Abstract
Transient receptor potential vanilloid type 1 (TRPV1) channels are structurally related, nonselective cation channels that exhibit a high permeability to calcium. Sensory nerve endings expressing TRPV1 channels play a prominent role in regulating the cardiac sympathetic afferent reflex and contribute to cardiac remodeling and dysfunction in chronic heart failure. However, the precise expression of TRPV1 channels in cardiomyocytes vs. non-cardiomyocytes remains debated. Here we utilized a tdTomato-GFP reporter mouse crossed with a mouse line expressing Cre recombinase under the control of the TRPV1 promoter to map the TRPV1 expression pattern in heart. In this model, TRPV1-negative cells express tdTomato protein (red), whereas TRPV1-positive cells express GFP protein (green). As we expected, substantial GFP expression was found in many small and medium diameter dorsal root ganglia neurons in heterozygous TRPV1-Cre +/−, tdTomato flox/flox +/− male mice, suggesting that this heterozygous model is sufficient for labeling TRPV1-positive cells. Furthermore, these results showed that GFP green staining was not detectable in cardiomyocytes. Instead, we found strong GFP green staining in cardiac blood vessels—thought to be arterioles—in the heart. We also observed strong GFP signals on PGP9.5-positive cardiac nerve endings in the epicardium. In summary, this study does not support the concept that TRPV1 channels are strongly expressed in mouse cardiomyocytes. We conclude that TRPV1 channels in mouse heart are mostly expressed on non-cardiomyocyte cells including cardiac nerve endings and vessels. These data have important implications for the modulations of cardiogenic reflexes.
Keywords: tdTomato reporter, cardiac spinal afferents, dorsal root ganglia, autonomic nerve system, resiniferatoxin
1. Introduction
The transient receptor potential vanilloid type 1 (TRPV1) channel is a nonselective cation channel with high calcium permeability activated by heat, protons, vanilloid compounds such as capsaicin, reseniferitoxin (RTX), and membrane-derived lipids including anandamide [1]. It is well established that TRPV1 protein is expressed in a subset of sensory afferent nerves with their cell bodies located in the dorsal root, trigeminal, and nodose ganglia [4, 15].
In the cardiovascular system, cardiac afferent fibers expressing TRPV1 are the sensory arm of the cardiac sympathetic afferent reflex (CSAR) [22] and contribute to elevated sympathetic tone [17–20] in chronic heart failure (CHF). Recent studies have reported that selective cardiac afferent denervation by epicardial application of a potent TRPV1 agonist, RTX, reduced CSAR-evoked sympatho-excitation and mitigate the deleterious effects after myocardial infarction in both rats and pigs [16, 21]. In these studies, epicardial application of RTX was chosen since the majority of TRPV1-positive cardiac afferents travel through the superficial layer of heart [16, 21]. While this treatment depletes a large portion of cardiac spinal afferents in the heart, it brings with it a potential risk to cause mild to moderate epicardium damage if cardiomyocytes also express TRPV1 receptors. To date, it is still debated whether cardiomyocytes express TRPV1 receptors. Evidence from Zahner et al. and from Facer et al. support the notion that TRPV1 receptors are primarily located on cardiac afferents from the epicardial surface and are expressed to very limited degree in ventricular cardiomyocytes from both rats and humans [6, 22]. On the other hand, contradictory results revealed extensive protein expression of TRPV1 in ventricular cardiomyocytes [1, 7, 8]. This discrepancy may be due to the non-specificity of commercial TRPV1 antibodies. Given the importance of cardiac TRPV1 afferents as a potential therapeutic target in CHF and the need for further insights into the contribution of TRPV1 receptors on cardiomyocytes, it is critical to resolve the expression pattern of TRPV1 receptors in the heart with a more reliable technique.
Here, we utilized a tdTomato-GFP reporter mouse and crossed it with a mouse line expressing Cre recombinase under the control of the TRPV1 promoter to map the TRPV1 expression pattern in the heart. In this transgenic mouse, cells expressing TRPV1 should be specifically visualized as GFP fluorescence. We aimed to investigate: 1) the expression profile of TRPV1 in sensory neurons in order to confirm the accuracy and sensitivity of this model; 2) the expression pattern of TRPV1 in ventricular cardiomyocytes, cardiac vessels, and nerve terminals.
2. Methods and materials
2.1. Generation of transgenic animals
All mouse experiments were approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center and were carried out under the guidelines of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The TRPV1-cre mice (B6.129-Trpv1tm1(cre)Bbm/J, stock#017769) and tdTomato reporter mice (Gt(ROSA)26Sortm4(ACTB-tdTomato-EGFP)Luo, stock#007676) were purchased from Jackson Laboratory (Bar Harbor, ME, USA). We crossed these two mouse strains and generated an F1 reporter mouse strain TRPV1Cre+/−:tdTomato+/− (TRPV1cre-tdTomato+/−) for visualization of TRPV1 cells. At approximately 21 d of age, pups were weaned, tails were clipped, and DNA was extracted. The DNA of tails were amplified by polymerase chain reaction (PCR). Two pairs of primers including wild type pair 13922 5′-TTC AGG GAG AAA CTG GAA GAA-3′, 13923 5′-TAG TCC CAG CCA TCC AAA AG-3′, and Mutant pair oIMR1084 5′-GCG GTC TGG CAG TAA AAA CTA TC-3′, oIMR1085 5′-GTG AAA CAG CAT TGC TGT CAC TT-3′ were used to detect wild type and TRPV1-cre allele, respectively. Three primers including 12177 5′-CTT TAA GCC TGC CCA GAA GA-3′, 30297 5′-TAG AGC TTG CGG AAC CCT TC-3′, and 30298 5′-AGG GAG CTG CAG TGG AGT AG-3′ were used to detect mutant tdTomato locus. The primers were used in the reaction mix (94°C/2 min; 10 cycles of 94°C/20 sec, 65°C/15 sec, and 68°C/10 sec; 28 cycles of 94°C/15 sec, 60°C/15 sec, 72°C/10 sec; 72°C/10 min). TRPV1cre-tdTomato+/− male mice (n=7) and tdTomato control male mice (n=5) at 8–10 weeks old were used in the further experiments.
2.2. Immunostaining
The tdTomato, TRPV1cre-tdTomato+/− mice were anesthetized with an intraperitoneal injection of urethane (800 mg/kg) and α-chloralose (40 mg/kg). Then, mice were perfused through the apex of left ventricle, first with Phosphate Buffered Saline (PBS, PH=7.4) followed by 4% paraformaldehyde (4% PFA in 0.1M PBS). The hearts and the thoracic dorsal root ganglia (T1-T4 DRGs) were immediately removed and immersed in 4% PFA overnight. Next, the tissues were rinsed with PBS and dehydrated with 30% sucrose in PBS overnight at 4 °C, and afterwards sectioned at 14 μm using a Leica cryostat (−20°C).
2.2.1. DAPI staining
The slides from T1-T4 DRGs and the heart were rinsed with PBS for 5 min. The slides were then mounted with an aqueous mounting medium containing DAPI (sc24941, Ultracruz, Dallas, TX, USA) and sealed with a coverslip.
