Abstract
Numerous studies have implicated changes in the Y chromosome in male cancers, yet few have investigated the biological importance of Y chromosome non-coding RNA. Here we identify a group of Y chromosome-expressed long non-coding RNA (lncRNA) that are involved in male non-small cell lung cancer (NSCLC) radiation sensitivity. Radiosensitive male NSCLC cell lines demonstrated a dose-dependent induction of linc-SPRY3-2/3/4 following irradiation, which was not observed in radioresistant male NSCLC cell lines. Cytogenetics revealed the loss of chromosome Y (LOY) in the radioresistant male NSCLC cell lines. Gain- and loss-of-function experiments indicated that linc-SPRY3-2/3/4 transcripts affect cell viability and apoptosis. Computational prediction of RNA binding proteins (RBP) motifs and UV-Crosslinked Immunoprecipitation (CLIP) assays identified IGF2BP3, an RBP involved in mRNA stability, as a binding partner for linc-SPRY3-2/3/4 RNA. The presence of linc-SPRY3-2/3/4 reduced the half-life of known IGF2BP3 binding mRNA, such as the anti-apoptotic HMGA2 mRNA, as well as the oncogenic c-MYC mRNA. Assessment of Y chromosome in NSCLC tissue microarrays and expression of linc-SPRY3-2/3/4 in NSCLC RNAseq and microarray data revealed a negative correlation between the loss of the Y chromosome or linc-SPRY3-2/3/4 and overall survival. Thus, linc-SPRY3-2/3/4 expression and LOY could represent an important marker of radiation therapy in NSCLC.
Keywords: Non-small cell lung cancer, radiation therapy, Y chromosome, lncRNA
Graphical Abstract
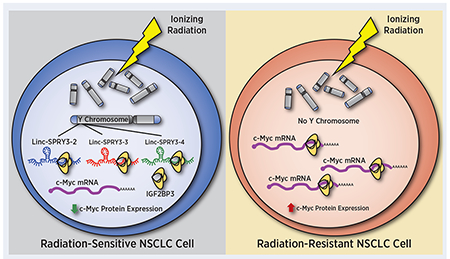
Overexpression of Y chromosome-encoded linc-SPRY3 RNAs after irradiation increases radiation sensitivity of male non-small cell lung cancer (NSCLC) cells by reducing stability of oncogenic mRNAs.
INTRODUCTION
Lung cancer is the most commonly diagnosed cancer worldwide and is the number one cause of cancer-related mortality (1). While there have been great strides in advancing treatment modalities for NSCLC, radiation therapy (RT) resistance remains a significant challenge. Previous studies have demonstrated mechanisms of radioresistance that include alterations in proteins involved in cellular response to DNA damage, the repair of DNA, apoptosis, hypoxia, and cell cycle regulators, among others (2,3). Regulatory non-coding RNAs have also been implicated in the development of radioresistance in NSCLC (4).
Regulatory non-coding RNAs include mature microRNAs (miRNAs) of around 22 nucleotides and long non-coding RNAs (lncRNAs) of >200 nucleotides (5). Studies have shown that most LncRNAs do not contain an open reading frame and could regulate a wide variety of processes through diverse mechanisms, including acting as signals (e.g. enhancers of transcriptional regulation), guides (e.g. recruitment of chromatin modifiers enzymes), or competing endogenous RNAs (e.g. sequestering miRNAs or RNA binding proteins) (6). The dysregulation of the expression and function of lncRNAs has been implicated in numerous cancer types, suggesting their potential use as biomarkers, prognostic indicators, or therapeutic targets (7). In NSCLC, lncRNAs have been associated with nearly every facet of cancer cell biology (8). Several lncRNAs have been associated with the development of resistance to radiotherapy (9). Examples of this include MALAT1 and HOTAIR, which are responsible for inhibiting apoptosis and promoting epithelial to mesenchymal transition thereby aiding in radioresistance (10).
Non-coding RNA genes, including those coding for lncRNAs have been mapped to every human chromosome, including the X and Y chromosomes (11). LncRNAs that map to the X chromosome include XIST, which is critical for the initiation of X-inactivation. The dysregulation of XIST has been shown in multiple cancers (7,12). Other X chromosome lncRNAs include TSIX and XACT, which also function in regulating X chromosome inactivation (12). In contrast, few studies have considered the possibility that the Y chromosome contains genes encoding regulatory RNAs (13–15). Only 27 distinct protein-coding genes are present on the Y chromosome, most of which function in sex determination and male fertility (16,17). The remaining are considered potential non-coding genes and pseudogenes (18). This suggests the Y chromosome is an untapped source of non-coding information, and could potentially provide new insights into numerous diseases - including NSCLC.
Here we report, for the first time, the importance of the lncRNAs linc-SPRY3 (also known as lnc-BPY2C): linc-SPRY3-2, linc-SPRY3-3, linc-SPRY3-4 (linc-SPRY3-2/3/4) in male NSCLC radiation response. Gain and loss of function experiments demonstrated that linc-SPRY3-2/3/4 are involved in radiation vulnerability of male NSCLC cells and tumor samples containing chromosome Y or linc-SPRY3-2/3/4. Together our findings suggest a role of male-specific lncRNAs in NSCLC radiation sensitivity.
MATERIALS AND METHODS
Cell Culture
Non-small cell lung cancer cell lines H460, H820, H157, H1299, A549, and WVU-Ma-0005 were cultured with DMEM (Sigma-Aldrich, St. Louis, MO, USA, D7777). H1650 and Ma-ALK-0001 cells were grown in RPMI (Corning, Corning, NY, USA, 50-020-PC). DMEM and RPMI media was supplemented with 10% fetal bovine serum (Gemini Bio-products, Sacramento, CA, USA, 100-106), L-glutamine (Gibco-LifeTech, Grand Island, NY, USA, 25030-081), HEPES (GE Healthcare Life Sciences, Logan, UT, USA, SH30237.01), penicillin–streptomycin (Gibco, 15140-122), and amphotericin B (Gibco, 15290-026). Human bronchial epithelial cells (HBEC) were cultured with keratinocyte-SFM supplemented with bovine pituitary extract, human recombinant epidermal growth factor, and L-glutamine (Gibco, 10724-011). All cells were grown in a humidified incubator at 37°C in the presence of 5% CO2. Cell lines HBEC, H460, H820, H157, A549, H1299, and H1650 were kindly provided by Dr. Erik A. Bey (Indiana University) and patient derived cell lines WVU-Ma-0005 and Ma-ALK-0001 were kindly provided by Dr. Patrick C. Ma (Pennsylvania State University). Experiments conducted in these cell lines were performed within 2 weeks after thawing from liquid nitrogen (2-3 passages). All cell lines were tested for mycoplasma and authenticated by ATCC using the mycoplasma testing service and the Short Tandem Repeat (STR) profiling service.
Microarray analysis
For global lncRNA expression, we used the GeneChip® Human Transcriptome Array 2.0 (ThermoFisher Scientific, Affymetrix, Santa Clara, CA, USA) that assesses 22,829 non-protein coding human genes. Microarray processing was carried out in the West Virginia University Genomics Core Facility. Briefly, 1 μg of total RNA was labeled using the Flashtag RNA labeling kit (Genisphere, Hatfield, PA, USA) according to the manufacturer’s instructions. Each sample was hybridized to the array at 48°C and 60 rpm for 16 hours then washed and stained on Fluidics Station 450 (Fluidics script FS450_0003) and finally scanned on a GeneChip® Scanner 3000 7G (Affymetrix, Santa Clara, CA, USA). Feature intensities were extracted by using GeneChip® Human Transcriptome Array 2.0 library files. Array data was deposited into the NCBI Gene Expression Omnibus (GEO) database (GEO accession #GSE147708).
Quantitative RT-PCR
Total RNA was extracted using Trizol Reagent (Ambion, Austin, TX, USA, 15596026) per manufacturer’s instructions, then treated with Turbo DNAfree DNase (Invitrogen, Carlsbad, CA, USA, AM1907) for 25 minutes at 37ºC. RNA concentrations were determined with a Nanodrop 2000 Spectrophotometer. 0.5 – 1 μg of total RNA was converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA, 170-6891), followed by qRT-PCR using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, 172-5271) and several pair of primers to specifically amplify the genes of interest in this study (Supplementary Table S1). Relative expression was calculated using the double delta CT method (relative expression = 2-ΔCT; where ΔCT =CT (Target RNA) – CT (endogenous control RNA)), where the endogenous control for lncRNA and mRNA was GAPDH and/or UBC.
