Abstract
Although substantial evidence supports aspirin’s efficacy in colorectal cancer chemoprevention, key molecular mechanisms are uncertain. An untargeted metabolomics approach with high-resolution mass spectrometry was used to elucidate metabolic effects of aspirin treatment in human colon tissue. We measured 10,269 metabolic features in normal mucosal biopsies collected at colonoscopy after approximately three years of randomized treatment with placebo, 81 mg/day or 325 mg/day aspirin from 325 participants in the Aspirin/Folate Polyp Prevention Study. Linear regression was used to identify aspirin-associated metabolic features and network analysis was used to identify pathways and predict metabolite identities. Poisson regression was used to examine metabolic features associations with colorectal adenoma risk. We detected 471 aspirin-associated metabolic features. Aside from the carnitine shuttle, aspirin-associated metabolic pathways were largely distinct for 81 mg aspirin (e.g., pyrimidine metabolism) and 325 mg (e.g., arachidonic acid metabolism). Among aspirin-associated metabolic features, we discovered three that were associated with adenoma risk and could contribute to the chemopreventive effect of aspirin treatment, and which have also previously been associated with colorectal cancer: creatinine, glycerol 3-phosphate and linoleate. The last two of these are in the glycerophospholipid metabolism pathway, which was associated with 81 mg aspirin treatment and provides precursors for the synthesis of eicosanoids from arachidonic acid upstream of cyclooxygenase inhibition by aspirin. Conversely, carnitine shuttle metabolites were increased with aspirin treatment and associated with increased adenoma risk. Thus, our untargeted metabolomics approach has identified novel metabolites and pathways that may underlie the effects of aspirin during early colorectal carcinogenesis.
INTRODUCTION
Chemoprevention has the potential to reduce the global burden of cancer and aspirin is one of the most promising agents on the horizon, but a deeper understanding of the molecular underpinnings is central to moving forward (1,2). Substantial evidence from pre-clinical, observational, and clinical studies supports the use of aspirin as a preventive agent for colorectal cancer (3,4). Aspirin has shown efficacy in reducing the risk of both preinvasive adenomas and colorectal cancer. This evidence basis recently led to the US Preventive Services Task Force recommendation for the use of aspirin to prevent colorectal cancer in addition to cardiovascular disease among those at increased risk of cardiovascular disease (5). Despite this, the key molecular mechanisms responsible for aspirin’s chemopreventive effects are not clearly understood (3,6–8). Aspirin incorporates two bioactive components in one molecule, a reactive acetyl moiety and a salicylate group, and is well known for its pleotropic effects (9). Its best-characterized pharmacologic activity is the irreversible acetylation and inhibition of the cyclooxygenases (COX-1 and COX-2), which are responsible for the initial step in the conversion of arachidonic acid into prostaglandins and related eicosanoids. However, there is also evidence for COX-independent mechanisms that could modify carcinogenesis (7,8,10,11). The identification of key targets in colorectal tissue that mediate aspirin’s protective effects may help to optimize its use for colorectal cancer chemoprevention.
We previously conducted the Aspirin/Folate Polyp Prevention Study, a randomized trial of aspirin (81 or 325 mg/day) for the prevention of colorectal adenomas among individuals with a history of adenomas (12). In this trial, the 81 mg/day dose reduced risk of any adenoma by 19% and advanced lesions by 41%, whereas there were statistically non-significant risk reductions for the 325 mg/day dose (4% and 17%, respectively) (12). In prior targeted analyses in this study population, we were unable to link aspirin-induced changes in inflammation markers in blood plasma or eicosanoid products of COX-1/COX-2 in urine to its chemopreventive effects in the colorectum (13,14). In the present work, we used an untargeted high-resolution metabolomics approach to investigate the effects of aspirin on low molecular weight molecules (metabolites) in normal colon tissue biopsies to identify key metabolic features and pathways involved in the chemopreventive effects of aspirin in the colorectum. While previous targeted analyses of eicosanoids have been performed, to our knowledge this is the first untargeted metabolomics analysis of aspirin’s effects in normal human colonic mucosa.
METHODS
Clinical Trial Study Population and Design
The Aspirin/Folate Polyp Prevention Study was a multicenter, double-blind, placebo-controlled, randomized trial to assess the chemopreventive effects of aspirin and folic acid in individuals with a recent history of colorectal adenomas, as described in detail previously (NCT00272324) (12,15). The study was approved by institutional review boards at all participating institutions, and all study participants provided written informed consent. Participants were enrolled from eight clinical centers in the United States and one in Canada. Recruitment began in July 1994 and ended in March 1998. Eligible participants were 21 to 80 years old with no history of colorectal cancer or familial colorectal cancer syndromes but with a recent colorectal adenoma and no remaining polyps after a complete colonoscopy within 3 months before study enrollment. At enrollment, participants completed a questionnaire regarding demographic and health factors and were asked to avoid using aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs) for the duration of study treatment. Following a 3-month run-in period, eligible participants who took at least 80% of their study pills were randomized in a 3 × 2 factorial design to aspirin (placebo, 81 mg/day, or 325 mg/day) and folic acid (placebo or 1 mg/day). Every four months during the study, participants completed questionnaires regarding adherence to study pill taking, medications used, and medical events. Treatment ended and adenoma occurrence was assessed at a surveillance colonoscopy approximately three years from the pre-enrollment colonoscopy.
All lesions removed from the large bowels of study participants were reviewed by a single study pathologist who was blinded to treatment assignments. Advanced adenomas were defined as those with cancer, high-grade dysplasia, more than 25% villous features, or an estimated diameter of at least 1 cm as assessed by the endoscopist. High-risk findings were defined as the occurrence of at least one advanced adenoma or multiple (3 or more) adenomas of any type.
Collection and Selection of Colon Tissue Biopsies
Normal colon mucosal biopsies were collected at the year three (end-of-treatment) colonoscopy from a sub-set of study participants who provided additional informed consent for the purpose of prior research, as described previously (16,17). Before their colonoscopy, participants were instructed to discontinue study treatment (aspirin or placebo) for one week to minimize bleeding risk per standard of care. Using biopsy forceps, tissue specimens were taken from the normal mucosa of the proximal colon, approximately 5 cm above the ileocecal valve. The specimens (approximately 10 mg) were put into freezer tubes and immediately frozen in liquid nitrogen or an ethanol/dry ice slurry prior to storage at ≤−70°C. Within a year of collection, specimens were shipped on dry ice to the Dartmouth study biorepository for storage at ≤−70°C and were never thawed prior to the present analysis. Subsequently, frozen tissue samples were shipped on dry ice to the Emory Department of Medicine Clinical Biomarkers Laboratory and maintained at ≤−70°C prior to metabolomics analysis.
