Abstract
Mechanisms regulating nuclear organization control fundamental cellular processes, including the cell and chromatin organization. Their disorganization, including aberrant nuclear architecture, has been often implicated in cellular transformation. Here, we identify Lamin A, among proteins essential for nuclear architecture, as SPANX, a cancer testis antigen previously linked to invasive tumor phenotypes, interacting protein in melanoma. SPANX interaction with Lamin A was mapped to the immunoglobulin fold-like domain, a region critical for Lamin A function, which is often mutated in laminopathies. SPANX down-regulation in melanoma cell lines perturbed nuclear organization, decreased cell viability and promoted senescence-associated phenotypes. Moreover, SPANX knockdown in melanoma cells promoted proliferation arrest, a phenotype mediated in part by IRF3/IL1A signaling. SPANX-knockdown in melanoma cells also prompted the secretion of IL1A, which attenuated the proliferation of naive melanoma cells. Identification of SPANX as a nuclear architecture complex component provides an unexpected insight into the regulation of Lamin A and its importance in melanoma.
Keywords: Melanoma, Lamin A/C, SPANX, Nuclear Architecture, IRF3
INTRODUCTION
Melanoma is the most aggressive form of skin cancer due to its propensity to metastasize and colonize distant organs (1). Understanding molecular mechansims underlying melanoma progression still represents a major knowledge gap, one that hinders effective clinical treatment. The capacities of tumors to grow, metastasize and resist treatment are often linked to their success in surviving harsh environmental conditions, including the ability to counter mechanical pressure generated as tumors grow in a constrained environment, either in the primary tumor, in narrow spaces during metastasis or when subjected to shear stress in the circulation (2). Protection of nuclei from mechanical constriction is provided by the lamina, a filamentous layer located at the inner nuclear membrane and composed mostly of lamins. In mammals, A-type lamins (mainly lamins A and C) are encoded by the LMNA gene, while B-type lamins are encoded by LMNB1 and LMNB2. Lamins are type V-intermediate filament proteins composed of an N-terminal head domain, a central rod domain required for polymerization, and a long C-terminal tail containing an immunoglobulin-like domain that functions in protein/protein interactions (3). While B-type lamins confer elasticity to nuclei, A-type lamins provide stiffness that renders nuclei resistant to deformation (4). Lamins also function in numerous cellular process including chromatin organization, DNA repair and the cell cycle (3).
Deregulation of LMNA function by mutation, as seen in laminopathies, leads to premature aging and death. These disorders are marked by altered chromatin organization and changes in gene expression, DNA damage and impaired DNA repair (4), as well as aberrantly-shaped nuclei and the appearance of nuclear blebs and micronuclei (5). Such changes promote cell cycle arrest and senescence and partially depend on changes in TP53/p53 activity (6). Lamin A has been also implicated in the control of cell division by controlling RB1 (Retinoblastoma-associated protein) or PCNA (Proliferating Cell Nuclear Antigen) (7,8), as in the control of mitotic spindle assembly and positioning (9) or nuclear envelope assembly at the end of mitosis (3).
Replication stress, nuclear envelope rupture and appearance of micronuclei—all seen in laminopathic and cancer cells—can also activate innate immunity responses (10). Indeed, ssDNA or cytosolic chromatin are detected by protein sensors that activate TBK1, resulting in activation of the NF-κB pathway or IRF3 (Interferon Regulatory Factor 3) (11). Replication stress in laminopathic cells also induces an interferon-like response, as reflected by increased STAT1 phosphorylation (12). In cancer, mitotic stress leads to formation of micronuclei (13), which activate inflammatory pathways (14). Interestingly, activation of innate immunity potentiates the response to anti-CTLA4 therapy in a melanoma model (15). However, activation of these pathways is also linked with the propensity to metastasize (16).
In mammals, the Sperm Protein Associated with the Nucleus on the X chromosome (SPANX) gene family includes five highly homologous members (SPANX-A1, -A2, -B1, -C and -D) (17) that differ in subcellular localization and relative expression in normal versus transformed cells (18,19). SPANX-A1, -A2, -C and -D constitute the A-type family and consist of 97 amino acid showing 94% identity, making them hardly differentiable. As the only member of the B-family, SPANXB1 contains a unique six amino acid insertion within its N-terminal domain (17). Hereafter, we use the term “SPANX” to refer to A-type SPANX genes, which under physiological conditions, are expressed only in testis and considered “cancer testis antigens” (20). However, in pathological conditions, such as cancers including melanoma, hematological malignancies and breast cancer, SPANX is expressed in non-testis tissues. Among different tumor types, SPANX-A1 and -A2 levels are particularly high in melanoma (20,21). Moreover, SPANX expression reportedly increases with melanoma stage (22,23). While some report that SPANX expression antagonizes the epithelial to mesenchymal transition in a lung cancer model (24), others suggest that SPANX expression promotes invasion of breast cancer cells (25). Herein, we identify SPANX as a lamin A/C interacting protein in melanoma and report that decreasing SPANX expression alters nuclear architecture in a manner that restricts melanoma cell proliferation.
