Abstract
Immunotherapy has revolutionized cancer treatment, yet most patients do not respond. While tumor antigens are needed for effective immunotherapy, a favorable tumor immune microenvironment is also critical. In this review, we discuss emerging evidence that tumor cells exploit cellular plasticity and dedifferentiation programs to avoid immune surveillance, which in turn drives metastatic dissemination and resistance to immunotherapy. A deeper understanding of these programs may provide novel opportunities to enhance the efficacy of existing immunotherapies.
Keywords: Tumor immunology, immune evasion, tumor dedifferentiation, cell plasticity
Tumor immunotherapy and resistance mechanisms
Studies over the past two decades have uncovered a crucial role for the immune system in tumor biology (1). Immune cells interact with and functionally influence tumor cells at every stage of tumor development and metastatic dissemination (1–3). Therapeutic interventions enhancing immune cell functions – including chimeric antigen receptor (CAR) T cell-based treatments and immune checkpoint blockade (ICB) – have revolutionized the clinical care of cancer patients with various types of malignancies (1,4–6). Despite these remarkable successes, most cancers are refractory to immunotherapy as a result of immune evasion of tumor cells (7–11). Moreover, immune evasion is an important step in the colonization of disseminated and dormant tumor cells in distant organs (2,3,12). Because tumor immune evasion leads to poor clinical outcomes by promoting therapy resistance and metastatic outgrowth, a deeper understanding of the underlying molecular and cellular mechanisms is needed.
Tumor cells use various strategies to evade immune surveillance (13,14). These include: (i) down-regulation of the antigen presentation machinery (15–18), (ii) silencing the expression of tumor associated antigens (19,20), (iii) dysregulation of tumor cell intrinsic interferon signaling pathways (21–25), (iv) recruitment of immunosuppressive cells (e.g. regulatory T cells and suppressive myeloid cells) to establish an “immune-privileged” microenvironment (26–29), (v) upregulation of immune suppressive molecules (e.g. PD-L1) (30–32), and (iv) metabolic activity of tumor cells (e.g. production of prostaglandin E2) (33–38). Conveniently (for the tumor cells), many of these immune-evading adaptations are driven by the very oncogenic signaling pathways that provide tumors with their enhanced growth and proliferation properties (e.g. Wnt, mTOR, MYC, and Kras signaling) (13,14,32,39–46). This, in turn, begs an obvious question: What biological properties link oncogenic signaling with immune regulation? One answer to this question, supported by several recent studies, is that such signaling can simultaneously alter cellular differentiation states of tumor cells as well as their recognition by the immune system.
Tumor cell plasticity and dedifferentiation drives immune evasion
Cellular plasticity – defined as a dramatic shift in cellular phenotype – is commonly observed in various types of malignancies, where it contributes to tumor progression and resistance to therapeutic interventions (47–51). One manifestation of plasticity in tumors is dedifferentiation, in which tumor cells lose their specialized properties and take on less differentiated phenotypes reminiscent of early embryonic development or regenerative processes (52). Loss of differentiation is known to be associated with increased tumor cell invasiveness and drug resistance (49–51), but there is growing evidence that tumor cell dedifferentiation is also coupled to immune surveillance.
Studies in melanoma, for example, have shown that tumor cell dedifferentiation – and adoption of a stem- or progenitor-like phenotype – leads to an escape of immune recognition by adoptively transferred T cells in both preclinical mouse models and patients (20,53). Two factors contribute to this immune-privileged state: (i) a loss of differentiation-associated antigens (20,53) and (ii) dedifferentiation-associated transcriptional changes that result in the recruitment of immunosuppressive myeloid cells (54). Moreover, a study published earlier this year linked dedifferentiation of melanoma cells with resistance to ICB in both preclinical mouse models and cancer patients (55). Thus, there is ample data to support a connection between tumor cell dedifferentiation and immune evasion in this lethal form of skin cancer.
There is also evidence for such a connection in other tumor types. In squamous cell carcinoma, for example, dedifferentiated tumor cells acquire stem-like properties and express the immune modulating molecule CD80, leading to escape from immune attack (56). Similarly, dedifferentiated tumor initiating cells can evade immune surveillance by dysregulation of PD-L1 or NKGD2 (57,58). Moreover, single cell analysis has revealed that lung cancer cells possess heterogeneous differentiation states that correspond to various stages of lung development. These distinct states are associated with different sensitivities to immune surveillance and metastatic colonization capacity, a consequence of differential expression of natural killer cell and T cell recognition and regulatory molecules (59). Interestingly, one recent study showed that metastasis-initiating colorectal cancer cells possess molecular features of regenerative epithelial cells (60). These studies highlight potential molecular connections linking tissue regeneration and immune surveillance, given that tissue damage, compensatory regeneration, and the associated inflammatory response may all promote tumor progression. Such an idea is in line with studies from multiple tumor types that epithelial mesenchymal transition or EMT (another form of altered cellular differentiation) leads to immune evasion through various molecular mechanisms, including increases in immune inhibitory molecules and decreases in the antigen presentation machinery (49,61–66). It is also important to note that dedifferentiation of tumor cells may cause increased expression of developmental antigens such as cancer testis antigens, which may have an opposing effect on immune surveillance (67–69). Together, these studies indicate that tumor cell dedifferentiation promotes immune evasion through distinct, but related, molecular mechanisms.