2.2.2. Neurofilament (NF200), Troponin I (Tn I) and protein gene product 9.5 (PGP9.5) immunofluorescence
All the slides were taken out and recovered to room temperature. After being blocked in 10% normal donkey serum for 1 h, T1-T4 DRGs were incubated with mouse anti-Neurofilament 200 (NF200, 1:100, N5389, Sigma-Aldrich, St. Louis, MO, USA) while the heart sections were treated with either rabbit anti-Tn I (1:100, ab47003, Abcam, Cambridge, MA, USA) or rabbit anti-protein gene product 9.5 (PGP9.5, 1:500, GTX109637, GeneTex, Irvine, CA, USA). Heart and DRG sections were left overnight at 4°C. Sections were washed and incubated with secondary antibodies of Alexa 405-conjugated goat anti-mouse or Alexa 405-conjugated goat anti-rabbit (1:200, Invitrogen, Carlsbad, CA, USA) for 1 h at room temperature. Slides were sealed with a coverslip.
All slides were observed on a Leica laser confocal microscope under 10X and 40X objective and images were captured using a digital camera system. Z-Stacks were acquired for NF200 immunostained DRG slides.
2.3. Data analysis and Statistics
For quantification of TRPV1 positive neurons, every fifth section (5 sections per mice) was used for consecutive cell quantification analyses using Image J software (https://imagej.nih.gov/ij/download.html). The number of GFP positive primary sensory neurons and total neurons in T1-T4 DRGs was counted, calculated as the percent of TRPV1 positive neurons, and then averaged as per group. The final percent of TRPV1 positive neurons in T1-T4 DRGs are expressed as mean ± SEM. Statistical analysis was carried out using GraphPad Prism 8 (San Diego, CA).
3. Results
3.1. Generation of the F1 TRPV1cre-tdTomato+/− mice
To examine the profile of TRPV1 expression in the heart, we generated TRPV1cre-tdTomato mice by crossing the previously characterized TRPV1-cre driver mice with tdTomato reporter mice (Figure 1A). The tdTomato mice have a two-colored fluorescent cassette (tdTomato: red; enhanced green fluorescent protein (EGFP): green) that can be differentially activated by Cre recombinase (Muzumdar et al., 2007). Normally, the tdTomato red fluorescence is constitutively expressed in the plasma membrane of all cells. When crossing with TRPV1cre mice, TRPV1 promoter elements drive expression of Cre recombinase to delete the tdTomato complementary DNA with a transcriptional stop cassette flanked by LoxP sites, and the GFP cassette is subsequently expressed. The littermates were genotyped for both TRPV1cre and tdTomato sequences (Figure 1B). TRPV1cre-tdTomato+/− and tdTomato (no cre) control male mice were used for the further experiments.
Figure 1.
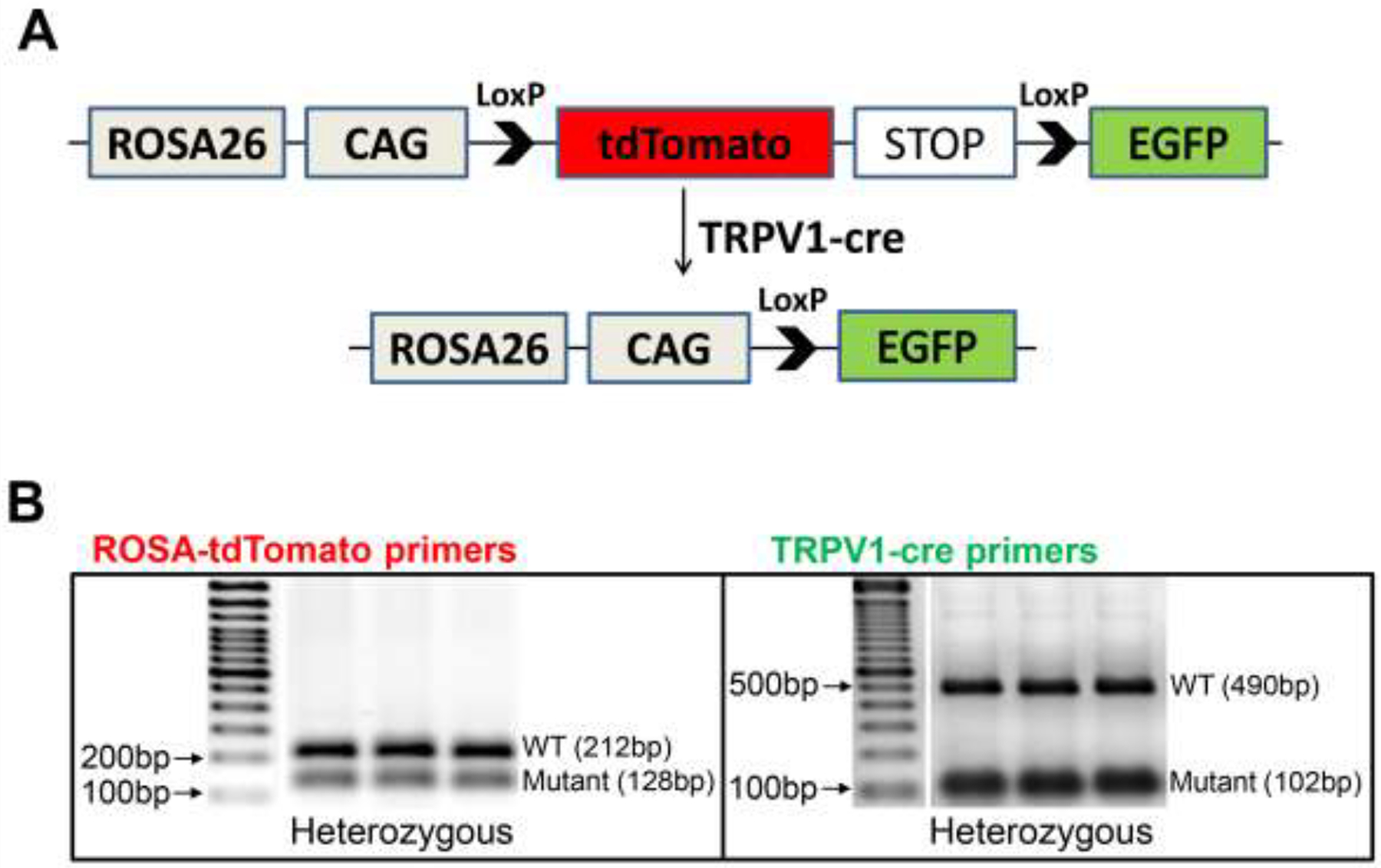
Generation of TRPV1cre-tdTomato reporter mice. (A) Schematic diagram of the gene targeting strategy to cross tdTomato mice with TRPV1cre mice. (B) PCR assay using primers to detect the on-target editing in DNA isolated tails tissue of F1 TRPV1cre-tdTomato+/− mice. ROSA-tdTomato: mutant, 128 base pairs (bp); wild type (WT) band, 212 bp. TRPV1cre: mutant, 102 bp; WT band, 490 bp.