Virtual Northern Blot
Total RNA (60µg) from 8Gy treated H460 cells was resolved on a 1.2% formaldehyde denaturing agarose gel overnight. The gel was imaged (to serve as ladder) and then fragmented into 25 fractions of equal size and RNA was recovered from each fraction by gel extraction. The recovered RNA was then used for qRT-PCR analysis of each individual lncRNA. The values generated by qRT-PCR were plotted as a fraction of the total RNA. Plotted values were a mean of three technical replicates per band.
Clonogenic Cell Survival Assay
Cells were seeded in triplicate 60mm gridded dishes containing 4 mL of media at 250 cells per dish. Cells were treated with sham or a single dose of radiation (2Gy, 4Gy, or 8Gy), and then colonies were allowed to form for 10-15 days. Colonies were fixed and stained with crystal violet in 33% methanol, and manually counted using a microscope. Surviving fraction values relative to the sham controls were plotted as a function of radiation dose.
Viral Transduction of shRNAs
HEK-293T cells were transfected with the lentiviral plasmid pLKO-shScramble (Addgene #1864) or pLKO.1-TRC cloning vector (Addgene #10878) containing shRNA sequences against linc-SPRY3-2, linc-SPRY3-3, or linc-SPRY3-4 and lentiviral packaging plasmids (PsPax2 Addgene #12260, and VSV-G Addgene #8454) using calcium phosphate transfection. Lentivirus was collected after 48 hours, filtered (0.45 µm) and supplemented with polybrene (1 μg/mL, MilliporeSigma, Burlington, MA, USA, TR-1003-G). WVU-Ma-0005 or H460 cells were exposed to lentivirus for 16-72 hours and allowed to recover for 48 hours post infection. Infected cells were stably selected via puromycin treatment (1.0-2.5 μg/mL) for 72 hours. Efficiency of shRNA knockdown was measured using qRT-PCR.
Nucleofection of Y Chromosome BAC
The bacterial artificial chromosome (BAC) RP11-88F4 clone, from the RPCI-11 Human Male BAC Library (containing a 95,171bp insert from ChrY:58,819,440-58,914,611 [GRCh37/hg19]), was purchased from the Children’s Hospital Oakland Research Institute and introduced to cells by nucleofection. A549 cells (4×106 cells/cuvette) were briefly resuspended in nucleofection buffer, and 16 μg of RP11-88F4 or pBACe3.6 (control) plasmid DNA was added. The cuvette was placed on the Amaxa Nucleofector II device and the appropriate program was selected (Nucleofector® Program X-001). Immediately after the Nucleofection process ended, cells were resuspended in rescue media and transferred into 6-well plates and incubated at 37ºC under 5% CO2. Expression of linc-SPRY3-2/3/4 was measured 48 hours post transfection by qRT-PCR.
Flow Cytometry
Cell viability and apoptosis were evaluated via Guava Eascyte HT Flow Cytometer (Millipore). Guava Viacount (Luminex, Austin, TX, 4000-0040) reagent was used for analysis of cell viability and Guava Nexin (Luminex, 4500-0455) reagent for apoptosis per manufacturer’s instructions. Cells were plated at equal densities in a 6-well plate, then sham treated or treated with a single dose of radiation (8Gy). Cells were trypsinized and collected 24, 48 and 72 hours after treatment. Parameters for the flow cytometer were set using sham controls.
Tumor Growth Delay Assay (TGD)
shControl or shlinc-SPRY3-2 (sh1) cells were trypsinized and suspended at 5 × 106 cells/mL in matrigel. 1 × 106 cells were injected subcutaneously into the right flanks of male nude mice (Nu/Nu-088, Charles River, Wilmington, MA, USA). Mice were housed in the West Virginia University Animal Facility under pathogen-free conditions with the approved Institutional Animal Care and Used Committee (IACUC) protocol number: 1606003213. Once tumors reached ±150 mm3, RT was initiated. Treatment groups (3 animals each) included sham treated control (0Gy) and radiation (20Gy, 5 fractions). Tumor volume and depth was measured using ultrasound imaging (Vevo 2100, FUJIFILM VisualSonics Inc.). Measurements were quantified using the Vevo LAB 2.2.0 software. Tumors were assessed until they reached >800mm3, day 21 after initiation of treatment, or mice showed evidence of morbidity. Relative tumor volume was determined by normalizing measured volumes to the starting volume (Day 0) of each respective animal.
UV Crosslinking and Immunoprecipitation (CLIP)
CLIP was performed according to the original protocols with some modifications (19,20). The detailed method is described in the Supplementary data.
RNA Stability Assay
Cells were treated with 3µg/mL of Actinomycin D and then collected at 0 hours, 3 hours, 6 hours and 9 hours post treatment. RNA was then extracted and used for subsequent qRT-PCR analysis.
C. elegans RNA was added as an exogenous spike for qRT-PCR (70 ng/µL) to amplify the worm specific gene Ama-1. Each time point was normalized to the 0 hour control.
siRNA Transfection
Cells were seeded at 3.5×105 cells per well in a 6-well dish. siRNAs were transfected utilizing the RNAiMax Lipofectamine reagent according to the manufacturer’s standard transfection protocol. 300pmol of scramble control siRNA (Horizon ON-TARGETplus Non-targeting Control siRNA #2, D-001810-02-05) was used. For the targeted lncRNA siRNAs, 100pmol of each linc-SPRY3 specific siRNA was used to create a pool with a total concentration of 300pmol.
Irradiation
Radiation protocols were carried out on an Xstrahl XenX small animal radiation research platform (Suwanee, GA, USA). The XenX was commissioned following the AAMP Task Group 61 protocol for clinical irradiators of 40-300 kV (11439485). Dosage of experimental design was confirmed using Gafchromic EBT3 film (31198412, 29923495). Mice were anesthetized via isoflurane inhalation for the duration of each treatment. Each tumor was treated to a total dose of 20Gy over 5 fractions given every other day with a 1.2 cm circular fixed collimator at a dose rate of 3.62Gy/min corrected for tumor volume per mouse. For cells, a similar confirmation of dose rate for the experimental setup was determined by Gafchromic EBT3 film. Cells were treated in single fraction doses of 2Gy, 4Gy, or 8Gy with a variable collimator at a dose rate of 1.7Gy/min.
DNA FISH AND Tissue Array Analysis
DNA FISH in cell lines was performed and quantified by the WVU Cytogenetics Laboratory. DNA FISH for the tissue arrays was performed as previously described with the following modifications (21). Slides were first baked at 60°C before proceeding through the protocol. Slides were deparaffinized with Skipdewax (Insitus Biotechnologies, Albuquerque, NM, USA, #T213) for 20 minutes at 80°C followed by three washes with distilled water for 2 minutes each. This process was repeated three times. The SRY probe (Cytocell Ltd., Tarrytown, NY, USA, LPU 026-A) was allowed to hybridize for 22 hours. All tissue arrays were purchased from US Biomax, Inc. (Derwood, MD, USA, Lung: OD-CT-RsLung01-009, Testis: T231a, Cervix: T103). Imaging for representative images for the tissue array FISH was performed via epifluorescent microscopy on a Nikon A1R/N SIM-E microscope. Imaging for quantification of Y chromosome in the lung tissue array was performed on an Olympus VS120 Slide Scanner microscope. Quantification of Y chromosome FISH in the lung TMA was done using IMARIS software. Kaplan-Meier method was used to estimate the survival curves and log-rank test was used to assess the difference of survival curves between groups. TMA data for each patient was evaluated by percentage of Y chromosome positive cells out of the total cells examined, where the cut-off point to define high and low was obtained from the classification and regression tree.
RNA Seq and microarray Analysis from NSCLC patient data
RNA-Seq data for patients diagnosed with NSCLC were obtained from the Gene Expression Omnibus (GEO) database (accession #GSE81089) (22). The expression of linc-SPRY3-2 was measured by TPM (transcripts per millions reads) by salmon (23). A TPM value below 0.1 was deemed as not-expressed. The threshold was determined by comparing to the values obtained from female patients, where linc-SPRY3-2 should not express. The survival period was estimated as days between surgery and vital date (i.e., day of death or latest contact per the definition from GSE81089). For the microarray data, the expression of linc-SPRY3-2 from male Early Stage Lung Squamous Cell Carcinomas patients were obtained from GEO database (accession #GSE74777) based on Affymetrix Human Transcriptome Array 2.0. The survival analysis was done by comparing the top 10% high expressing patients of linc-SPRY3-2. The survival curves were made with the survival R package, and p-value was calculated by log-rank test.