MATERIALS AND METHODS
Cell culture
Human HEK293T cells were obtained from the American Type Culture Collection (ATTC). A375 were obtained from ATCC and WM1366 from the Wistar Institute, UACC612 from the university of Arizona Cancer Center. These cells were grown in DMEM as outlined (26). MM150922 were derived from melanoma patient and grown in RPMI as described earlier (27). All lines were maintained at 37°C in a humidified atmosphere with 5% CO2. Cell lines were regularly checked for mycoplasma contamination using a luminescence-based kit (Lonza) and were kept in culture for up to 6 weeks. Melanoma cell lines were authenticated at SBP core genomic facility.
Reagents and antibodies
Puromycin, blasticidin and doxycyclin were purchased from Merck. Agarose beads conjugated to anti-FLAG or anti-HA antibodies were purchased from Merck (catalog # A2220) or Roche (catalog # 11 815 016 001), respectively. Antibodies were purchased as follows: SPANX (Abcam; ab47252 diluted at 1/1000 for western blot and ab119280 diluted at 1/200 for immunofluorescence), GAPDH (Abcam; ab8245, diluted at 1/1000), FLAG (Merck; F1804, diluted at 1/1000), HA (Biolegend; 16B12, diluted at 1/1000), LMNA (Santa Cruz Biotechnology; sc376248, diluted at 1/1000 and 1/200 for western blot and immunofluorescence respectively), V5 (Cell signaling Technology; D3H8Q, diluted at 1/1000), H3 (Santa Cruz Biotechnology; sc8654, diluted at 1/200), and IRF3 (Cell Signaling Technology; 10949, diluted at 1/200).
Plasmids
To generate SPANXA and lamin A WT and mutant expression constructs, 1ng of appropriate cDNA derived from A375 cells was PCR-amplified and subcloned into pre-digested ApaI and EcoRI-cut pcDNA3 vector using a NEBuilder® HiFi DNA Assembly kit (New England Biolabs). The same protocol was used to generate doxycycline-inducible SPANXA and SPANXC overexpression vectors, except that PCR-amplified cDNA was subcloned into EcoRI-cut pLVX TetOne-puro plasmid (Clontech). Primer sequences used to generate these constructs are available upon request. Lentiviral pLKO.1 vectors expressing SPANX-specific shRNAs were obtained from the La Jolla Institute for Immunology RNAi Center (La Jolla, CA, USA). Doxycycline-inducible lentiviral pLKO.1 vectors expressing IRF3 or IL1A-specific shRNAs were generated by amplification of the shRNA sequence obtained by chemical synthesis (Merck) and subcloned into EcoRI-linearized lentiviral pLKO.1 vectors expressing blasticidine resistance.
Cell transfection and transduction
For siRNA and plasmid transfection, cells were transfected according to the manufacturer’s instructions using Jetprime transfection reagent (Polyplus transfection) or CalFectin (SignaGen Laboratories). For lentiviral transduction, lentiviral particles were prepared by transfection of HEK293T cells with shRNA- or expression vectors and the second-generation packaging plasmids ΔR8.2 and Vsv-G (Addgene). Virus-containing supernatants were collected 36h later and added with polybrene (Merck) to pre-seeded melanoma cells. After 24h, cells were transferred to 10-cm culture dishes for selection in appropriate antibiotics. For cells transduced with doxycycline-inducible vectors, cells were treated with 500ng/mL of doxycycline prior to experiments.
Patient-derived xenografts (PDX)
PDX models of human melanoma were established at the Wistar Institute as previously described (28). When the PDX tumor was harvested, tissue pieces were snap-frozen in liquid N2 for RNA extraction. Clinical information is provided in Table S1.
