Abstract
Background:
Lung cancer remains the leading cause of cancer death worldwide with 15–20% occurring in never-smokers. To assess genetic determinants for prognosis among never smokers, we conducted a genome-wide investigation in the International Lung Cancer Consortium(ILCCO).
Methods:
Genomic and clinical data from 1569 never-smoking lung cancer patients of European ancestry from 10 ILCCO studies were included. Hazard ratios(HRs) and 95% confidence intervals of overall survival were estimated. We assessed whether the associations were mediated through mRNA expression based 1553 normal lung tissues from the Lung expression quantitative trait loci(eQTL) dataset and GTEx. For cross-ethnicity generalization, we assessed the associations in a Japanese study(N=887).
Results:
One locus at 13q22.2 was associated with lung adenocarcinoma survival at genome-wide level, with carriers of rs12875562-T allele exhibiting poor prognosis(HR=1.71(1.41–2.07), p=3.60×10−8), and altered mRNA expression of LMO7DN in lung tissue(GTEx, p=9.40×10−7; Lung eQTL dataset, p=0.003). Furthermore, two of 11 independent loci that reached the suggestive significance level(p<10−6) were significant eQTL affecting mRNA expression of nearby gene in lung tissues, including CAPZB at 1p36.13 and UBAC1 at 9q34.3. One locus encoding NWD2/KIAA1239 at 4p14 showed associations in both European(HR=0.50(0.38–0.66), p=6.92×10−7) and Japanese populations(HR=0.79(0.67–0.94), p=0.007).
Conclusions:
Based on the largest genomic investigation on the lung cancer prognosis of never smokers to date, we observed that lung cancer prognosis is affected by inherited genetic variants.
Impact:
We identified one locus near LMO7DN at genome-wide level and several potential prognostic genes with cis-effect on mRNA expression. Further functional genomics work is required to understand their role in tumor progression.
Keywords: Never smokers, LMO7DN, eQTL, Genetics of Cancer Risk and Outcome, Lung Cancer
INTRODUCTION
With over 1 million deaths each year, lung cancer continues to be the leading cause of cancer mortality worldwide, and the five-year survival rate remains low at only 10 to 20%(1,2). While it is well established that tobacco smoking is the primary cause of lung cancer, inherited genetic variations has also been established as etiological factors through genome-wide association studies (GWAS), which identified susceptibility loci including CHRNA3/5, TERT-CLPTM1L, the HLA/MHC region, CHEK2 and more in the last decade(3–9).
Approximately 15 to 20% of lung cancer cases occur in individuals who are lifelong never smokers (10,11). Many studies have shown significant differences in the etiology and clinical characteristics between never and ever smokers, and lung cancer in never smokers is being recognized as a distinct disease entity. Most notably, smokers and never smokers have different histological presentation with adenocarcinoma being the main histological type among never smoking patients (10), and never smokers have a higher prevalence of EGFR mutations and those with EGFR mutations show longer survival after treatment with EGFR inhibitors than ever smokers do. Additional features that distinguish lung cancer in never smokers and ever smokers are differences in their somatic mutations and methylation profiles (12,13).
Inherited genetic variation has been hypothesized to influence lung cancer survival and several genome-wide association studies (GWAS) were performed with a focus on overall survival (14); in early stage lung cancer patients (15,16), patients who received platinum based chemotherapies (17–19), and advanced non-small cell lung cancer (20,21), although most studies have relatively modest sample sizes ranging from 100 to 400 lung cancer patients. Moreover, we hypothesize that there are distinctive genetic factors contribute to lung cancer prognosis in smokers and never smokers, and analyzing never smokers separately would provide a greater insight on the genetic components of lung cancer survival for this specific population. To increase our power for genomic discovery, we conducted a meta-analysis of ten GWAS with clinical prognosis data based on a total of 1569 never-smoking lung cancer patients of European ancestry in a two-stage analysis. The generalizability of the candidate association across ethnicity was tested in the Japanese non-smoking population in the second stage. The potential functional significance of the genetic regions related to prognosis was investigated using expression quantitative trait loci (eQTL) analysis based on four independent studies from the Universities of Laval, University of British Columbia and Groningen, and the Genotype-Tissue Expression (GTEx) data (22,23).
MATERIALS and METHODS
Description of participating studies
A total of 12 studies in the International Lung Cancer Consortium (ILCCO) participated in this analysis, including 10 lung cancer GWAS of European populations and 2 studies in Japanese populations to asses for generalizability. Never smokers were defined as individuals who smoked fewer than 100 cigarettes during their lifetime, with the exception of Liverpool Lung Project in which the definition was individuals who smoked 10 cigarettes per week regularly (among those 98.5% also fit under the former definition). All participants provided written informed consent and research protocols of all studies were reviewed and approved by the local institutional review boards of each participating study. Information of each study is summarized in Table 1, and included in the Supplementary Materials.
Table 1.