Cell Fractionation
Cells were pelleted and immediately processed for cell fractionation to extract RNA (protocol adapted from previous publication (24). Pelleted cells were disrupted using an autoclaved glass Dounce homogenizer. Homogenate was centrifuged to pellet the nuclear fraction and the cytoplasmic fraction (supernatant) was collected. Each fraction was then treated with DNAseI (New England Biolabs) for 20 minutes at 37ºC and then RNA was extracted using Trizol Reagent (Life Technologies) per manufacturer’s instructions. Once the RNA was extracted it was treated with Turbo DNAfree DNase (Ambion) for 20 minutes at 37ºC. RNA concentrations were obtained using a Nanodrop 2000 Spectrophotometer (Thermo Scientific). qRT-PCR for cell fractionations was normalized using an exogenous spike of C. elegans RNA (70 ng/µL) and the worm specific gene Ama-1.
Statistical Analysis
Data are presented as mean ±SD, ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. The statistical significance between experimental groups was determined by ANOVA followed by Tukey’s multiple comparisons test. Analyses were performed using GraphPad Prism V8 and Microsoft Excel 2013.
RESULTS
Identification of radiation-responsive lncRNAs expressed from the Y chromosome
To identify lncRNAs potentially involved in radiation response of NSCLC cells, we profiled the expression of over 20,000 human lncRNAs in the male NSCLC H460 cell line irradiated with 7 Gy of ionizing radiation (IR), compared to unirradiated (0 Gy) H460 cells 24 hours post-treatment. Using a change in gene expression of 1.5-fold or more, we identified 106 lncRNAs as up-regulated following exposure to radiation, while 59 lncRNAs exhibited down-regulation (Supplementary Table S2, Supplementary Fig. S1). Among the highest upregulated lncRNAs, we observed changes in transcripts annotated as linc-SPRY3-2, linc-SPRY3-3, and linc-SPRY3-4, also known as lnc-BPY2C-4, lnc-BPY2C-25, and lnc-BPY2C-2, respectively. Surprisingly, the linc-SPRY3-2/3/4 transcripts map to a region of the Y chromosome heterochromatic block known as the DYZ1 region that contains a single H3K27Ac mark suggesting transcription in this region (Fig. 1A). The DYZ1 region has been described as genetically inert since it is composed of heterochromatin and, due to its highly repetitive nature, remains poorly sequenced (16). Previously attempted sequence analyses of the human Y chromosome have revealed numerous copies of the pentanucleotide repeat motif 5’-TTCCA-3’ in the DYZ1 region (25,26). Only one previous publication has shown expression of two non-coding RNAs (AK128024.1 and AY598346.2) from the DYZ1 region in a testis-specific manner (27). The linc-SPRY3-2/3/4 sequence fragments obtained from the online database LNCipedia (28) contain several 5’-TTCCA-3’ repeat motifs (Fig. 1B) and partially overlap with AK128024.1 and AY598346.2 (Supplementary Fig. S2), further supporting that these lncRNAs are transcribed from the DYZ1 region of the Y chromosome. Using the linc-SPRY3-2/3/4 sequence fragments, we designed primers to specifically amplify each linc-SPRY3 transcript in order to perform virtual northern blot analysis. Using RNA from 8Gy treated H460 cells, we found two peaks of amplification migrating around 5Kb and 1Kb regardless of which linc-SPRY3 primers were used. This suggests linc-SPRY3-2/3/4 could be partial sequences of a longer lincRNA transcript(s). (Supplementary Fig. S3A–C).
Figure 1: Microarray analysis reveals a family of Y chromosome, radiation inducible long non-coding RNAs.
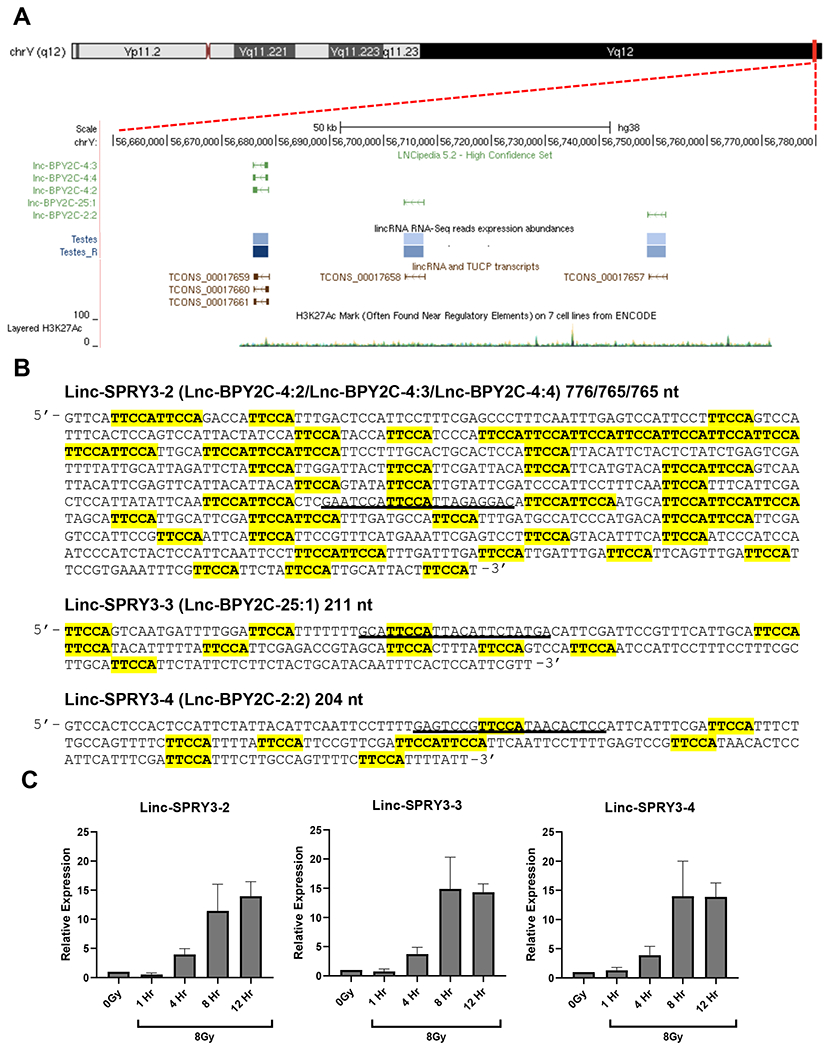
(A) Y chromosome schematic obtained from the UCSC Genome Browser demonstrating the genomic location of the three linc-SPRY3 transcripts in the DYZ1 region (also known as lnc-BPY2C). (B) Linc-SPRY3 transcripts sequences. Highlighted sections indicate pentanucleotide repeats. The black underline regions show the sequences target by the shRNAs and siRNAs used in this study (C) qRT-PCR analysis of the linc-SPRY3 family members validating the radiation induced expression in H460 cells. Time course expression following a single dose of 8Gy IR of linc-SPRY3-2/3/4. GAPDH mRNA was used to normalize qRT-PCR analysis. Error bars represent SD from the mean (n=3).
We next developed qRT-PCR assays of the linc-SPRY3 transcripts to assess their expression in H460 cells following exposure to a single dose of IR (8Gy) at different time points post-treatment versus non-irradiated controls. We observed a 5-fold induction in the expression of the linc-SPRY3-2/3/4 transcripts four hours post-exposure to radiation, that increased to almost 15-fold after eight hours and remained elevated for at least twelve hours after treatment (Fig. 1C).
Linc-SPRY3-2/3/4 expression reveals a dose responsive signature to IR
In order to expand our findings, we examined the expression of the linc-SPRY3 transcripts in several male NSCLC cell lines in addition to H460: the radiosensitive cell lines H820 and H157, and three radioresistant lines: H1299, A549 and H1650 (29–33). We also included two early passage male patient-derived NSCLC cell lines: WVU-Ma-0005 and Ma-ALK-0001. Clonogenic cell survival assays were performed to determine and verify the radiation response of these NSCLC cell lines. Our clonogenic assay data confirmed the radiosensitivity of H460 and H820 as well as the radioresistance of A549 and H1299, which were used as controls for the characterization of the response of the patient-derived cell lines. Our data showed that the WVU-Ma-0005 cell line is radiosensitive and the Ma-ALK-0001 cell line is radioresistant (Supplementary Fig. S4). Next, cells were treated with a single dose of IR (2Gy, 4Gy, or 8Gy) and collected 72 hours post-treatment. Controls were established by treating the cells identically in parallel and omitting the irradiation step. We observed that human bronchial epithelial cells (HBEC), which served as a normal control, the radiosensitive cell lines H460, H820, WVU-Ma-0005 and H157 demonstrated a dose-dependent increase in expression of the linc-SPRY3 RNAs, (Fig. 2A–D, Supplementary Fig. S5A–F). In contrast, the radioresistant cell lines A549, H1299, Ma-ALK-0001 and H1650 showed minimal to no expression of these lncRNAs when compared to no retro-transcription (RT) controls (Fig. 2E–G and Supplementary Fig. S5G–K). To confirm that this expression profile is specific to male radiosensitive NSCLC cells, we investigated the expression of the linc-SPRY3 transcripts in two established female NSCLC cell lines H1975 (radiosensitive) and H1819 (radioresistant) (29). qRT-PCR revealed no amplification signal above background in the female cell lines, a finding consistent with the Y-specific mapping of the linc-SPRY3 genes (Supplementary Fig. S6A–B). Finally, we assessed the coding potential and cellular location of the linc-SPRY3 RNAs in H460 and WVU-Ma-0005 cells. In both cell lines, we detected linc-SPRY3 RNA species exclusively in the nucleus (Supplementary Fig. S7A and S7B) and we found no coding probability in any of the lncRNA transcripts (Supplementary Fig. S7C) supporting our hypothesis of their potential non-coding function.
