were performed and DHT-induced fold-change of the expression for the identified directly AR-regulated genes was plotted [center: median; box: 25th to 75th interquartile range (IQR); whiskers: 1.5x IQR; outliers: individual data points]. f, qRT-PCR for PSA/NKX3.1 expression in LNCaP cells pretreated with LSD1 inhibitors for 4h (S2101/OG-L002 at 50μM and C12 at 5μM) and then treated with/out DHT (10nM, 24h). g, ChIP-qPCR for AR binding in LNCaP stable cells expressing doxycycline-inducible LSD1-WT (LN-LSD1WT) or LSD1-K661A (LN-LSD1K661A). h, RT-PCR for KLK3 and NKX3.1 mRNA expression in LNCaP cells stably overexpressing LSD1-WT or LSD1-K661A mutant treated with/out DHT (0–1nM, 24h). i, LN-LSD1WT cells treated with different doses of GSK2879552 (6 days) and with/out doxycycline were subjected to the measurement of cell density (mean±SD). j, ChIP-qPCR for FOXA1 at KLK3 enhancer in LNCaP cells treated with GSK2879552 (1μM, 4h). k, GSEA showing top-ranked pathways that were repressed or enhanced by GSK2879552 (1μM for 2 weeks versus 50μM for 48h). Note: Experiments described in this figure were all done under hormone-depleted conditions.
Extended Data Fig. 5|. LSD1 regulates AR-V7 chromatin binding and activity.
a, ChIP-qPCR for FOXA1 binding at AR-regulated enhancers in LNCaP stable cells expressing doxycycline- inducible ARV7 (LNCaP-tetARV7) treated with/out GSK2879552 (50μM, 4h). b, Immunoblotting for V5 (AR-V7) in LNCaP-tetARV7 cells treated with vehicle, doxycycline only, doxycycline plus GSK2879552 (10μM), doxycycline plus S2101 (10μM), or doxycycline plus ORY-1001 (2.5μM) for 48h. c, qRT-PCR for the expression of AR-V7-regulated genes in CWR22-RV1 cells treated with GSK2879552 (0–2.5 μM for 48h). d, Immunoblotting for AR-V7 in CWR22-RV1 cells treated with GSK2879552 (0–10 μM, 48h). e, qRT-PCR for AR-FL/V7-regulated genes in the LSD1-KO line versus the control line with/out 10nM DHT treatment. Note: Experiments described in this figure were all done under hormone-depleted conditions.
Extended Data Fig. 6|. Identification of methylated lysine 270 in FOXA1 as an LSD1 substrate.
a, FOXA1 immunoprecipitation was performed in LNCaP cells treated with different LSD1 inhibitors (S2101 at 50μM and C12 at 5μM), followed by immunoblotting for methyl-lysine (images cropped from the same blot). b, FOXA1 immunoprecipitation was performed in LN-LSD1WT or LN-LSD1K661A cells, followed by immunoblotting for methyl-lysine. c, Mass-spectrometry analysis on immunoprecipitated V5-FOXA1 in LNCaP cells stably overexpressing V5-tagged FOXA1. Covered residues are in yellow. Residues with detected post-translational modifications are indicated in green. d, LNCaP stable cells expressing doxycycline-inducible V5-tagged FOXA1- WT (LNCaP-tetFOXA1WT) or FOXA1-K270R (LNCaP-tetFOXA1K270R) were generated. V5-FOXA1 expression induced by doxycycline treatment (0–0.1μg/ml) was confirmed by immunoblotting. e, ChIP-qPCR for FOXA1-WT or K270R binding (anti-V5) at AR-regulated enhancers in these stable cells (doxycycline supplemented) treated with S2101 (50μM, 24h). f, g, CWR22-RV1 cells stably expressing doxycycline-inducible FOXA1-WT (CWR22RV1-tetFOXA1WT) or K270R mutant (CWR22RV1-tetFOXA1K270R) were established. (f) Immunoblotting for LSD1 in those stable cells (doxycycline supplemented) transfected with siRNA against LSD1 (siLSD1) or non-target control (siNTC) and (g) ChIP-qPCR for V5-FOXA1 binding were performed. Note: Experiments described in this figure were all done under hormone-depleted conditions.