Patient characteristics of the participating studies with European and Asian ancestry after quality control filters
| European ancestry | Asian ancestry Japan | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OncoArray | Total | |||||||||||
| Characteristics No (%) | Toronto | MDACC | Mayo Clinic | Harvard | Central Europe | MSH-PMH | HLCS | CAPUA | MDACC | LLP | ||
| Subtotal | 208 | 327 | 349 | 92 | 59 | 88 | 240 | 42 | 71 | 93 | 1569 | 887 |
| MFT** | 101 | 87 | 126 | 101.9 | 124 | 38.4 | 83 | 72.7 | 25.6 | 84.8 | 86.8 | 56.8 |
| MST* | 20.1 | 27.7 | 54.3 | 49.6 | 13 | 87 | 53.5 | 10.09 | 42 | 14.7 | 34.6 | 23.9 |
| Vital status | ||||||||||||
| Dead | 146 (70) | 217 (66) | 219 (63) | 54 (59) | 53 (90) | 31 (35) | 143 (60) | 41 (98) | 25 (35) | 70 (75) | 999 (65) | 326 (37) |
| Alive | 62 (30) | 110 (34) | 130 (37) | 38 (41) | 6 (10) | 57 (64) | 97 (40) | 1 (2) | 46 (65) | 23 (25) | 570 (35) | 561 (63) |
| Age, | 62.4 (12.6) | 61.4 (13.4) | 62.1 (13.4) | 64.2 (12.9) | 63.9 (8.5) | 65.4 (12.4) | 61.8 (11.9) | 70.1 (9.3) | 60.7 (11.1) | 67.2 (8.5) | 62.8 (12.5) | 59.1 (9.1) |
| Mean (SD) | ||||||||||||
| Gender | ||||||||||||
| Male | 69 (33) | 104 (32) | 117 (34) | 61 (66) | 11 (19) | 20 (23) | 82 (34) | 6 (14) | 31 (44) | 57 (61) | 558 (36) | 133 (15) |
| Female | 139 (67) | 223 (68) | 232 (66) | 31 (34) | 48 (81) | 68 (77) | 158 (66) | 36 (86) | 40 (56) | 36 (39) | 1012 (64) | 754 (85) |
| Clinical stage | ||||||||||||
| I-IIIA | 74 (36) | 183 (56) | 244 (70) | 55 (60) | 10 (42) | 50 (57) | 112 (48) | 10 (26) | 19 (28) | 43 (60) | 804(53) | 638 (72) |
| IIIB-IV | 134 (64) | 144 (44) | 105 (30) | 37 (40) | 14 (58) | 37 (43) | 122 (52) | 28 (74) | 49 (72) | 29 (40) | 695(46) | 249 (28) |
| Histology | ||||||||||||
| Adenocarcinoma | 138 (69) | 244 (75) | 237 (68) | 57 (62) | 30 (51) | 79 (90) | 149 (62) | 27 (64) | 66 (93) | 38 (41) | 1065(68) | 871 (98) |
| Squamous cell | 14 (7) | 26 (8) | 16 (5) | 7 (8) | 10 (17) | 5 (5) | 9 (4) | 5 (12) | 1 (1) | 36 (39) | 129(8) | 10 (1) |
| Other | 48 (24) | 57 (17) | 96 (27) | 28 (30) | 19 (32) | 4 (6) | 82 (34) | 10 (24) | 4 (6) | 19 (20) | 367(24) | 6 (1) |
| Surgery | ||||||||||||
| Yes | 67 (32) | 135 (41) | 131 (38) | 50 (54) | n/a | n/a | n/a | n/a | n/a | n/a | 383 (41) | 639 (71) |
| No | 141 (68) | 192 (59) | 218 (62) | 42 (46) | n/a | n/a | n/a | n/a | n/a | n/a | 551 (59) | 248 (28) |
| Chemotherapy | ||||||||||||
| Yes | 107 (51) | 197 (60) | 107 (31) | 37 (40) | 7 (12) | n/a | n/a | n/a | n/a | n/a | 455 (48) | 222 (25) |
| No | 101 (49) | 130 (40) | 242 (69) | 55 (60) | 23 (39) | n/a | n/a | n/a | n/a | n/a | 496 (52) | 665 (75) |
| Radiation | ||||||||||||
| Yes | 89 (43) | 88 (27) | 17 (5) | 24 (26) | 17 (29) | n/a | n/a | n/a | n/a | n/a | 235 (25) | 26 (3) |
| No | 119 (57) | 239 (73) | 332 (95) | 68 (74) | 13 (22) | n/a | n/a | n/a | n/a | n/a | 703 (75) | 861 (97) |
MFT = median follow-up time, months
MST = median survival time, months; values in parentheses represent percentages based on column totals.
Genotyping and Imputation
Genotyping in each study was conducted using Illumina HumanHap300K, 370K, 610K, 660K, OmniExpress or OncoArray. In general, the quality control procedures were similar across studies with exclusion of variants based on low call rate (<90%), and low minor allele frequency (<1%). Individuals with high missing rate (>5 or 10%), gender discrepancies, unexpected duplicates or relatedness were excluded. Details of genotyping and quality control procedures as applied to the lung cancer OncoArray project have been previously published (24). After applying quality control steps and restricting to genotyped individuals of European ancestry with no smoking history and complete clinical follow-up information, data were available on a total of 1,569 never smoking lung cancer patients, including 208 from Toronto, 327 from MDACC, 349 from Mayo, 59 from Central Europe, 92 from Harvard study and 534 in the five studies genotyped in the OncoArray project. The key characteristics of all participating studies are summarized in Table 1.
To facilitate the meta-analysis across genotyping platforms, genotype imputation was conducted in each study based on the March 2012 release of the 1000-Genomes Project. The Toronto and Mayo Clinic studies were imputed using IMPUTE2 (25–27), and the IARC-Central Europe and Harvard studies were imputed using MaCH software (28). Variants that were not present in any genotyping array, or with sub-optimal imputation quality were excluded from the analysis based on IMPUTE2 Info < 0.3 and MACH RSQR < 0.3. After applying quality control filters, 629,283 SNPs were available for the meta-analysis. For the Japanese GWAS study, the 887 lung cancer patients from National Cancer Center Hospital and Aichi Cancer Centre were genotyped using Illumina HumanOmini1-Quad and Illumina 660W. The Japanese study was imputed using IMPUTE2. Standard quality control steps applied to remove potential errors and biases have been previously described (29). Briefly, individuals with gender discrepancies, low call rates (<98%) and first-degree relatives were excluded, and variants with Hardy-Weinberg Equilibrium (P < 10−6) were removed.
Statistical analysis
Study-specific analysis of GWAS data
Overall survival time was defined as the time from date of lung cancer diagnosis to date of death or the last known date alive. Cox proportional-hazards model was applied to assess marginal effects of patient characteristics on lung cancer survival. Genomic inflation factor was estimated by comparing observed and expected p-values. Quantile-quantile (Q-Q) plots were used to assess the extent to which the observed distribution of the test statistic follows the expected distribution for each study. OncoArray project data were pooled and analysed as one study as they were all genotyped and processed at the same time. The analytical process was summarized in Supplementary Figure 1.
For each variant that passed QC procedures as described, multivariate Cox proportional hazards regression was used to assess the association of lung cancer survival within each study. The probabilistic genotype dosage model was used for the main analysis and included potential confounding factors that might influence patient survival including age (as a continuous variable), sex (male or female), clinical stage (IA-IIIA, IIIB-IV), and where available, treatment information. To limit inflation of the calculated test-statistics due to population sub-structure, each study was independently adjusted by the top two to six principal components (PCs) (30). The Japanese studies were adjusted by the top five PCs. Hazard Ratios (HR) and their corresponding 95% confidence intervals (95% CI) for survival were computed based on cox regression models.