3.2. GFP proteins were expressed in small- and medium-sized DRG neurons in the TRPV1cre-tdTomato+/− mice
As TRPV1 channels are primarily expressed in sensory neurons, we first determined the GFP expression pattern between TRPV1cre-tdTomato+/− and tdTomato control mice in T1-T4 DRGs that innervate the heart. As shown in Figure 2A, TRPV1cre-tdTomato+/− mice robustly expressed GFP proteins in T1-T4 DRGs, most of which are small and medium sized. Moreover, we co-stained the DRG slides with NF200 (Figure 2B), which is a large diameter, myelinated A-fiber neuronal marker. Our results showed the majority of GFP-positive DRG neurons in TRPV1cre-tdTomato+/− mice were not co-localized with NF200. As for the percentage of TRPV1 neurons in the T1-T4 DRGs, our data suggested that 55% of neurons were GFP-positive (Figure 2C) in TRPV1cre-tdTomato+/− mice. However, no GFP-positive neurons were observed in the tdTomato control mice. These results demonstrate that our TRPV1cre-tdTomato+/− mice model is sensitive enough to detect the TRPV1-positive cells.
Figure 2.
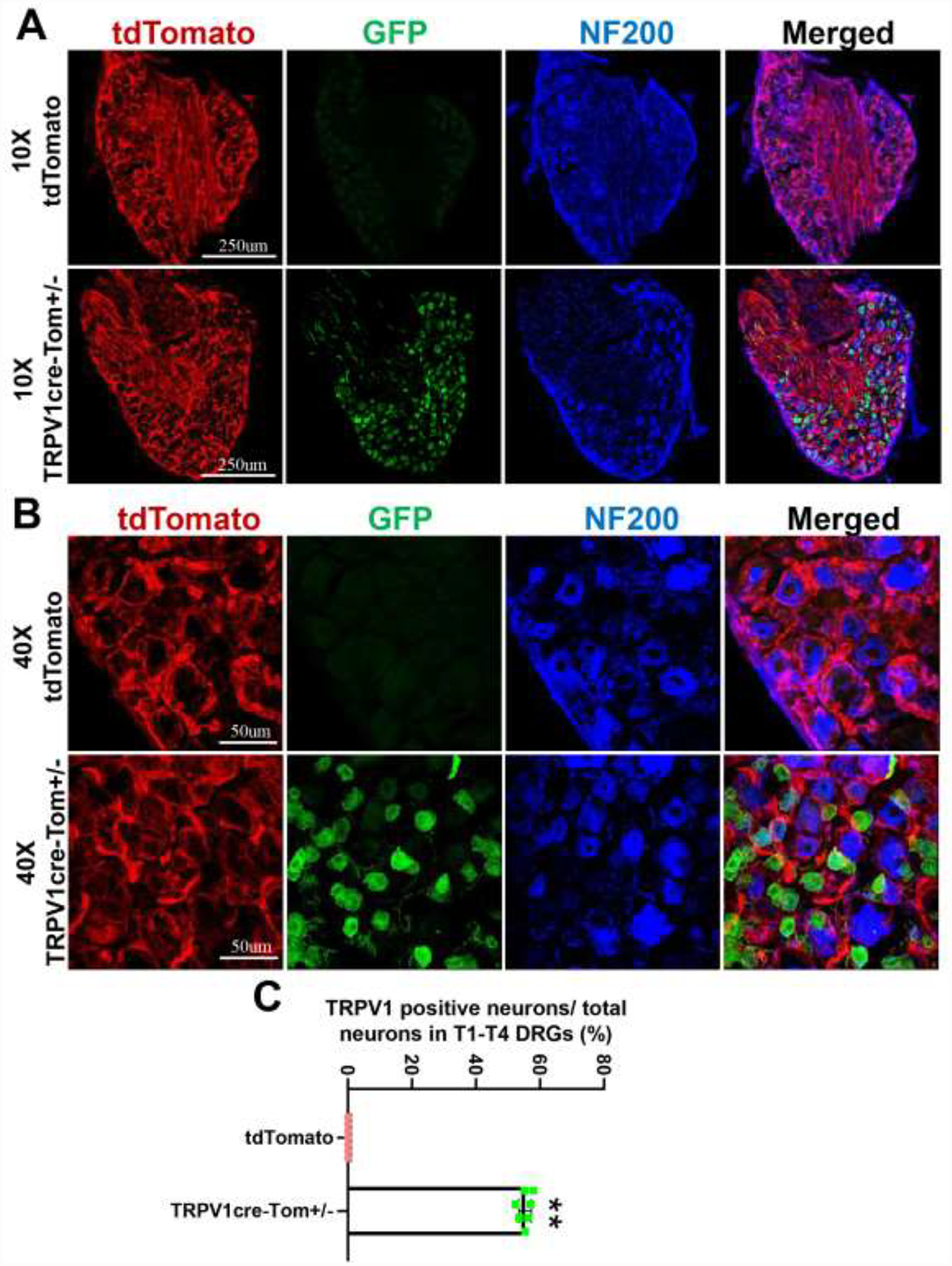
Representative images showing the expression of GFP positive T1-T4 dorsal root ganglions (DRGs) neurons under 10x objective (A) and 40x objective (B) in TRPV1cre-tdTomato+/− (TRPV1cre-Tom+/−) and tdTomato mice. NF200: neurofilament 200, a myelinated A fiber neuron marker. n=5/tdTomato group, n=7/TRPV1cre-tdTomato group. (C) Mean data showing that the percentage of TRPV1 positive neurons in T1-T4 DRGs of TRPV1cre-tdTomato+/− mice. Data are expressed as mean±SE. n=7/TRPV1cre-tdTomato+/− group.
3.3. GFP signals were not detected in cardiomyocytes in the TRPV1cre-tdTomato+/− mice
We further examined GFP fluorescence in the ventricles of both tdTomato mice and TRPV1cre-tdTomato+/− mice. We were not able to detect any significant GFP fluorescence in the ventricular cardiomyocytes of either tdTomato control or TRPV1cre-tdTomato+/− mice (Figure 3A&B). We repeated the experiments by co-staining the heart slides with the cardiomyocyte marker Tn I [10]. GFP fluorescence was not detected in the cardiomyocytes in either group (Figure 3C). Similarly, GFP fluorescence was not detected in atrial cardiomyocytes (Figure 4).
Figure 3.