Extended Data Fig. 7|. K270R mutation of FOXA1 enhances the chromatin binding of FOXA1 and subsequently stabilizes AR recruitment.
a, LNCaP-tetFOXA1WT cells treated with GSK2879552 (50μM) were fractionated into the soluble nuclear fraction and the insoluble chromatin-bound fraction, followed by immunoblotting for V5 and histone 3 (H3). b, CWR22RV1-tetFOXA1WT or CWR22RV1-tetFOXA1K270R cells were fractionated into the soluble nuclear fraction and the insoluble chromatin-bound fraction, followed by immunoblotting for V5 and H3. c, LNCaP-tetFOXA1WT or LNCaP-tetFOXA1K270R cells were treated with/out doxycycline for 48h, and DHT or vehicle for 4h. ChIP-qPCR for AR binding at AR-regulated enhancers was shown. d, ChIP-qPCR for AR binding in LNCaP-tetFOXA1WT or LNCaP-tetFOXA1K270R cells treated with DHT (10nM) versus DHT plus enzalutamide (10μM). e, qRT-PCR for androgen-induced gene expression in response to enzalutamide treatment in CWR22RV1-tetFOXA1WT or CWR22RV1-tetFOXA1K270R cells (in the presence of 10nM DHT). Note: Experiments described in this figure were all done under hormone-depleted conditions.
Extended Data Fig. 8|. LSD1 inhibition impairs FOXA2 binding in PC-3 cells.
a, The amino acid sequences at the wing2 region of FOXA1 and FOXA2 were aligned. The red arrow indicates the K270 of FOXA1. b, Immunoblotting for FOXA2 in the indicated PCa cell lines. c, d, FOXA2 ChIP-seq was performed in PC-3 cells treated with vehicle or GSK2879552 (50μM, 4h). (c) Heatmap view for FOXA2 ChIP- seq peak intensity and (d) the mean of FOXA2 ChIP-seq signals at FOXA2 binding sites (Veh vs GSK-4h: p=1.6×10–6) were shown. e, HOXB13 ChIP-seq was performed in LNCaP cells treated with vehicle or GSK2879552 (50μM, 4h). Heatmap view for HOXB13 ChIP-seq peak intensity was shown. f, ChIP-qPCR for HOXB13 binding at indicated enhancer sites in LNCaP cells treated with/out GSK2879552 (50μM, 4h). Note: Experiments described in this figure were all done under hormone-depleted conditions.
Extended Data Fig. 9|. LSD1 inhibition suppresses tumor growth of the CWR22-RV1 CRPC model.
a, The correlation between LSD1 and FOXA1 mRNA expression levels in TCGA PCa cohort. b, LSD1 and FOXA1 expressions in normal prostate versus PCa tumors using TCGA PCa dataset (center: median; box: 25th to 75th IQR; whiskers: 1.5x IQR; outliers: individual data points). c, FOXA1 mRNA expression levels in cell lines derived from different diseases using data from cancer cell line encyclopedia, CCLE (center: median; box: 25th to 75th IQR; whiskers: 1.5x IQR; outliers: individual data points). d, CWR22-RV1 cells (hormone depleted condition) were treated with different doses of GSK2879552, and cell density was measured after 2 days of treatments (mean±SD). e, Castrated SCID male mice bearing CWR22-RV1 xenograft tumors received DMSO or GSK2879552 treatment and the body weight was measured at the endpoint. f, Immunoblotting for H3K4me2 in the vehicle versus GSK2879552 treated group.
Extended Data Fig. 10|. LSD1 inhibition suppresses tumor growth of FOXA1-positive CRPC PDX models.
a, The mRNA expression of AR-V7 was examined in tumor samples from vehicle-treated xenograft tumors in comparison with C4–2 (AR-V7 negative) derived xenograft tumors. b, c, LuCaP35CR tumors were established in castrated male SCID mice and treated with LSD1 inhibitors. Immunoblotting for H3K4me2 in LuCaP35CR (b) treated with vehicle versus GSK2879552 or (c) treated with vehicle versus ORY-1001. d, Bodyweight for mice bearing LuCaP35CR treated with vehicle versus ORY-1001. e, f, Castrated SCID male mice bearing LuCaP35CR xenograft tumors received daily DMSO or ORY-1001 (0.06mg/kg) via intraperitoneal injection (n=7 independent tumors). (e) The tumor volume was measured at the indicated time. (f) After the mice were sacrificed, tumor samples were subjected to RT-PCR analysis for indicated AR-FL/V7 regulated genes. g, LuCaP77CR tumors were established in castrated male SCID mice and treated with LSD1 inhibitors. Immunoblotting for H3K4me2 in LuCaP77CR tumors treated with vehicle versus GSK2879552. h, LNCaP and DU145 cell lines were treated with 0–200μM GSK2879552 for 4 days and the cell density was measured (mean±SD). i, CWR22-RV1 cells (in hormone depleted condition) were treated with DMSO, GSK2879552 (5μM), enzalutamide (10μM), or combination of GSK2879552 and enzalutamide for 2d, and then cell density was measured.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by grants from NIH (R00 CA166507 and R01 CA211350 to C.C., U54 CA156734 to C.C. and J.A.M., and P01 CA163227 to S.P.B.), DOD (W81XWH-15-1-0554 and W81XWH-19-1-0777 to S.G., W81XWH-16-1-0445 and W81XWH-19-1-0361 to C.C.), CIHR (142246, 152863, 152864, and 159567 to H.H.H.), Prostate Cancer Canada (RS2016-1022 and TAG2018-2061 to H.H.H.), NSERC (498706 to H.H.H.), Terry Fox Program Project Grants (1093 to H.H.H.), and Princess Margaret Cancer Foundation (to H.H.H.). H.H.H. holds a Joey and Toby Tanenbaum Brazilian Ball Chair in Prostate Cancer Research. We thank Dr. John Asara and Min Yuan from Beth Israel Deaconess Medical Center and James Lee from Dana Farber Cancer Institute for the work on mass spectrometry analyses.