Survival rates were estimated using the Kaplan-Meier method and median survival times were calculated based on diagnosis and death dates. Log-rank tests were used to examine for differences between survival estimates of genotypes pooled across studies.
Meta-Analysis of GWAS data
A fixed-effects meta-analysis was performed to combine study-specific hazard ratios (HR) of sequence variants using an inverse variance-based weighting method implemented in the METAL program (31). The combined estimates were only computed for those variants observed in at least three studies. I2 statistic was calculated to assess the proportion of the total variation due to heterogeneity, and I2 > 75% and PHET < 0.05 were applied to filter out variants with high study heterogeneity (32).
Given the biological heterogeneity across lung cancer histological types, we have also conducted additional analysis restricted to 1,065 adenocarcinoma patients, as this is the predominant histological type of never smokers. We did not consider a subgroup analysis for other histology types due to small sample size. For genetic variants with p-value of less than 10−6, we assessed the generalizability based on the Japanese study. All statistical tests were two-sided.
Functional Significance
Genetic variants with combined p-value less than 10−6 for lung cancer survival were followed up for potential functional significance through an expression quantitative trait loci (eQTL) investigation based on the Lung eQTL dataset, which includes 3 independent studies and GTEx data. All data sources have been described previously (22,23). Briefly for the Lung eQTL dataset, whole-genome gene expression profiling in the lung was performed on a custom Affymetrix array (GPL10379). Microarray pre-processing and quality controls were conducted as previously described (22). Genotyping was carried on the Illumina Human 1M-Duo BeadChip array. Only cis-eQTL were considered in this study, testing probe sets located within 1 Mb up and downstream of the SNPs associated with lung cancer survival. Genotypes and gene expression were available in a total of 1038 individuals including 409 from Laval University, 287 patients from the University of British Columbia (UBC), and 342 from University of Groningen(33). Association tests were carried in each cohort and then meta-analyzed using Fisher’s method. Expression QTL analyses were performed adjusted for age, sex and smoking status. In addition, the Genotype-Tissue Expression (GTEx) database (http://www.gtexportal.org/home/) of RNAseq analysis was queried to examine the functional association between candidate variants and expressions of nearby genes in 515 human lung tissues (Release V7).
RESULTS
Study Population Characteristics
The baseline characteristics of 1,569 never-smoking lung cancer patients with European ancestry and 887 Asian lung cancer patients from Japan are shown in Table 1. All studies had similar age distribution with mean age at diagnosis of approximately 62 years across all studies with European ancestry. As expected, approximately two-thirds of the patients were females, and lung adenocarcinoma was the primary histological type in all studies. Median follow-up time (MFT) ranged from 26 months in the MDACC-OncoArray study to 126 months in the Mayo Clinic study. Overall, 53% of patients were diagnosed with localized stage (I-IIIA) and the remaining 47% with advanced stage (IIIB-IV). The association between key patient characteristics and survival is shown in Supplementary Table 1. As expected, clinical stage is the most prominent factor associated with survival. Treatment information was available in five of the studies as surgery, chemotherapy or radiation.
Genetic variants associated with lung cancer survival
A total of 629,283 single nucleotide variants (SNVs) were included in the combined analysis after quality control filtering procedures previously described. The distribution of the bottom 95% of P-values was similar to the expected distribution, and the genomic control parameter was 1.02 based on the combined analysis. The associations between genetic variants and lung cancer overall survival for all lung cancer and adenocarcinoma patient across chromosomes are shown in Manhattan plots (Supplementary Figure 2). The main findings for overall survival among all lung cancer patients and adenocarcinoma patients are summarized in Table 2. For lung cancer overall, no regions reached genome-wide significance and four variants at 1p22.3, 8q2.3, 9q31.3 and 10p14 were associated with overall survival at p-value less than 10−6 (Supplementary Figure 3A–3D).
Table 2.
The associations of sentinel variants representing each locus and overall survival of all lung cancer and adenocarcinoma
| European ancestry | Japanese ancestry | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Lead SNP | Gene¥ | Effect | Ref | MAF | HR* (95% CI) | P | I2 | P(HET) | MAF | HR* (95% CI) | P |
| Lung cancer | ||||||||||||
| 1p22.3 | rs2911600 | LINC01461 | T | C | 0.033 | 2.02(1.53,2.66) | 6.41E-07 | 36.2 | 0.180 | 0.066 | 0.88 (0.64,1.21) | 0.438 |
| 8q23.3 | rs7822185 | LINC00536 | A | G | 0.035 | 2.71(1.83,4.00) | 6.45E-07 | 49.7 | 0.113 | 0.063 | 1.05 (0.77,1.45) | 0.729 |
| 9q31.3 | rs4978466 | Intergenic | A | G | 0.292 | 0.76(0.69,0.85) | 4.78E-07 | 38.6 | 0.164 | 0.083 | 0.99 (0.74,1.32) | 0.976 |
| 10p14 | rs17143938 | Intergenic | T | C | 0.058 | 0.58(0.47,0.72) | 5.99E-07 | 0 | 0.714 | n/a | n/a | n/a |
| Adenocarcinoma | ||||||||||||
| 1p36.13 | rs214346 | CAPZB | A | G | 0.424 | 0.72(0.63,0.82) | 5.86E-07 | 29.5 | 0.225 | 0.366 | 1.01 (0.85,1.19) | 0.900 |
| 4p14 | rs17603438 | NWD2/KIAA1239 | A | G | 0.100 | 0.50(0.38,0.66) | 6.92E-07 | 0 | 0.588 | 0.452 | 0.79 (0.67,0.94) | 0.007 |
| 5q11.2 | rs10041935 | Intergenic | A | C | 0.267 | 0.73(0.65,0.83) | 6.84E-07 | 38.2 | 0.152 | 0.275 | 0.89 (0.75,1.06) | 0.199 |
| 7p21.3 | rs10247578 | Intergenic | A | G | 0.403 | 1.38(1.23,1.55) | 7.69E-08 | 32.7 | 0.191 | 0.203 | 0.88 (0.72,1.09) | 0.258 |
| 9q34.3 | rs6569 | UBAC1 | A | C | 0.481 | 0.72(0.63,0.82) | 5.94E-07 | 0 | 0.991 | 0.489 | 1.00 (0.87,1.16) | 0.922 |
| 10p15.3 | rs17158233 | Intergenic | A | G | 0.024 | 3.68(2.23,6.09) | 3.66E-07 | 0 | 0.628 | 0.090 | 1.22 (0.96,1.56) | 0.100 |
| 11q14.1 | rs17148028 | DLG2 | T | G | 0.061 | 0.48(0.36,0.64) | 5.66E-07 | 0 | 0.968 | 0.125 | 1.26 (0.97,1.65) | 0.081 |
| 13q22.2 | rs12875562 | Intergenic | T | C | 0.122 | 1.71(1.41,2.07) | 3.60E-08 | 0 | 0.959 | 0.112 | 1.11(0.87,1.42) | 0.372 |
Hazard ratios (HRs) are from a meta-analysis of Cox-PH models adjusted by age, sex, clinical stage, principal components and when available by treatment. Ref: reference allele; Effect: the allele being evaluated; MAF : Minor allele frequency of the single nucleotide polymorphism; P : P-value; I2 : the I-squared statistic described the percentage of variation across studies that is due to heterogeneity; P(HET): the p-value for heterogeneity.
