Abstract
The cytosolic pattern recognition receptor NLRP3 senses host-derived danger signals and certain microbe-derived products in both humans and rodents. NLRP3 activation assembles an inflammasome complex, which contains the adapter protein ASC and caspase-1, whose activation triggers the maturation and release of the proinflammatory cytokines IL-1β and IL-18. Serine 5 (S5) phosphorylation of NLRP3 prevents its oligomerization and activation, while dephosphorylation of this residue by the phosphatase PP2A allows NLRP3 activation. However, the protein kinase that mediates NLRP3 S5 phosphorylation is unknown. Here, we show that AKT associates with NLRP3 and phosphorylates it on S5, limiting NLRP3 oligomerization. This phosphorylation event also stabilizes NLRP3 by reducing its ubiquitination on lysine 496, which inhibits its proteasome mediated degradation by the E3 ligase Trim31. Pharmacologic manipulation of AKT kinase activity reciprocally modulates NLRP3 inflammasome mediated IL-1β production. Inhibition of AKT reduced IL-1β production following the intraperitoneal injection of LPS into mice. We propose that AKT, Trim31 and PP2A together modulate NLRP3 protein levels and tendency to oligomerize, thereby setting a tightly regulated threshold for NLRP3 activation.
Keywords: AKT, NLRP3, TRIM31, inflammasome, phosphorylation, ubiquitination, oligomerization
Introduction
Inflammasomes are large protein complexes that mediate rapid immune responses against pathogens and sterile stressors (1–3). NLRP3 (NOD-, LRR-, and pyrin domain-containing protein 3) is a pattern recognition receptor (PRR) that upon activation constitutes the NLRP3 inflammasome through the recruitment of ASC and caspase-1 (4). Activation of the NLRP3 inflammasome requires two signals. The first is a priming signal (experimentally triggered by LPS) that results in the transcriptional upregulation of inflammasome components and induces post-translational modifications on NLRP3 to license NLRP3 inflammasome assembly (5). The second signal is experimentally induced by extracellular ATP, crystalline uric acid, pore forming toxins (e.g. nigericin), or other cell stress inducers and promotes formation of an active inflammasome complex (6–9). This occurs via NLRP3 self-oligomerization, PYD-PYD domain dependent helical assembly of the adaptor protein ASC, and subsequent activation of pro-caspase-1. Activated caspase-1 then cleaves pro-IL-1β and pro-IL-18 into their mature forms, which are important in initiating the inflammatory cascade (5).
NLRP3 is subject to several post-translational modifications including phosphorylation, ubiquitination, and oligomerization, all of which regulate NLRP3 activation (10–12). For instance, c-Jun N terminal kinase 1 (JNK1) phosphorylates human NLRP3 at S198, which is required for NLRP3 deubiquitination and self-oligomerization (13, 14). Conversely, protein kinase A (PKA) mediated phosphorylation of NLRP3 at S295 blocks NLRP3 activation by promoting NLRP3 ubiquitination (15). A recent study reported that NLRP3 phosphorylation at S5 during the priming stage decreases NLRP3 oligomerization and subsequent cytokine secretion. This process is reversed by phosphatase PP2A mediated NLRP3 S5 dephosphorylation, which mitigates electrostatic repulsion of the NLRP3 PYD domain and facilitates NLRP3 self-oligomerization and ASC recruitment (16). Though the unique function of NLRP3 phosphorylation at S5 is defined, the kinase responsible for phosphorylation of this site remains unknown.
AKT is a well-studied serine/threonine kinase that functions downstream of phosphoinositide 3-kinase (PI3K); it exists as three isoforms and has a physiological role in nearly every organ system (17). In monocytes and macrophages, AKT is activated by LPS stimulation, and appears to be immunosuppressive as it negatively regulates the immune response downstream of LPS (18–20). Here, we extend the immunosuppressive function of AKT by characterizing it as a dual regulator of the NLRP3 inflammasome. We show that AKT phosphorylates NLRP3 at S5, preventing NLRP3 and ASC oligomerization and reducing inflammatory cytokine production. We also show that AKT mediated phosphorylation of S5 enhances NLRP3 stability during the LPS priming phase, which occurs via inhibition of TRIM31 ubiquitination and degradation of NLRP3. Finally, we show that inhibition of AKT enhances inflammasome activity in vivo, demonstrating a physiological role for AKT as a regulator of NLRP3.
Material and Methods
Cell isolation and culture
Primary bone marrow-derived macrophages (BMDMs) were prepared as previously described (21). In short, bone marrow was isolated from femurs of 8-week-old C57BL/6 mice. Bone marrow was suspended in sterile ACK lysis buffer for red blood cell lysis and the remaining cells plated in DMEM (Gibco) supplemented with L929-cell conditioned media (30% vol/vol). Loosely adherent cells were removed on day 4 and fresh medium added; macrophages were used on day 6–8 of culture. THP-1, L929, and HEK293T cells were obtained from ATCC. HEK293T and L929 cells were grown in DMEM, while THP-1 cells were grown in RPMI (Gibco) with 4.5 g/l glucose, 1 mmol/l sodium pyruvate and 20 μM HEPES. Prior to use, THP-1 cells were differentiated into macrophages using 250 nM Phorbol-12-myristate-13-acetate (PMA) for 24 h. All media was supplemented with 10% fetal calf serum (FCS, Gibco), penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were maintained in a 5% CO2 incubator (Thermo) at 37°C.
Reagents, plasmids, siRNA, and transfection
MK2206 (HY-10358) and SC79 (HY-18749) were purchased from MedChemExpress. E. Coli LPS (0111:B4) was purchased from Sigma. The Myc-NLRP3 construct and its truncated mutants were a kind gift from Dr. Yong-Jun Liu (Baylor Institute for Immunology Research), and Myc-NLRP3 truncated constructs were made using primers to delete portions of the open reading frame. The Flag-NLRP3 plasmid was a kind gift from Dr. Gabriel Nunez (The University of Michigan Medical School). Flag-NLRP3 S5A and Flag-NRLP3 K496R plasmids were made using mutagenic primers; mutant plasmids were sequenced to verify fidelity prior to use. Plasmids encoding AU-Trim31, Flag-TRIM31, and Flag-TRIM31ΔRING were a kind gift from Dr. Wei Zhao (Shandong University School of Medicine). Plasmids encoding a control vector and bacterial flagellin were kind gifts from Dr. Russel Vance (University of California Berkley). The pCMV6-Flag-NLRP3, pCMV6-ASC, and pCMV6-pro-caspase-1 plasmids have been previously described (22). The pCMV5 hemagglutinin (HA)-Akt wild type, HA-Myr-AKT (constitutively active) HA-AKT-K179M (Akt kinase-dead form) were kindly provided by Dr P. N. Tsichlis (Kimmel Cancer Institute, Thomas Jefferson University). Plasmids were transfected using X-tremeGene HP DNA Transfection Reagent (Roche) following the manufacturers protocol. For THP-1 cells, cells were seeded in 12 well plates at 5×105 cells/well using OPTI-MEM during transfection. OPTI-MEM media was replaced by culture media after 6 hrs of transfection to ensure cell recovery. AKT1/2, Trim31, and scrambled control siRNAs were purchased from Santa Cruz Biotechnology. siRNA was prepared at 40 nM with 2ul transfection reagent and transfected overnight prior to assay. Efficient siRNA knockdown was validated at the RNA or protein level.
Inflammasome activation and cytokine detection
Cells were seeded in 12-well culture plates (primary BMDMs: 1×106/well; THP-1 cells: 5×105/well) or 96 well plates (primary BMDMs and THP-cell: 1× 105/well) and cultured overnight. Cells were primed with 200 ng/ml LPS for 6 h in opti-MEM (Gibco) unless otherwise noted. The NLRP3 inflammasome was activated with 5 μM nigericin for 45 min or 2 mM ATP for 1 h. For rapid NLRP3 activation, BMDMs were stimulated with 1 μg/ml LPS for 3 h and 7.5 mM Nigericin for 30 min. To activate the AIM2 inflammasome, 2 μg/ml poly(dA:dT) was transfected into BMDMs with X-tremeGene HP for 6 hrs. For NLRC4 inflammasome activation, 400 ng of a flagellin encoding vector was transfected using X-treme GENE HP for 6 hrs. In order to detect cytokine secretion after inflammasome activation, we isolated protein from the culture media after inflammasome activation (23). In short, 0.5 mL of media from each sample was collected and mixed with 500 μl methanol and 125 μl chloroform. The resultant mixture was vortexed and centrifuged at 13,000 rpm for 5 min at 4°C. The upper phase was discarded, while 500 μl methanol was added to the lower phase followed again by centrifugation. The supernatant was again decanted, and the remaining protein pellet dried for 5 min at 50°C. The protein pellet was dissolved in 60 μl sample buffer with 0.1M DTT, heated at 95°C for 15 min, and loaded onto an SDS-PAGE gel for separation and detection via immunoblotting. Alternatively, samples were analyzed for mouse IL-1β (88–7013, Invitrogen), IL-6 (431406, BioLegend), and TNF-α (430906, BioLegend) by ELISA according to the manufacturer’s protocol.