Figure 2: Radiation sensitive and radiation resistant NSCLC cell lines show significant difference in the expression of the linc-SPRY3 family due to Y chromosome loss.
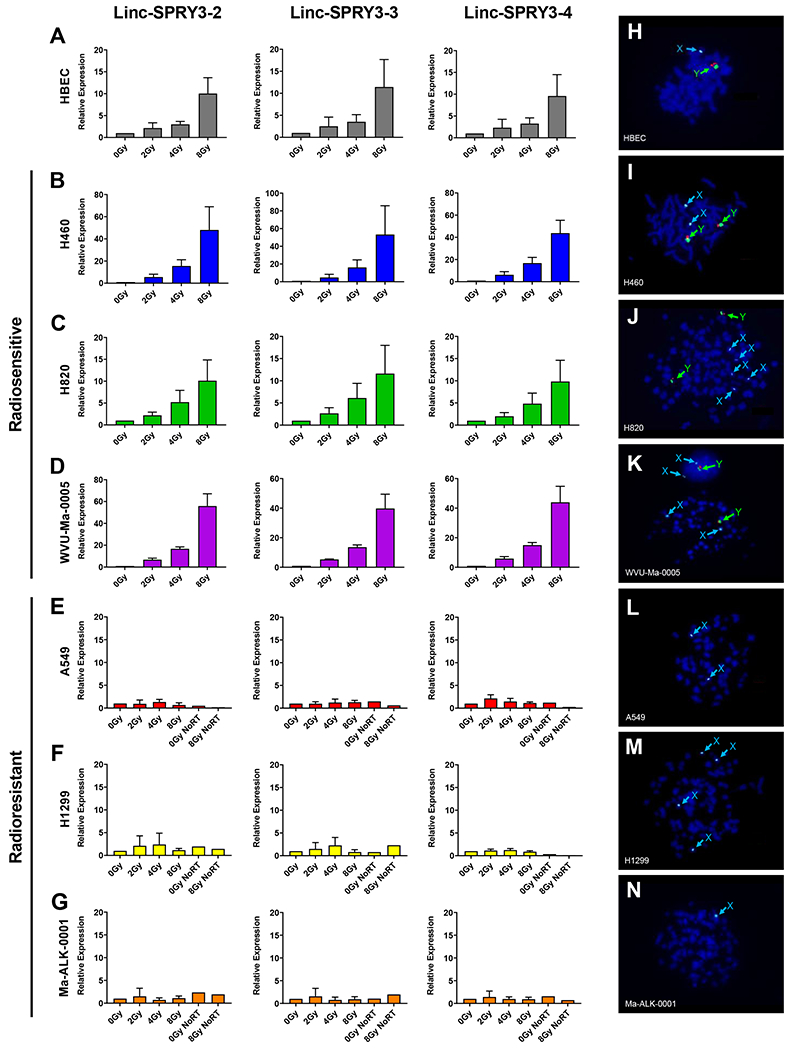
qRT-PCR analysis of (A) HBEC and radiosensitive cell lines (B) H460, (C) H820, and (D) WVU-Ma-0005, demonstrates a dose dependent response in linc-SPRY3 expression. qRT-PCR analysis of radioresistant cell lines, (E) A549, (F) H1299, (G) Ma-ALK-0001, shows little to no change in linc-SPRY3 expression. Values are relative to untreated control (0Gy) of the same collection day. GAPDH mRNA was used to normalize qRT-PCR analysis. Error bars represent SD from the mean (n=3). (H-N) Cytogenetic analysis via DNA-FISH reveals complete loss of the Y chromosome in the radioresistant cell lines while HBECs and radiosensitive cell lines retain their Y chromosome. Images are a representative from 200 counted cells.
Loss of the linc-SPRY3 transcripts in the radioresistant cell lines is due to loss of the Y chromosome
The differences in the expression of the linc-SPRY3 transcripts in radiosensitive and radioresistant NSCLC cell lines prompted us to hypothesize that radioresistant lines harbored genetic changes involving chromosome Y. There are reports of changes in the Y chromosome in cancers dating back several decades that include evidence for the mosaic loss of chromosome Y (LOY) (21,34–41). LOY involves the loss of the entire Y chromosome and occurs in human male tissues as a consequence of aging, however, only recent studies have begun validating a correlation between LOY and cancer biology (18,21,36–38,40–45). Using DNA-FISH, we investigated the sex chromosome status of our panel of cell lines. As expected, the immortalized HBECs showed a normal male genotype containing a single copy of the X and Y chromosomes (Fig. 2H). The radiosensitive cell lines (H460, H820, and H157) possessed multiple copies of both the X and Y chromosomes, while the patient-derived cell line WVU-Ma-0005 contained a single copy of the X and Y chromosomes (Fig. 2I–K and Supplementary Fig. S8A). Interestingly, the radioresistant cell lines A549, H1299, Ma-ALK-0001 and H1650 showed complete loss of the Y chromosome (Fig. 2L–N and Supplementary Fig. S8B). Cytogenetic analysis and quantification of LOY in all the NSCLC cell lines verified our DNA-FISH analysis, (Supplementary Table S3) suggesting that loss of the linc-SPRY3 transcripts in radioresistant cell lines is due to LOY, and that these noncoding RNAs and potentially other genes mapping to the Y chromosome could influence the response of cells to radiation.
The DYZ1 region of the long arm of the Y chromosome restores radiosensitivity
As described above, the repetitive nature of the sequences around the gene(s) coding for the linc-SPRY3 transcript(s) (Fig. 1B) means no complete sequence(s) for these RNAs exists hindering gene-specific gain-of-function studies. However, we were interested to determine if a bacterial artificial chromosome (BAC, RP11-88F4) encompassing the DYZ1 region of the long arm of the human Y chromosome where the linc-SPRY3 transcript(s) are located (a 95,171bp insert from ChrY:58,819,440-58,914,611 [GRCh37/hg19]) could alter the response of the Y chromosome negative, radioresistant NSCLC cell line A549 to IR. After 72 hours, we extracted DNA from the nucleofected cells and performed PCR for the BAC plasmid (pBACe3.6) to determine efficiency of the nucleofection (Supplementary Fig. S9). Next, we detected the expression of the three linc-SPRY3 transcripts in the BAC-transfected A549 cells (A549_88F4) by RT-PCR, but not in the A549 BAC control cells (A549_Ctrl) (Fig. 3A).
Figure 3: Linc-SPRY3 RNAs are important for cell survival following 8Gy irradiation.