Host or closest gene
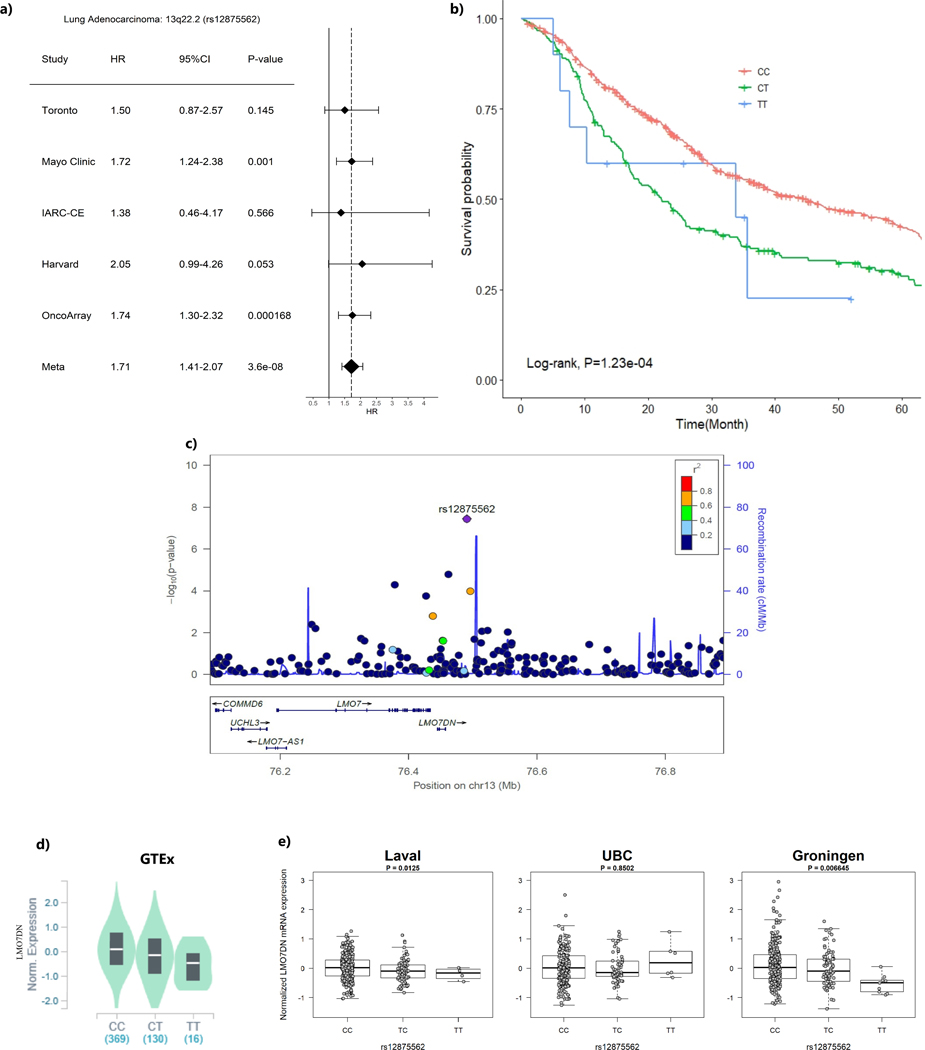
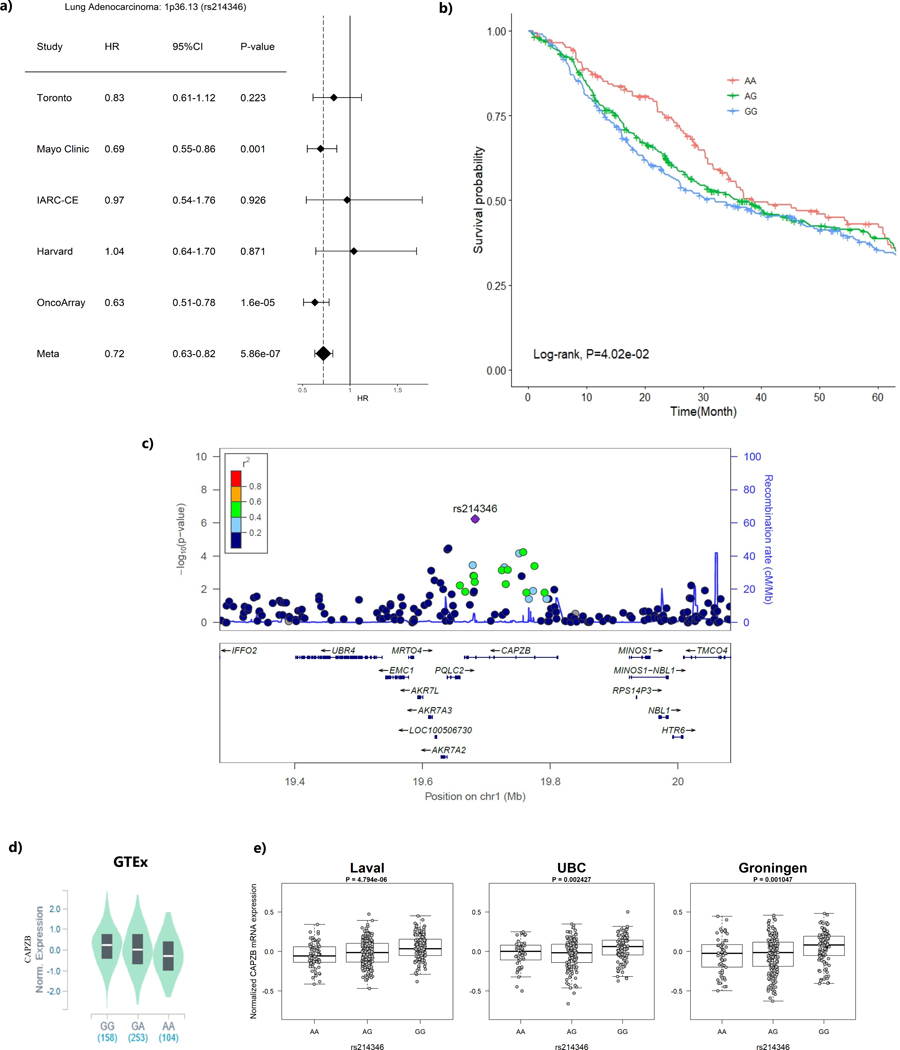
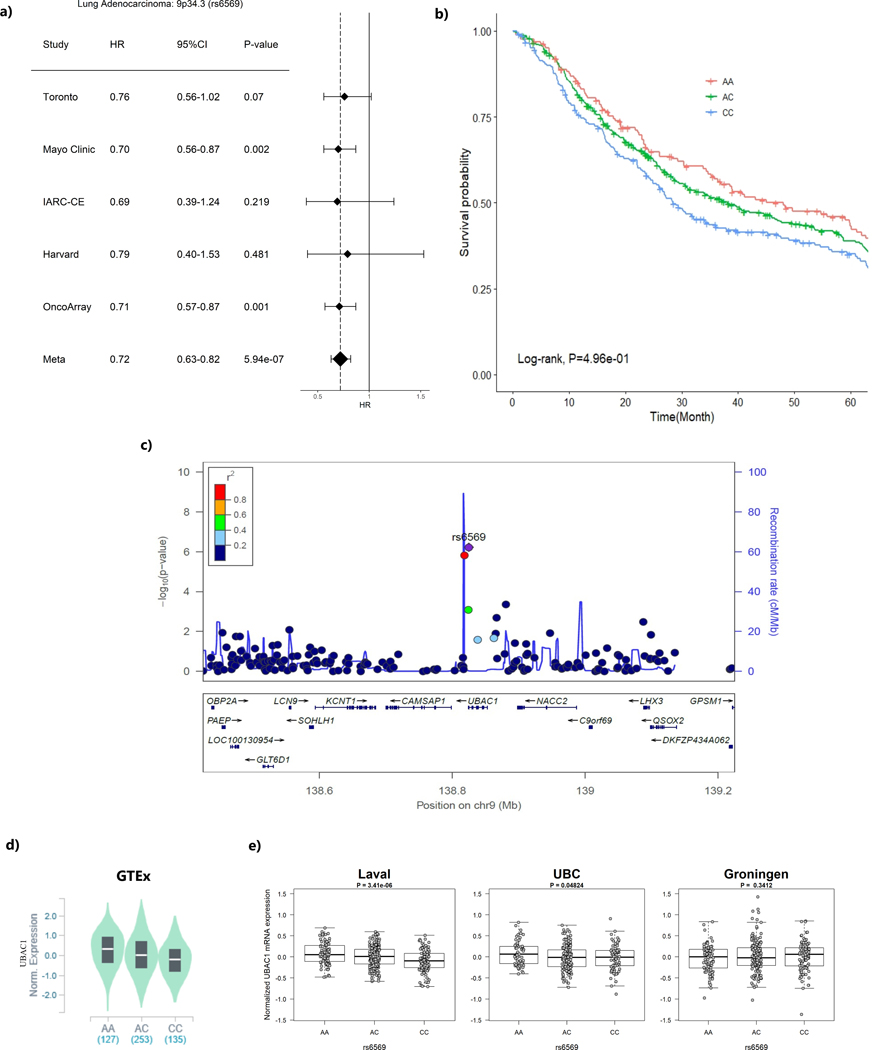
When restricting the analysis to 1,065 lung adenocarcinoma patients, the intergenic region at 13q22.2 (represented by rs12875562) reached significance GWAS level with T allele was associated with shorter survival time (HR = 1.71, 95%CI=1.41–2.07, P=3.60×10−8) (Table 2, Figure 1). In addition, seven other loci had suggestive evidence of association with overall survival at p-value ≤ 10−6(Table 2). Among those loci that showed suggestive evidence, it was worthwhile to mention that loci encoding CAPZB gene at 1p36.13 (represented by rs214346), and encoding UBAC1 gene at 9q34.3 (represented by rs6569) both conferred consistent association across studies with HR of 0.72 (95%CI=0.63–0.82, p=5.86×10−7) and 0.72 (95%CI= 0.63–0.82, p=5.94×10−7), respectively (Figure 2a and Figure 3a). The genetic locus that conferred the most distinctive survival patterns by genotype is located in 11q14 (represented by rs17148028) encoding DLG2 (Table 2 and Supplementary Figure 3i) with HR of 0.48 (0.36–0.64) with carriers of T allele exhibiting better prognosis. The forest plots and regional plots of the remaining loci are shown in the Supplementary Figure 3.
Figure 1. The region ch13q22.2 (represented by rs12875562) and lung adenocarcinoma survival.
(a) Forest plots for study-specific hazard ratios (HR) and 95% confidence intervals (CI)
(b) Kaplan-Meier plot by genotype of the sentinel variant over 5-year time period.
(c) Regional plot that include the 1Mb around the sentinel variant rs12875562. X-axis represent the chromosome position and Y-axis represent –log10 (p-value) with color representing the linkage disequilibrium with the sentinel variant in r2
(d) eQTL p-value from the GTEx data based on normal lung tissues
(e) eQTL P-values from the Lung eQTL dataset: Laval University, University of British Columbia (UBC) and University of Groningen. The left y axis represents gene expression levels in the lung adjusted for age, sex and smoking status. The x axis represents genotyping groups for rs12875562.
Figure 2. The region ch1p36.13 (represented by rs214346) and lung adenocarcinoma survival.
(a) Forest plots for study-specific hazard ratios (HR) and 95% confidence intervals (CI)
(b) Kaplan-Meier plot by genotype of the sentinel variant over 5-year time period.