Cell Lysis and Immunoprecipitation
For standard immunoblotting, cells were lysed in Triton lysis buffer [20mM HEPES (pH 7.5), 50mM β-Glycerol phosphate, 2mM EGTA, 1% Triton-X 100, and protease inhibitor cocktail (Roche)] on ice for 20 minutes. Cell lysates were cleared by centrifugation, the supernatant collected and diluted in 4× NuPage LDS sample buffer (Thermo), and the samples heated at 100 °C for 10 min. For immunoprecipitation experiments, cells were lysed in CHAPS buffer [20mM HEPES (pH 7.5), 50mM β-Glycerol phosphate, 2mM EGTA, 1% CHAPs, and protease inhibitor cocktail (Roche)] at 4°C for 30 min. Lysates were centrifuged at 7,500 rpm at 4°C for 10 min, and the supernatants collected and incubated at 4°C for 1.5 h with anti-HA antibody (C29F4, Cell signaling) or overnight with either anti-NLRP3 antibody (D4D8T, Cell Signaling) or anti-Flag M2 beads (Sigma). When appropriate, immunoprecipitates were isolated by protein G pulldown, and isolated immunoprecipitates washed 6 times with lysis buffer. Proteins were released by the addition of DTT, 2-mercaptoethanol, LDS Sample Buffer, and heating at 100 °C for 10 minutes.
Immunoblotting
Samples were loaded onto a 4–20% Novex Tris-Glycine gel, separated by SDS-PAGE, and transferred to a nitrocellulose membrane using the iBLOT Gel Transfer System (Invitrogen) on setting P2 for 7 minutes. Immunoblots were analyzed using the following antibodies: rabbit monoclonal anti-NLRP3 (1:1000, D4D8T, Cell Signaling), rabbit polyclonal anti-hCaspase-1 (1:1000, #3866, Cell Signaling), mouse monoclonal anti-IL-1β (1:1000, D4T2D, Cell Signaling), rabbit monoclonal anti-AKT1 (1:1000, C73H10, Cell Signaling), rabbit monoclonal anti-pAKT S473 (1:1000, D9E, Cell Signaling), anti-AKT substrate (1:500, #9614, Cell Signaling) rabbit polyclonal anti-hTrim31 (1:,000, #12543–1-AP, Proteintech), rabbit monoclonal anti-ASC (1:1000, F-9, Santa Cruz), mouse monoclonal anti-Flag (1:1000, M2, Sigma), rabbit polyclonal anti-HA (1:1000, SAB4300603, Sigma), mouse monoclonal anti-myc (1:1000, 9E10, Biolegend), HRP-anti-GAPDH (1:10000, HRP-60003, Proteintech). When appropriate, species specific secondary antibodies conjugated to HRP (Cell Signaling) were used to detect protein bands by enhanced chemiluminescence (ECL, Bio-Rad). Images were obtained using a Thermo iBright FL1000, and quantification of band intensity was performed using ImageJ (NIH). All immunoblotting experiments were independently performed at least three times with similar results and representative figures shown. Bands are normalized to its loading control and presented as fold change of the control for the immunoblot shown.
Reconstitution of NLRP3 inflammasome in HEK293T cells
For reconstitution of the NLRP3 inflammasome in HEK293T cells, cells were cultured in 12-well plates at a density of 1.5×105 cells/well overnight prior to transfection. Cells were co-transfected with plasmids for pCMV6-Flag-NLRP3 (10 ng/well), pCMV6-ASC (5 ng/well) and pCMV6-pro-caspase-1 (10 ng/well) using X-treme GENE HP (Roche). Cell lysates were collected using standard procedures and the maturation of caspase-1 p20 assessed by immunoblotting.
RNA isolation and quantitative PCR
Total RNA was isolated using Trizol (Thermo) according to the manufacturer’s protocol. RNA concentration was assessed by absorbance at 260 and 280 nm, and complementary DNA (cDNA) synthesis performed using Omniscript Reverse Transcription kit (Qiagen) following the manufacturer’s instructions using 2ug isolated RNA as template. Quantitative real-time (qRT-PCR) was performed using SYBR GREEN Master Mix (Takara) on a QuantStudio 3 Real-Time PCR system (Thermo) and primers for the genes of interest. The RT-PCR program used an initial enzyme activation step (10 min, 95°C), followed by 45 cycles consisting of a denaturing (95°C, 10 s) and annealing/extension (60°C, 1 mi). Measurements were performed in triplicate, and the results were normalized to GAPDH and relative mRNA levels calculated using the 2−ΔΔCt method. The primers used for qRT-PCR were as follows:
ACTIN: F’:TCCTGTGGCATCCACGAAACT, R’: GGAGCAATGATCCTGATCTTC;
NLRP3: F’: GGTCCTCTTTACCATGTGCTTC, R’: AAGTCATGTGGCTGAAGCTGTA;
ASC: F’: AAAAGTTCAAGATGAAGCTGCTG, R’: CTCCTGTAAGCCCATGTCTCTAA;
CASP-1: F’: TTTCAGTAGCTCTGCGGTGT, R’: TTTCTTCCTGATTCAGCACTCTC;
IL-1β: F’: CCCAAGCAATACCCAAAGAAGAAG, R’: TGTCCTGACCACTGTTGTTTCC;
IL-6: F’: ACAAGAAAGACAAAGCCAGAGTCC, R’: CTAGGTTTGCCGAGTAGATCTC;
TNF-α: F’: ATGAGCACAGAAAGCATGATC, R’: ATGAGCACAGAAAGCATGATC.
ASC speck staining
BMDMs were plated onto cover slips and primed with 1 μg/ml LPS for 3 hrs, followed by stimulation with PBS or 5 μM Nigericin for 15min. Cells were fixed with 4% paraformaldehyde followed by permeabilization with 0.5% Triton X-100 at 4°C for 30 min. Slides were blocked for 30 minutes with 5% BSA and stained with anti-ASC antibody (F-9, Santa Cruz) overnight. The following day, cells were washed, stained with a TRITC-conjugated secondary antibody, and nuclei countered stained using 4’, 6’-diamidino-2-phenylindole (DAPI, 1μg/ml). Imaging was performed using a Zeiss 880 confocal microscope with minimal laser power.
ASC oligomer cross-linking
BMDMs were primed with 1 μg/ml LPS for 3 h followed by stimulation with PBS (non-treated) or 5 μM nigericin for 15 min. Cells were lysed with Triton lysis buffer at 4°C for 30 min, and 3% of the lysates collected as whole cell lysates. The remaining lysate was mixed with the same volume of a CHAPS containing buffer [20mM HEPES-KOH (PH=7.5), 5 mM MgCl2, 0.5 mM EGTA, 0.1% CHAPS] and centrifuged at 8000×g at 4°C for 15 min. The resultant pellet was considered the Triton insoluble fraction. For ASC cross-linking, the Triton insoluble fraction was washed, re-suspended in PBS, and cross-linked for 30 min at 37°C with 4mM disuccinimidyl suberate (DSS). The pellet was collected after centrifugation for 8 min at 5000×g and dissolved in SDS loading buffer for immunoblot analysis.
In vitro kinase assay
Lysate from cells expressing Flag-NLRP3 (or its indicated mutant) was added to immunoprecipitated and purified HA-AKT (or its mutants) using a kinase reaction buffer [50 mM HEPES (pH=7.5), 25 mM β-glycerolphosphate, 20 mM MgCl2, 200 μM ATP, 4 mM DTT, 10 mM β-Me] at room temperature for 1 hr. The reaction was stopped by adding LDS loading buffer, and the samples analyzed by gel electrophoresis and immunoblotting.