Conserved immune evasion mechanisms in normal cells
Why might acquisition of a stem- or progenitor-like state – the phenotypic consequence of dedifferentiation – lead to immune evasion? One possibility is that tumor cells are simply recapitulating an evolutionarily conserved program that protects stem cells from immune attack. It is known that certain quiescent tissue-resident stem cells evade detection and killing by the innate and adaptive immune system by downregulating molecules involved in antigen presentation (70). Interestingly, one recent study provided evidence that disseminated pancreatic tumor cells in the liver can evade T cell mediated immune surveillance when they reside in a quiescent state (71). Likewise, other studies have highlighted the function of slow cycling and less differentiated cancer stem cells in shaping the tumor immune microenvironments (72). Collectively, these findings indicate that dedifferentiation-associated changes in cell proliferation may regulate the interaction between tumor cells and surrounding immune cells.
Recently, an evolutionarily conserved molecular mechanism for immune invasion was described in which epigenetic regulation by the polycomb repressive complex 2 (PRC2) robustly repressed the expression of antigen presentation molecules in embryonic stem cells, tissue specific progenitor cells, and cancer cells (15). Thus, immune privilege may be a feature of cells that normally exist in a dedifferentiated state: tissue resident stem and progenitor cells. By extension, cancer cells – responding to oncogenic signals that promote dedifferentiation – may simply be exploiting such evolutionarily conserved mechanisms to evade immune surveillance.
Important questions to be answered
This concept of differentiation-associated tumor immune regulation raises several unanswered questions: (i) How does tumor cell plasticity and dedifferentiation mechanistically lead to immune evasion? While several possible mechanisms have been described, additional molecular links between tumor differentiation status and immune system activity remain to be elucidated; (ii) Do epigenetic changes couple tumor dedifferentiation to immune evasion? Epigenetic regulators have been demonstrated to drive tumor cell plasticity and dedifferentiation (49). Given recent studies highlighting the important role of epigenetic factors in regulating antigen presentation on tumor cells (15), tumor cell identity may be directly coupled to immune escape through the action of epigenetic regulators; (iii) Do different tumor types use distinct mechanisms to evade immune surveillance? Studies to date have revealed divergent mechanisms of immune evasion in various tumor types, ranging from reduced antigen presentation by tumor cells to heightened immunosuppression in the tumor microenvironment. Further study is needed to determine whether tumors arising in different tissues-of-origin prefer certain immune-evasive strategies or whether a variety of mechanisms are available for a given tumor regardless of lineage. Likewise, given the fact that features of the host tissue also contribute to tumor immunity (73), tactics for immune evasion in metastases may track with either the primary tumor’s lineage, or with the site of dissemination; (iv) Does cellular plasticity associated with tissue injury-regeneration confer immune protective effects to incipient cancers? Cellular plasticity is a feature of normal tissues subjected to injury or inflammation, as is commonly observed in premalignant states of metaplasia (49). Thus, the mechanisms underlying plasticity in these inflammatory states may provide incipient tumors with additional immuno-protective properties; (v) Does “redifferentiation” increase the susceptibility of a tumor to immune surveillance? The less differentiated a tumor is, the more aggressive its behavior. Hence, therapeutic approaches that promote tumor cell redifferentiation can provide clinical benefit (74). An additional benefit of such approaches might be an increased susceptibility of tumor cells to immune surveillance, thereby enhancing the efficacy of existing immunotherapies such as CAR-T cells and checkpoint blockade; and (vi) What are the implications for stem cell biology? A further understanding of immune-evasive mechanisms in cancer may inform strategies for preserving stem cell viability and longevity in normal tissues by protecting these self-renewing cells from aging-dependent immune-mediated attrition.