Footnotes
DATA AVAILABILITY
All data generated or analyzed during this study are included in this published article. The GEO accession for ChIP-seq and RNA-seq data is GSE149007. Readers are welcome to comment on the online version of the paper. Correspondence and requests for materials should be addressed to C.C. (changmeng.cai@umb.edu).
CODE AVAILABILITY STATEMENT
This study did not generate any unpublished code, software, or algorithm. All data analyses were performed using cited software with the parameters indicated in the Methods section.
COMPETING INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Gao N. et al. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol 17, 1484–507 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Carroll JS et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Jozwik KM & Carroll JS Pioneer factors in hormone-dependent cancers. Nat Rev Cancer 12, 381–5 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Yang YA & Yu J Current perspectives on FOXA1 regulation of androgen receptor signaling and prostate cancer. Genes Dis 2, 144–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–53 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Shi YJ et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell 19, 857–64 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Metzger E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–9 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Wissmann M. et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol 9, 347–53 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Metzger E. et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation. Nat Cell Biol 10, 53–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzger E. et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature 464, 792–6 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Cai C. et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep 9, 1618–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupien M. et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–70 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swinstead EE et al. Steroid Receptors Reprogram FoxA1 Occupancy through Dynamic Chromatin Transitions. Cell 165, 593–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzger E. et al. Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nat Struct Mol Biol 23, 132–9 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Sehrawat A. et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci U S A 115, E4179–E4188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mimasu S. et al. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry 49, 6494–503 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Mohammad HP et al. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 28, 57–69 (2015). [DOI] [PubMed] [Google Scholar]
- 18.He HH et al. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res 22, 1015–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin HJ, Zhao JC, Wu L, Kim J & Yu J Cooperativity and equilibrium with FOXA1 define the androgen receptor transcriptional program. Nat Commun 5, 3972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheng W. et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell 174, 549–563 e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonarakis ES et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 371, 1028–38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z. et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res 20, 1590–600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharp A. et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest 129, 192–208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu R. et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 69, 16–22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y. et al. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res 71, 2108–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J. et al. p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–8 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Wang J. et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41, 125–9 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Kontaki H & Talianidis I Lysine methylation regulates E2F1-induced cell death. Mol Cell 39, 152–60 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S & Carroll JS FOXA1 Directs H3K4 Monomethylation at Enhancers via Recruitment of the Methyltransferase MLL3. Cell Rep 17, 2715–2723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbieri CE et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44, 685–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grasso CS et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parolia A. et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J. et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 580, 93–99 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Iwafuchi M. et al. Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nat Genet (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scher HI et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 367, 1187–97 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Park JW, Lee JK, Witte ON & Huang J FOXA2 is a sensitive and specific marker for small cell neuroendocrine carcinoma of the prostate. Mod Pathol 30, 1262–1272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brechka H, Bhanvadia RR, VanOpstall C & Vander Griend DJ HOXB13 mutations and binding partners in prostate development and cancer: Function, clinical significance, and future directions. Genes Dis 4, 75–87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maes T. et al. ORY-1001, a Potent and Selective Covalent KDM1A Inhibitor, for the Treatment of Acute Leukemia. Cancer Cell 33, 495–511 e12 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Pishas KI et al. Therapeutic Targeting of KDM1A/LSD1 in Ewing Sarcoma with SP-2509 Engages the Endoplasmic Reticulum Stress Response. Mol Cancer Ther 17, 1902–1916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corey E. et al. LuCaP 35: a new model of prostate cancer progression to androgen independence. Prostate 55, 239–46 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Nguyen HM et al. LuCaP Prostate Cancer Patient-Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease an--d Serve as Models for Evaluating Cancer Therapeutics. Prostate 77, 654–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Bono JS et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q. et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138, 245–56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics 28, 1838–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsi-Yang Fritz M, Leinonen R, Cochrane G & Birney E Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res 21, 734–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu LJ et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramirez F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Wang S & Li W RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–5 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Wang S. et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc 8, 2502–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D & Carroll JS FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43, 27–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y. et al. Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat Genet 48, 1003–13 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.