LPS-induced systemic inflammation in vivo
Twenty-eight 8-week-old female C57BL/6J mice (Jax Stock No 000664) were used in this study (7 mice per group). Mice were pre-treated with DMSO or MK2206 (80 mg/kg) intraperitoneally for 2 h, followed by intraperitoneal injection of LPS (5 mg/kg). Mice were sacrificed 4 h after injection, and both 1.5 mL intraperitoneal lavage samples in PBS and blood samples collected. Serum was isolated from blood by use of a serum filter and centrifugation at 1500rpm for 15 min. Cytokine levels were measured in both serum and lavage samples using ELISA. All animal experiments and protocols were approved by the National Institute of Allergy and Infectious Diseases Animal Care and Use Committee at the National Institutes of Health.
Quantification and statistical analysis
Statistical analysis was performed using Prism 6 (GraphPad). A standard two-tailed unpaired Student’s t test, one-way ANOVA, or Mann-Whitney test was used for statistical analysis. P <0.05 was considered statistically significant.
Results
AKT kinase activity negatively regulates NLRP3 inflammasome activation
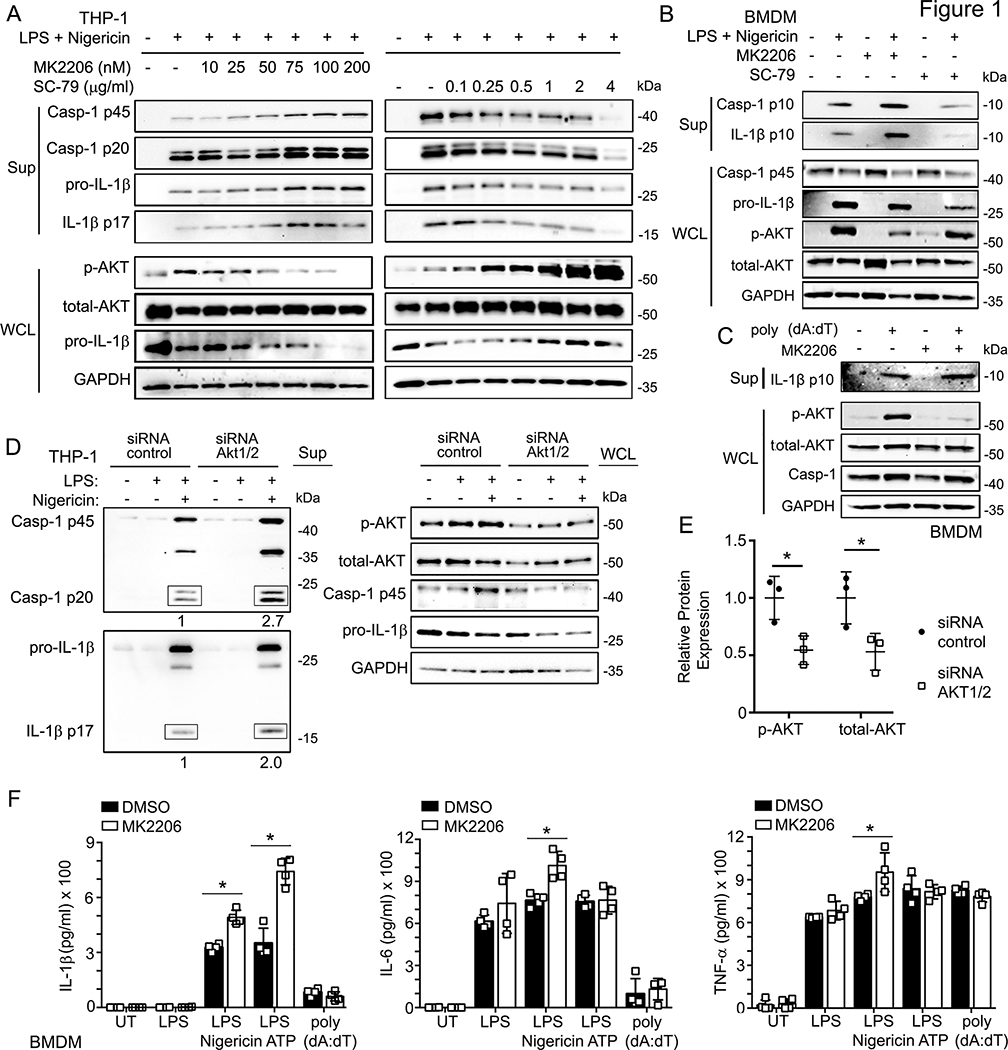
Previous studies indicate that NLRP3 inflammasome activation enhances AKT activity in several cell types including murine bone marrow derived macrophages (BMDMs), the human monocytic THP-1 cell line, and the alveolar epithelial A549 cell line (24, 25). In this study, we tested the impact of modulating AKT activity on inflammasome activity in THP-1 cells by LPS-priming and nigericin treating cells with the AKT inhibitor MK2206 and the AKT activator SC-79. We found that a graded MK2206 treatment gradually increased the amount of cleaved caspase-1 and IL-1β secretion after NRLP3 inflammasome activation with nigericin, while escalating amounts of SC-79 caused a progressive decline in cleaved caspase-1 and IL-1β secretion. Antibodies that detect the phosphorylation status of AKT S473 were used as an internal control to monitor AKT activation (26). LPS plus nigericin stimulation expectedly increased p-AKT, which was blunted by the of addition MK2206 and exacerbated by SC-79 (Fig. 1A). We verified these results in primary cells using LPS primed and nigericin treated mouse BMDMs (Fig. 1B). In contrast, when we treated LPS-primed BMDMs with poly(dA:dT) to activate AIM2 inflammasomes or flagellin to activate NLRC4 inflammasomes MK2206 did not affect IL-1β secretion suggesting that the effects of AKT are NLRP3 specific (Fig. 1C, Fig. S1A).
Figure 1.
AKT is involved in NLRP3-mediated inflammasome activation.
(A) Immunoblots of culture supernatants and the whole cell lysates of LPS-primed THP-1 cells treated with indicated doses of MK2206 (AKT inhibitor) or SC-79 (AKT activator) for 1 h, followed by LPS (100 ng/ml, 6 hrs), and Nigericin treatment (5 μM, 45 min).
(B) Immunoblots of the culture supernatants (Sup) and whole cell lysates (WCL) of BMDMs treated with indicated stimuli: MK2206 (AKT inhibitor, 100 nM, pre-treated 1 h), SC-79 (AKT activator, 1 μg/ml, 1.5 hrs), and/or LPS (100 ng/ml, 6 hrs), Nigericin (5 μM, 45 min).
(C) Immunoblots of the culture supernatants (Sup) and whole cell lysates (WCL) of BMDMs transfected with poly (dA:dT) (2 μg/ml, 6 hrs), and/or MK2206 (AKT inhibitor, 100 nM, pre-treated 1 h).
(D) Immunoblots of the supernatant and the whole cell lysates of the control-siRNA or Akt1/2-siRNA transfected THP-1 cells for 24 hours, followed by LPS (100 ng/ml, 6 h) and/or Nigericin (5 μM, 45 min) treatment.
(E) Quantification of the p-AKT and total-AKT protein levels in the control-siRNA or Akt1/2-siRNA transfected THP-1 cells.
(F) Elisa assays of IL-1β (left), IL-6 (middle), and TNF-α (right) in supernatants from the BMDMs treated with the indicated stimuli: MK2206 (AKT inhibitor, 100 nM, pre-treated 1 h); LPS (100 ng/ml, 6 h), following by Nigericin (5 μM, 45 min) or ATP (2 mM, 1 h); 2μg/ml poly (dA:dT) transfected for 6 hours. UT- untreated. Immunoblots shown are representative of at least 3 independent experiments and band quantification is shown for the representative blot. All experiments were repeated at least three times, and bars represent the means ± SEM. *p<0.05, **p<0.01, ***p<0.001, as determined by student’s t test.
To verify that the effects of MK2206 and SC-79 on NLRP3 inflammasome activity were specific to AKT, we assayed cleaved caspase-1 and IL-1β secretion after knockdown of AKT1/2 in NLRP3 activated THP-1 cells. AKT1/2 depleted macrophages showed a 2 to 3-fold increase in cleaved caspase-1 and IL-1β secretion compared to controls, further implicating AKT as a negative regulator of NLRP3 (Fig. 1D). The efficacy of AKT knockdown was confirmed at the protein level and found to be approximately 50% (Fig. 1D and E).