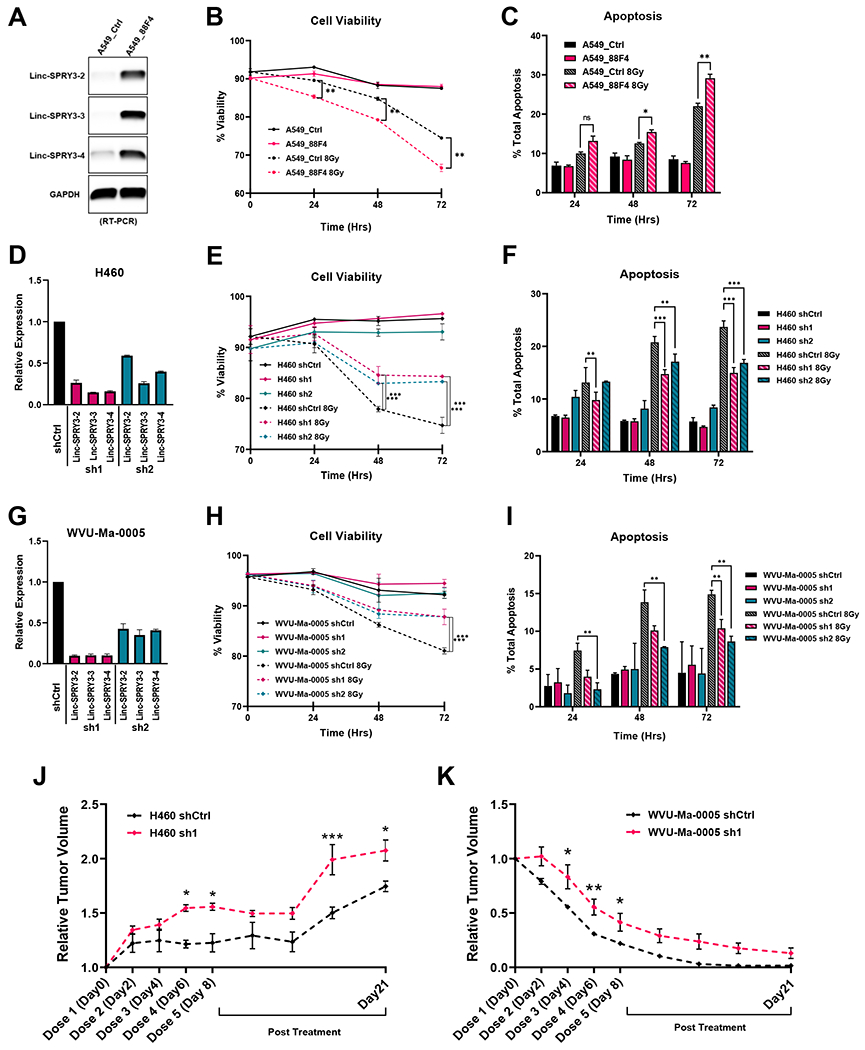
(A) RT-PCR analysis shows expression of the linc-SPRY3 family in BAC RP11-88F4 nucleofected A549 cells. (B-C) Overall cell viability and apoptosis were measured by flow cytometry. Y chromosome BAC nucleofection results in lower cell viability and increased apoptosis compared to control in radioresistant A549 cells after 8Gy IR. (D, G) qRT-PCR analysis reveals effective knockdown of the linc-SPRY3 RNAs in radiosensitive cell lines H460 and WVU-Ma-0005. (E-F, H-I) Knockdown of the linc-SPRY3 family in both H460 and WVU-Ma-0005 cells using two different shRNAs against linc-SPRY3 family (sh1 and sh2) resulted in better overall cell viability and increased resistance to apoptosis when compared with shControl (shCtrl) after 8Gy IR. Untreated controls are provided to demonstrate baseline cell viability and apoptosis. (J-K) Tumor growth delay assay (n = 3) shows higher resistance to radiation of the sh1 tumors relative to shCtrl tumors in both H460 and WVU-Ma-0005 animal experiments. Mean relative tumor volumes are plotted. Error bars represent SD from the mean (n=3) (A-I). Error bars represent SD from the mean of 3 mice in each group (J-K). Untreated tumor controls are represented in Supplementary Fig. S9. ANOVA was used to determine statistical significance.
We measured cell viability and apoptosis in the A549_Ctrl and the A549_88F4 cells after a single dose of 8Gy IR. Notably, the A549_88F4 cells showed significantly worse cell viability when compared to control cells in response to treatment (Fig. 3B). Additionally, A549_88F4 cells showed increased apoptosis compared to control cells after IR with this difference reaching statistical significance at 48 and 72 hours (Fig. 3C). These experiments suggest that the DYZ1 region of the long arm of the Y chromosome harbors genes that influence the response to IR, which includes the linc-SPRY3 family. To obtain further evidence to support this hypothesis, we conducted a series of loss-of-function studies using shRNAs to reduce the levels of the linc-SPRY3 transcripts in NSCLC cells.
First, we designed and validated two shRNAs; one targeting a linc-SPRY3-2 sequence (sh1) and one a linc-SPRY3-3 sequence (sh2) (Fig. 1B and Supplementary Table S4). Interestingly, we observed a significant reduction in the levels of all three linc-SPRY3 RNAs in H460 and WVU-Ma-0005 cells (Fig. 3D and 3G), consistent with our hypothesis that linc-SPRY3 transcripts may be a longer lincRNA transcript. Both H460 and WVU-Ma-0005 cells expressing the linc-SPRY3 shRNAs exhibited increased survival and reduced apoptosis after 8Gy IR in comparison to control cells (Fig. 3E–F & 3H–I); consistent with a loss of linc-SPRY3 transcripts expression resulting in resistance to radiation.
We next sought to determine if reducing the expression of the linc-SPRY3 transcripts would result in IR resistance in vivo. Briefly, 6-8 week old mice were injected with H460 or WVU-Ma-0005 cells harboring either the shCtrl or sh1 constructs. Once tumors reached a volume of 150mm3, mice were treated with fractionated IR to a total dose of 20 Gy, mimicking the clinical procedures for patients with NSCLC (Supplementary Fig. S10A). Ultrasound imaging was used to ensure accurate measurement of both tumor depth and volume, the appropriate settings for IR administration, as well as the tracking tumor growth (Supplementary Fig. S10B–C). Untreated mice showed rapid development of tumors regardless of the type of shRNA used (Supplementary Fig. S10D–E). As expected, IR treated mice exhibited slower tumor progression, while sh1 tumors showed larger tumor volumes both during and after IR compared to shCtrl tumors. This suggests that loss of linc-SPRY3 transcripts results in tumors with higher resistance to RT (Fig. 3J–K and Supplementary Fig. S10D–E).
The linc-SPRY3 transcripts interact with the RNA binding protein IGF2BP3 and affect RNA stability in HMGA2 and c-MYC mRNAs
In an attempt to identify a potential mechanism for the linc-SPRY3-2/3/4 RNAs we utilized A daTabase of RNA binding proteins and AssoCiated moTifs (ATtRACT) software (46) to predict RNA binding proteins (RBPs) potentially interacting with the linc-SPRY3 transcripts. The analysis revealed 13 potential RBPs interacting with the linc-SPRY3 transcripts but the largest number of predicted binding sites across all three family members was the RBP Insulin Like Growth Factor 2 mRNA Binding Protein 3 (IGF2BP3, also known as IMP3) (Fig. 4A). To confirm the predicted association of IGF2BP3 with the linc-SPRY3 RNAs, we performed UV crosslinking and immunoprecipitation (CLIP) for IGF2BP3 in irradiated H460 cells. RT-PCR of the recovered RNA revealed that IGF2BP3 does indeed directly interact with all three linc-SPRY3 family members as well as the known binding partner HMGA2 mRNA (47) but not with the control GAPDH (Fig. 4B). IGF2BP3 is a known RBP whose functions include stabilizing mRNAs. Two such targets include c-Myc and HMGA2 (48). These two targets were of specific interest to us, since previous studies have shown that downregulation of c-Myc and HMGA2 results in an increase in apoptosis in multiple cancers including lung cancer (49–54).
Figure 4: The linc-SPRY3 family act as molecular sponges for IGF2BP3 preventing it from stabilizing HMGA2 and c-Myc.