Growth assays
For the purpose of monitoring growth rate, 5×103 melanoma cells were seeded in white 96-well clear-bottomed plates (Corning) and growth was monitored every 24h using CellTiter Glo kit (Promega) according to the manufacturer’s recommendations. Luminescence intensity was measured using an Infinite 2000 Pro reader (Tecan). Alternatively, 5 × 105 cells transfected with siRNA were seeded per well, in 6-well plates. After 24h, cells were fixed and stained using a ready-to-use solution of crystal violet (Merck). Plates were pictured using a scanner (Epson). Growth was quantified by solubilization of crystal violet in 10% acetic acid and monitoring optical density at 595nm using a Clariostar spectrophotometer (BMG Labtech).
Western blot analysis
Cells were lysed by addition of lysis buffer [100 mM Tris-HCl pH 7.5, 5% sodium dodecyl sulfate (SDS)]. Proteins were visualized using a chemiluminescence method (Image-Quant LAS400, GE Healthcare).
RT-qPCR
RNA extraction, cDNA synthesis and qPCR were performed as previously described (26). Primer sequences are listed in the Supplementary Material and Methods. Primer efficiency was measured in preliminary experiments, and amplification specificity was confirmed by dissociation curve analysis.
Colony formation
A375 (1000 cells) and HEK293T (1500 cells) cells were seeded in 6-well plates. Seven to 10 days later, colonies were fixed and stained using a ready-to-use solution of crystal violet (Merck). Plates were pictured using a scanner (HP ScanJet G4010), and colonies were analyzed using ImageJ software.
Spheroid formation
Spheroids were formed using the hanging drop method. Briefly, 500 cells diluted in 20 μL were seeded on microplates (Nunc). Eight days later, spheroids were pictured using a fluorescence microscope (Olympus 1X71S1F-2). Sphere area was measured using ImageJ software.
Cell cycle analysis
Cell were harvested using accutase (Merck) and washed twice with PBS. Cells were then fixed with ice-cold 70% ethanol at least overnight at 4°C. Following fixation, cells were washed twice with ice-cold PBS and stained with propidium iodide (PI)-staining buffer (50μg/mL PI, 0.1% Triton X-100, 0.1% sodium citrate and 10μg/mL RNase) for 30min before analysis by FACS (BD LSRFortessa™, BD Biosciences).
β-galactosidase staining
5 × 105 cells were seeded on 6-well plates and β-galactosidase staining was performed using a kit (Cell Signaling Technology). Cells were imaged using a DMi8 microscope (Leica). The percentage of β-galactosidase-positive cells was assessed using Image J software.
Immunoprecipitation
Cells were lysed in lysis buffer [50mM Tris-HCl pH7.5, 120mM NaCl. 12mM EDTA pH8, 0.5% Nonidet P-40 (NP-40) and 1mM dithiothreitol (DTT)] supplemented with 1X protease inhibitor cocktail (Merck) and 1X phosphatase inhibitor cocktail (Roche) and then centrifuged for 10 min at 17,000 g. The supernatant was incubated overnight at 4˚ C with beads conjugated with the appropriate antibody. Beads were pelleted by centrifugation, washed 5 times with lysis buffer, and boiled in Laemmli buffer to elute proteins. Finally, proteins were resolved on SDS-PAGE and subjected to western blotting as described above.
Mass spectrometry
A375 cells transduced with empty pLVX or pLVX-SPANXA-FLAG were subjected to immunoprecipitation as described above, except that elution from beads was performed by incubation with 3XFlag peptide (150 μg/mL, Merck) for 1 h at 4° C and eluates were subjected to tryptic digestion followed by LC-MS/MS, as described previously (29). Raw data were analyzed using MaxQuant (v1.6.0.1) with default settings. Fold changes were calculated by dividing protein intensity of the FLAG immunoprecipitate (SPANXA IP) by protein intensity of the control immunoprecipitate (control IP). Thresholds were set based on the disctribution of the log2 transformed ratio, as previously described (29). The mass spectrometry data have been deposited (MassIVE, MSV000084617).