Concluding remarks
In summary, there is mounting evidence to suggest that tumor cells hijack immune evasive mechanisms from normal somatic stem and progenitor cells. As cells become less differentiated during tumor progression, they employ both cell autonomous and non-cell autonomous mechanisms to change their susceptibility to immune recognition and destruction (Figure 1). However, there are many remaining questions that need to be explored. Recent development of transcriptional and epigenetic profiling techniques that could examine molecular features of tumor cells at single cell resolution will facilitate a detailed picture of interactions between dedifferentiated tumor cells and the immune microenvironment. In addition, establishment of improved in vitro organoid systems will be helpful, as they could allow high throughput unbiased screens to differentiation-promoting agents which could simultaneously slow tumor growth and improve immune recognition. In conclusion, elucidation of the mechanisms by which tumor cells evade immune destruction, and their links to evasive mechanisms utilized by normal somatic stem and progenitor cells, may provide novel therapeutic opportunities for enhancing the efficacy of existing immunotherapies. At the same time, such knowledge may broaden our understanding of interactions between immune cells and stem cells in other biological contexts.
Figure 1. Dedifferentiation of tumor cells leads to immune evasion.
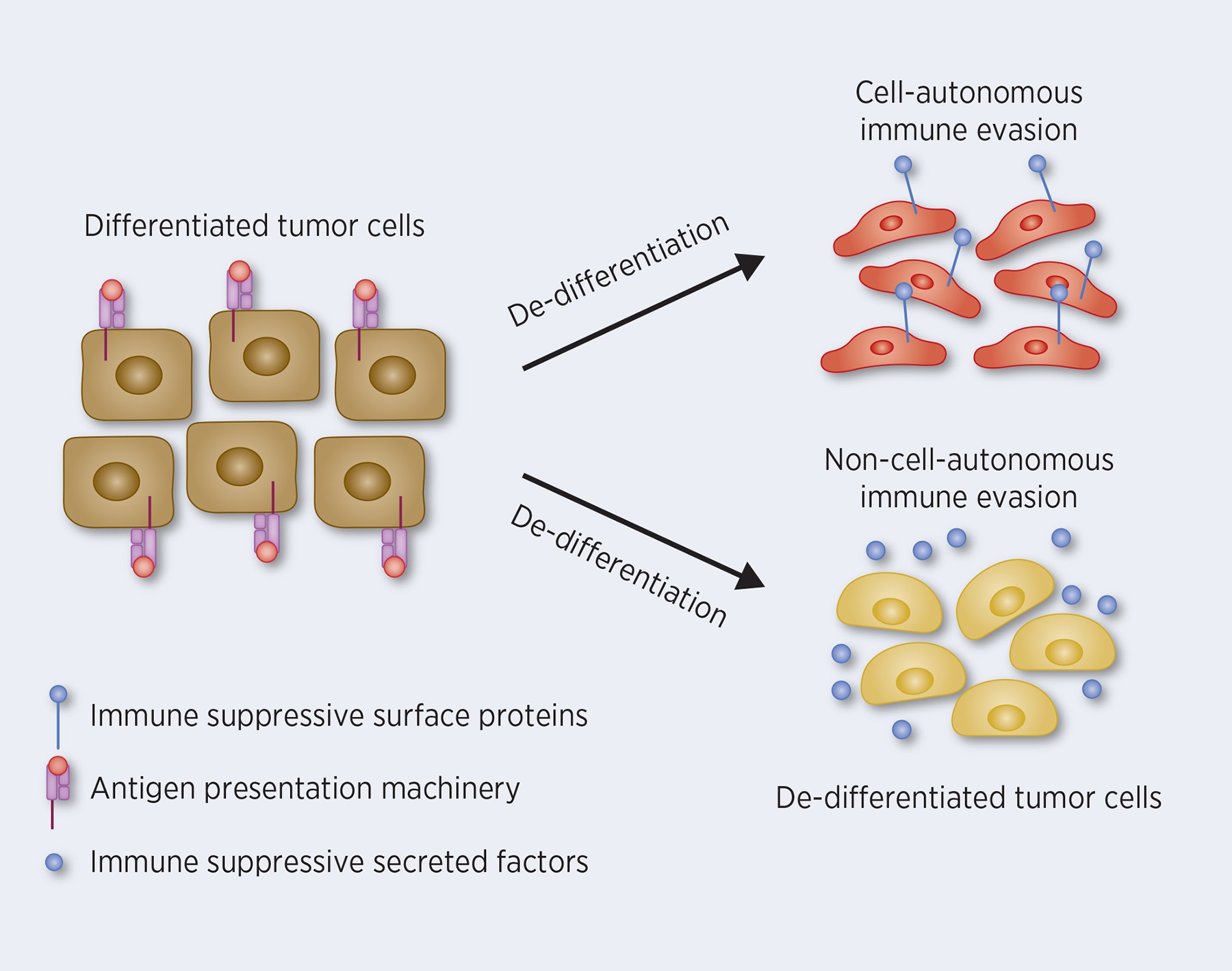
Diagram showing how dedifferentiation of tumor cells induces immune evasion through cell-autonomous mechanisms (e.g. loss of differentiation-associated antigens, decreased expression of antigen presentation molecules, and increased expression of immune suppressive molecules, such as PD-L1) and non-cell-autonomous mechanisms (e.g. expression of suppressive myeloid cells recruiting chemokines and growth factors).