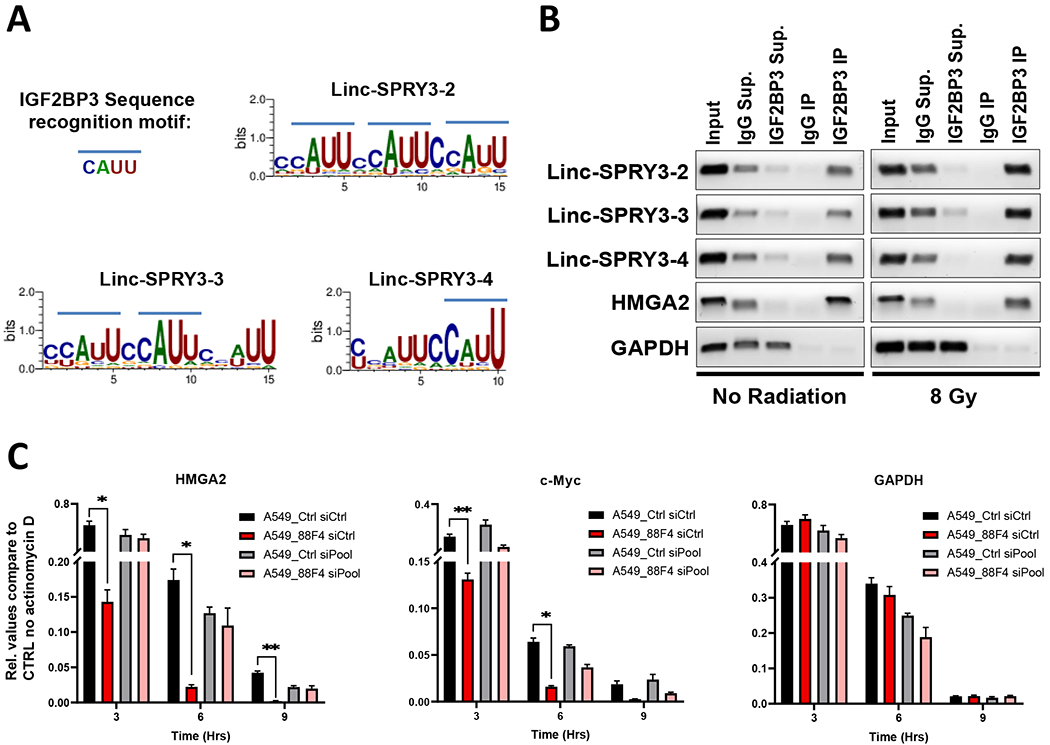
(A) Predicted binding motifs (using WebLogo3) of IGF2BP3 in each linc-SPRY3 transcript. (B) RT-PCR amplification of the recovered RNA from IGF2BP3 CLIP assays of untreated (No Radiation) or treated (8 Gy) H460 cells. The lanes are as follows (from left to right): Input RNA, IgG Supernatant, IGF2BP3 Supernatant, IgG Immunoprecipitation, IGF2BP3 Immunoprecipitation. HMGA is provided as a positive control and GAPDH as a negative control for IGF2BP3 interaction. Gel images are a representative of duplicate experiments. (C) Actinomycin D (ActD) RNA stability assays show rapid degradation of HMGA2 and c-Myc mRNA when the linc-SPRY3 RNAs are present (A549_88F4 siCtrl). siRNA knockdown rescues mRNA stability of HMGA2 and c-Myc (A549_88F4 siPool). GAPDH is provided as a negative control. Error bars represent SD from the mean (n=3)
In order to confirm that the association of IGF2BP3 with the linc-SPRY3 family members affects its ability to target and stabilize c-Myc and HMGA2, we performed RNA stability assays using Actinomycin D (ActD) to stop global transcription in A549 cells with or without the expression of the linc-SPRY3 transcripts (A549_Ctrl and A549_88F4 cells) as well as the knockdown of the linc-SPRY3 transcripts by a pool of siRNAs (siPool). qRT-PCR analysis revealed more rapid degradation of HMGA2 and c-Myc mRNAs in the A549_88F4 siCtrl cells when compared to A549_Ctrl siCtrl cells but GAPDH mRNA was not affected (red bar compared to black bar) (Fig. 4C). Furthermore, we observed rescue of c-Myc and HMGA2 mRNAs stabilization in the A549_88F4 siPool cells after the knockdown of the linc-SPRY3 transcripts (Fig. 4C and Supplementary Fig. S11). Overall, these data suggest that when the linc-SPRY3 RNAs are present they could potentially act as competing endogenous RNAs (ceRNAs) for IGF2BP3, thereby preventing it from stabilizing its mRNA targets c-Myc and HMGA2 affecting apoptosis response.
Y chromosome loss and loss of the linc-SPRY3 RNAs are potential poor prognostic indicators for male NSCLC
To assess the potential clinical relevance of the presence or absence of the Y chromosome and radiation sensitivity and resistance, we performed DNA-FISH for the Y chromosome on a lung cancer tissue microarray and quantified LOY for 30 male cores as well as a testicular cancer core (positive control) and a cervical cancer core (negative control). Representative images taken of male lung cancer cores and a testicular cancer core reveal numerous Y chromosome positive cells, while images taken of female lung cancer cores and a cervical cancer core reveal no detectable presence of the Y chromosome DYZ1 (Supplementary Fig. S12A–H). Using images from male cores of the NSCLC tissue array, we applied a custom image analysis pipeline to determine the percentage Y chromosome positivity. The representative images demonstrate the DNA-FISH of a primary Y chromosome negative core (Fig. 5A, inset is absent of punctate dots) and a heavily Y chromosome positive core (Fig. 5C, arrows in inset indicate punctate dots). Figures 5B and 5D show the result of the image analysis in which Y chromosome negative cells are artificially colored purple and Y chromosome positive cells are artificially colored red. We next used the supplied clinical data to perform Kaplan-Meier analysis. Our analysis revealed a trend towards a negative relationship between Y chromosome loss and patient survival. Though not statistically significant, the trend revealed worse survival for those patients with < 25% Y chromosome positivity (Fig. 5E).
Figure 5: The DYZ1 region of the Y chromosome and the linc-SPRY3 family show negative correlations with patient survival.
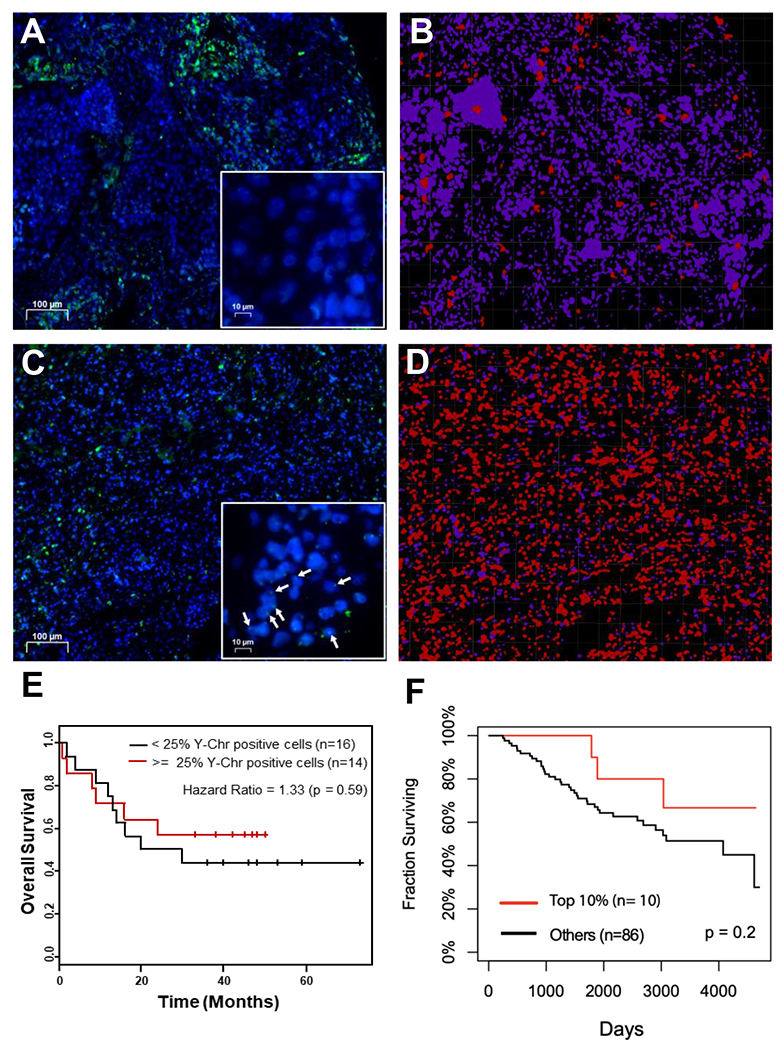
(A-D) Representative images from the imaging analysis performed on 30 cores from a lung cancer tissue array to determine the percentage of Y chromosome DYZ1 positive cells. Y chromosome positive cells are red and Y chromosome negative cells are purple. (E) Kaplan-Meier analysis of the 30 cores analyzed for Y chromosome DYZ1 positivity. (F) Survival curves for NSCLC male patients (GSE74777).
Finally, we accessed the publicly available microarray data (GSE74777) containing 107 Early Stage Lung Squamous Cell Carcinomas (96 male) and the NSCLC short-read RNAseq dataset (GSE81089) containing 198 samples (95 male) to assess the expression of the linc-SPRY3-2 transcript and clinical outcomes (the expression of the other two lncRNAs was only detected in limited number of patients in the RNAseq dataset and therefore were not considered). Interestingly, male patients with high linc-SPRY3 transcripts expression (top 10%) from the microarray data showed a better surviving outcome than the rest of the patients (Fig. 5F). Furthermore, in the RNA-seq study, male patients with tumors expressing low levels of linc-SPRY3-2 exhibited a worse overall survival than those in which their tumor did express linc-SPRY3-2 (Supplementary Fig. S12I). Further studies are needed to validate these initial findings (statistically not significant), but the trends observed in two independent studies suggest that the presence of the Y chromosome and the expression of linc-SPRY3 RNAs warrants further analysis for their functions in radiation response of male NSCLC tumors.