Previous work has shown that AKT inhibitors repress NF-κB dependent transcription (27). To determine if AKT regulates NLRP3 inflammasome activation via modulation of NF-κB, we stimulated untreated or MK2206 treated BMDMs with LPS, LPS plus nigericin, LPS plus ATP, or LPS plus poly (dA:dT) and measured the secretion of NF-κB dependent cytokines IL-6 and TNF-α in addition to IL-1β by ELISA. As before, AKT inhibition increased IL-1β levels after NLRP3 specific stimulation (nigericin and ATP) but not with LPS alone or AIM2 activation (Fig. 1F). Notably, there was no increase in IL-6 or TNF-α after LPS alone, LPS + ATP, or LPS + poly (dA:dT), but a slight increase in IL-6 and TNF-α after LPS + nigericin. Similarly, we found no transcriptional differences in IL-6, TNF-α, NLRP3, or ASC in THP-1 cells following LPS stimulation in the presence or absence of MK2206 (Fig. S1B). This data indicates that AKT negatively regulates inflammasome activation in an NF-κB-independent, NLRP3-specific manner in human and murine cells.
NLRP3 LRR domain interacts with the AKT kinase domain
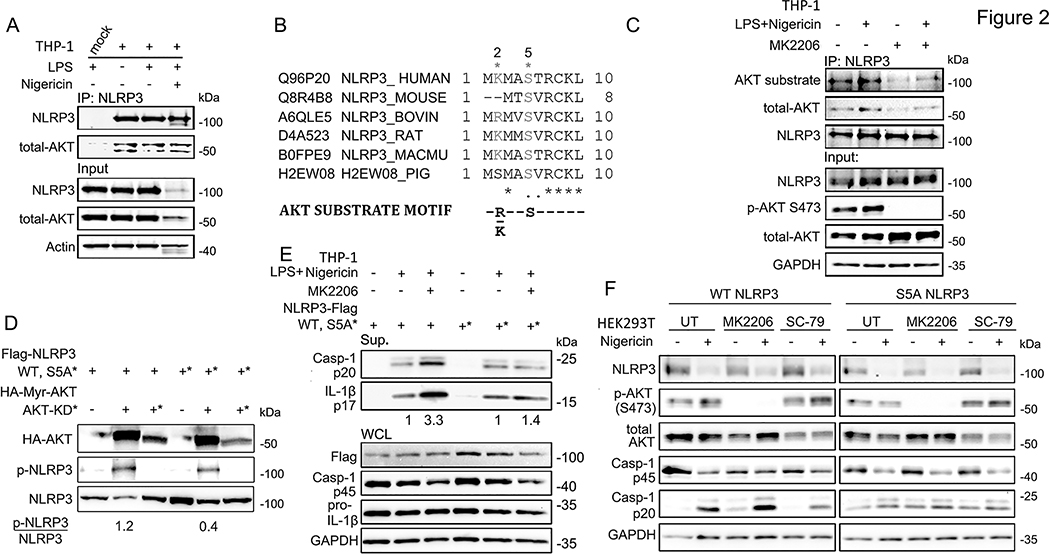
To investigate the mechanism through which AKT inhibits NLRP3 inflammasome activation, we first examined whether AKT co-immunoprecipitates with NLRP3. We assessed resting, LPS-primed, and nigericin-stimulated THP-1 cells and found endogenous AKT and NLRP3 co-immunoprecitated under each condition suggesting a constitutive interaction between the two proteins (Fig. 2A). Structurally, NLRP3 contains a pyrin domain (PYD), a NACHT domain, and leucine-rich repeats (LRRs). To identify the domain of NLRP3 that mediates its interaction with AKT, we used a series of Myc-tagged NLRP3 truncated mutants, including the NRLP3 PYD domain, NLRP3 PYD domain deletion mutant (ΔPYD), NLRP3 LRR domain, and NLRP3 LRR deletion mutant (ΔLRR) (Fig. S2A). Co-immunoprecipitation experiments in HEK293T cells with these truncated mutants showed that NLRP3 PYD and ΔLRR mutants lost their ability to bind AKT1 while NLRP3 ΔPYD and LRR mutants did not (Fig. S2B), indicating that the LRR domain of NLRP3 is required for the NLRP3-AKT interaction. AKT1 contains an N-terminal pleckstrin homology domain (PH, aa 1–108), a central kinase domain (aa 150–408), and a C-terminal regulatory domain (aa 409–480). To determine the domain through which AKT1 interacts with NLRP3, we created four HA-tagged truncated mutants including HA-AKT1 1–149 (containing the PH domain), HA-AKT1 150–480 (PH domain deletion mutant), HA-AKT1 120–233 (containing a partial kinase domain), and HA-AKT1 1–408 (a regulatory domain deletion mutant) (Fig. S2C). Co-immunoprecipitation experiments in HEK293T cells showed that Flag-NLRP3 interacted with HA-AKT1 WT, HA-AKT1 150–480, HA-AKT1 120–233, and HA-AKT1 1–408, but not with HA-AKT1 1–149 (Fig. S2D). These results suggest that AKT interacts with NLRP3 via its kinase domain.
Figure 2.
AKT interacts with NLRP3 and phosphorylates Serine 5.
(A) Immunoprecipitation assay showing that endogenous NLRP3 interacts with AKT in THP-1 cells with or without inflammasome activation.
(B) Uniprot alignment of NLRP3 orthologs surrounding Serine 5 site compares with motif of AKT-phosphate substrate.
(C) Immunoprecipitation assay showing the interaction of NLRP3 with AKT in THP-1 cells after treated with the indicated stimuli: MK2206 (100 nM, pre-treated 1 h), LPS (100 ng/ml, 6 hrs), followed by Nigericin (5 μM, 45 min).
(D) In vitro kinase assay showing AKT phosphorylation of NLRP3 S5.
(E) Immunoblots of culture supernatants and the whole cell lysates of Flag-NLRP3 WT or S5A mutant overexpressed THP-1 cells with the indicated stimuli: MK2206 (100 nM, pre-treated 1 h), LPS (100 ng/ml, 6 hrs), Nigericin (5 μM, 45 min).
(F) Immunoblots of the NLRP3 inflammasome reconstitution in HEK293T cells. To reconstitute the NLRP3 inflammasome, Flag-tagged ASC, and Flag-tagged pro-caspase-1 were transfected with Flag-NLRP3 WT or S5A mutant in HEK293T cells, and then treated with indicated stimuli: MK2206 (100 nM, pre-treated 1 h), SC-79 (1 μg/ml, 2 hrs), and/or Nigericin (5 μM, 45 min). Immunoblots shown are representative of at least 3 independent experiments and band quantification is shown for the representative blot.
NLRP3 phosphorylation at Serine 5 is mediated by AKT
Several studies have shown that phosphorylation of NLRP3 regulates its activation (28). The evolutionarily conserved NLRP3 S5 is one such site and prevents NLRP3 activation upon phosphorylation by regulating charge-charge interactions between PYD domains required for NLRP3 and ASC oligomerization (16). While it known that the phosphatase PP2A licenses NLRP3 activation by dephosphorylating this site, the kinase involved in its phosphorylation remains unknown (16). The known motif for AKT substrates is RXXS/T (or KXXS/T) (29), and the amino acid sequence surrounding serine 5 on human NLRP3 matches that of known AKT substrates (Fig. 2B). Thus, we hypothesized that AKT regulates NLRP3 activation via direct phosphorylation of NLRP3 at S5. To test this hypothesis, we immunoprecipitated endogenous NLRP3 from nigericin activated THP-1 cells and immunoblotted with an anti-phospho-AKT substrate (RXXS*/T*) antibody. Activation of the NLRP3 inflammasome increased the phosphorylation of NLRP3 at AKT-substrate sites, while treatment with MK2206 both decreased basal phosphorylation at these sites and prevented further phosphorylation after NLRP3 activation (Fig. 2C). These results suggest that phosphorylation of NLRP3 after its activation are AKT dependent. Next, we performed an in vitro phosphorylation assay to test whether AKT directly phosphorylates NLRP3 at S5. Immunoprecipitated HA-Myr-AKT (constitutively active) or HA-AKT-K179M (kinase dead) was mixed with HEK293T cell lysates expressing either Flag-NLRP3 WT or its non-phosphorylated mutant Flag-NLRP3 S5A. Phosphorylation of WT NLRP3 was induced by incubation with constitutively active AKT, while no phosphorylation was seen after incubation with kinase dead AKT (Fig. 2D). Importantly, the NLRP3 S5 mutant showed a three-fold reduction in phosphorylation compared to NLRP3 WT (Fig. 2D), further suggesting AKT directly phosphorylates NLRP3 on S5.