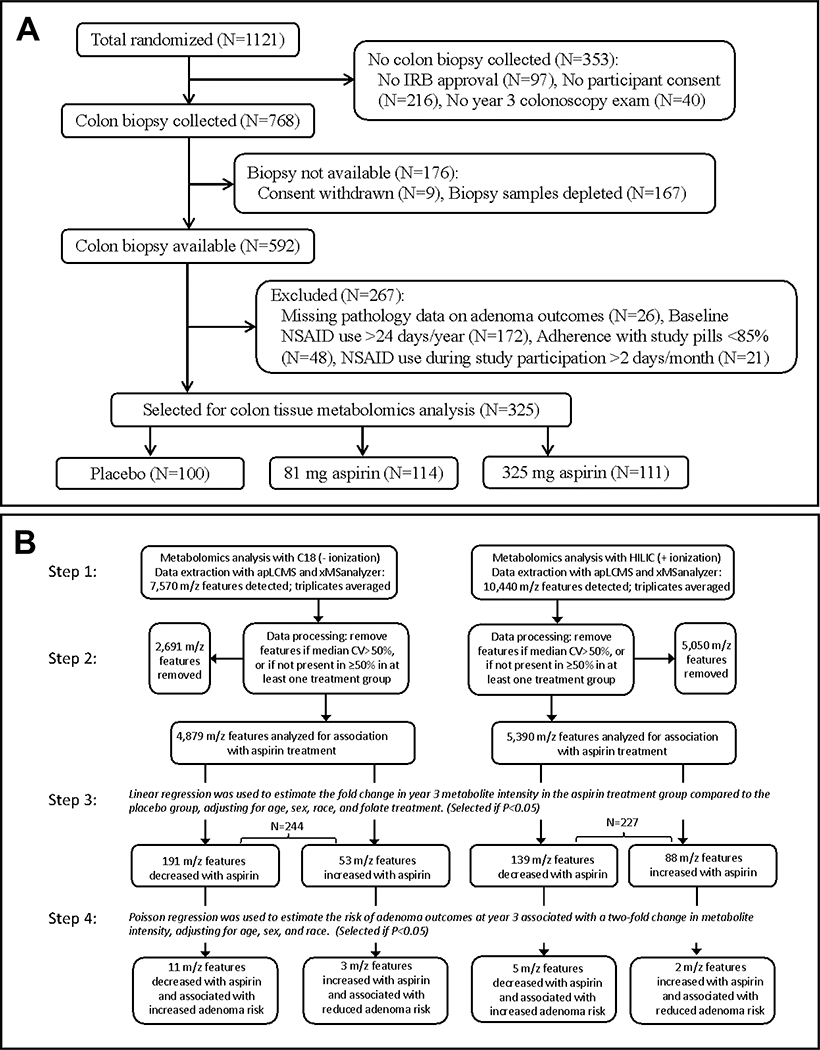
Selection of participants with colon biopsies for inclusion in the current work was as follows (Figure 1A). Among 1,121 randomized participants, normal tissue biopsies were collected from 768 participants. For the present research, of 592 participants with biopsies still available, we excluded 26 due to missing data on adenoma outcomes. Furthermore, to maximize the separation between the placebo control and aspirin treatment groups, we excluded participants with NSAID use >24 days/year at baseline or >2 days/month during study participation, or with <85% adherence to study pills, leaving 325 participants for analysis in the work described here.
Figure 1.
Flow Charts. A, Selection of participants for inclusion in colon tissue metabolomics analysis. B, Workflow for metabolomics data analysis. In step 3, separate linear models were run for metabolic features associated with low (81 mg) or high (325 mg) doses of aspirin treatment vs. placebo, and total numbers of metabolic features associated with either dose aspirin are shown. In step 4, separate Poisson regression models were run for three types of adenoma outcomes (any adenoma, advanced adenoma or high-risk findings) and total numbers of metabolic features associated with any of these three outcomes are shown in the last row. These metabolic features (N=21 in total) are detailed in Table 3 and may contribute to the chemopreventive effect of aspirin treatment to reduce adenoma outcomes.
High Resolution Metabolomics Analyses and Data Processing
The global metabolic effects of aspirin in colon tissue were assessed using dual liquid chromatography coupled with high-resolution mass spectrometry with dual ionization to maximize detection of diverse metabolites. Methods for sample preparation and metabolomics analyses were similar to those described previously (18–20). Briefly, batches of 40 experimental samples were prepared daily by the addition of 50 μl water, 200 μl acetonitrile and 5 μl of a mixture containing 14 stable isotope internal standards. Samples were homogenized for a few seconds using an Active Motif EpiShear probe sonicator, incubated on ice for 30 minutes, and centrifuged at 14,000 rpm at 4°C for 10 minutes. The resulting supernatant (100 μl) was removed and six technical replicates (10 μl each) were analyzed for each sample: three using C18 chromatography (Higgins Analytical, 50 × 2.1 mm column) and three using HILIC chromatography (Waters Xbridge BEH Amide, 50 × 2.1 mm column). Mass spectral data were collected with a 5-min gradient on a Dionex UliMate 3000 rapid separation liquid chromatography system coupled with a Thermo Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Diego, CA) at a resolution of 120,000 and operated in negative or positive electrospray ionization mode with C18 or HILIC chromatography, respectively. Each of the colon tissue samples were identified only with an eight-digit barcode and analyzed in random order by a metabolomics analyst blinded to each sample’s treatment classification. Reference samples analyzed included pooled human plasma samples from the National Institute of Standards and Technology (NIST, Gaithersburg, MD; SRM #1950) and from Equitech Bio (Kerrville, TX). In addition, a colon tissue reference sample was made using normal tissue removed at surgery from an anonymous donor. Reference samples of each type were included at the beginning, middle and end of each analytical batch for quality control purposes including evaluating instrument stability. Average pairwise Pearson correlation coefficients for all features across all pooled plasma and colon reference samples were, respectively, 0.96 and 0.98 using HILIC+ and 0.94 and 0.84 using C18- analyses.
Raw data files were extracted using apLCMS v6.3.3 (21) with modification by xMSanalyzer v2.0.7 (22), with each unique metabolic feature (ion) defined by accurate mass (±5 ppm) mass-to-charge ratio (m/z) and retention time. Batch correction was performed using Combat (23) and ion intensity values were averaged across triplicates. Extraction of mass spectral data initially yielded 7,570 and 10,440 metabolic features in the C18 and HILIC datasets, respectively (Figure 1B, step 1). Samples with mean overall pairwise Pearson correlation <70% between replicates were excluded from the analyses: 4 and 0 samples were excluded from the C18 and HILIC datasets, respectively. Metabolic features were excluded due to high variability between triplicates (median coefficient of variation >50%) or low abundance (undetectable in >50% of samples in all three treatment groups), leaving a total of 10,269 features to be analyzed, 4,879 and 5,390 in the C18 and HILIC datasets, respectively (Figure 1B, step 2).
Detection of Aspirin Catabolites in Blood Plasma Samples
To validate adherence to randomized aspirin treatments, raw intensities of two common aspirin catabolites (salicylic acid and salicyluric acid) were assessed in participants’ baseline and year three plasma samples collected as described previously (16,17). Notably, the colon tissue biopsies could not be used for this purpose because they were collected a week after treatment cessation, as described above. The year three blood samples analyzed here were collected while participants were still on study treatment a median of 17 days (interquartile range 11–35 days) prior to their year three end-of-treatment colonoscopy. Briefly, non-fasting blood collected in 7-ml EDTA Vacutainer brand tubes was immediately put on ice and then centrifuged at 1,100×g for 10 min at 4°C. The plasma fraction was removed and stored frozen at ≤−20°C for up to 12 months prior to shipment on dry ice to the Dartmouth study biorepository for storage at ≤−70°C. Of the 325 participants selected for the tissue metabolomics analysis as described above, 293 had paired baseline and year three plasma samples for this analysis of aspirin catabolites. Of those 586 samples, 25% (149) had never been thawed previously, 73% (430) had been thawed once, and 1% (7) had been thawed twice. Samples selected were shipped on dry ice to the Emory Department of Medicine Clinical Biomarkers Laboratory and maintained at ≤−70°C prior to metabolomics analysis.
Methods for plasma sample preparation and metabolomics analyses were similar to those described previously (18–20). Briefly, batches of 20 experimental samples were prepared daily by thawing on ice and removal of 50 μl and then addition of 100 μl of acetonitrile containing 2.5 μl of internal standards. The samples were kept on ice for 30 minutes followed by centrifugation at 14,000 rpm at 4°C for 10 minutes. The resulting supernatant (100 μl) was removed, and three technical replicates (10 μl each) were analyzed for each sample using C18 chromatography (Thermo Accucore C18, 100 × 2.1 mm column). Mass spectral data were collected with a Thermo QExactive High Field mass spectrometer (Thermo Fisher Scientific, San Diego, CA) operated in positive electrospray ionization mode at a resolution of 120,000. Each of the plasma samples were identified with an eight-digit barcode and paired sets of baseline and year 3 samples from the same participant were analyzed in a single batch in random order by a metabolomics analyst blinded to their treatment classification. Data files were extracted as described for the tissue data. Identities of aspirin catabolites were confirmed at level 1 per the Metabolomics Standards Initiative (MSI) (24) by co-elution using tandem mass spectrometry (MS/MS) relative to authentic standards: m/z = 139.0389, salicylic acid [M+H] and m/z = 196.0604, salicyluric acid [M+H]. For each of the three treatment groups, the differences in aspirin catabolite intensities between paired baseline and year three plasma samples were assessed using Wilcoxon Signed-Rank tests and visualized with box plots.