Acknowledgements
JL acknowledges support from the Blavatnik Family Fellowship program. BZS acknowledges support from the NCI (R01CA229803).
References
- 1.Galon J, Bruni D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity. 2020;52:55–81. [DOI] [PubMed] [Google Scholar]
- 2.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goddard ET, Bozic I, Riddell SR, Ghajar CM. Dormant tumour cells, their niches and the influence of immunity. Nat Cell Biol. 2018;20:1240–9. [DOI] [PubMed] [Google Scholar]
- 4.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–5. [DOI] [PubMed] [Google Scholar]
- 5.Sanmamed MF, Chen L. A Paradigm Shift in Cancer Immunotherapy: From Enhancement to Normalization. Cell. 2018;175:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown CE, Mackall CL. CAR T cell therapy: inroads to response and resistance. Nat Rev Immunol. 2019;19:73–4. [DOI] [PubMed] [Google Scholar]
- 9.Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol. 2019;16:372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiong HQ. Molecular targeting therapy for pancreatic cancer. Cancer Chemother Pharmacol. 2004;196:430–41. [DOI] [PubMed] [Google Scholar]
- 12.Kitamura T, Qian B-Z, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15:73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;35:S185–98. [DOI] [PubMed] [Google Scholar]
- 14.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell. 2019;36:385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulson KG, Voillet V, McAfee MS, Hunter DS, Wagener FD, Perdicchio M, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun. 2018;9:3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicklin DJ, Wang Z, Arienti F, Rivoltini L, Parmiani G, Ferrone S. beta2-Microglobulin mutations, HLA class I antigen loss, and tumor progression in melanoma. J Clin Invest. 1998;101:2720–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–6. [DOI] [PubMed] [Google Scholar]
- 21.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017;7:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:397–404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan D, Kobayashi A, Jiang P, De Andrade LF, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature. 2019;565:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, et al. Tumor-Derived Granulocyte-Macrophage Colony-Stimulating Factor Regulates Myeloid Inflammation and T Cell Immunity in Pancreatic Cancer. Cancer Cell. 2012;21:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Huang Q, Luster TA, Hu H, Zhang H, Ng WL, et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cancer Discov. 2020;10:270–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, et al. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity. 2018;49:178–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discov. 2016;6:80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorand RD, Nthale J, Myers JT, Barkauskas DS, Avril S, Chirieleison SM, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markosyan N, Li J, Sun YH, Richman LP, Lin JH, Yan F, et al. Tumor cell-intrinsic EPHA2 suppresses anti-tumor immunity by regulating PTGS2 (COX-2). J Clin Invest. 2019;129:3594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böttcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172:1022–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zelenay S, Van Der Veen AG, Böttcher JP, Snelgrove KJ, Rogers N, Acton SE, et al. Cyclooxygenase-Dependent Tumor Growth through Evasion of Immunity. Cell. 2015;162:1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W-R, Tian M-X, Yang L-X, Lin Y-L, Jin L, Ding Z-B, et al. PKM2 promotes metastasis by recruiting myeloid-derived suppressor cells and indicates poor prognosis for hepatocellular carcinoma. Oncotarget. 2015;6:846–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coleman MF, Cozzo AJ, Pfeil AJ, Etigunta SK, Hursting SD. Cell Intrinsic and Systemic Metabolism in Tumor Immunity and Immunotherapy. Cancers (Basel). 2020;12:852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imbert C, Montfort A, Fraisse M, Marcheteau E, Gilhodes J, Martin E, et al. Resistance of melanoma to immune checkpoint inhibitors is overcome by targeting the sphingosine kinase-1. Nat Commun. 2020;11:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodir NM, Kortlever RM, Barthet VJA, Campos T, Pellegrinet L, Kupczak S, et al. Myc instructs and maintains pancreatic adenocarcinoma phenotype. Cancer Discov. 2020;10:588–607. [DOI] [PubMed] [Google Scholar]
- 40.Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell. 2019;35:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-Induced GM-CSF Production Promotes the Development of Pancreatic Neoplasia. Cancer Cell. 2012;21:836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. [DOI] [PubMed] [Google Scholar]
- 43.Welte T, Kim IS, Tian L, Gao X, Wang H, Li J, et al. Oncogenic mTOR signalling recruits myeloid-derived suppressor cells to promote tumour initiation. Nat Cell Biol. 2016;18:632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–5. [DOI] [PubMed] [Google Scholar]