We then tested whether AKT regulated NLRP3 activity via its phosphorylation at NLRP3 S5 by transiently transfecting NRLP3 WT or its S5A mutant into PMA differentiated THP-1 cells and activating the NLRP3 inflammasome in the presence or absence of MK2206. MK2206 increased cleaved caspase-1 and IL-1β after inflammasome activation in the presence of WT NLRP3, but not its S5A mutant (Fig. 2E), suggesting AKT inhibits the NLRP3 inflammasome via activity at NLRP3 serine 5. Finally, we used an inflammasome reconstitution system in HEK293T cells to validate our findings. As expected, AKT inhibitor MK2206 increased cleaved caspase-1 levels while the AKT activator SC-79 decreased cleaved caspase-1 after WT NLRP3 activation, but neither inhibitor affected caspase-1 cleavage after activation of NLRP3 S5A (Fig. 2F). Taken together, our results demonstrate that AKT inhibits NLRP3 inflammasome activation by direct phosphorylation of NLRP3 S5.
AKT regulates NLRP3 oligomerization and degradation during activation of the NLRP3 inflammasome
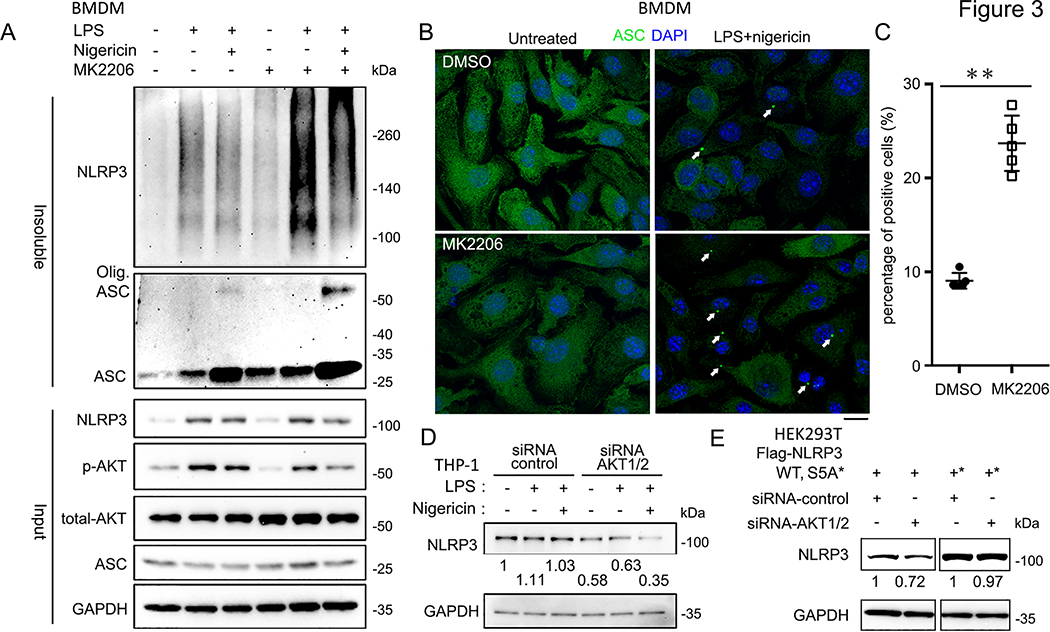
Activation of the NLRP3 inflammasome results in its PYD domain dependent auto-oligomerization and subsequent recruitment/oligomerization of the adaptor protein ASC, resulting in the formation of a prion like complex which is visible as ASC specks by immunofluorescence (30, 31). Previous studies have shown that phosphorylation of NLRP3 S5 negatively regulates this process due to electrostatic repulsion between PYD domains, which inhibits NLRP3 self-oligomerization (16). In order to show that AKT mediated changes in cleaved caspase-1 and IL-1β following NLRP3 activation are due to its phosphorylation of NLRP3 S5, we tested whether AKT affects the oligomerization of NLRP3 and ASC by isolating the triton insoluble fraction of LPS and nigericin treated BMDMs. As expected, activation of the NLRP3 inflammasome with high dose LPS (1 μg/ml for 3 h) or LPS (1 μg/ml for 3 h) + nigericin (15 min) increased the amount of oligomerized NLRP3 detected in the triton insoluble fraction compared to the controls. Total ASC and its oligomerized form were also increased in the triton insoluble fraction after treatment with LPS plus nigericin (Fig. 3A). Pretreatment with the AKT inhibitor MK2206 further increased NLRP3 oligomerization both after treatment with LPS alone and after LPS plus nigericin (Fig. 3A). To confirm these findings, we used confocal fluorescent microscopy to assay ASC speck formation in BMDMs. We consistently found enhanced ASC speck formation in MK2206 treated BMDMs after activation of the NLRP3 inflammasome (Fig. 3B, C). Taken together, this data shows that AKT kinase activity, presumably via phosphorylation on NLRP3 S5, inhibits NLRP3 and ASC oligomerization after activation of the NLRP3 inflammasome. Furthermore, this process may begin during the priming stage of inflammasome activation, as inhibition of AKT enhances NLRP3 oligomerization after treatment with LPS alone.
Figure 3.
AKT is involved in NLRP3 oligomerization and degradation with inflammasome activation.
(A) Immunoblots of DSS cross-linked insoluble fraction of DMSO- or MK2206-treated BMDMs after treating with the indicated stimuli: MK2206 (100 nM, pre-treated 1 h), LPS (1 μg/ml, 3 hrs) and Nigericin (5 μM, 15 min).
(B) Immunofluorescence images of BMDMs treated with indicated stimuli: MK2206 (100 nM, pre-treated 1 h), LPS (1 μg/ml, 3 hrs) and Nigericin (5 μM, 15 min), labeled with anti-ASC antibodies (green). The nuclei were labeled by DAPI (blue). Arrows indicate the ASC speck-like structure in the cells.
(C) Qualification of the percentage of the BMDMs with ASC speck like structure from (B). Experiments were repeated three times, more than 100 cells were qualified for each replicate, and bars represent the means ± SEM. **p<0.01, as determined by student’s t test.
(D) Immunoblots of the whole cell lysates of control-siRNA or AKT1/2-siRNA transfected THP-1 cells treated with the indicated stimuli: LPS (100 ng/ml, 6 hrs), Nig. (5 μM, 45 min). Expressions of endogenous NLRP3 protein level were normalized to untreated group.
(E) Immunoblots of the whole cell lysates of Flag-NLRP3 WT, S5A, or S5E overexpressed in control-siRNA transfected or AKT 1/2 depleted HEK293T cells. Blotted NLRP3 protein levels were normalized to control-siRNA transfected samples, respectively. Immunoblots shown are representative of at least 3 independent experiments and band quantification is shown for the representative blot.
In addition to phosphorylation, NLRP3 activation is regulated by ubiquitin dependent degradation (32). Thus, we next evaluated whether AKT regulation of NLRP3 degradation contributes to the AKT dependent inflammasome phenotypes observed. Surprisingly, knockdown of AKT1/2 in THP-1 cells resulted in a 3-fold decrease in NLRP3 protein levels after priming with LPS (Fig. 3D). To evaluate whether AKT phosphorylation of NLRP3 S5 is involved its regulation of NLRP3 protein levels, we transfected NLRP3 WT or the NLRP3 S5A mutant into HEK293T cells while knocking down AKT1/2. Knockdown of AKT1/2 decreased NLRP3 WT levels but had no effect on the protein levels of NLRP3 S5A (Fig. 3E). These results suggest that AKT phosphorylation of NRLP3 S5 prevents the degradation of NLRP3 during the LPS priming phase. Notably, AKT mediated phosphorylation of NLRP3 S5 dually and opposingly regulates NLRP3 activation, inhibiting its assembly but preventing its degradation.