(c) Regional plot that include the 1Mb around the sentinel variant rs214346. X-axis represent the chromosome position and Y-axis represent –log10 (p-value) with color representing the linkage disequilibrium with the sentinel variant in r2
(d) eQTL p-value from the GTEx data based on normal lung tissues
(e) eQTL P-values from the Lung eQTL dataset: Laval University, University of British Columbia (UBC) and University of Groningen. The left y axis represents gene expression levels in the lung adjusted for age, sex and smoking status. The x axis represents genotyping groups for rs214346
Figure 3. The region ch9q34.3 (represented by rs6569) and lung adenocarcinoma survival.
(a) Forest plots for study-specific hazard ratios (HR) and 95% confidence intervals (CI)
(b) Kaplan-Meier plot by genotype of the sentinel variant over 5-year time period.
(c) Regional plot that include the 1Mb around the sentinel variant rs6569. X-axis represent the chromosome position and Y-axis represent –log10 (p-value) with color representing the linkage disequilibrium with the sentinel variant in r2
(d) eQTL p-value from the GTEx data based on normal lung tissues
(e) eQTL P-values from the Lung eQTL dataset: Laval University, University of British Columbia (UBC) and University of Groningen. The left y axis represents gene expression levels in the lung adjusted for age, sex and smoking status. The x axis represents genotyping groups for rs6569
Among the total 12 loci, the locus encoding NWD2/KIAA1236 located at 4p14 (represented by rs17603438) also showed an association with lung cancer survival in the Japanese cohort. The major allele A was correlated with longer survival time in both the European cohorts (HR = 0.50, 95%CI= 0.38–0.66, P=6.92×10−7) and in the Japanese study (HR = 0.79, 95%CI= 0.67–0.94, P=0.0072). No other loci showed generalizable association across ethnic groups.
Supplementary Table 2 summarizes the result of genetic variants previously reported to be associated with lung prognosis based on two of the studies included in this analysis (34). Three of the eight loci remained to be nominally significant (Supplementary Table 2) at p-value of 10−2 to 10−4 based on ten studies. No follow-up analyses were performed on these variants given the weak level of evidence.
Functional Characterization
To investigate whether the variants associated with lung cancer survival may modulate the mRNA expression in the lung tissues, we conducted eQTL analysis for the top 12 loci identified by overall and adenocarcinoma only analysis in a total of four independent studies, including three lung microarray studies and GTEx, based on a total of 1553 lung tissues. Variants that were shown to have significant cis-effect on the mRNA expression of a nearby gene across all four studies are shown in Table 3 (Table 3).
Table 3:
Expression quantitative trait loci (eQTL) identified in both Lung MicroArray project and GTEx database
| Lung MicroArray eQTL | GTEx eQTL for lung tissue d | |||||
|---|---|---|---|---|---|---|
| Location | Lead SNP | Genea | Target Geneb | P-valuec | Target Geneb | P-value |
| 1p36.13 | rs214346 | CAPZB | CAPZB | 2.78 × 10−10 | CAPZB | 1.10 × 10−9 |
| 9q34.3 | rs6569 | UBAC1 | UBAC1 | 1.14 × 10−3 | UBAC1 | 9.20 × 10−14 |
| 13q22.2 | rs12875562 | Intergenic | LMO7DN | 0.003 | LMO7DN | 9.40 × 10−7 |
based on variant location
gene with the most significant eQTL association within a 1 Mb up and downstream of the transcription probe set.
eQTL P-value is based on meta-analysis of Laval University, University of British Columbia, University of Groningen studies.
Genotype-Tissue Expression (GTEx) database (http://www.gtexportal.org/home/) of RNAseq analysis in 515 lung human tissues (Release V7)
CAPZB, Capping Actin Protein Of Muscle Z-Line Subunit Beta; UBAC1, UBA Domain Containing 1; LMO7DN, LIM Domain 7 Downstream Neighbour
Three loci demonstrated consistent cis-effects across all 4 eQTL studies, including the only GWAS level significant variant, rs12875562 located at 13q22.2 with significant eQTL effect on LMO7 Downstream Neighbour (LMO7DN) gene expression with p-value of 9.40×10−7 in GTEx and 3.44×10−3 in Lung eQTL dataset and 9.40×10−7 in GTEx (Table 3 and Figure 1). Patients with the minor allele T had a poor survival and lower LMO7DN expressions. The strongest eQTL signal came from rs214346 located in CAPZB at 1p36.13, which showed a consistent association with increased expression of CAPZB with in Lung eQTL dataset (p-value of 2.78×10−10) and in GTEx lung tissue (p=1.10×10−9) (Table 3 and Figure 2). Patients with minor A allele have better survival and lower CAPZB expression in lung tissue. Finally, rs6569, located at 9q34.3 in UBAC1 gene was found to be associated with decreased expression of UBAC1 in both Lung eQTL dataset (p= 1.14×10−3) and GTEx (p=9.20×10−14) (Table 3 and Figure 3). The minor allele A of this variant was associated with longer survival (HR=0.71, 95%CI=0.62–0.81, P=5.94×10−7) and a higher expression of UBAC1.
DISCUSSION
Based on the largest GWAS on lung cancer prognosis for never smokers conducted to date, we identified one locus that reached GWAS significance level at ch13q22 for patients with lung adenocarcinoma, and 11 loci that provided suggestive evidence; among those ch4p14 was also associated lung cancer prognosis in Japanese population. Three of the top 12 loci were shown to affect mRNA expression in nearby genes as cis-eQTL across multiple studies, including the region with top signal in ch13q22, which provided additional level of evidence of the association related prognosis.
The variant at the 13q22 locus is located adjacent to the LMO7 gene and LMO7 Downstream Neighbour (LMO7DN). LMO7 encodes a fibrous actin-binding protein that is commonly expressed in many human tissues, but particularly high in the lung epithelial cells. It was suggested to be involved in the maintenance of epithelial architecture (35), and is considered to act as tumor suppresser gene, as LMO7 knock-out mice were shown to develop spontaneous lung adenocarcinoma (36). LMO7 expression was shown to be associated with lung cancer prognosis, but the direction of effect is not yet conclusive, which can be attributed to histological types included in the studies as well as other related genes in the same regulatory pathway (37,38). We did not observe a consistent eQTL association with LMO7 per se, but with its downstream neighbour (LMO7DN) instead, with carriers of T allele exhibited poor prognosis for lung adenocarcinoma and lower LMO7DN expression. This suggests that LMO7DN might play a more important role in lung tumor progression. This is the first time LMO7DN is identified as a gene associated with lung cancer prognosis at the genome-wide level.