- 45.Kitajima S, Thummalapalli R, Barbie DA. Inflammation as a driver and vulnerability of KRAS mediated oncogenesis. Semin Cell Dev Biol. 2016;58:127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 2017;6:1301–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagers AJ, Weissman IL. Plasticity of Adult Stem Cells. Cell. 2004;116:639–48. [DOI] [PubMed] [Google Scholar]
- 48.Merrell AJ, Stanger BZ. Adult cell plasticity in vivo: de-differentiation and transdifferentiation are back in style. Nat Rev Mol Cell Biol. 2016;17:413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan S, Norgard RJ, Stanger BZ. Cellular Plasticity in Cancer. Cancer Discov. 2019;9:837–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hölzel M, Bovier A, Tüting T. Plasticity of tumour and immune cells: a source of heterogeneity and a cause for therapy resistance? Nat Rev Cancer. 2013;13:365–76. [DOI] [PubMed] [Google Scholar]
- 51.Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2020;19:39–56. [DOI] [PubMed] [Google Scholar]
- 52.Friedmann‐Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15:244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mehta A, Kim YJ, Robert L, Tsoi J, Comin-Anduix B, Berent-Maoz B, et al. Immunotherapy Resistance by Inflammation-Induced Dedifferentiation. Cancer Discov. 2018;8:935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D, et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun. 2015;6:8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pérez-Guijarro E, Yang HH, Araya RE, El Meskini R, Michael HT, Vodnala SK, et al. Multimodel preclinical platform predicts clinical response of melanoma to immunotherapy. Nat Med. 2020;1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miao Y, Yang H, Levorse J, Yuan S, Polak L, Sribour M, et al. Adaptive Immune Resistance Emerges from Tumor-Initiating Stem Cells. Cell. 2019;177:1172–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paczulla AM, Rothfelder K, Raffel S, Konantz M, Steinbacher J, Wang H, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572:254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castagnoli L, Cancila V, Cordoba-Romero SL, Faraci S, Talarico G, Belmonte B, et al. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene. 2019;38:4047–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallée V-P, et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020;26:259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganesh K, Basnet H, Kaygusuz Y, Laughney AM, He L, Sharma R, et al. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat Cancer. 2020;1:28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, et al. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res. 2017;77:3982–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tripathi SC, Peters HL, Taguchi A, Katayama H, Wang H, Momin A, et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc Natl Acad Sci. 2016;113:E1555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noman MZ, Janji B, Abdou A, Hasmim M, Terry S, Tan TZ, et al. The immune checkpoint ligand PD-L1 is upregulated in EMT-activated human breast cancer cells by a mechanism involving ZEB-1 and miR-200. Oncoimmunology. 2017;6:e1263412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn Y-H, Byers LA, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014;5:5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lou Y, Diao L, Cuentas ERP, Denning WL, Chen L, Fan YH, et al. Epithelial–Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin Cancer Res. 2016;22:3630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res. 2016;22:609–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gjerstorff MF, Andersen MH, Ditzel HJ, Gjerstorff MF, Andersen MH, Ditzel HJ. Oncogenic cancer/testis antigens: prime candidates for immunotherapy. Oncotarget. 2015;6:15772–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gordeeva O Cancer-testis antigens: Unique cancer stem cell biomarkers and targets for cancer therapy. Semin Cancer Biol. 2018;53:75–89. [DOI] [PubMed] [Google Scholar]
- 69.Gibbs ZA, Whitehurst AW. Emerging Contributions of Cancer/Testis Antigens to Neoplastic Behaviors. Trends in cancer. 2018;4:701–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Agudo J, Park ES, Rose SA, Alibo E, Sweeney R, Dhainaut M, et al. Quiescent Tissue Stem Cells Evade Immune Surveillance. Immunity. 2018;48:271–285.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pommier A, Anaparthy N, Memos N, Kelley ZL, Gouronnec A, Yan R, et al. Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science. 2018;360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prager BC, Xie Q, Bao S, Rich JN. Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell. 2019;24:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salmon H, Remark R, Gnjatic S, Merad M. Host tissue determinants of tumour immunity. Nat Rev Cancer. 2019;19:215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Thé H Differentiation therapy revisited. Nat Rev Cancer. 2018;18:117–27. [DOI] [PubMed] [Google Scholar]