DISCUSSION
In this study the unbiased expression profiling of lncRNAs, following the irradiation of a NSCLC cell line, revealed the induced expression of three annotated lncRNAs that map to the highly repetitive, heterochromatic DYZ1 region of the Y chromosome: linc-SPRY3-2, linc-SPRY3-3, and linc-SPRY3-4. Further investigation revealed expression of this group of lncRNAs is IR dose dependent and begins approximately 4 hours after administration of IR and maintained through 72 hours (Fig. 1). Interestingly, expression of these lncRNAs was only detected in radiosensitive cell lines, but not in radioresistant cell lines, regardless of their oncogenic driver mutations (Supplementary Table S3). Cytogenetic analysis of all cell lines showed loss of the Y chromosome in the radioresistant cell lines, while the radiosensitive cells lines retained their Y chromosome, explaining the differences seen in linc-SPRY3-2/3/4 expression (Fig. 2). Gain-of-function and Loss-of-function experiments demonstrated that the linc-SPRY3 RNAs are important for cellular response to IR as statistically significant changes were seen in cell viability and apoptosis (Fig. 3). In vivo experiments further confirmed these observations as knockdown of the lncRNAs promoted radioresistance in the NSCLC cellular phenotype.
Additionally, we have provided significant evidence that these lncRNAs could act as competing endogenous RNAs (ceRNAs) for the RNA binding protein IGF2BP3 (also known as IMP3) (Fig. 4). In normal tissues, IGF2BP3 is primarily expressed during embryonic development, but its overexpression has been associated with numerous cancer types and as such has been classified as an oncogenic protein (48,55). More specifically, it has been previously shown that IGF2BP3 is upregulated across all NSCLC subtypes and that its loss leads to increased apoptosis (56). As a RBP, IGF2BP3 regulates several genes primarily by binding and stabilizing target mRNAs in order to increase translation efficiency (48). Two such targets, as mentioned previously, are c-Myc and HMGA2. Interestingly, previous studies have shown that downregulation of c-Myc results in increased radiosensitivity by promoting apoptosis (49,50). Additionally, while there is little to no previous evidence of a role for HMGA2 in radiation sensitivity, there are multiple studies characterizing its relationship to apoptosis (52–54). Moreover, HMGA2 expression is controlled by the transcription factor c-Myc (57) which suggests a potential feedback loop interaction. Based on our data, we propose the hypothesis that the presence of the linc-SPRY3-2/3/4, following IR, sequester IGF2BP3 and inhibit the stabilization of c-Myc and HMGA2 which results in downregulation of their proteins making the cells more vulnerable to apoptosis following IR. On the other hand, we cannot discard the possibility that the linc-SPRY3 transcripts could regulate the expression of c-MYC and HMGA2 at the transcriptional level. Lastly, DNA-FISH analysis, microarray analysis, and RNA-seq revealed a negative correlation between loss of the Y chromosome and loss of linc-SPRY3-2 expression and overall patient survival, respectively (Fig. 5). This establishes, for the first time, a connection between the Y chromosome, the linc-SPRY3 transcripts, and how male NSCLC cells respond to radiation.
In conclusion, we show for the first time the importance of a Y chromosome derived group of lncRNAs in regulating male NSCLC radiation response. We provide significant evidence that the linc-SPRY3-2/3/4 transcripts are potentially tumor suppressive and their loss via LOY contributes to radioresistance in NSCLC cells. We also provide evidence that these lncRNAs directly bind IGF2BP3, preventing it from stabilizing its targets HMGA2 and c-Myc which we believe contributes to the radiosensitive phenotype. Moreover, we show a trending negative correlation between Y chromosome and linc-SPRY3 loss and patient survival. We do, however, acknowledge the limitations of this study. More data is needed to fully validate a clinically relevant relationship of both the Y chromosome and linc-SPRY3-2/3/4 family in NSCLC patient survival. It would also be interesting to investigate the behavior of these lncRNAs in an acquired resistance model, utilizing isogenic radioresistant cell lines generated from radiosensitive parental cells like H460 and WVU-Ma-0005 cell lines.
Considering the dose dependent response in expression observed in the radiosensitive cell lines, we speculate that these nuclear lncRNAs could be involved in apoptosis and/or the DNA damage response and repair pathway. We also hypothesize that linc-SPRY3 transcripts and the Y chromosome could eventually be used as biomarkers for cancer radiotherapy. Overall, we believe that more attention should be given to the Y chromosome, as many of its non-coding regions remain uncharted. This has left a vast untapped resource in the realm of cancer cell biology that has yet to be explored.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This study describes previously unknown Y chromosome-expressed lncRNA regulators of radiation response in male NSCLC and show a correlation between loss of chromosome Y and radioresistance.
ACKNOWLEDGMENTS
This work was supported in part by WVU Cancer Institute, National Cancer Institute (NCI) Plan (2V882), American Cancer Society IRG Internal Pilot Funding (IRG-09-061-04), WVCTSI Award (NIH/NIGMS U54GM104942), and Tumor Microenvironment (TME) CoBRE Grant (NIH/NIGMS P20GM121322). Dr. Karen Hayes was supported in part by The Ladies Auxiliary to the VFW of the United States Cancer Research Fellowship (CK003229). Dr. Gangqing Hu was supported with Bioinformatic Core grants NIH-NIGMS U54 GM-104942 and P20 GM103434. We thank Elisabeth Seftor (WVU Cancer Institute) for her assistance with flow cytometry analysis. We thank Dr. Malcolm Mattes (WVU Cancer Institute) for lending his expertise in radiation oncology. We thank Jamie Senft of the WVU Medicine Clinical Cytogenetics Laboratory for performing the DNA-FISH and providing the images and analysis for all of our cell lines. We thank Sarah Mclaughlin and the WVU Animal Models Imaging Facility (AMIF), as well as the WVU Microscope Imaging Facility (MIF) and their funding sources (NIH Grants P30GM103488, S10RR026378, U54GM104942, P20GM103434) for their assistance with animal and cell imaging. We thank Dr. Alexey Ivanov for his suggestions. We thank the staff of the WVUCI Preclinical Tumor Models core facility for their assistance with the in vivo study. We would also like to thank Dr. Natasha Caplen of the National Cancer Institute (National Institutes of Health, Bethesda, MD) for her assistance in editing this work.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34 [DOI] [PubMed] [Google Scholar]
- 2.Willers H, Azzoli CG, Santivasi WL, Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J 2013;19:200–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim BM, Hong Y, Lee S, Liu P, Lim JH, Lee YH, et al. Therapeutic Implications for Overcoming Radiation Resistance in Cancer Therapy. Int J Mol Sci 2015;16:26880–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Q Predictive relevance of ncRNAs in non-small-cell lung cancer patients with radiotherapy: a review of the published data. Biomark Med 2018;12:1149–59 [DOI] [PubMed] [Google Scholar]
- 5.Calore F, Lovat F, Garofalo M. Non-coding RNAs and cancer. Int J Mol Sci 2013;14:17085–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol 2019;21:542–51 [DOI] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016;29:452–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei MM, Zhou GB. Long Non-coding RNAs and Their Roles in Non-small-cell Lung Cancer. Genomics Proteomics Bioinformatics 2016;14:280–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Chen S, Yang B, Mao W, Yang X, Cai J. Molecular Mechanisms of lncRNAs in Regulating Cancer Cell Radiosensitivity. Biosci Rep 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi HC, Tsai CY, Tsai MM, Yeh CT, Lin KH. Roles of Long Noncoding RNAs in Recurrence and Metastasis of Radiotherapy-Resistant Cancer Stem Cells. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigo R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet 2018;19:535–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jegu T, Aeby E, Lee JT. The X chromosome in space. Nat Rev Genet 2017;18:377–89 [DOI] [PubMed] [Google Scholar]