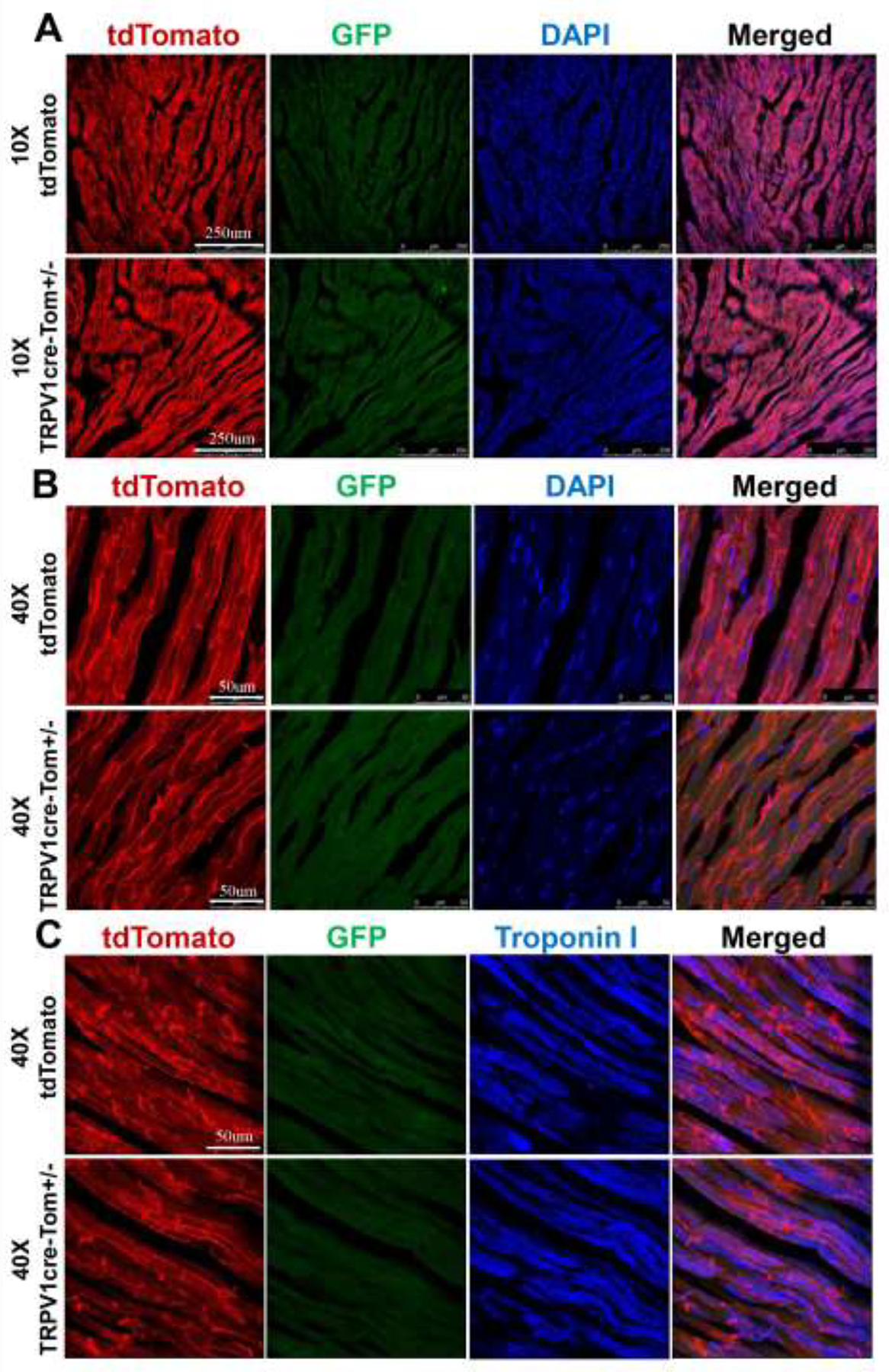
Representative images showing that GFP positive fluorescence is absent in ventricular cardiomyocytes under 10x objective (A) and 40x objective (B and C) in both TRPV1cre-tdTomato+/− (TRPV1cre-Tom+/−) and tdTomato mice. DAPI: 4,6-diamidino-2-phenylindole, a cell nucleus marker; Troponin I, a marker for cardiomyocytes. n=5/tdTomato group, n=7/TRPV1cre-tdTomato+/− group.
Figure 4.
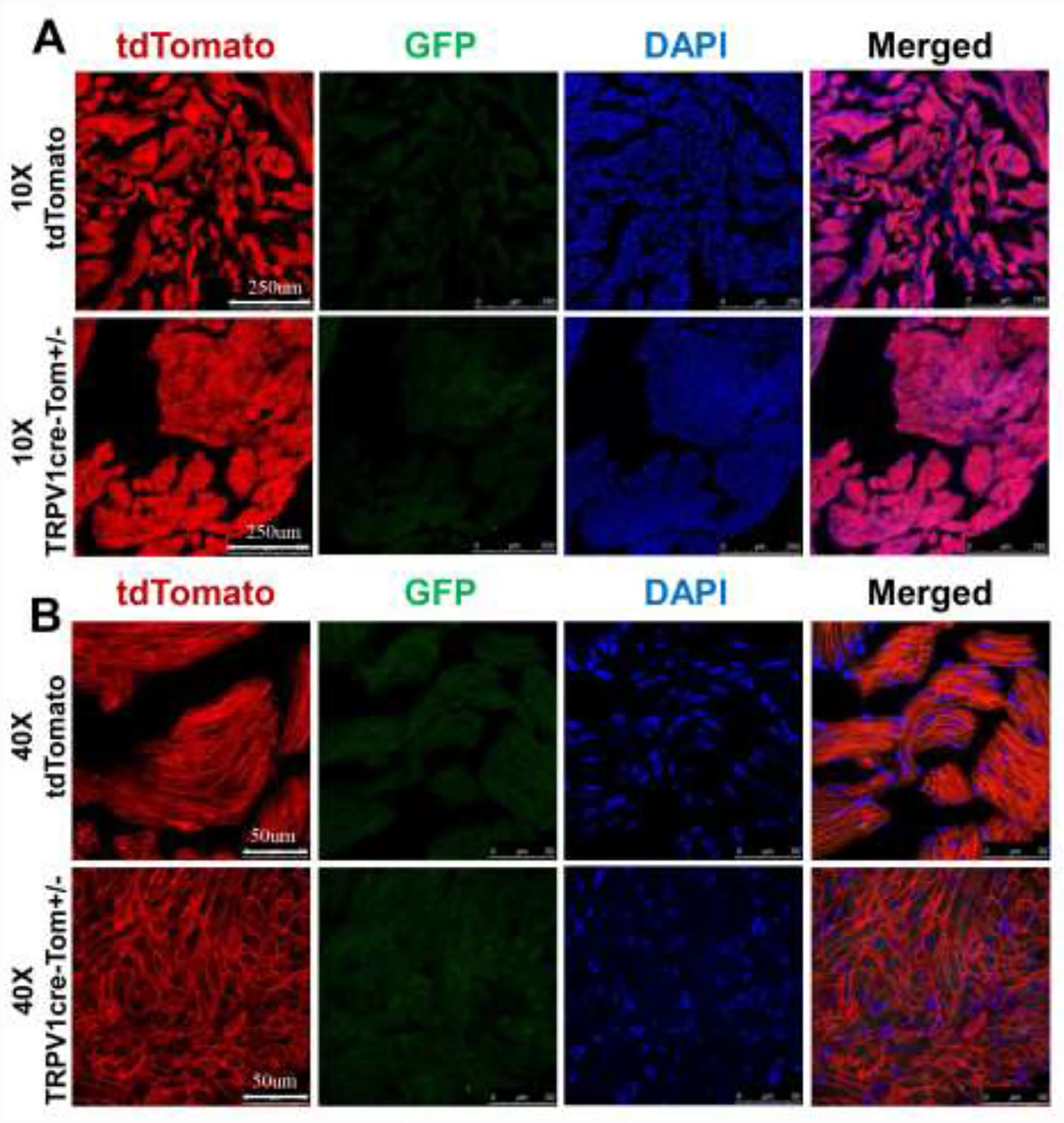
Representative images showing that GFP positive fluorescence is absent in atrial cardiomyocytes under 10x objective (A) and 40x objective (B) in both TRPV1cre-tdTomato+/− (TRPV1cre-Tom+/−) and tdTomato mice. DAPI: 4,6-diamidino-2-phenylindole, a cell nucleus marker. n=5/tdTomato group, n=7/TRPV1cre-tdTomato+/− group.