Statistical Analyses
Data from tissue samples was normalized by dividing the ion intensity of each metabolic feature by the total ion intensity for all features in the sample. Zero intensity values were replaced by the minimum value for that feature across all samples divided by 2 prior to normalization. Normalization naturally leads to extremely small values for the ion intensities that may cause instability in analyses when these are log2-transformed prior to regression modelling. Hence, the ion intensity values were multiplied by an arbitrary large constant 109, to ensure that all log2 values were greater than 0. The multiplication has no impact on the resulting p-values.
To identify metabolic features in colon tissue associated with aspirin treatment we used multivariable linear regression with the log2-transformed ion intensity as the dependent variable and the randomized aspirin treatment assignment as the predictor variable with adjustment for age, sex, race (coded as a binary variable: non-Hispanic white vs. other) and folate treatment assignment. Separate models were used for assessing associations with 81 mg/day aspirin vs. placebo or 325 mg/day aspirin vs. placebo in intention-to-treat analyses. Results were visualized using Manhattan and volcano plots. Metabolic pathways associated with aspirin treatment were identified using Mummichog v2.0.6 (25), a set of algorithms specifically designed for high-throughput metabolomics that utilizes the collective power in metabolic networks to help resolve the ambiguity in metabolite prediction in a data-driven analysis.
Aspirin-associated metabolic features were subsequently assessed for their associations with adenoma outcomes using multivariable Poisson regression models for binary data with common outcomes to estimate risk ratios (RR) and 95% confidence intervals (CIs) for associations with two-fold changes in ion intensity, adjusting for age, sex, and race. Three separate models were used to assess associations with risk of at least one adenoma, advanced adenoma (those with cancer, high-grade dysplasia, more than 25% villous component, or an estimated diameter of at least 1 cm) or high-risk findings (at least one advanced adenoma or at least 3 adenomas of any type).
The number of independent estimates in our analysis is unknown because multiple metabolic features detected on our platform can map to one metabolite, and metabolites in a pathway are correlated. False discovery rate (FDR) q-values were computed using the Benjamini-Hochberg method with a 0.2 threshold for significance (26). In regression analyses, aspirin-associated metabolic features were selected and prioritized using a two-sided P-value threshold of <0.05 without adjustment for multiple testing to avoid type II error. This approach, common in metabolomics studies, supports the discovery of novel metabolites associated with aspirin treatment and enriches input information for pathway analyses (25,27). P-values in pathway analyses were calculated using permutation tests with sampling from Gamma distributions to account for clustered data; enriched metabolic pathways were selected using a Mummichog scoring threshold of P<0.05 to avoid type I error (25,27). In all other analyses (e.g., changes in aspirin catabolite intensities in plasma), two-sided P<0.05 were considered statistically significant. Analyses were conducted in R v3.5.1 and SAS v9.4.
Metabolite Annotation and Identification
Metabolite identification was performed by comparing accurate mass and retention time to authentic standards in an in-house library run under identical conditions using MS/MS. Additional tentative annotations were assigned using Mummichog v2.0.6 (25) and xMSannotator v1.3.2 (28) with Human Metabolome Database v3.5. xMSannotator uses a multi-step approach based on m/z, retention time, isotopes, adducts, correlation across samples, and network and pathway associations to assign database matches into different categories: high, medium, or low confidence. Metabolite identities were classified per MSI criteria (24): level 1 (confirmed by MS/MS and co-elution with authentic standards), level 2 (tentative annotation using xMSannotator with medium or high confidence), and level 3 (tentatively characterized chemical class), and level 4 (accurate mass or no database match). Features that were redundant (multiple adducts), implausible, or with suspect retention time were excluded.
RESULTS
Population Characteristics
Characteristics of the 325 participants with colon tissue biopsies included in the present analyses were similar across the three aspirin treatment groups (Table 1). The mean age at enrollment was approximately 58 years, 34% were female, and 87% were non-Hispanic whites. Risk factors for colorectal neoplasia were similar across the treatment groups, including body mass index, smoking status, alcohol use, and family history of colorectal cancer. Overall, 53% of these participants were randomly assigned to receive folate supplementation. Study treatment lasted 32.7 ± 2.4 months on average. Overall, 37% of participants had at least one adenoma at their year three colonoscopy, 8% had at least one advanced adenoma, and 11% had high-risk findings.
Table 1.
Characteristics of Randomized Participants Selected for Colon Tissue Metabolomics Study, by Aspirin Treatment Group (N=325)
| Placebo (N=100) | 81 mg Aspirin (N=114) | 325 mg Aspirin (N=111) | Pd | |
|---|---|---|---|---|
| Baseline Characteristics: | ||||
| Age at Enrollment, Mean ± SD | 57.8 ± 9.2 | 57.7 ± 8.1 | 58.7 ± 8.6 | 0.61 |
| Female, N (%) | 37 (37%) | 37 (33%) | 37 (33%) | 0.76 |
| Race/ethnicity, N (%) | 0.49e | |||
| White, Non-Hispanic | 86 (86%) | 97 (85%) | 100 (90%) | |
| Black, Non-Hispanic | 4 (4.0%) | 5 (4.4%) | 1 (0.9%) | |
| Hispanic | 6 (6.0%) | 7 (6.1%) | 3 (2.7%) | |
| Other | 4 (4.0%) | 5 (4.4%) | 7 (6.3%) | |
| Body-Mass Index, kg/m2, mean ± SD | 27.2 ± 4.6 | 27.3 ± 4.1 | 27.9 ± 4.8 | 0.53 |
| Current Smoker, N (%) | 13 (13%) | 11 (9.7%) | 14 (13%) | 0.69 |
| Alcohol Use, N (%) | 62 (66%) | 71 (65%) | 72 (67%) | 0.95 |
| Family History of Colorectal Cancer, N (%) | 33 (42%) | 35 (39%) | 37 (45%) | 0.71 |
| Study and Follow-up Characteristics: | ||||
| Allocated to Folate Treatment, N (%) | 48 (48%) | 63 (55%) | 60 (54%) | 0.53 |
| Study Follow-Up Time, Mean ± SDa | 32.7 ± 2.6 | 32.6 ± 2.3 | 32.7 ± 2.3 | 0.95 |
| ≥1 Adenoma at Year 3 Colonoscopy | 33 (33%) | 39 (34%) | 47 (42%) | 0.30 |
| ≥1 Advanced Adenoma at Year 3 Colonoscopyb | 6 (6.0%) | 8 (7.0%) | 13 (11.7%) | 0.27 |
| High-risk Findings at Year 3 Colonoscopyc | 9 (9.0%) | 11 (9.7%) | 16 (14.4%) | 0.38 |
| Paired Baseline and Year 3 Plasma Samples Analyzed for Aspirin Metabolites, N (%) | 90 (90%) | 105 (92%) | 98 (88%) | 0.63 |
Note: N’s for missing data for alcohol use: 6 placebo, 4 aspirin (81 mg), 3 aspirin (325 mg); for family history of colorectal cancer: 21 placebo, 24 aspirin (81 mg), 29 aspirin (325 mg).
Time in months from enrollment to year 3 end of treatment colonoscopy.
Advanced adenomas are those with cancer, high-grade dysplasia, >25% villous component, or diameter ≥1 cm.
High-risk refers to ≥1 advanced adenomas or ≥3 adenomas of any type.
P-values for comparisons among the three treatment groups: Pearson chi-square tests for categorical variables and analysis of variance likelihood ratio chi-square tests for continuous variables.
P-value using Fisher’s exact test.
Plasma Levels of Aspirin Catabolites: Salicylic Acid and Salicyluric Acid
Among the 325 participants with colon biopsy samples, 293 (90%) also had paired baseline and year three plasma samples (Table 1) that were used to validate adherence to randomized treatment by analysis of two common aspirin catabolites: salicylic acid and salicyluric acid (Supplementary Figure 1S). Intensities of salicylic acid and salicyluric acid statistically significantly increased between baseline and year three among participants in the two aspirin treatment groups (P=1.9 X 10−6 and P=4.9 X 10−12, respectively for 81 mg aspirin; P=7.9 X 10−13 and P=2.2 X 10−17, respectively, for 325 mg aspirin) but not among participants in the placebo group. There was a small decrease in salicyluric acid (P=0.02) in the placebo group at year three compared to baseline that was likely due to modest use of aspirin pre-enrollment (although we excluded those with baseline NSAID use >24 days/year) and subsequent avoidance of personal aspirin use during study participation.