NLRP3 S5 phosphorylation prevents Trim31-mediated ubiquitination of NLRP3 at Lysine 496
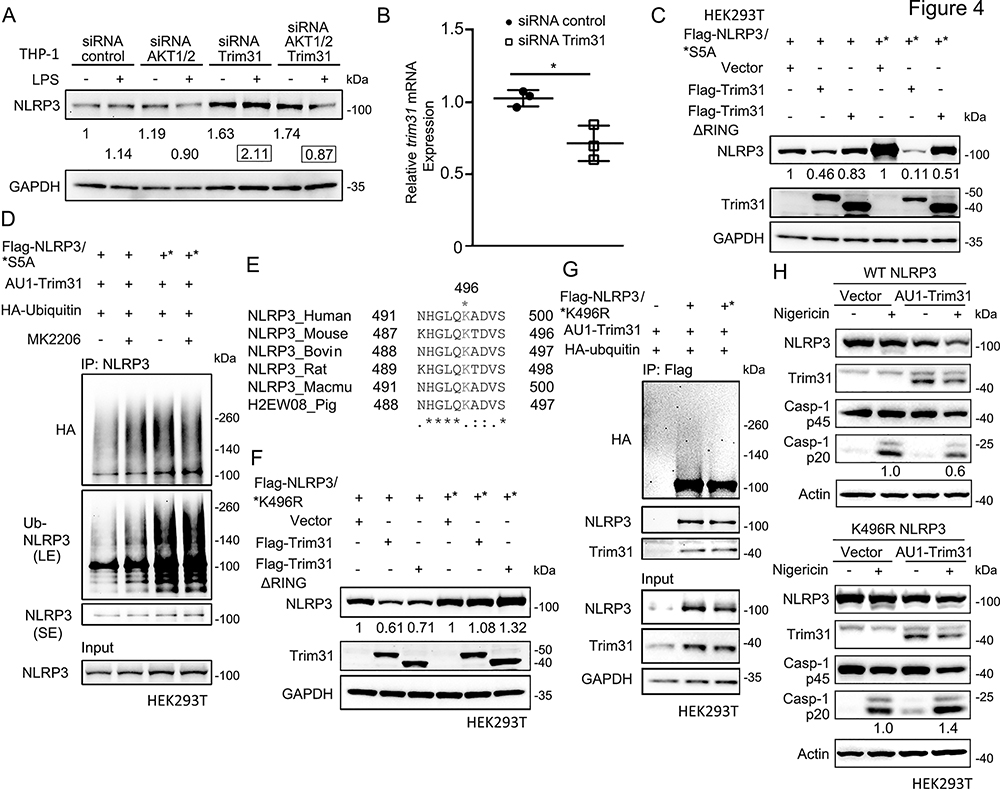
TRIM31 functions as an E3 ligase that induces NLRP3 ubiquitin-proteasome degradation (32). To investigate TRIM31’s involvement in AKT mediated regulation of NLRP3 protein levels, we first reduced the expression of AKT1/2, TRIM31, or both in THP-1 cells and assayed NLRP3 protein levels. While we did not observe a consistent change in NLRP3 levels after AKT knockdown in the unprimed state, knockdown of TRIM31 and simultaneous AKT1/2 and TRIM31 knockdown increased NLRP3 protein levels 1.63 and 1.74-fold respectively (Fig. 4A). In LPS primed THP-1 cells, AKT1/2 knockdown consistently decreased NLRP3 levels while TRIM31 knockdown increased NLRP3 levels; however, simultaneous knockdown of AKT1/2 and TRIM31 markedly decreased NLRP3 levels compared to the control (Fig. 4A). Adequate knockdown of TRIM31 was verified by qRT-PCR (Fig. 4B). This data indicates that after LPS priming, AKT knockdown promotes NLRP3 degradation either through a pathway parallel to TRIM31 or by sensitizing NLRP3 to TRIM31 mediated degradation.
Figure 4.
S5 phosphorylation prevents Trim31-mediated ubiquitination of NLRP3 at K496 site.
(A) Immunoblots of the whole cell lysates of control-siRNA-, AKT1/2-siRNA-, or/and Trim31-siRNA-transfected LPS-primed THP-1 cells. The endogenous NLRP3 protein levels were normalized to control-siRNA transfected untreated sample (showed in fold).
(B) Quantification of the Trim31 mRNA level in control-siRNA- or Trim31-siRNA-treated THP-1 cells. Control-siRNA or Trim31-siRNA was transfected into THP-1 cells for 48 hrs, and the trim31 mRNA level was tested from real-time PCR. Experiments were repeated three times, and bars represent the means ± SEM. *p<0.05, as determined by student’s t test.
(C) Immunoblots of the whole cell lysates of the HEK293T cells transfected with Flag-NLRP3 (WT or S5A mutant), and Flag-Trim31 (WT or ΔRing mutant). Expressions of transfected NLRP3 protein level were normalized to vector-transfected groups, respectively.
(D) Immunoblots showing the ubiquitination of NLRP3. AU1-Trim31 and HA-Ubiquitin were co-transfected into HEK293T cells with Flag-NLRP3 WT or S5A mutant, followed by treatment of MK2206 (100 nM, 6 hrs.) and MG132 (20 μM, 4 hrs). The whole cell lysates were immunoprecipitated with anti-NLRP3 and immunoblotted with indicated antibodies.
(E) Uniprot alignment of NLRP3 orthologs surrounding Lysine 496 site.
(F) Immunoblots of the whole cell lysates of the HEK293T cells transfected with Flag-NLRP3 (WT or K496R mutant), and Flag-Trim31 (WT or ΔRing mutant). Expressions of transfected NLRP3 protein level were normalized to vector-transfected groups, respectively.
(G) Immunoblots showing the ubiquitination of NLRP3. AU1-Trim31 was co-transfected into HEK293T cells with Flag-NLRP3 WT or K496R mutants, followed by treatment of MG132 (20 μM, 4 hrs.). The whole cell lysates were immunoprecipitated with anti-Flag and immunoblotted with indicated antibodies.
(H) Immunoblots of the NLRP3 inflammasome reconstitution in HEK293T cells. To reconstitute the NLRP3 inflammasome, Flag-tagged ASC, Flag-tagged pro-caspase-1, and AU1-tagged Trim31 were transfected with Flag-NLRP3 WT or K496R mutant in HEK293T cells, and then respectively treated with Nigericin (5 μM, 45 min) before collection. Immunoblots shown are representative of at least 3 independent experiments and band quantification is shown for the representative blot.
We next expressed NLRP3 WT or its S5A mutant with TRIM31 WT or its E3-ubiquitin ligase loss of function mutant (TRIM31ΔRING) in HEK293T cells. Expression of TRIM31, but not TRIM31ΔRING decreased protein levels of NLRP3 WT while NLRP3 S5A was degraded more efficiently by expression of TRIM31 (Fig. 4C), suggesting non-phosphorylated NLRP3 is more sensitive to TRIM31 mediated degradation. Consistently, in HEK293T cells inhibition of AKT enhanced the ubiquitination of NLRP3 WT after co-expression of TRIM31, while the NLRP3 S5A mutant showed higher levels of basal phosphorylation and was not affected by the expression of TRIM31 (Fig. 4D). In sum, these experiments show that AKT mediated phosphorylation of NLRP3 S5 negatively regulates TRIM31 mediated NLRP3 degradation.
While it is known that TRIM31 drives NLRP3 ubiquitin-proteasome degradation, the ubiquitination site on NLRP3 remains unknown (32). Using the PhosphoSitePlus database (https://phosphosite.org/), we found that Lysine 496 (K496) is a previously identified evolutionarily conserved ubiquitination site based on mass spectrometry (Fig. 4E). We constructed a non-ubiquitinated NLRP3 K496R mutant and expressed it with TRIM31 in HEK293T cells. While NLRP3 WT protein levels are reduced upon expression of TRIM31, NLRP3 K496R protein levels are unaffected by expression of TRIM31 (Fig. 4F). Consistently, TRIM31 induced NLRP3 ubiquitination is reduced in the NLRP3 K496R mutant compared to NLRP3 WT (Fig. 4G), suggesting that NLRP3 K496 ubiquitinated by TRIM31. We then confirmed a regulatory role for NLRP3 K496 in inflammasome activation using a reconstitution system in HEK293T cells. Overexpressed TRIM31 only impaired caspase-1 activation after nigericin treatment in NLRP3 WT expressing cells, but not in NLRP3 K496R expressing (Fig. 4H). Taken together, our data demonstrates that the AKT-NLRP3 axis facilitates the stability of NLRP3 by phosphorylating NLRP3 S5, which prevents TRIM31 ubiquitination at K496 and its subsequent ubiquitin-proteasome degradation.