3.4. GFP signals were expressed in cardiac blood vessels in the TRPV1cre-tdTomato+/− mice
Although we were not able to detect GFP fluorescence in cardiomyocytes, we did observe striated GFP fluorescence running in the extracellular space between ventricular cardiomyocytes in TRPV1cre-tdTomato+/− mice and not in tdTomato control mice (Figure 5A). This circumferential banding pattern is characteristic of blood vessels [12]. The location of GFP fluorescence was restricted to certain small- to medium diameter vessels (50 μm~250 μm). Given their size and location, the vessels are most likely arterioles. By further co-staining with the cardiomyocyte marker Tn I (Figure 5B), we confirmed that the GFP-positive fluorescence observed in the blood vessel-like structures was not co-localized with Tn I-positive cardiomyocytes. However, a small proportion of the blood vessels’ components in the TRPV1cre-tdTomato mice possessed tdTomato fluorescence, indicating that TRPV1 composed only a proportion of blood vessel cells. All blood vessels in the ventricle of tdTomato mice were completely GFP-negative. Overall, our data confirmed that TRPV1 was not expressed in the ventricular cardiomyocytes.
Figure 5.
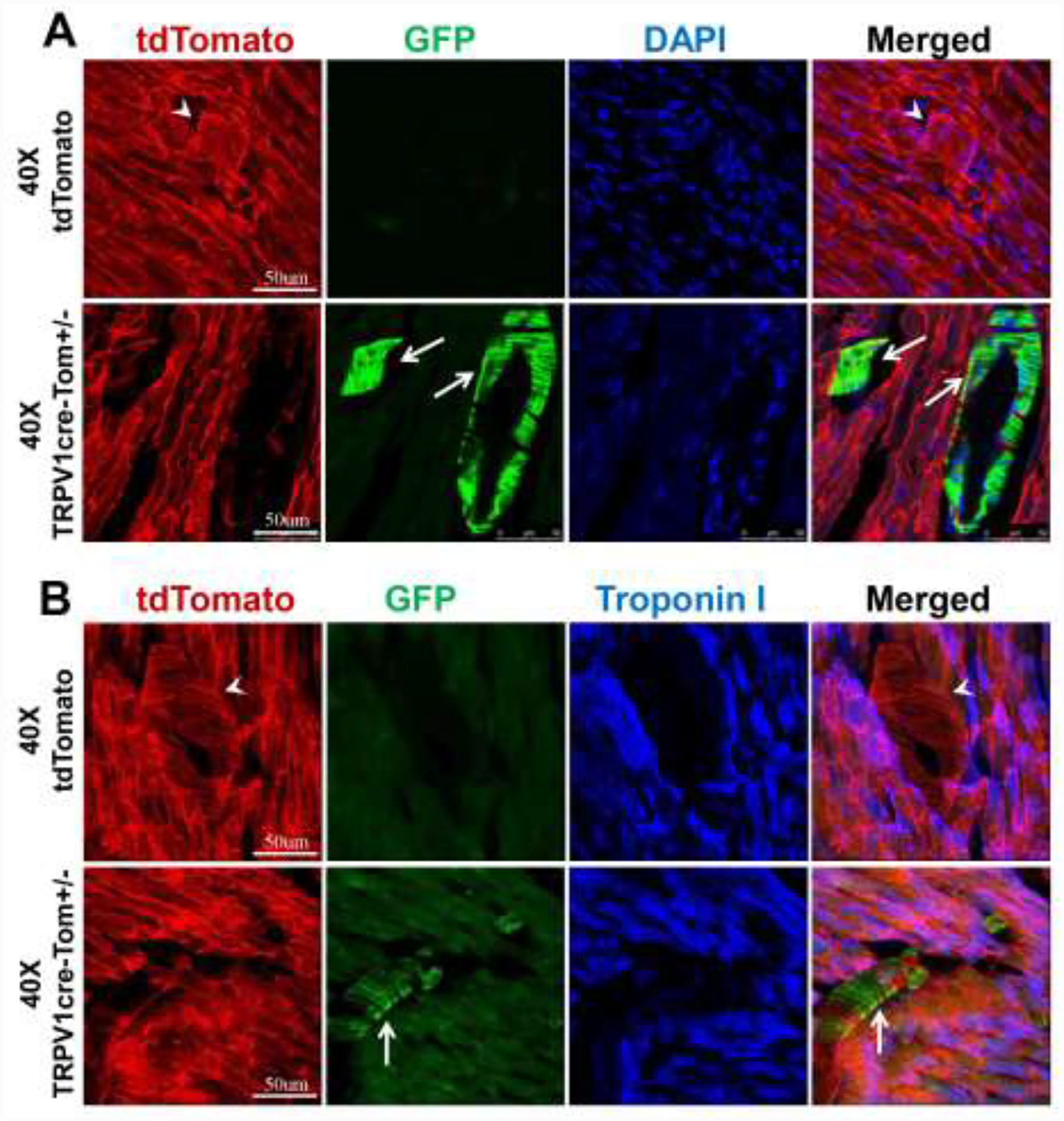
Representative images showing that non-cardiomyocyte structures such as blood vessels are GFP positive under 40x objective (A and B) in the ventricles of TRPV1cre-tdTomato+/− (TRPV1cre-Tom+/−) mice. White arrows point to the GFP-positive blood vessels. DAPI: 4,6-diamidino-2-phenylindole, a cell nucleus marker; Troponin I, a marker for cardiomyocytes. n=5/tdTomato control group, n=7/TRPV1cre-tdTomato+/− group.
3.5. GFP signals were expressed in cardiac nerve endings in the TRPV1cre-tdTomato+/− mice
Since previous evidence has shown that the heart is innervated by TRPV1-expressing afferent nerves and these afferent nerves are essential for the cardiac sympathetic afferent reflex (CSAR) during myocardial ischemia, we immunostained the ventricle slides from both tdTomato and TRPV1cre-tdTomato+/− mice with a nerve endings marker PGP9.5 [2] to examine the TRPV1 afferent innervation in the heart. Figure 6 shows a few thin nerve fibers and small areas positive for PGP9.5 between cardiomyocytes near the epicardial surface in both tdTomato mice and TRPV1cre-tdTomato+/− mice. The tdTomato control mice showed no colocalization of GFP and PGP9.5 positive areas, while the colocalization was observed in the TRPV1cre-tdTomato+/− mice. Collectively, these data suggest that TRPV1 nerve endings are located densely in the epicardial surface, which is consistent with former studies [6, 22].
Figure 6.