Colon Tissue Metabolic Features and Pathways Associated with Aspirin Treatment
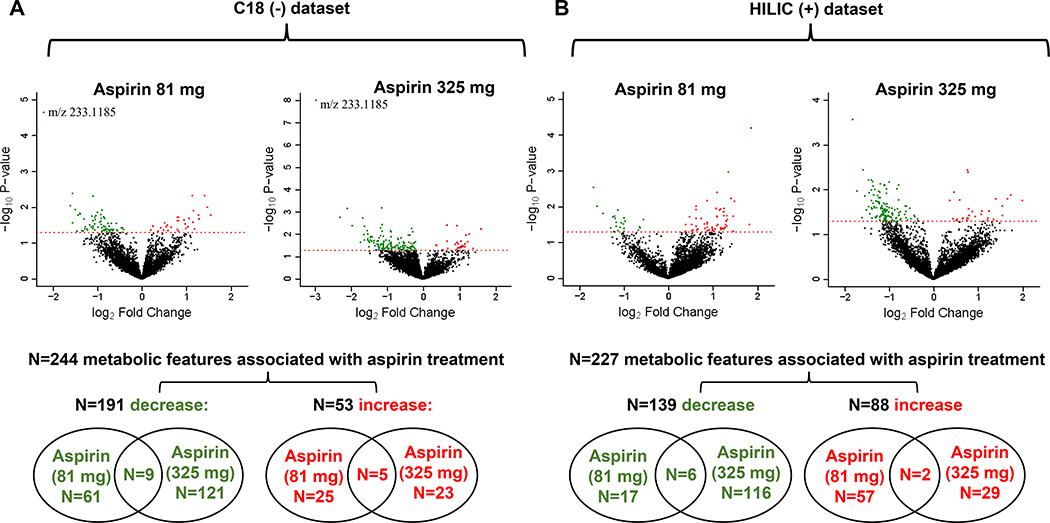
Of 10,269 metabolic features analyzed, 471 were associated with aspirin treatment (81 or 325 mg/day): 244 and 227 in the C18 and HILIC datasets, respectively (Figure 1B, step 3; Figure 2). The majority of metabolic features decreased with aspirin treatment: 330 (70%) (191 and 139 in the C18 and HILIC datasets, respectively; Figure 2). Only 22 features (4.7%) were associated with both doses of aspirin treatment (14 and 8 in the C18 and HILIC datasets, respectively; Figure 2). Following correction for multiple testing, only one feature was statistically significantly associated with both low and high dose aspirin treatment: m/z 233.1185 (q=0.11 and 9.8 X 10−5, respectively). This was annotated as multiple forms of Pterosin, compounds detected in some types of vegetables.
Figure 2.
Colon tissue metabolic features associated with aspirin treatment. A, C18 (–) dataset. B, HILIC (+) dataset. Linear regression was used to estimate the change in year 3 ion intensities in the aspirin treated group compared to the placebo group, adjusting for age, sex, race, and folate treatment. Volcano plots depict for each metabolic feature the magnitude of the change on the x-axis (log2fold change with aspirin treatment vs. placebo) and the statistical significance on the y-axis (-log10p-value for the association with aspirin treatment). Color-coded metabolic features statistically significantly increased (red) or decreased (green) with aspirin treatment (P<0.05). One feature significant at a false discovery rate (q<0.2) is indicated in the C18 volcano plots as m/z 233.1185. Venn diagrams indicate overlap of significant metabolic features across the two aspirin treatment groups (81 or 325 mg) for those that either decrease or increase with treatment.
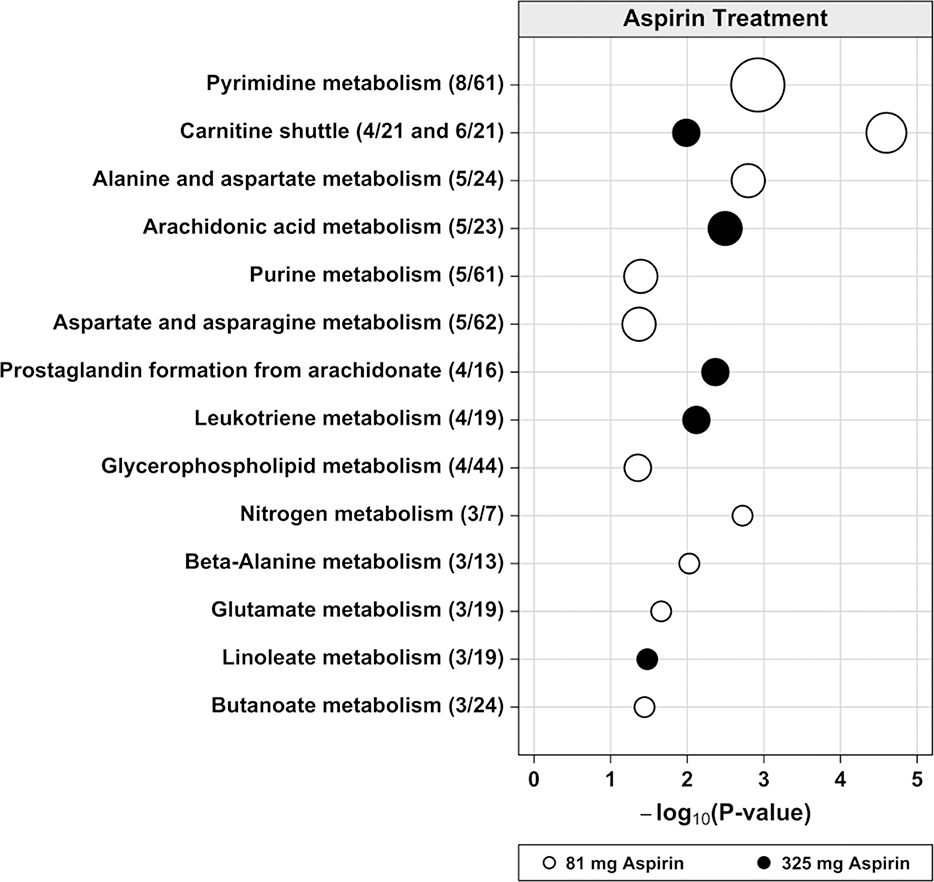
Metabolic pathways associated with 81 mg versus 325 mg aspirin treatment were largely distinct (Figure 3). The three pathways with the lowest p-values associated with 81 mg aspirin were involved in energy metabolism from fatty acids (carnitine shuttle), nucleotide (pyrimidine) metabolism, and amino acid (alanine and aspartate) metabolism. In contrast, the three pathways with the lowest p-values associated with 325 mg aspirin were related to arachidonic acid metabolism, including prostaglandin formation from arachidonate, and leukotriene metabolism. Only the carnitine shuttle pathway was associated with both doses of aspirin treatment. Putative or confirmed identities for 20 metabolic features in the aspirin-associated pathways along with their fold changes with aspirin treatment are shown in Table 2. As expected, levels of several putative arachidonic acid metabolites were substantially reduced with aspirin treatment. Metabolic features with the largest fold changes with aspirin treatment were: an 80% decrease (0.20 fold change) in the putative prostaglandin metabolite alpha-hydroxy-9,15-dioxoprostanoate with 325 mg aspirin (m/z=351.2180, P=0.002) and a 2.68 fold increase in the confirmed carnitine shuttle metabolite arachidyl carnitine with 81 mg aspirin (m/z=456.4043, P=0.006).
Figure 3.
Dysregulated metabolic pathways associated with aspirin treatment in colon tissue. The vertical axis represents the pathways (circles) with the radius representing the number of hits (significant metabolite features). The horizontal axis represents the negative log10 of the gamma adjusted P-values for each pathway with at least 3 hits. The solid circles are for 81 mg aspirin and the open circles are for 325 mg aspirin treatment. In parentheses next to each pathway name is the number of hits divided by the pathway size (total number of features detected in the pathway).
Table 2.