The region in ch1p36.13 encodes CAPZB gene, whose mRNA expression is affected by the variant with A allele associated with lower level of expression. CAPZB is a regulator of actin filament length that determines the mitotic cortex thickness during cell cycle progression, and it is associated with cell growth and motility in epithelioid sarcoma (39,40). Variants within the same locus were previously associated with total platelet mass (41), autoimmune related disorders such as Crohn’s disease and psoriasis (42,43). Other variants of the same gene have also been shown to be related to lung function (44,45), although the exact mechanism of how this gene is associated with lung cancer prognosis in never smokers is not clear.
The region in ch9q34.3 encodes UBAC1, which is associated with innate immune system and Class I MHC mediated antigen processing. The sequence variants in this gene have been shown to be associated with interleukin-4 and interleukin-6 levels (46). It has not been previously reported to be associated with cancer risk or prognosis. It is biologically plausible that this gene may modulate tumor progression through the innate immune pathway. Finally, genetic variants of DLG2 located at 11q14.1 were previously shown to be associated with lung function (47) and familial squamous cell lung carcinoma (48), as well as anthropometric measurement such as body fat mass, fat and body mass index (49), which support a role of this gene in lung carcinogenesis and prognosis.
Although previous studies have identified several genetic loci associated with lung cancer overall survival, primarily in smoking related lung cancer and early stage non-small cell lung cancer (14,17–19), we did not observe the association with those previously reported loci in our study of never smokers. This is not a surprise as we expect distinctive genetic architecture contribute to lung cancer survival in smokers and never smokers.
For cross-ethnic generalizability, we observed only one association in ch4p14, encoding NWD2/KIAA1239 gene that was potentially generalizable between European and Japanese populations. This could be due to differences in the genetic background or different linkage disequilibrium patters underlying causal variants between European and Japanese population. In addition, the Japanese study has different characteristics such as distribution of sex, stage, treatment modality and overall survival, which could also contribute to limited generalizability across different ethnicities. Interestingly, the 4p14 locus was previous reported to be associated with smoking behaviour(50).
Our study has several limitations – First, potential heterogeneity between patient characteristics across studies may not be fully accounted for. In particular treatment information was not available for studies included in the OncoArray project and therefore could not be adjusted for in the model. However, it is expected that adjustment for clinical stage would mitigate the potential confounding effect of treatment as these two factors are highly correlated. Similarly, we do not have information on somatic mutations such as EGFR or ALK, which would affect prognosis. It is likely that the differences we observed across ethnic population are due to the differences in these mutations, which are markedly different by ethnic groups. Second, our sample size might be underpowered due to relatively rare occurrence of lung cancer in never smokers, which can lead to potential false negative results. Nevertheless, this is the largest genome-wide analysis for lung cancer prognosis among never smokers conducted to date, and the results have high relevance considering the percentage of never smokers are increasing among lung cancer patients.
In summary, we identified one locus at genome-wide significant level at 13q22 which was consistent with known role of genetic region encoding LMO7/LMO7DN in cancer pathology, along with several potential loci that would require further validations. The integrated evidence from both the associations with lung cancer survival and eQTL are complementary and provide support of our hypothesis that inherited genetic factors affect lung cancer survival in never smokers. Functional genomic experiments to assess the effect of altered gene regulation would be required to further understand the therapeutic potential of these biologically plausible genes.
Supplementary Material
ACKNOWLEDGEMENT
Dr. Amos is a Research Scholar of the Cancer Prevention Research Institute of Texas and part of his effort is supported by RR170048. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
The Mayo Clinic Study is supported by NIH-grants CA77118/CA80127/CA84354 and Mayo Foundation. Support was provided by the Mayo Clinic Shared Resources (PI: Ping Yang).
Funding sources: The genetic data generation was supported by U.S. NIH U19 CA148127 and the analysis was supported by the U19 CA203654 and CIHR FDN 167273. R.J.H. holds Canada Research Chair in Integrative Molecular Epidemiology.
Footnotes
Conflict of Interest Statement: The authors declare no potential conflicts of interest.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394–424 doi 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Thun MJ, Henley SJ, Burns D, Jemal A, Shanks TG, Calle EE. Lung cancer death rates in lifelong nonsmokers. J Natl Cancer Inst 2006;98(10):691–9 doi 10.1093/jnci/djj187. [DOI] [PubMed] [Google Scholar]
- 3.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008;40(5):616–22 doi 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKay JD, Hung RJ, Han Y, Zong X, Carreras-Torres R, Christiani DC, et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nature genetics 2017;49(7):1126–32 doi 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452(7187):633–7 doi 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 6.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet 2009;85(5):679–91 doi 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet 2008;40(12):1404–6 doi 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452(7187):638–42 doi 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet 2008;40(12):1407–9 doi 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer 2007;7(10):778–90 doi 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 11.Thun MJ, Henley SJ, Travis WD. Lung Cancer In: Thun MJ, Linet MS, Cerhan JR, Haiman CA, Schottenfeld D, editors. Cancer Epidemiology and Prevention, 4th Edition New York: Oxford University Press; 2018. p 519–52. [Google Scholar]
- 12.Subramanian J, Govindan R. Molecular genetics of lung cancer in people who have never smoked. Lancet Oncol 2008;9(7):676–82 doi 10.1016/s1470-2045(08)70174-8. [DOI] [PubMed] [Google Scholar]
- 13.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150(6):1121–34 doi 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosse Y, Amos CI. A Decade of GWAS Results in Lung Cancer. Cancer Epidemiol Biomarkers Prev 2018;27(4):363–79 doi 10.1158/1055-9965.EPI-16-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YT, Heist RS, Chirieac LR, Lin X, Skaug V, Zienolddiny S, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol 2009;27(16):2660–7 doi 10.1200/jco.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang S, Pan Y, Wang Y, Hu L, Cao S, Chu M, et al. Genome-wide association study of survival in early-stage non-small cell lung cancer. Ann Surg Oncol 2015;22(2):630–5 doi 10.1245/s10434-014-3983-0. [DOI] [PubMed] [Google Scholar]