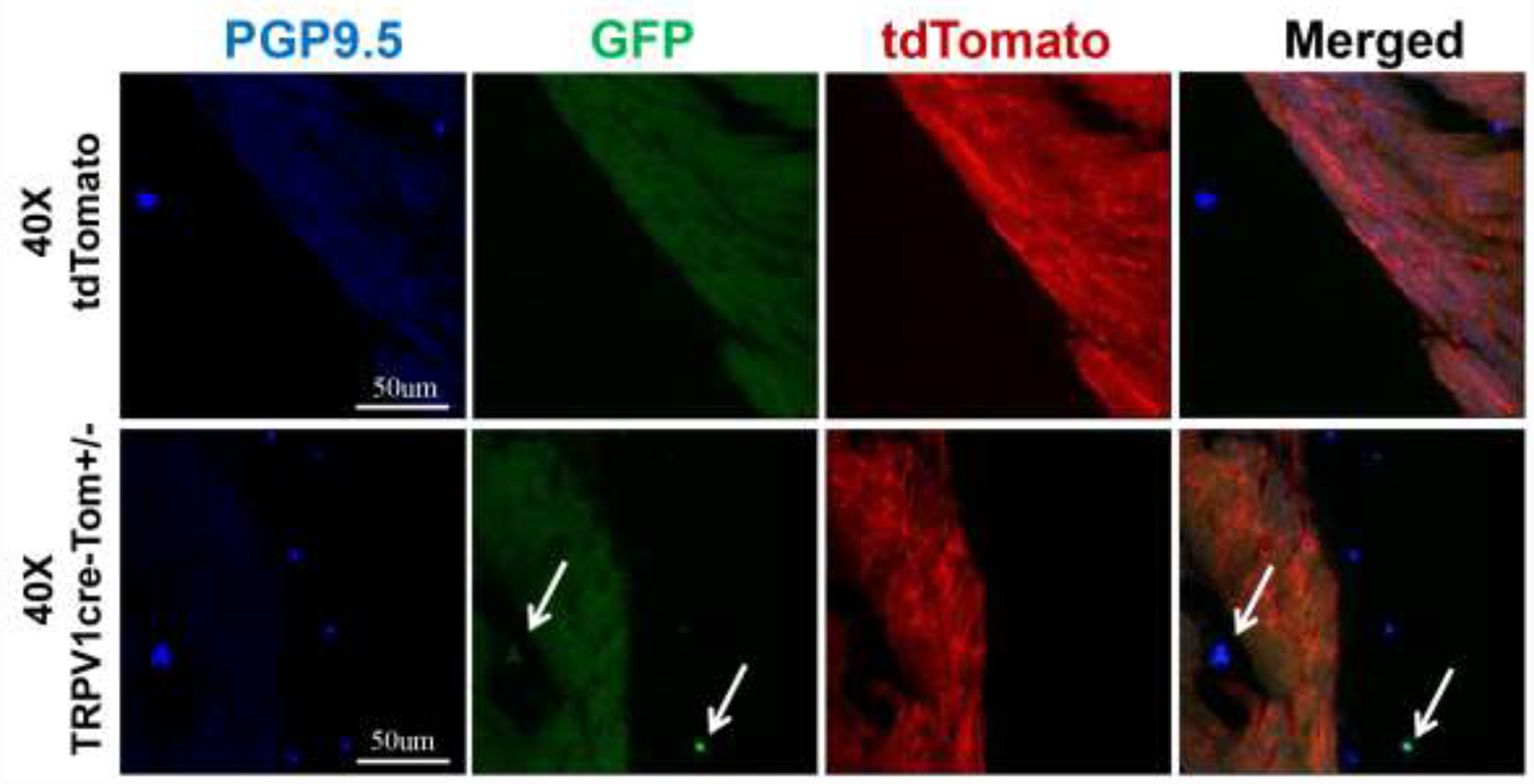
Representative images showing TRPV1 sensory nerve endings are expressed on the ventricular epicardial surface under 40x objective in TRPV1cre-tdTomato+/− (TRPV1cre-Tom+/−) mice. White arrows point to GFP-positive TRPV1 sensory nerve endings. PGP9.5: protein gene product 9.5, a specific marker for neurons and nerve endings. n=5/tdTomato control group, n=7/TRPV1cre-tdTomato+/− group.
4. Discussion
In order to more definitively resolve the disparate issue concerning TRPV1 expression in cardiomyocytes, we utilized a TRPV1cre-tdTomato recombination mouse model using a cre-loxP strategy to visualize TRPV1-positive cells as GFP positive fluorescence. Compared with traditional techniques, this approach has several advantages including enhanced sensitivity and greater signal to background used to localize TRPV1. The primary results include: 1) tdTomato control mice display no GFP-positive neurons in the T1-T4 DRGs, while GFP-positive neurons are widely distributed in the T1-T4 DRGs of TRPV1cre-tdTomato+/− mice and account for 55% of the total primary neuron population. 2) Histological evidence showed that cardiomyocytes in the ventricle were GFP negative in both tdTomato and TRPV1cre-tdTomato+/− mice. Further co-staining with the cardiomyocyte marker, TnI, produced similar results. Atrium mapping of TRPV1cre-tdTomato+/− mice also showed a similar phenomenon. 3) We detected strong GFP fluorescence in some blood vessel-like structures in the TRPV1cre-tdTomato+/− mice. 4) By co-staining with nerve ending marker PGP9.5, we found intense PGP9.5 positive nerve endings near the epicardial surface and GFP positive TRPV1 nerve endings colocalized with these PGP9.5 positive nerve endings in TRPV1cre-tdTomato+/− mice.
Cre expression in our mouse line faithfully corresponds with the expression of endogenous TRPV1 [5]. Persistent visualization of transient gene expression of TRPV1 was allowed after crossing it with tdTomato mice [5]. The observed GFP fluorescence of TRPV1 receptors is further supported by immunofluorescence data performed by other groups. First, GFP positive neurons occupied almost 55% of the total neurons and most were small and medium size C fiber primary afferent neurons, consistent with previous well-established studies [3, 4, 11, 15]. Second, our results showed that TRPV1 was expressed in the sensory nerve endings of the epicardial surface. Zahner et al. reported that TRPV1 receptors revealed an intricate network of thin and tortuous fibers in the myocardium, predominantly near the epicardial surface [22]. Facer et al. further showed that TRPV1 nerve fibers were densely expressed on the human ventricular surface [6]. Third, TRPV1 channels in arteriolar smooth muscle cells and their roles in the regulation of vascular tone has been reported [5, 9, 12], which is consistent with our finding that GFP fluorescence showed a characteristic circumferential banding pattern on blood vessels. In addition, the location of GFP fluorescence was restricted to certain small- to medium diameter vessels (50 μm~250 μm), given their size and location, are most likely arterioles. Future experiments need to be done to confirm whether TRPV1 is restricted to smooth muscle cells or endothelial cells in arterioles. Collectively, our strategy permits high sensitivity and accuracy in the tissue localization of TRPV1 protein.
Discrepancies still exist concerning the TRPV1 expression in cardiomyocytes. Some studies reported dense expression of TRPV1 throughout the endocardium, myocardium and epicardium [1, 7, 8] whereas the others reported very faint or no TRPV1 immunostaining in the myocardium [6, 22]. Such differences may be due to different sensitivities of antibodies used in immunocytochemical approaches or inappropriate signal to noise ratio. Pharmacological strategies were also used to suggest a functional contribution of this channel in isolated cardiomyocytes or fibroblasts [13, 14, 23]. Regarding these studies, it is important to note that antibody specificity for western blotting was not always tested and the purity of cardiomyocytes or fibroblast preparations have not always been reported. Despite using what we consider to be a sensitive technique for detecting TRPV1, we did not detect any GFP fluorescence in ventricular cardiomyocytes of TRPV1cre-tdTomato+/− mice. Although one could argue that the TRPV1 gene might be expressed in cardiomyocytes at very low levels or in a specific cellular compartment so that GFP signals were too low to detect, our current data does not support this scenario.