Metabolic Features in Pathways Associated with Aspirin Treatment: Fold changes with Aspirin and Associations with Adenoma Outcomes
| m/z | RT | Col | FC (95% CI)a 81 mg Aspirin | FC (95% CI)a 325 mg Aspirin | RR (95% CI)b Any Adenomac | RR (95% CI)b Advancedc | RR (95% CI)b High-Riskc | Putative Identityd | Adductd | MSI leveld |
|---|---|---|---|---|---|---|---|---|---|---|
| A. Metabolic features that decreased with aspirin treatment: | ||||||||||
| 88.012 | 19.3 | C18− | 0.74 (0.56–0.98) | 0.87 (0.74–1.03) | 0.94 (0.91–1.02) | 0.95 (0.83–1.12) | 0.89 (0.83–1.02) | Pyruvate | M(C13)−H | 1 |
| 103.0591 | 18.7 | C18− | 0.74 (0.55–1.00) | 1.07 (0.91–1.25) | 0.94 (0.91–1.01) | 1.17 (0.93–1.35) | 0.87 (0.83–0.99) | 2-aminobutyrate | M(C13)−H | 1 |
| 156.0288 | 26.4 | C18− | 0.49 (0.26–0.90) | 0.68 (0.39–1.17) | 1.02 (0.98–1.05) | 0.97 (0.91–1.05) | 0.96 (0.92–1.03) | N-Acetyl-L-aspartate | M−H2O−H | 1 |
| 171.0069 | 18.4 | C18− | 0.57 (0.34–0.96) | 0.78 (0.51–1.21) | 1.02 (0.97–1.06) | 1.10 (1.00–1.14) | 1.12 (1.02–1.15) | Glycerol 3-phosphate | M−H | 1 |
| 173.0569 | 24.2 | C18− | 0.56 (0.33–0.97) | 0.79 (0.50–1.25) | 1.00 (0.97–1.03) | 0.93 (0.89–1.02) | 0.92 (0.90–1.00) | 5,6-Dihydrouracil | M+CH3COO | 2 |
| 191.0679 | 19.9 | C18− | 0.54 (0.29–0.99) | 0.86 (0.51–1.44) | 1.02 (0.98–1.05) | 1.04 (0.94–1.12) | 0.99 (0.93–1.06) | L-Asparagine | M+CH3COO | 1 |
| 231.1358 | 20.2 | C18− | 0.38 (0.18–0.83) | 1.24 (0.65–2.36) | 1.01 (0.98–1.03) | 1.03 (0.96–1.08) | 1.02 (0.97–1.07) | Spermic acid 2 | M−H | 2 |
| 319.2253 | 35 | HILIC+ | 0.77 (0.43–1.38) | 0.48 (0.24–0.96) | 1.00 (0.98–1.03) | 1.00 (0.93–1.08) | 0.97 (0.93–1.03) | Leukotriene A4 | M+H | 2 |
| 335.2205 | 30.2 | HILIC+ | 0.81 (0.46–1.43) | 0.43 (0.21–0.84) | 0.98 (0.96–1.01) | 1.04 (0.96–1.10) | 0.97 (0.93–1.03) | Prostaglandin C2e | M+H | 2 |
| 336.2241 | 25.6 | HILIC+ | 0.76 (0.42–1.38) | 0.42 (0.21–0.83) | 0.99 (0.97–1.02) | 1.06 (0.96–1.12) | 1.01 (0.95–1.07) | 12,13-DHOME | M+Na | 2 |
| 351.2180 | 23 | C18− | 0.66 (0.27–1.63) | 0.20 (0.08–0.54) | 0.98 (0.97–1.01) | 0.98 (0.94–1.03) | 0.96 (0.94–1.01) | Alpha-Hydroxy-9,15-dioxoprostanoatee,f | M−H | 2 |
| 370.2323 | 21.2 | C18− | 0.34 (0.16–0.70) | 0.31 (0.15–0.64) | 1.00 (0.98–1.03) | 1.01 (0.94–1.07) | 1.00 (0.95–1.06) | Thromboxane B2f | M(C13)−H | 2 |
| B. Metabolic features that increased with aspirin treatment: | ||||||||||
| 261.2224 | 244.8 | C18− | 1.35 (1.02–1.78) | 0.92 (0.61–1.38) | 0.95 (0.93–1.00) | 0.92 (0.85–1.04) | 0.94 (0.87–1.06) | Linoleate | M−H2O−H | 1 |
| 400.3402 | 24.8 | HILIC+ | 1.30 (0.92–1.83) | 1.36 (1.01–1.84) | 1.22 (1.06–1.24) | 1.41 (0.92–1.75) | 1.36 (0.95–1.60) | L-Palmitoylcarnitine | M+H | 1 |
| 426.3568 | 24.8 | HILIC+ | 1.39 (0.95–2.04) | 1.50 (1.08–2.10) | 1.18 (1.04–1.20) | 1.23 (0.90–1.48) | 1.25 (0.95–1.44) | Octadecenoyl carnitinee | M+H | 2 |
| 428.3716 | 25 | HILIC+ | 1.24 (0.91–1.70) | 1.38 (1.08–1.76) | 1.22 (1.04–1.26) | 1.43 (0.94–1.75) | 1.44 (0.99–1.67) | Stearoylcarnitine | M+H | 1 |
| 448.3429 | 23.9 | HILIC+ | 1.83 (1.06–3.15) | 1.03 (0.53–2.00) | 1.05 (0.99–1.08) | 1.00 (0.92–1.09) | 1.04 (0.94–1.12) | C20:4 carnitinee | M+H | 2 |
| 450.3572 | 24.5 | HILIC+ | 2.49 (1.18–5.26) | 1.58 (0.71–3.51) | 1.02 (0.99–1.04) | 1.03 (0.95–1.09) | 1.03 (0.96–1.09) | Dihomo-gamma-linolenyl carnitinee | M+H | 2 |
| 456.4043 | 25.3 | HILIC+ | 2.68 (1.34–5.35) | 1.65 (0.79–3.46) | 1.03 (0.99–1.05) | 0.99 (0.93–1.06) | 1.01 (0.95–1.06) | Arachidyl carnitine | M+H | 1 |
| 476.3729 | 24.5 | HILIC+ | 2.30 (1.04–5.07) | 2.64 (1.21–5.78) | 1.01 (0.99–1.03) | 1.08 (0.99–1.13) | 1.06 (0.98–1.10) | Adrenyl carnitine | M+H | 2 |
NOTE: Only metabolic features from pathways associated with aspirin treatment in Figure 3 are included.
Abbreviations: RT, retention time in seconds; Col, chromatography column (C18 or HILIC) and ionization mode (− or +); FC; fold change; RR, risk ratio; CI, confidence interval; MSI, Metabolomics Standards Initiative; 12,13-DHOME, 12,13-hydroxyoctadec-9(Z)-enoate.
Linear regression was used to estimate the fold change in year 3 ion intensities in the aspirin treatment group compared to the placebo treatment group, adjusting for age, sex, race, and folate treatment. Bolding indicates statistically significant (P<0.05).
Poisson regression was used to assess the association of risk of adenoma outcomes with a two-fold change in ion intensity, adjusting for age, sex, and race. Bolding indicates statistically significant (P<0.05).
Any adenoma refers to ≥1 adenoma of any type; advanced refers to adenomas with cancer, high-grade dysplasia, >25% villous component, or diameter ≥1 cm; high-risk refers to ≥1 advanced adenoma or ≥3 adenomas of any type.
For putative identities shown, the adduct form and MSI identification confidence level are indicated in the columns to the right.
For these metabolic features, an additional adduct form was detected but is not shown to reduce redundancy in the table.
In pathway analysis, two metabolic features in the Prostaglandin Formation from Arachidonate pathway were associated with any aspirin treatment (combined analysis of 81 and 325 mg), but not either dose alone.
Association of Colon Tissue Metabolic Features with Adenoma Risk
Aspirin-associated metabolic features were assessed for their associations with adenoma risk to identify those that could contribute to the chemopreventive effects of aspirin (Figure 1B, step 4; Table 3). There are only two categories of metabolic features that could contribute to aspirin’s chemopreventive effects: A) those that increased with aspirin treatment and were also associated with a reduction in adenoma risk, or B) those that decreased with aspirin treatment and were also associated with an increase in adenoma risk. In total, we discovered 21 metabolic features meeting these criteria, of which seven have putative or confirmed identities (Table 3, Supplementary Table S1).