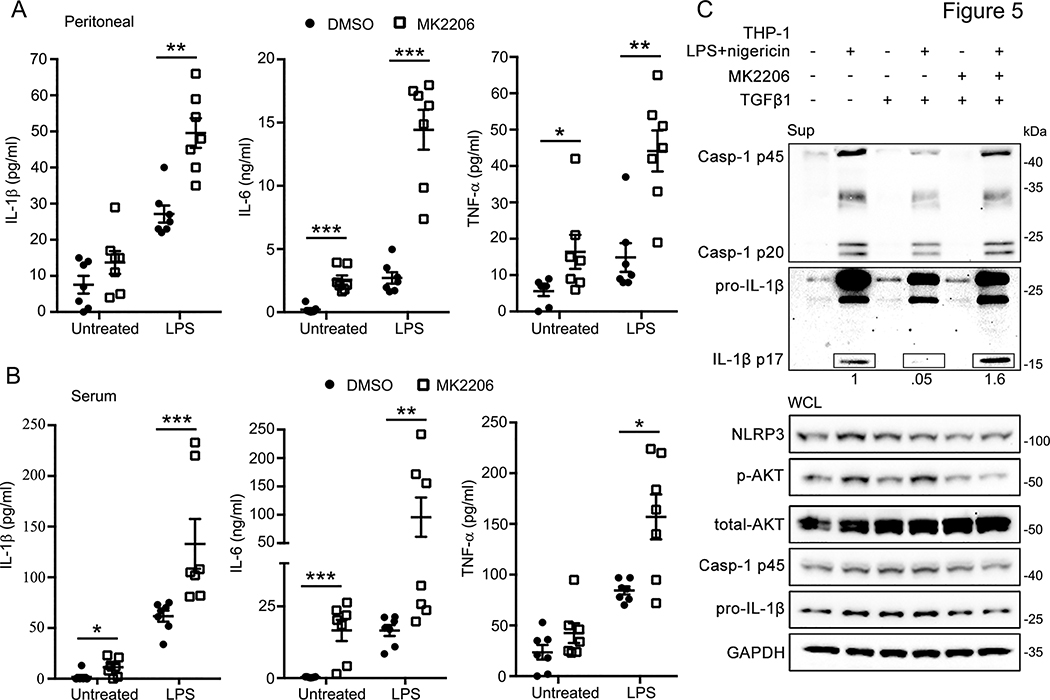
AKT regulates inflammation in vivo and is required for TGF-β mediated inflammasome activation
Finally, we pursued two lines of experiments to establish the physiological relevance of our findings. To characterize whether AKT regulates inflammation in vivo, we used intraperitoneal low dose LPS (0.5 mg/kg) to induce inflammation in mice in the presence or absence of MK2206 (80 mg/kg). Consistent with our in vitro data, IL-1β was increased in both peritoneal lavage samples and blood serum of MK2206 treated mice compared to the control (Fig. 5A, B). The results also show increased IL-6 and TNF-α in peritoneal lavage and serum samples of MK2206 treated mice (Fig. 5A&B), potentially due to enhanced inflammation downstream of NLRP3 activation. Given the concordance between the in vivo and in vitro data, it is likely that AKT regulates NLRP3 inflammasome activation in vivo. Second, we investigated whether AKT plays a role in the physiological inhibition of NLRP3 inflammasome activity by TGF-β. TGF-β is a broadly immunosuppressive cytokine known both to rapidly induce phosphorylation of AKT (33) and to restrict spontaneous activation of NLRP3 (34, 35). In nigericin activated THP-1 cells, TGF-β reduced the secretion of cleaved caspase-1 and mature IL-1β as previously reported. However, pre-incubation with the AKT inhibitor MK2206 attenuated the inhibitory activity of TGF-β after NLRP3 inflammasome activation (Fig. 5C), implicating AKT downstream of TGF-β in NLRP3 inflammasome inhibition. These data provide evidence that AKT plays a physiological role in the regulation of NLRP3 inflammasome activation.
Figure 5.
Inhibition of p-AKT can promote inflammasome activation in vivo and recover the reduction of inflammasome by TGFβ1.
(A-B) Elisa assays of IL-1β, IL-6, and TNF-α in peritoneal lavage (A) or serum (B) from C57BL/6J mice after treated with MK2206 (80 mg/kg, 2 hrs., i.p.), and followed by LPS (0.5 mg/kg, 4 hrs., i.p.). n = 7 mice/group with ELISAs performed in triplicate. Bars represent the mean ± SEM, *p<0.05, **p<0.01, ***p<0.001 as determined by Mann-Whitney test.
(C) Immunoblots of culture supernatants and the whole cell lysates of THP-1 cells treated with the indicated stimuli: MK2206 (100 nM, pre-treated 2 hrs.), TGFβ1 (10 ng/ml, pre-treated 1 h), LPS (100 ng/ml, 6 hrs.), and Nigericin treatment (5 μM, 45 min). Immunoblots shown are representative of at least 3 independent experiments and band quantification is shown for the representative blot.
Discussion
Our results delineate a model through which AKT, TRIM31, and PP2A function together to regulate the NLRP3 inflammasome. During the priming phase (signal 1), AKT mediated phosphorylation of NLRP3 S5 enhances NLRP3 stability via inhibition of TRIM31 mediated ubiquitination and degradation. In response to signal 2, AKT phosphorylation of NLRP3 S5 prevents excess NLRP3 oligomerization and reduces IL-1β production. Previous studies indicate that dephosphorylation of NLRP3 S5 by PP2A is important in NLRP3 activation (16), suggesting AKT and PP2A function together to tightly regulate the threshold for NLRP3 activation. Notably, AKT directly acts on NLRP3 (and not ASC), as modulation of AKT activity does not affect activation of the AIM2 and NLRC4 inflammasomes (ASC is a component of both).
Several kinases are known to modify NLRP3. JNK1 is a positive regulator of NLRP3 assembly via phosphorylation of NLRP3 S194 (13), while PKA is a negative regulator of NLRP3 assembly via phosphorylation of NLRP3 S295 (15). Here, we identify AKT as the kinase responsible for phosphorylating NLRP3 S5. Previous studies show that NLRP3 S5 phosphorylation hinders homotypic NLRP3 PYD interactions and heterotypic NLRP3/ASC PYD interactions, thus preventing accidental NLRP3 assembly. Consistently, we show that AKT inhibits NLRP3 oligomerization during LPS priming and reduces cytokine secretion after NLRP3 activation via phosphorylation of NLRP3 S5. We delineate additional mechanisms by which AKT modulates NLRP3, showing NLRP3 S5 phosphorylation inhibits TRIM31 mediated NLRP3 ubiquitination at K496 and subsequent degradation.
Based our observations and previous investigations (16), there is considerable evidence that AKT contributes to the tight regulation of NLRP3 seen physiologically. We propose that upon LPS priming, AKT is activated to phosphorylate NLRP3 S5. This first prevents TRIM31 mediated NLRP3 K496 ubiquitination and subsequent degradation. Concomitantly, AKT inhibits NLRP3 oligomerization thereby preventing excess inflammasome activity in the absence of a second signal. Upon NLRP3 activation by a second signal, PP2A mediated dephosphorylation of NLRP3 S5 licenses NLRP3 oligomerization and enables NLRP3 inflammasome assembly (16). This dephosphorylation likely enables TRIM31-mediated ubiquitination and degradation of NLRP3, which may play a role in preventing excessive inflammation following NLRP3 inflammasome activation.
AKT has several immunosuppressive functions in monocytes and macrophages; it decreases monocyte activation after LPS stimulation (18), skews macrophages towards an M2 (anti-inflammatory) phenotype (36), and functions downstream of the immunosuppressive TGF-β pathway (33). Our data extends the mechanisms through which AKT exerts its anti-inflammatory effects to include regulation of NLRP3. Interestingly, PP2A is known to dephosphorylate AKT at T308, leading to kinase inactivation (37). Thus, in addition to directly dephosphorylating NLRP3 S5 during inflammasome activation (16), PP2A may indirectly decrease S5 phosphorylation by terminating AKT activation. While further investigation is needed to determine the molecular details, our study firmly implicates AKT in TGF-β mediated NLRP3 inhibition and provides in vivo evidence of AKT mediated NLRP3 inhibition.
Finally, in addition to implicating AKT as a regulator of NLRP3, we provide evidence that NLRP3 K496 is a bona fide TRIM31 ubiquitination site. Previous studies have shown that several ubiquitin E3 ligases, including FBXL2, PARKIN, TRIM31 or MARCH7, function as negative regulator of NLRP3 by promoting its proteasome or autophagy dependent degradation (11, 32, 38, 39). Despite the identification of several E3 ligases that mediate the ubiquitination of NLRP3, the precise ubiquitination sites have not been identified. Further studies are needed to determine whether other E3 ligases target NLRP3 K496, which we have shown to be important in controlling NLRP3 stability.