Compared with traditional immunofluorescence procedures, the cre-loxP recombination genetic approach clearly provides a better resolution for detecting the expression pattern of the TRPV1 receptor, in both the DRGs and in the heart. Since cre recombinase was able to specifically delete DNA sequences of the tdTomato-stop codon that are flanked by two loxP sites, which subsequently allowed for GFP-green visualization of TRPV1 expressing cells, the GFP and tdTomato fluorescence would never colocalize. For instance, our data show that GFP positive-TRPV1 primary afferent neurons, which were expressed throughout the T1-T4 DRGs, were devoid of tdTomato fluorescence in T1-T4 DRGs of TRPV1cre-tdTomato+/− mice. Moreover, TRPV1-positive neurons were largely restricted to the small and medium diameter C fibers. GFP-positive blood vessels in the extracellular space of ventricular cardiomyocytes in TRPV1cre-tdTomato+/− mice were never found to be associated with cardiomyocytes that were immunoassayed for the cardiomyocyte marker Tn I and never colocalized with tdTomato fluorescence. The greater signal-to-noise ratio and minimal background using TRPV1cre-tdTomato+/− mice is particularly advantageous compared to studies using TRPV1 antibodies. Thus, our data unequivocally demonstrates that, throughout the adult mouse left ventricle and atrium, TRPV1 expression was not detected within any cardiomyocyte. One potential limitation of this study is that only male mice were used to detect the distribution of TRPV1 in cardiomyocytes and non-cardiomyocytes in the heart. A potential sex difference in the expression pattern of TRPV1 in heart need to be identified in the future study.
Pharmacological strategies were used to suggest a functional contribution of this channel in CHF. The cardiac afferent fibers expressing TRPV1 play an important role in the CSAR [22], which is activated by agonists applied to the epicardial surface and contributes to elevated sympathetic tone in CHF [17–20]. A previous study from our lab demonstrated that chronic and selective cardiac sympathetic afferent denervation by epicardial application of the potent TRPV1 receptor agonist RTX markedly reduced the CSAR-evoked sympatho-excitation and had beneficial effects on cardiac remodeling and dysfunction in CHF [16]. A study in pigs also showed that percutaneous epicardial depletion of TRPV1 sensory afferents with RTX was a viable upstream target to mitigate chronic cardiac sympatho-excitation and its deleterious effects after myocardial infarction [21]. These studies indicate that selective cardiac sympathetic afferent deafferentation by RTX by application on the epicardial surface may offer a clinical benefit to improve cardiac and autonomic function in CHF. However, the rationale for epicardial RTX application is based on the fact that a majority of TRPV1 protein is expressed in cardiac spinal afferent nerve fibers on the surface or superficial layers of the ventricles but not in the cardiomyocytes. If TRPV1 was expressed in cardiomyocytes, toxic irreversible activation of TRPV1 by RTX in cardiomyocytes would worsen cardiac function after myocardial infarction, instead of improving it. Here, our data clearly demonstrate that TRPV1 was not detected in cardiomyocytes. Potential cardiomyocyte side effects of RTX treatment for CHF would be minimal. Moreover, TRPV1 receptors compose a large proportion of the nerve endings near the epicardial surface, which further supports the beneficial effect of functional ablation of epicardial TRPV1 nerve endings in CHF. Future experiments are needed to confirm that there is no functional TRPV1 expression by measuring the sensitivity of isolated ventricular cardiomyocytes to highly selective pharmacological agonists/antagonists.
5. Conclusion
In conclusion, our study reveals no expression of TRPV1 channels in ventricular cardiomyocytes. We propose that the absence of TRPV1 expression in cardiomyocytes may contribute to the beneficial effects of application of the potent TRPV1 agonist, RTX and its use as a potential clinical intervention in CHF.
Figure 7.
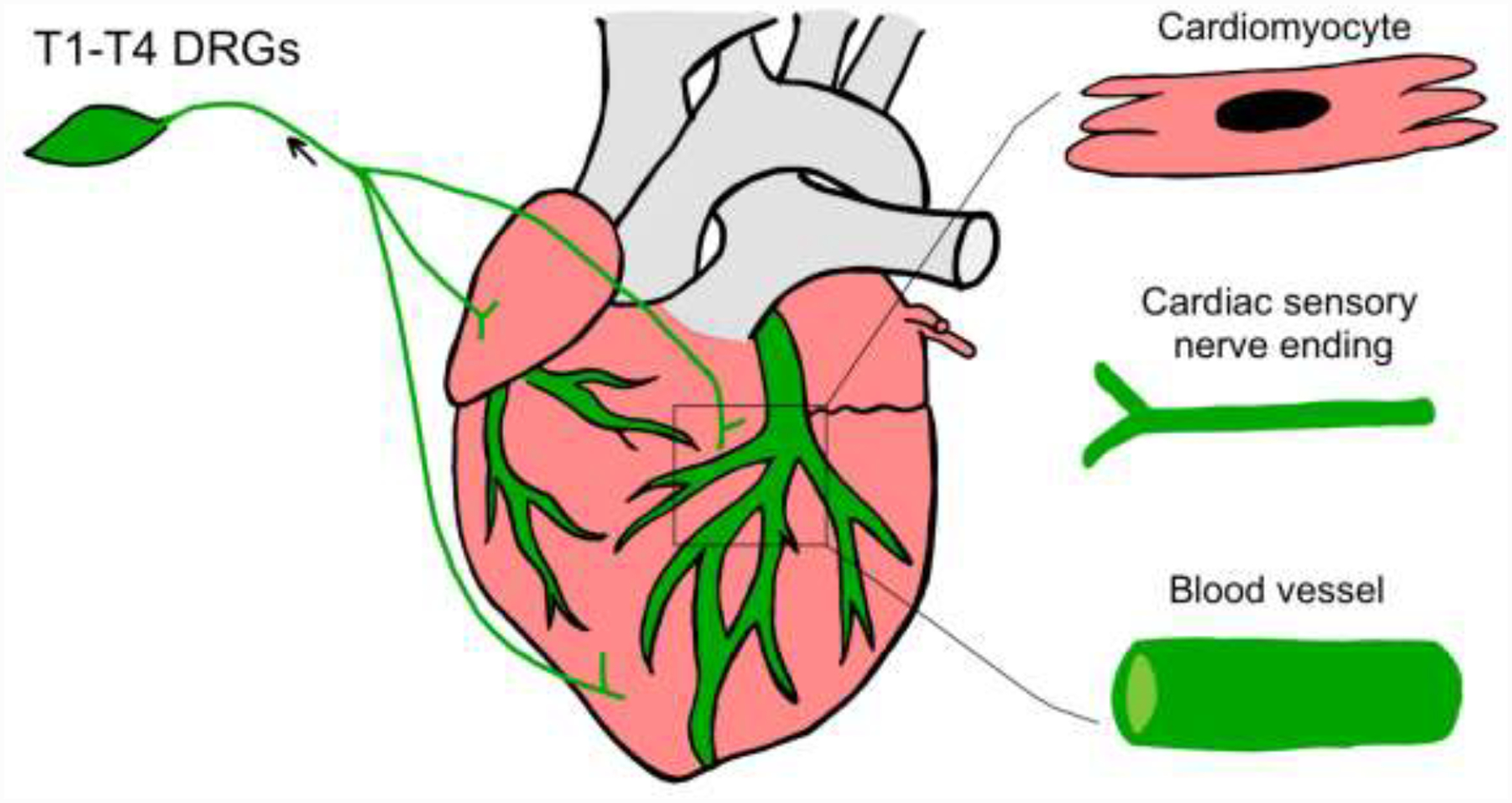
A schematic diagram showing cardiac expression pattern of TRPV1 receptors using a Transgenic Reporter Mouse Model. Green color in T1-T4 DRGs, cardiac nerve endings and blood vessels indicate a TRPV1-positive innervation.
Highlights.