Table 3.
Metabolic Features That May Contribute to the Chemopreventive Effects of Aspirin Treatment: Fold Changes with Aspirin and Associations with Adenoma Outcome
| m/z | RT | Col | FC (95% CI)a 81 mg Aspirin | FC (95% CI)a 325 mg Aspirin | RR (95% CI)b Any Adenomac | RR (95% CI)b Advancedc | RR (95% CI)b High-Riskc | Putative Identityd | Adductd | MSI Leveld |
|---|---|---|---|---|---|---|---|---|---|---|
| A: Metabolic features that decreased with aspirin treatment compared to placebo, and were also associated with increased risk of adenoma outcomes: | ||||||||||
| 133.9746 | 290.8 | HILIC+ | 0.90 (0.55–1.45) | 0.48 (0.26–0.89) | 1.03 (0.99–1.06) | 1.15 (1.03–1.19) | 1.11 (0.99–1.16) | |||
| 138.5112 | 108.8 | HILIC+ | 0.64 (0.42–0.97) | 0.50 (0.31–0.82) | 1.01 (0.97–1.05) | 1.18 (1.01–1.24) | 1.15 (1.02–1.19) | |||
| 171.0069 | 18.4 | C18− | 0.57 (0.34–0.96) | 0.78 (0.51–1.21) | 1.02 (0.97–1.06) | 1.10 (1.00–1.14) | 1.12 (1.02–1.15) | Glycerol 3-phosphate | M−H | 1 |
| 197.1899 | 20.8 | HILIC+ | 0.75 (0.54–1.04) | 0.58 (0.39–0.86) | 1.07 (0.99–1.10) | 1.19 (1.02–1.25) | 1.21 (1.05–1.24) | |||
| 201.1247 | 19.9 | C18− | 0.55 (0.32–0.95) | 1.04 (0.70–1.55) | 1.02 (0.97–1.05) | 1.10 (1.01–1.13) | 1.11 (1.02–1.13) | Leucyl-Alanine | M+Na−2H | 2 |
| 398.2864 | 242.7 | C18− | 0.89 (0.44–1.80) | 0.46 (0.22–0.97) | 1.05 (1.01–1.06) | 1.11 (1.01–1.14) | 1.08 (1.00–1.12) | |||
| 435.313 | 215.3 | C18− | 0.61 (0.37–0.99) | 1.20 (0.81–1.78) | 1.06 (0.99–1.09) | 1.28 (1.05–1.33) | 1.08 (0.94–1.19) | Varanic acid | M−H | 2 |
| 439.2982 | 219.1 | C18− | 0.68 (0.48–0.99) | 0.92 (0.70–1.22) | 1.10 (0.97–1.17) | 1.48 (1.05–1.64) | 1.41 (1.05–1.52) | |||
| 460.2833 | 208.1 | C18− | 0.67 (0.48–0.96) | 0.89 (0.76–1.03) | 1.10 (0.99–1.16) | 1.46 (1.02–1.66) | 1.31 (0.99–1.47) | LysoPE (C18:1) | M−H2O−H | 2 |
| 516.0552 | 16.3 | C18− | 0.77 (0.39–1.54) | 0.46 (0.22–0.94) | 1.05 (1.01–1.06) | 1.03 (0.96–1.09) | 1.01 (0.96–1.07) | |||
| 527.3094 | 198.2 | C18− | 0.56 (0.34–0.91) | 1.16 (0.91–1.48) | 1.07 (0.98–1.11) | 1.18 (1.02–1.24) | 1.04 (0.92–1.15) | |||
| 589.5188 | 291.4 | C18− | 0.84 (0.62–1.15) | 0.68 (0.47–0.97) | 0.99 (0.95–1.05) | 1.28 (1.01–1.40) | 1.06 (0.92–1.18) | Polyoxyethylene dioleate | M−H | 2 |
| 591.8592 | 253 | C18− | 0.61 (0.26–1.41) | 0.34 (0.15–0.81) | 1.04 (1.00–1.05) | 1.03 (0.96–1.08) | 1.04 (0.98–1.08) | |||
| 634.3312 | 222.3 | C18− | 0.82 (0.56–1.19) | 0.62 (0.40–0.95) | 1.00 (0.95–1.05) | 1.26 (1.00–1.39) | 1.20 (1.01–1.28) | |||
| 714.3432 | 50.3 | HILIC+ | 0.78 (0.41–1.50) | 0.48 (0.24–0.99) | 1.05 (1.00–1.06) | 1.08 (0.98–1.14) | 1.07 (0.99–1.11) | |||
| 816.4432 | 295.7 | HILIC+ | 0.98 (0.43–2.22) | 0.43 (0.19–0.99) | 1.04 (1.00–1.05) | 0.99 (0.94–1.05) | 0.99 (0.95–1.04) | |||
| B. Metabolic features that increased with aspirin treatment compared to placebo, and were also associated with decreased risk of adenoma outcomes: | ||||||||||
| 115.0632 | 35.9 | HILIC+ | 1.17 (0.94–1.46) | 1.29 (1.05–1.59) | 0.91 (0.88–1.00) | 0.88 (0.78–1.07) | 0.90 (0.79–1.08) | Creatinine | M+H | 1 |
| 118.0589 | 17.3 | C18− | 1.43 (1.04–1.95) | 1.15 (0.81–1.63) | 0.92 (0.90–0.98) | 0.97 (0.84–1.14) | 0.97 (0.87–1.10) | |||
| 167.0379 | 175.1 | C18− | 1.74 (1.08–2.81) | 1.91 (1.24–2.96) | 0.97 (0.94–1.02) | 0.88 (0.86–0.98) | 0.89 (0.87–0.97) | |||
| 261.2224 | 244.8 | C18− | 1.35 (1.02–1.78) | 0.92 (0.61–1.38) | 0.95 (0.93–1.00) | 0.92 (0.85–1.04) | 0.94 (0.87–1.06) | Linoleate | M−H2O−H | 1 |
| 681.9263 | 281 | HILIC+ | 1.29 (0.50–3.34) | 2.93 (1.17–7.34) | 0.97 (0.96–1.00) | 0.99 (0.95–1.04) | 0.99 (0.95–1.04) | |||
NOTE: Only metabolic features associated with at least one dose of aspirin treatment (P<0.05, bold type) and also with risk of at least one of the adenoma outcomes (P<0.05, bold type) in the opposing direction are presented.
Abbreviations: RT, retention time in seconds; Col, chromatography column (C18 or HILIC) and ionization mode (− or +); FC; fold change; RR, risk ratio; CI, confidence interval; MSI, Metabolomics Standards Initiative; LysoPE, lysophosphatidylethanolamine.
Linear regression was used to estimate the fold change in year 3 ion intensity in the aspirin treatment group compared to the placebo group, adjusting for age, sex, race, and folate treatment.
Poisson regression was used to assess the association of risk of adenoma outcomes at year 3 with a two-fold change in ion intensity, adjusting for age, sex, and race.
Any adenoma refers to ≥1 adenoma of any type. Advanced refers to adenomas with cancer, high-grade dysplasia, >25% villous component, or diameter ≥1 cm. High-risk refers to ≥1 advanced adenoma or ≥3 adenomas of any type.
For putative identities shown, adduct form and MSI identification confidence level are indicated in the columns to the right.
The 20 metabolic features in aspirin-associated pathways in Table 2 were also assessed for their associations with adenoma risk (Table 2). Findings for glycerol 3-phosphate (m/z=171.0069) and linoleate (m/z=261.2224) are included in this table (in addition to Table 3) because they were identified as components of the glycerophospholipid metabolism pathway. However, there was no evidence that any of the other metabolic features from the pathway analyses could contribute to the chemopreventive effects of aspirin because either: 1) they were not statistically significantly associated with adenoma risk, or 2) they exhibited associations that were in a direction that would increase rather than reduce adenoma risk with aspirin treatment. These include putative arachidonate pathway metabolites (e.g., prostaglandin C2, leukotriene A4) that decreased with aspirin treatment but were not associated with adenoma risk. Also, some putative or confirmed carnitine shuttle metabolites (e.g., L-palmitoylcarnitine, octadecenoyl carnitine, stearoylcarnitine) increased with aspirin treatment and were associated with increased, rather than reduced, adenoma risk.