- 17.Sato Y, Yamamoto N, Kunitoh H, Ohe Y, Minami H, Laird NM, et al. Genome-wide association study on overall survival of advanced non-small cell lung cancer patients treated with carboplatin and paclitaxel. J Thorac Oncol 2011;6(1):132–8 doi 10.1097/JTO.0b013e318200f415. [DOI] [PubMed] [Google Scholar]
- 18.Wu C, Xu B, Yuan P, Miao X, Liu Y, Guan Y, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res 2010;70(23):9721–9 doi 10.1158/0008-5472.can-10-1493. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Ye Y, Rosell R, Amos CI, Stewart DJ, Hildebrandt MA, et al. Genome-wide association study of survival in non-small cell lung cancer patients receiving platinum-based chemotherapy. J Natl Cancer Inst 2011;103(10):817–25 doi 10.1093/jnci/djr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y, Yoon KA, Joo J, Lee D, Bae K, Han JY, et al. Prognostic implications of genetic variants in advanced non-small cell lung cancer: a genome-wide association study. Carcinogenesis 2013;34(2):307–13 doi 10.1093/carcin/bgs356. [DOI] [PubMed] [Google Scholar]
- 21.Hu L, Wu C, Zhao X, Heist R, Su L, Zhao Y, et al. Genome-wide association study of prognosis in advanced non-small cell lung cancer patients receiving platinum-based chemotherapy. Clin Cancer Res 2012;18(19):5507–14 doi 10.1158/1078-0432.CCR-12-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, Laviolette M, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet 2012;8(11):e1003029 doi 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348(6235):648–60 doi 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol Biomarkers Prev 2017;26(1):126–35 doi 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 2012;44(8):955–9 doi 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1(6):457–70 doi 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010;11(7):499–511 doi 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010;34(8):816–34 doi 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nature genetics 2012;44(8):900–3 doi 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 30.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics 2006;38(8):904–9 doi 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 31.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26(17):2190–1 doi 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003;327(7414):557–60 doi 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosse Y, Li Z, Xia J, Manem V, Carreras-Torres R, Gabriel A, et al. Transcriptome-wide association study reveals candidate causal genes for lung cancer. Int J Cancer 2019. doi 10.1002/ijc.32771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Wang L, Ye Y, Aakre JA, Pu X, Chang GC, et al. Genome-wide association study of genetic predictors of overall survival for non-small cell lung cancer in never smokers. Cancer Res 2013;73(13):4028–38 doi 10.1158/0008-5472.can-12-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holaska JM, Rais-Bahrami S, Wilson KL. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum Mol Genet 2006;15(23):3459–72 doi 10.1093/hmg/ddl423. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka-Okamoto M, Hori K, Ishizaki H, Hosoi A, Itoh Y, Wei M, et al. Increased susceptibility to spontaneous lung cancer in mice lacking LIM-domain only 7. Cancer Sci 2009;100(4):608–16 doi 10.1111/j.1349-7006.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsson T, Kvarnbrink S, Holmlund C, Botling J, Micke P, Henriksson R, et al. LMO7 and LIMCH1 interact with LRIG proteins in lung cancer, with prognostic implications for early-stage disease. Lung Cancer 2018;125:174–84 doi 10.1016/j.lungcan.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura H, Hori K, Tanaka-Okamoto M, Higashiyama M, Itoh Y, Inoue M, et al. Decreased expression of LMO7 and its clinicopathological significance in human lung adenocarcinoma. Exp Ther Med 2011;2(6):1053–7 doi 10.3892/etm.2011.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chugh P, Clark AG, Smith MB, Cassani DAD, Dierkes K, Ragab A, et al. Actin cortex architecture regulates cell surface tension. Nat Cell Biol 2017;19(6):689–97 doi 10.1038/ncb3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukaihara K, Suehara Y, Kohsaka S, Kubota D, Toda-Ishii M, Akaike K, et al. Expression of F-actin-capping protein subunit beta, CAPZB, is associated with cell growth and motility in epithelioid sarcoma. BMC Cancer 2016;16:206 doi 10.1186/s12885-016-2235-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGraw KL, Cheng CH, Chen YA, Hou HA, Nilsson B, Genovese G, et al. Non-del(5q) myelodysplastic syndromes-associated loci detected by SNP-array genome-wide association meta-analysis. Blood Adv 2019;3(22):3579–89 doi 10.1182/bloodadvances.2019000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng S, Lin S, Zou J, Wang Y, Ji Q, Lv Z. Association between rs12045440 Polymorphism in the CAPZB Intron and Serum TSH Concentrations in Chinese Thyroid Tumor Patients. Int J Endocrinol 2015;2015:250542 doi 10.1155/2015/250542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhan M, Chen G, Pan CM, Gu ZH, Zhao SX, Liu W, et al. Genome-wide association study identifies a novel susceptibility gene for serum TSH levels in Chinese populations. Hum Mol Genet 2014;23(20):5505–17 doi 10.1093/hmg/ddu250. [DOI] [PubMed] [Google Scholar]
- 44.Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet 2019;51(3):481–93 doi 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L, et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet 2017;49(3):416–25 doi 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahola-Olli AV, Wurtz P, Havulinna AS, Aalto K, Pitkanen N, Lehtimaki T, et al. Genome-wide Association Study Identifies 27 Loci Influencing Concentrations of Circulating Cytokines and Growth Factors. Am J Hum Genet 2017;100(1):40–50 doi 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am J Hum Genet 2019;104(1):65–75 doi 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Byun J, Schwartz AG, Lusk C, Wenzlaff AS, de Andrade M, Mandal D, et al. Genome-wide association study of familial lung cancer. Carcinogenesis 2018;39(9):1135–40 doi 10.1093/carcin/bgy080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tachmazidou I, Suveges D, Min JL, Ritchie GRS, Steinberg J, Walter K, et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am J Hum Genet 2017;100(6):865–84 doi 10.1016/j.ajhg.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SL, Carmella SG, Chen M, Patel Y, Stram DO, Haiman CA, et al. Mercapturic Acids Derived from the Toxicants Acrolein and Crotonaldehyde in the Urine of Cigarette Smokers from Five Ethnic Groups with Differing Risks for Lung Cancer. PLoS One 2015;10(6):e0124841 doi 10.1371/journal.pone.0124841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.