The TRPV1cre-tdTomato reporter mouse model was sensitive to detect the TRPV1 cells.
The reporter mouse model suggested that TRPV1 channels were not expressed in cardiomyocytes.
TRPV1 channels were expressed in cardiac blood vessels and cardiac nerve endings.
This study has important implications for studying the TRPV1 channels in heart.
Funding Statement
This study was supported by NIH grant 1R01 HL-152160 and partially supported by 2R01 HL126796 and 1R01 HL-121012. Dr. Hanjun Wang is also supported by Margaret R. Larson Professorship in Anesthesiology. Dr. Irving H. Zucker is supported in part by the Theodore F. Hubbard Professorship for Cardiovascular Research.
Abbreviations:
- TRPV1
Transient receptor potential vanilloid type 1
- RTX
reseniferitoxin
- CSAR
cardiac sympathetic afferent reflex
- DRG
dorsal root ganglia
- CHF
chronic heart failure
- NF200
Neurofilament
- TnI
Troponin I
- PGP9.5
protein gene product 9.5
- GFP
green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no financial conflict of interest.
References
- [1].Andrei SR, Sinharoy P, Bratz IN, Damron DS, TRPA1 is functionally co-expressed with TRPV1 in cardiac muscle: Co-localization at z-discs, costameres and intercalated discs, Channels 10 (2016) 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brady CM, Apostolidis AN, Harper M, Yiangou Y, Beckett A, Jacques TS, Freeman A, Scaravilli F, Fowler CJ, Anand P, Parallel changes in bladder suburothelial vanilloid receptor TRPV1 and pan-neuronal marker PGP9.5 immunoreactivity in patients with neurogenic detrusor overactivity after intravesical resiniferatoxin treatment, BJU international 93 (2004) 770–776. [DOI] [PubMed] [Google Scholar]
- [3].Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, Julius D, Impaired nociception and pain sensation in mice lacking the capsaicin receptor, Science 288 (2000) 306–313. [DOI] [PubMed] [Google Scholar]
- [4].Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D, The capsaicin receptor: a heat-activated ion channel in the pain pathway, Nature 389 (1997) 816–824. [DOI] [PubMed] [Google Scholar]
- [5].Cavanaugh DJ, Chesler AT, Jackson AC, Sigal YM, Yamanaka H, Grant R, O’Donnell D, Nicoll RA, Shah NM, Julius D, Basbaum AI, Trpv1 reporter mice reveal highly restricted brain distribution and functional expression in arteriolar smooth muscle cells, The Journal of neuroscience : the official journal of the Society for Neuroscience 31 (2011) 5067–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Facer P, Punjabi PP, Abrari A, Kaba RA, Severs NJ, Chambers J, Kooner JS, Anand P, Localisation of SCN10A gene product Na(v)1.8 and novel pain-related ion channels in human heart, International heart journal 52 (2011) 146–152. [DOI] [PubMed] [Google Scholar]
- [7].Gao F, Liang Y, Wang X, Lu Z, Li L, Zhu S, Liu D, Yan Z, Zhu Z, TRPV1 Activation Attenuates High-Salt Diet-Induced Cardiac Hypertrophy and Fibrosis through PPAR-delta Upregulation, PPAR research 2014 (2014) 491963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hurt CM, Lu Y, Stary CM, Piplani H, Small BA, Urban TJ, Qvit N, Gross GJ, Mochly-Rosen D, Gross ER, Transient Receptor Potential Vanilloid 1 Regulates Mitochondrial Membrane Potential and Myocardial Reperfusion Injury, Journal of the American Heart Association 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kark T, Bagi Z, Lizanecz E, Pasztor ET, Erdei N, Czikora A, Papp Z, Edes I, Porszasz R, Toth A, Tissue-specific regulation of microvascular diameter: opposite functional roles of neuronal and smooth muscle located vanilloid receptor-1, Molecular pharmacology 73 (2008) 1405–1412. [DOI] [PubMed] [Google Scholar]
- [10].Maynard SJ, Menown IB, Adgey AA, Troponin T or troponin I as cardiac markers in ischaemic heart disease, Heart 83 (2000) 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mickle AD, Shepherd AJ, Mohapatra DP, Sensory TRP channels: the key transducers of nociception and pain, Progress in molecular biology and translational science 131 (2015) 73–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Phan TX, Ton HT, Chen Y, Basha ME, Ahern GP, Sex-dependent expression of TRPV1 in bladder arterioles, American journal of physiology. Renal physiology 311 (2016) F1063–F1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Qi Y, Qi Z, Li Z, Wong CK, So C, Lo IC, Huang Y, Yao X, Tsang SY, Role of TRPV1 in the Differentiation of Mouse Embryonic Stem Cells into Cardiomyocytes, PloS one 10 (2015) e0133211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun Z, Han J, Zhao W, Zhang Y, Wang S, Ye L, Liu T, Zheng L, TRPV1 activation exacerbates hypoxia/reoxygenation-induced apoptosis in H9C2 cells via calcium overload and mitochondrial dysfunction, International journal of molecular sciences 15 (2014) 18362–18380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, Julius D, The cloned capsaicin receptor integrates multiple pain-producing stimuli, Neuron 21 (1998) 531–543. [DOI] [PubMed] [Google Scholar]
- [16].Wang HJ, Wang W, Cornish KG, Rozanski GJ, Zucker IH, Cardiac sympathetic afferent denervation attenuates cardiac remodeling and improves cardiovascular dysfunction in rats with heart failure, Hypertension 64 (2014) 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang W, Schultz HD, Ma R, Cardiac sympathetic afferent sensitivity is enhanced in heart failure, The American journal of physiology 277 (1999) H812–817. [DOI] [PubMed] [Google Scholar]
- [18].Wang W, Zucker IH, Cardiac sympathetic afferent reflex in dogs with congestive heart failure, The American journal of physiology 271 (1996) R751–756. [DOI] [PubMed] [Google Scholar]
- [19].Wang WZ, Gao L, Pan YX, Zucker IH, Wang W, AT1 receptors in the nucleus tractus solitarii mediate the interaction between the baroreflex and the cardiac sympathetic afferent reflex in anesthetized rats, American journal of physiology. Regulatory, integrative and comparative physiology 292 (2007) R1137–1145. [DOI] [PubMed] [Google Scholar]
- [20].Wang WZ, Gao L, Wang HJ, Zucker IH, Wang W, Interaction between cardiac sympathetic afferent reflex and chemoreflex is mediated by the NTS AT1 receptors in heart failure, American journal of physiology. Heart and circulatory physiology 295 (2008) H1216–H1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yoshie K, Rajendran PS, Massoud L, Mistry J, Swid MA, Wu X, Sallam T, Zhang R, Goldhaber JI, Salavatian S, Ajijola OA, Cardiac TRPV1 afferent signaling promotes arrhythmogenic ventricular remodeling after myocardial infarction, JCI insight 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zahner MR, Li DP, Chen SR, Pan HL, Cardiac vanilloid receptor 1-expressing afferent nerves and their role in the cardiogenic sympathetic reflex in rats, The Journal of physiology 551 (2003) 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang Y, Li L, Hua Y, Nunn JM, Dong F, Yanagisawa M, Ren J, Cardiac-specific knockout of ET(A) receptor mitigates low ambient temperature-induced cardiac hypertrophy and contractile dysfunction, Journal of molecular cell biology 4 (2012) 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]