There are certain limitations to our study which merit discussion. First, AKT and TRIM31 have feedback loops effecting NF-κB activation. We provide several lines of evidence suggesting AKT modulation of NLRP3 is largely NF-κB independent. For example, we observed no transcriptional differences in IL-6, TNF-α, NLRP3, or ASC in LPS stimulated THP-1 cells with or without MK2206 (Fig. S1B). Similarly, we see no differences in TNF-α and IL-6 secretion in BMDMs following LPS with or without MK2206 (Fig. 1F). Nevertheless, it remains possible that AKT or TRIM31 mediated NF-κB feedback loops contribute to the phenotypes observed. Furthermore, our in vivo data uses a broad inducer of inflammation (LPS) rather than NLRP3 specific activator. Given the increase in IL-1β in this model, it is likely that AKT modulation of NLRP3 explains some but not all of the phenotypes seen in in vivo. In sum, our studies provide insight into the molecular mechanisms governing NLRP3 inflammasome activation. Improved understanding of NLRP3 activation may result in the development of novel therapeutics targeting NLRP3.
Supplementary Material
Key Points.
AKT phosphorylates NLRP3 at S5, preventing self-oligomerization and IL-1β release.
NLRP3 S5 phosphorylation prevents TRIM31-mediated degradation during LPS priming.
Acknowledgements
Dr. Anthony Fauci for his longstanding support. This research was supported by the intramural program of NIAID, project number AI000739–25.
Footnotes
Conflict of interest: The authors declare no conflicts of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Martinon F, Burns K, and Tschopp J 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular cell 10: 417–426. [DOI] [PubMed] [Google Scholar]
- 2.Petrilli V, Dostert C, Muruve DA, and Tschopp J 2007. The inflammasome: a danger sensing complex triggering innate immunity. Current opinion in immunology 19: 615–622. [DOI] [PubMed] [Google Scholar]
- 3.Harris J, Lang T, Thomas JPW, Sukkar MB, Nabar NR, and Kehrl JH 2017. Autophagy and inflammasomes. Mol Immunol 86: 10–15. [DOI] [PubMed] [Google Scholar]
- 4.Broz P, and Dixit VM 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16: 407–420. [DOI] [PubMed] [Google Scholar]
- 5.Swanson KV, Deng M, and Ting JPY 2019. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nature Reviews Immunology 19: 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, and Dixit VM 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440: 228–232. [DOI] [PubMed] [Google Scholar]
- 7.Martinon F, Petrilli V, Mayor A, Tardivel A, and Tschopp J 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440: 237–241. [DOI] [PubMed] [Google Scholar]
- 8.Becker CE, and O’Neill LA 2007. Inflammasomes in inflammatory disorders: the role of TLRs and their interactions with NLRs. Seminars in immunopathology 29: 239–248. [DOI] [PubMed] [Google Scholar]
- 9.Freche B, Reig N, and van der Goot FG 2007. The role of the inflammasome in cellular responses to toxins and bacterial effectors. Seminars in immunopathology 29: 249–260. [DOI] [PubMed] [Google Scholar]
- 10.Gong T, Jiang W, and Zhou R 2018. Control of Inflammasome Activation by Phosphorylation. Trends in biochemical sciences 43: 685–699. [DOI] [PubMed] [Google Scholar]
- 11.Han S, Lear TB, Jerome JA, Rajbhandari S, Snavely CA, Gulick DL, Gibson KF, Zou C, Chen BB, and Mallampalli RK 2015. Lipopolysaccharide Primes the NALP3 Inflammasome by Inhibiting Its Ubiquitination and Degradation Mediated by the SCFFBXL2 E3 Ligase. The Journal of biological chemistry 290: 18124–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bednash JS, and Mallampalli RK 2016. Regulation of inflammasomes by ubiquitination. Cellular & molecular immunology 13: 722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song N, Liu ZS, Xue W, Bai ZF, Wang QY, Dai J, Liu X, Huang YJ, Cai H, Zhan XY, Han QY, Wang H, Chen Y, Li HY, Li AL, Zhang XM, Zhou T, and Li T 2017. NLRP3 Phosphorylation Is an Essential Priming Event for Inflammasome Activation. Mol Cell 68: 185–197 e186. [DOI] [PubMed] [Google Scholar]
- 14.Ren G, Zhang X, Xiao Y, Zhang W, Wang Y, Ma W, Wang X, Song P, Lai L, Chen H, Zhan Y, Zhang J, Yu M, Ge C, Li C, Yin R, and Yang X 2019. ABRO1 promotes NLRP3 inflammasome activation through regulation of NLRP3 deubiquitination. EMBO J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, Zheng M, Zhang X, Xia D, Ke Y, Lu L, and Wang D 2016. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity 45: 944. [DOI] [PubMed] [Google Scholar]
- 16.Stutz A, Kolbe CC, Stahl R, Horvath GL, Franklin BS, van Ray O, Brinkschulte R, Geyer M, Meissner F, and Latz E 2017. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med 214: 1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manning BD, and Toker A 2017. AKT/PKB Signaling: Navigating the Network. Cell 169: 381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guha M, and Mackman N 2002. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. The Journal of biological chemistry 277: 32124–32132. [DOI] [PubMed] [Google Scholar]
- 19.Luyendyk JP, Schabbauer GA, Tencati M, Holscher T, Pawlinski R, and Mackman N 2008. Genetic analysis of the role of the PI3K-Akt pathway in lipopolysaccharide-induced cytokine and tissue factor gene expression in monocytes/macrophages. J Immunol 180: 4218–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troutman TD, Hu W, Fulenchek S, Yamazaki T, Kurosaki T, Bazan JF, and Pasare C 2012. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proceedings of the National Academy of Sciences of the United States of America 109: 273–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vural A, Nabar NR, Hwang IY, Sohn S, Park C, Karlsson MCI, Blumer JB, and Kehrl JH 2019. Galphai2 Signaling Regulates Inflammasome Priming and Cytokine Production by Biasing Macrophage Phenotype Determination. J Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue Y, Nabar NR, Shi CS, Kamenyeva O, Xiao X, Hwang IY, Wang M, and Kehrl JH 2018. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis 9: 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi C-S, Nabar NR, Huang N-N, and Kehrl JH 2019. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discovery 5: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, and Dixit VD 2011. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nature medicine 17: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kong H, Zeng X, Liu W, Wang Z, Yan X, Wang H, and Xie W 2016. Activation of NLRP3 inflammasome enhances the proliferation and migration of A549 lung cancer cells. Oncology reports 35: 2053–2064. [DOI] [PubMed] [Google Scholar]
- 26.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, and Cohen P 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 7: 261–269. [DOI] [PubMed] [Google Scholar]
- 27.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, and Baldwin AS 2008. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes & development 22: 1490–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song N, and Li T 2018. Regulation of NLRP3 Inflammasome by Phosphorylation. Frontiers in immunology 9: 2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, and Comb MJ 2010. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal 3: ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, and Egelman EH 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156: 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, Hauenstein AV, Wu Z, Nunez G, Mao Y, and Wu H 2019. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, Han L, Jiang G, Zhang L, Gao C, and Zhao W 2016. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun 7: 13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkes MC, Mitchell H, Penheiter SG, Dore JJ, Suzuki K, Edens M, Sharma DK, Pagano RE, and Leof EB 2005. Transforming growth factor-beta activation of phosphatidylinositol 3-kinase is independent of Smad2 and Smad3 and regulates fibroblast responses via p21-activated kinase-2. Cancer Res 65: 10431–10440. [DOI] [PubMed] [Google Scholar]
- 34.Malireddi RKS, Gurung P, Mavuluri J, Dasari TK, Klco JM, Chi H, and Kanneganti TD 2018. TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. The Journal of experimental medicine 215: 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangan MS, and Latz E 2018. TAK1ng control: TAK1 restrains NLRP3 activation. The Journal of experimental medicine 215: 1007–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, and Tsatsanis C 2017. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J Immunol 198: 1006–1014. [DOI] [PubMed] [Google Scholar]
- 37.Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, and Chiang CW 2008. Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. The Journal of biological chemistry 283: 1882–1892. [DOI] [PubMed] [Google Scholar]
- 38.Mouton-Liger F, Rosazza T, Sepulveda-Diaz J, Ieang A, Hassoun SM, Claire E, Mangone G, Brice A, Michel PP, Corvol JC, and Corti O 2018. Parkin deficiency modulates NLRP3 inflammasome activation by attenuating an A20-dependent negative feedback loop. Glia 66: 1736–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, and Zhou R 2015. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 160: 62–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.