DISCUSSION
In this untargeted metabolomics analysis of colon tissue, we found that top metabolic pathways associated with 81 mg/day aspirin treatment primarily involved energy, nucleotide and amino acid metabolism, and were largely distinct from pathways associated with 325 mg/day aspirin treatment, which primarily involved pathways related to arachidonic acid metabolism, including prostaglandin and leukotriene metabolism. However, the carnitine shuttle pathway was associated with both doses of aspirin treatment. Based on associations with adenoma risk, we discovered several metabolites in colon tissue involved in energy metabolism that may contribute to the chemopreventive effects of aspirin during early colorectal carcinogenesis, including glycerol 3-phosphate, creatinine and linoleate. Conversely, we also discovered several carnitine shuttle metabolites that may be associated with increased risk of carcinogenesis with aspirin treatment.
Interestingly, the three confirmed metabolites linked to reductions in carcinogenesis with aspirin treatment have all previously been associated with colorectal cancer risk: glycerol 3-phosphate, creatinine and linoleate (Supplementary Table S1). In cross-sectional studies, linoleate in stool (29–33) and blood (34) and creatinine in urine (35) were inversely associated with cancer status. In studies of colorectal cancer vs normal mucosal tissue, creatinine was lower in tumors (30,36), whereas glycerol 3-phosphate was higher (37). The directions of these associations were all consistent with our findings for aspirin treatment and adenoma risk, i.e., aspirin treatment increased linoleate and creatinine levels, and reduced glycerol 3-phosphate levels, as would be expected given chemopreventive effects of aspirin. Furthermore, two of these metabolites (glycerol-3-phosphate and linoleate) are in the glycerophospholipid metabolism pathway, which was associated with treatment with 81 mg aspirin. Glycerophospholipids provide precursors for the synthesis of eicosanoids from arachidonic acid upstream of cyclooxygenase inhibition by aspirin (38). Also, linoleic acid in stool has been associated with specific gut microbial profiles (29,33), and the gut microbiome has been linked to colorectal cancer risk (39). Thus, a novel hypothesis is that the mechanism for the chemopreventive effect of aspirin may involve modification of the gut microbiome.
There have been a few previous studies of the metabolic effects of aspirin in humans. In one trial, 40 healthy adults were randomized to 325 mg aspirin treatment or placebo for 60 days in a crossover design (40). Among 363 metabolites that were analyzed in plasma, aspirin treatment reduced levels of 2-hydroxyglutarate, a putative oncometabolite associated with epigenetic dysregulation in myeloid malignancies. In our colon tissue specimens, although 325 mg aspirin treatment appeared to be associated with slightly reduced 2-hydroxyglutarate levels (fold change=0.88, P=0.08), 81 mg aspirin treatment was not (fold change=0.98, P=0.83) and changes in 2-hydroxyglutarate levels were also not significantly associated with adenoma outcomes. Also, in a cross-sectional study of 58 individuals, a targeted metabolomic analysis of the colon mucosa biopsied at routine colonoscopy showed decreased levels of eicosanoids with aspirin use (41), in agreement with our findings. Finally, in analyses of up to 156 participants in the Hereditary and Phenotype Intervention Heart Study who were treated with 81 mg aspirin for 14 days, serum levels decreased for 25 out of 30 oxylipids measured as well as linoleic acid and arachidonic acid (42–44). The decrease in linoleic acid detected in serum is opposite what we observed in colon tissue with long-term aspirin treatment and short-term cessation of treatment.
Our findings have a number of interesting implications. Modulation of energy metabolism appears to be a key factor in aspirin’s chemopreventive effects in colon tissue. It is well known that transformed cells reprogram energy metabolism to carry out aerobic glycolysis, a phenomena called the Warburg effect (45). Aspirin may be able to suppress this reprogramming via effects on the AMPK and mTOR signaling pathways (11). Also, the metabolic changes that we detected appear to be relatively long-lasting since they were detected in normal colon mucosal biopsies collected at colonoscopies a week after cessation of aspirin treatment.
Interestingly, aspirin-associated metabolic features and pathways differed for the two aspirin doses used (81 mg and 325 mg) and some of the metabolic changes caused by aspirin treatment might increase rather than decrease risk. These two observations may help to explain the lack of a typical dose-response relationship observed in our clinical trial where the lower 81 mg dose appeared more efficacious than the 325 mg dose in preventing adenomas (12). For example, increases in some carnitine shuttle metabolites appeared greater for the 325 mg dose as compared to the 81 mg dose, and these increases were associated with increased adenoma risk rather than reduced risk. Our findings are in agreement with recent evidence in cultured cells that aspirin increases mitochondrial fatty acid oxidation via stimulation of the carnitine shuttle (46). Potentially this increase in energy production from fatty acids may increase carcinogenesis. Ultimately, this finding may have important implications for optimizing the dose of aspirin used in chemoprevention. It is noteworthy that similar unusual dose responses were seen for associations of aspirin use with ovarian cancer risk in the Nurses’ Health Studies, where low dose (≤100 mg) aspirin was inversely associated with risk but the 325 mg dose was not associated with risk (47), and in another colorectal adenoma prevention trial (48).
Although we detected reductions in putative eicosanoids metabolites downstream of cyclooxygenase inhibition, especially for the 325 mg aspirin dose, we did not find evidence that these changes contribute to the chemopreventive effects of aspirin since changes in these metabolites were not associated with adenoma risk. This is in agreement with null findings from our prior targeted analysis in which the effect of aspirin in reducing adenoma risk was independent of urinary prostanoid levels (14). Thus, despite previous research implicating cyclooxygenase inhibition (4), our findings do not provide evidence for a role of these metabolites in the chemopreventive effects of aspirin in the normal mucosa at early stages of colorectal carcinogenesis. This is perhaps not surprising since COX-2 levels may not be elevated in non-neoplastic tissue.
This work has important strengths. We examined the effect of aspirin in individuals who were randomly assigned to daily treatment with two defined doses of aspirin (81 mg or 325 mg) or a placebo control for approximately three years. We analyzed the effects of aspirin in the relevant target tissue (colon). We linked our metabolomics findings to prospective data on adenoma outcomes. Finally, we carefully selected individuals for this metabolomics analysis in order to minimize aspirin exposure in the placebo control group.
There are also some limitations. Colon tissue biopsies were only collected at the end of treatment and a week after treatment cessation. However, we measured increases in two common aspirin catabolites (salicylate and salicyluric acid (9)) in plasma samples collected just prior to treatment cessation compared to those collected at baseline to confirm treatment adherence and lack of exposure in the placebo group. There was probably some variability in the amount of mucosal tissue in the biopsies, which were not weighed. However, data was normalized during statistical analyses to address this limitation. Also, 16 of 21 metabolic features discovered that may contribute to aspirin’s protective effects were unidentified, a common limitation inherent in current untargeted metabolomics analyses (49,50). Nevertheless, the 3 metabolites with confirmed identities have previously been associated with colorectal cancer, and 2 of these are involved in the biosynthesis of eicosanoids from arachidonic acid upstream of cyclooxygenase inhibition by aspirin. Finally, we did not adjust for multiple comparisons in the present exploratory analysis. However, the strength of the untargeted approach to discover novel aspirin-associated pathways and metabolites outweighs this limitation and identifying and validating the metabolic features discovered here should be a priority in future work.
In conclusion, our metabolomics analysis of normal colon mucosa from individuals who underwent long term aspirin treatment suggests that the modulation of energy metabolism and the effect of cyclooxygenase inhibition on arachidonic acid precursors may be key to aspirin’s chemopreventive efficacy in early colorectal carcinogenesis. Conversely, aspirin’s effects on carnitine shuttle metabolism may increase carcinogenesis and could be important for understanding dose-dependent effects of the drug.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the participants and staff of the Aspirin/Folate Polyp Prevention Study for their valuable contributions. This work was supported by NCI/NIH R01CA188038 to E. Barry and R01CA059005 to J. Baron. The study biorepository was supported by NIGMS/NIH P20GM104416. Aspirin and placebo tablets used in the Aspirin/Folate Polyp Prevention Study were provided by the Bayer Corporation.