- 13.Molina E, Chew GS, Myers SA, Clarence EM, Eales JM, Tomaszewski M, et al. A Novel Y-Specific Long Non-Coding RNA Associated with Cellular Lipid Accumulation in HepG2 cells and Atherosclerosis-related Genes. Sci Rep 2017;7:16710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai IL, Chang YS, Chan WL, Lee YT, Yen JC, Yang CA, et al. Male-Specific Long Noncoding RNA TTTY15 Inhibits Non-Small Cell Lung Cancer Proliferation and Metastasis via TBX4. Int J Mol Sci 2019;20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao G, Yao J, Kong D, Ye C, Chen R, Li L, et al. The Long Noncoding RNA TTTY15, Which Is Located on the Y Chromosome, Promotes Prostate Cancer Progression by Sponging let-7. Eur Urol 2018 [DOI] [PubMed] [Google Scholar]
- 16.Quintana-Murci L, Fellous M. The Human Y Chromosome: The Biological Role of a “Functional Wasteland”. J Biomed Biotechnol 2001;1:18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes JF, Page DC. The Biology and Evolution of Mammalian Y Chromosomes. Annu Rev Genet 2015;49:507–27 [DOI] [PubMed] [Google Scholar]
- 18.Maan AA, Eales J, Akbarov A, Rowland J, Xu X, Jobling MA, et al. The Y chromosome: a blueprint for men’s health? European Journal of Human Genetics 2017;25:1181–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol 2008;488:85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes KE, Barr JA, Xie M, Steitz JA, Martinez I. Immunoprecipitation of Tri-methylated Capped RNA. Bio Protoc 2018;8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollows R, Wei W, Cazier JB, Mehanna H, Parry G, Halford G, et al. Association between loss of Y chromosome and poor prognosis in male head and neck squamous cell carcinoma. Head Neck 2019;41:993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mezheyeuski A, Bergsland CH, Backman M, Djureinovic D, Sjoblom T, Bruun J, et al. Multispectral imaging for quantitative and compartment-specific immune infiltrates reveals distinct immune profiles that classify lung cancer patients. J Pathol 2018;244:421–31 [DOI] [PubMed] [Google Scholar]
- 23.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez I, Hayes KE, Barr JA, Harold AD, Xie M, Bukhari SIA, et al. An Exportin-1-dependent microRNA biogenesis pathway during human cell quiescence. Proc Natl Acad Sci U S A 2017;114:E4961–E70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakahori Y, Mitani K, Yamada M, Nakagome Y. A human Y-chromosome specific repeated DNA family (DYZ1) consists of a tandem array of pentanucleotides. Nucleic Acids Res 1986;14:7569–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman MM, Bashamboo A, Prasad A, Pathak D, Ali S. Organizational Variation of DYZ1 Repeat Sequences on the Human Y Chromosome and Its Diagnostic Potentials. DNA and Cell Biology 2004;23:561–71 [DOI] [PubMed] [Google Scholar]
- 27.Jehan Z, Vallinayagam S, Tiwari S, Pradhan S, Singh L, Suresh A, et al. Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2. Genome Res 2007;17:433–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res 2019;47:D135–D9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das AK, Sato M, Story MD, Peyton M, Graves R, Redpath S, et al. Non-small-cell lung cancers with kinase domain mutations in the epidermal growth factor receptor are sensitive to ionizing radiation. Cancer Res 2006;66:9601–8 [DOI] [PubMed] [Google Scholar]
- 30.Oweida A, Sharifi Z, Halabi H, Xu Y, Sabri S, Abdulkarim B. Differential response to ablative ionizing radiation in genetically distinct non-small cell lung cancer cells. Cancer Biol Ther 2016;17:390–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong X, Luo G, Zhou X, Luo W, Wu X, Zhong R, et al. Rad51 in regulating the radiosensitivity of non-small cell lung cancer with different epidermal growth factor receptor mutation status. Thorac Cancer 2016;7:50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang J, Kim W, Kwon T, Youn H, Kim JS, Youn B. Plasminogen activator inhibitor-1 enhances radioresistance and aggressiveness of non-small cell lung cancer cells. Oncotarget 2016;7:23961–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang J, Kim E, Kim W, Seong KM, Youn H, Kim JW, et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J Biol Chem 2013;288:27343–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Center R, Lukeis R, Vrazas V, Garson OM. Y chromosome loss and rearrangement in non-small-cell lung cancer. International Journal of Cancer 1993;55:390–3 [DOI] [PubMed] [Google Scholar]
- 35.Matturri L, Lavezzi AM. Recurrent chromosome alterations in non-small cell lung cancer. Eur J Histochem 1994;38:53–8 [PubMed] [Google Scholar]
- 36.Jordan JJ, Hanlon AL, Al-Saleem TI, Greenberg RE, Tricoli JV. Loss of the short arm of the Y chromosome in human prostate carcinoma. Cancer Genet Cytogenet 2001;124:122–6 [DOI] [PubMed] [Google Scholar]
- 37.Bianchi NO, Richard SM, Pavicic W. Y chromosome instability in testicular cancer. Mutat Res 2006;612:172–88 [DOI] [PubMed] [Google Scholar]
- 38.Minner S, Kilgue A, Stahl P, Weikert S, Rink M, Dahlem R, et al. Y chromosome loss is a frequent early event in urothelial bladder cancer. Pathology 2010;42:356–9 [DOI] [PubMed] [Google Scholar]
- 39.Patel R, Khalifa AO, Isali I, Shukla S. Prostate cancer susceptibility and growth linked to Y chromosome genes. Front Biosci (Elite Ed) 2018;10:423–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loftfield E, Zhou W, Yeager M, Chanock SJ, Freedman ND, Machiela MJ. Mosaic Y Loss Is Moderately Associated with Solid Tumor Risk. Cancer Res 2019;79:461–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caceres A, Jene A, Esko T, Perez-Jurado LA, Gonzalez JR. Extreme down-regulation of chromosome Y and cancer risk in men. J Natl Cancer Inst 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Center R, Lukeis R, Vrazas V, Garson OM. Y chromosome loss and rearrangement in non-small-cell lung cancer. Int J Cancer 1993;55:390–3 [DOI] [PubMed] [Google Scholar]
- 43.Wallrapp C, Hahnel S, Boeck W, Soder A, Mincheva A, Lichter P, et al. Loss of the Y chromosome is a frequent chromosomal imbalance in pancreatic cancer and allows differentiation to chronic pancreatitis. International Journal of Cancer 2001;91:340–4 [DOI] [PubMed] [Google Scholar]
- 44.Bianchi NO. Y chromosome structural and functional changes in human malignant diseases. Mutat Res 2009;682:21–7 [DOI] [PubMed] [Google Scholar]
- 45.Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet 2014;46:624–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giudice G, Sanchez-Cabo F, Torroja C, Lara-Pezzi E. ATtRACT-a database of RNA-binding proteins and associated motifs. Database (Oxford) 2016;2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheen YS, Liao YH, Lin MH, Chu CY, Ho BY, Hsieh MC, et al. IMP-3 promotes migration and invasion of melanoma cells by modulating the expression of HMGA2 and predicts poor prognosis in melanoma. J Invest Dermatol 2015;135:1065–73 [DOI] [PubMed] [Google Scholar]
- 48.Mancarella C, Scotlandi K. IGF2BP3 From Physiology to Cancer: Novel Discoveries, Unsolved Issues, and Future Perspectives. Front Cell Dev Biol 2019;7:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciccarelli C, Di Rocco A, Gravina GL, Mauro A, Festuccia C, Del Fattore A, et al. Disruption of MEK/ERK/c-Myc signaling radiosensitizes prostate cancer cells in vitro and in vivo. J Cancer Res Clin Oncol 2018;144:1685–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung J, Kim EJ, Chung HK, Park HJ, Jeong SY, Choi EK. c-Myc down-regulation is involved in proteasome inhibitor-mediated enhancement of radiotherapeutic efficacy in non-small cell lung cancer. Int J Oncol 2012;40:385–90 [DOI] [PubMed] [Google Scholar]
- 51.Cui F, Hou J, Huang C, Sun X, Zeng Y, Cheng H, et al. C-Myc regulates radiation-induced G2/M cell cycle arrest and cell death in human cervical cancer cells. J Obstet Gynaecol Res 2017;43:729–35 [DOI] [PubMed] [Google Scholar]
- 52.Shi Z, Wu D, Tang R, Li X, Chen R, Xue S, et al. Silencing of HMGA2 promotes apoptosis and inhibits migration and invasion of prostate cancer cells. J Biosci 2016;41:229–36 [DOI] [PubMed] [Google Scholar]
- 53.Gao X, Dai M, Li Q, Wang Z, Lu Y, Song Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac Cancer 2017;8:501–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao N, Wang FX, Wang G, Zhao QS. Targeting the HMGA2 oncogene by miR-498 inhibits non-small cell lung cancer biological behaviors. Eur Rev Med Pharmacol Sci 2018;22:1693–9 [DOI] [PubMed] [Google Scholar]
- 55.Lederer M, Bley N, Schleifer C, Huttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol 2014;29:3–12 [DOI] [PubMed] [Google Scholar]
- 56.Shi R, Yu X, Wang Y, Sun J, Sun Q, Xia W, et al. Expression profile, clinical significance, and biological function of insulin-like growth factor 2 messenger RNA-binding proteins in non-small cell lung cancer. Tumour Biol 2017;39:1010428317695928. [DOI] [PubMed] [Google Scholar]
- 57.Yang G, Xiong G, Feng M, Zhao F, Qiu J, Liu Y, et al. OLR1 promotes pancreatic cancer metastasis via increased c-Myc expression and transcription of HMGA2. Mol Cancer Res 2020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