Data Sharing: This data is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org, where it has been assigned Project ID PR000730. The data can be accessed directly via it’s Project DOI: 10.21228/M89X1C. This work is supported by NIH grant, U2C- DK119886.
Footnotes
Potential Conflict of Interest Disclosures: Together with the Trustees of Dartmouth College, John A. Baron holds a use patent, not currently licensed, for the chemopreventive use of aspirin for colorectal cancer. The other authors had no disclosures.
REFERENCES
- 1.William WN Jr., Heymach JV, Kim ES, Lippman SM. Molecular targets for cancer chemoprevention. Nature reviews Drug discovery 2009;8:213–25 [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Yurgelun MB, Alexandrov L, Rao A, Bejar R, Polyak K, et al. Precancer Atlas to Drive Precision Prevention Trials. Cancer Res 2017;77:1510–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan AT, Arber N, Burn J, Chia WK, Elwood P, Hull MA, et al. Aspirin in the chemoprevention of colorectal neoplasia: an overview. Cancer Prev Res (Phila) 2012;5:164–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drew DA, Cao Y, Chan AT. Aspirin and colorectal cancer: the promise of precision chemoprevention. Nat Rev Cancer 2016;16:173–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibbins-Domingo K, Force USPST. Aspirin Use for the Primary Prevention of Cardiovascular Disease and Colorectal Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med 2016;164:836–45 [DOI] [PubMed] [Google Scholar]
- 6.Thun MJ, Jacobs EJ, Patrono C. The role of aspirin in cancer prevention. Nature reviews Clinical oncology 2012;9:259–67 [DOI] [PubMed] [Google Scholar]
- 7.Alfonso L, Ai G, Spitale RC, Bhat GJ. Molecular targets of aspirin and cancer prevention. Br J Cancer 2014;111:61–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umar A, Steele VE, Menter DG, Hawk ET. Mechanisms of nonsteroidal anti-inflammatory drugs in cancer prevention. Semin Oncol 2016;43:65–77 [DOI] [PubMed] [Google Scholar]
- 9.Schror K Acetylsalicylic Acid. Wiley-Blackwell; 2016. [Google Scholar]
- 10.Gala MK, Chan AT. Molecular pathways: aspirin and Wnt signaling-a molecularly targeted approach to cancer prevention and treatment. Clin Cancer Res 2015;21:1543–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Feng Y, Liu X, Ma J, Li Y, Wang T, et al. Beyond a chemopreventive reagent, aspirin is a master regulator of the hallmarks of cancer. J Cancer Res Clin Oncol 2019;145:1387–403 [DOI] [PubMed] [Google Scholar]
- 12.Baron JA, Cole BF, Sandler RS, Haile RW, Ahnen D, Bresalier R, et al. A randomized trial of aspirin to prevent colorectal adenomas. N Engl J Med 2003;348:891–9 [DOI] [PubMed] [Google Scholar]
- 13.Ho GY, Xue X, Cushman M, McKeown-Eyssen G, Sandler RS, Ahnen DJ, et al. Antagonistic effects of aspirin and folic acid on inflammation markers and subsequent risk of recurrent colorectal adenomas. J Natl Cancer Inst 2009;101:1650–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedirko V, Bradshaw PT, Figueiredo JC, Sandler RS, Barry EL, Ahnen DJ, et al. Urinary metabolites of prostanoids and risk of recurrent colorectal adenomas in the Aspirin/Folate Polyp Prevention Study (AFPPS). Cancer Prev Res (Phila) 2015;8:1061–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole BF, Baron JA, Sandler RS, Haile RW, Ahnen DJ, Bresalier RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. Journal of the American Medical Association 2007;297:2351–9 [DOI] [PubMed] [Google Scholar]
- 16.Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, Hamdan R, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev 2009;18:1041–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace K, Grau MV, Levine AJ, Shen L, Hamdan R, Chen X, et al. Association between folate levels and CpG Island hypermethylation in normal colorectal mucosa. Cancer prevention research 2010;3:1552–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park YH, Lee K, Soltow QA, Strobel FH, Brigham KL, Parker RE, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology 2012;295:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013;9:S132–S43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go YM, Walker DI, Liang Y, Uppal K, Soltow QA, Tran V, et al. Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicol Sci 2015;148:531–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009;25:1930–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uppal K, Soltow QA, Strobel FH, Pittard WS, Gernert KM, Yu T, et al. xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27 [DOI] [PubMed] [Google Scholar]
- 24.Sumner WL, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007;3:211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, et al. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 2013;9:e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289–300 [Google Scholar]
- 27.Uppal K, Walker DI, Liu K, Li S, Go YM, Jones DP. Computational Metabolomics: A Framework for the Million Metabolome. Chem Res Toxicol 2016;29:1956–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uppal K, Walker DI, Jones DP. xMSannotator: An R Package for Network-Based Annotation of High-Resolution Metabolomics Data. Anal Chem 2017;89:1063–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013;8:e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phua LC, Chue XP, Koh PK, Cheah PY, Ho HK, Chan EC. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol Ther 2014;15:389–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, et al. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis 2014;35:2089–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, et al. Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One 2016;11:e0152126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Wang J, Rao B, Deng L. Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp Ther Med 2017;13:2848–54 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.May-Wilson S, Sud A, Law PJ, Palin K, Tuupanen S, Gylfe A, et al. Pro-inflammatory fatty acid profile and colorectal cancer risk: A Mendelian randomisation analysis. Eur J Cancer 2017;84:228–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Y, Xie G, Chen T, Qiu Y, Zou X, Zheng M, et al. Distinct urinary metabolic profile of human colorectal cancer. J Proteome Res 2012;11:1354–63 [DOI] [PubMed] [Google Scholar]
- 36.Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, et al. Metabolite profiling of human colon carcinoma--deregulation of TCA cycle and amino acid turnover. Mol Cancer 2008;7:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res 2009;69:4918–25 [DOI] [PubMed] [Google Scholar]
- 38.Hishikawa D, Hashidate T, Shimizu T, Shindou H. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J Lipid Res 2014;55:799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 2013;105:1907–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liesenfeld DB, Botma A, Habermann N, Toth R, Weigel C, Popanda O, et al. Aspirin Reduces Plasma Concentrations of the Oncometabolite 2-Hydroxyglutarate: Results of a Randomized, Double-Blind, Crossover Trial. Cancer Epidemiol Biomarkers Prev 2016;25:180–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gottschall H, Schmocker C, Hartmann D, Rohwer N, Rund K, Kutzner L, et al. Aspirin alone and combined with a statin suppresses eicosanoid formation in human colon tissue. J Lipid Res 2018;59:864–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yerges-Armstrong LM, Ellero-Simatos S, Georgiades A, Zhu H, Lewis JP, Horenstein RB, et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther 2013;94:525–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellero-Simatos S, Lewis JP, Georgiades A, Yerges-Armstrong LM, Beitelshees AL, Horenstein RB, et al. Pharmacometabolomics reveals that serotonin is implicated in aspirin response variability. CPT Pharmacometrics Syst Pharmacol 2014;3:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellero-Simatos S, Beitelshees AL, Lewis JP, Yerges-Armstrong LM, Georgiades A, Dane A, et al. Oxylipid Profile of Low-Dose Aspirin Exposure: A Pharmacometabolomics Study. J Am Heart Assoc 2015;4:e002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011;11:325–37 [DOI] [PubMed] [Google Scholar]
- 46.Uppala R, Dudiak B, Beck ME, Bharathi SS, Zhang Y, Stolz DB, et al. Aspirin increases mitochondrial fatty acid oxidation. Biochem Biophys Res Commun 2017;482:346–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnard ME, Poole EM, Curhan GC, Eliassen AH, Rosner BA, Terry KL, et al. Association of Analgesic Use With Risk of Ovarian Cancer in the Nurses’ Health Studies. JAMA Oncol 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole BF, Logan RF, Halabi S, Benamouzig R, Sandler RS, Grainge MJ, et al. Aspirin for the chemoprevention of colorectal adenomas: meta-analysis of the randomized trials. J Natl Cancer Inst 2009;101:256–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Mol Cell 2015;58:699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016;17:451–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.