RNA sequencing
RNA was prepared 7 days following SPANX knockdown, and triplicates were used for RNAseq analysis. Libraries were prepared from isolated total RNA using the QuantSeq 3’ mRNA-Seq Library Prep Kit FWD for Illumina from Lexogen, (Vienna, Austria). Barcoded libraries were pooled and single end sequenced (1X75) on the Illumina NextSeq 500 using the High output V2.5 kit (Illumina Inc., San Diego CA). Raw data in FASTQ files were generated by standstard illumina base calling pipeline and stored in illumina cloud service BaseSpace. Reads were mapped to the human reference genome (hg19) using STAR mapping algorithm (version 2.5.2a) after removing adapters and trimming low quality reads. FeatureCounts implemented in Subread (v1.50) was used to count the sequencing reads from mapped BAM files. Analyses of differentially expressed genes (DEGs) were subsequently performed using a negative binomial test method (edgeR) implemented using SARTools R Package. DEG analysis was performed using the following criteria: |log2 (Fold change)| >1 and adjusted p-value <0.01. IPA software (http://www.ingenuity.com) was used to identify the pathways associated with the DEG. The RNAseq data have been deposited in GEO repository with the following accession number: GSE141823.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature (RT), washed three times in PBS, permeabilized in 0.2% Triton X-100 for 5 min and blocked with 0.2% Triton X-100 and 10% FBS in PBS for 30 min. Primary antibodies were diluted in staining buffer (0.2% Triton X-100, 2% FBS in PBS) and incubated overnight at 4° C in a humidified chamber. Slides were then washed 3 times in staining buffer, and secondary antibodies (Life Technologies) diluted in staining buffer were added to the slides for 1 h at RT in a humidified chamber shielded from light. Finally, samples were stained by incubation with Hoechst 33342 1 μM for 20 min at RT, washed three times in staining buffer and mounted with VECTASHIELD® Antifade Mounting Medium (Vector laboratories). Alternatively, samples were directly mounted using VECTASHIELD® Antifade Mounting Medium with DAPI (Vector laboratories). Samples were observed using a Zeiss LSM800 confocal microscope with a 63x objective or using a fluorescence microscope DMi8 (Leica) with a 100X oil immersion objective. Images were processed using the 3D deconvolution tool from LASX software (Leica), and the same parameters were used to process all images. Super-resolution microscopy (Super-Resolution Radial Fluctuations; SRRF), was performed using a Zeiss LSM800 confocal microscope with a 63x 1.4NA objective. A region of interest of 9.3μm × 9.3μm was imaged using the Airyscan function. A series of 25 images was taken at 3.7fps; these were processed using the Airyscan algorithm using the ZEN blue software (Zeiss), followed by further processing using the SRRF algorithm in FIJI.
Electron microscopy
Five million cells were seeded in 100 mm plates, and one day later, cells were washed with preheated serum-free medium and fixed 1 h at RT in 2% glutaraldehyde/3% paraformaldehyde in 0.1 M sodium cacodylate buffer containing 5 mM CaCl2 and 3% sucrose. Cells were then washed with sodium cacodylate buffer, scraped and embedded in agarose. Next, samples were post-fixed and stained with 1% osmium tetraoxide, 0.5% potassium hexancyanaferrate and 0.5% potassium dichromate in 0.1 M cacodylate buffer, and in-block stained with 1% uranyl acetate for 1 h at RT. Cells were dehydrated in ethanol and embedded in Epon 812, and then 75 nm ultrathin sections were cut with a ultramicrotome UC7 (Leica) and transferred to copper grids for viewing using a Zeiss Ultra-Plus FEG-SEM equipped with a STEM detector at accelerating voltage of 30 kV.
Generation of SPANX antibody
SPANXA was PCR-amplified from pcDNA3-SPANXA-FLAG generated as described above, and subcloned into pGex-4T2 vector (Addgene). One liter of cultured BL21(DE3)pLysS competent cells (Promega) previously transformed by electroporation was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG) at 0.4 mM for 4 h at RT. Cells were washed twice with ice-cold PBS and centrifuged for 10 min at 5000 g. Cells were lysed in 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 20 mM β-mercaptoethanol and 1X EDTA-free protease inhibitor cocktail (Merck), sonicated and centrifuged 20 min at 14,000 g at 4° C. The supernatant was incubated with 5 mL of gluthatione sepharose beads (GE Healthcare Life Sciences), pre-washed twice with lysis buffer overnight at 4° C. Beads were then washed 5 times with Hepes 25 mM pH 7.6. Elution of SPANXA was performed by on-beads digestion at RT for 9 h with 100 units of thrombin. Samples were resolved by SDS-PAGE and stained with Coomassie blue (Biorad) according to the manufacturer’s intructions. The band corresponding to SPANXA was cut from the gel and used as antigen to immunize rabbits (ProSci Inc, CA). Bleeds obtained after 3–4 immunizations were used for Westerns and IP as noted.
In vivo tumor and Immunohistochemistry
All animal studies were conducted at the SBP animal facility in accordance with the Institutional Animal Care and Use Committee guidelines (approval #18–079). Six-week-old female nude mice were purchased from Charles River Laboratories (Wilmington, MA), and 7–10 days later inoculated with 1×106 of A375 cells . Following subcutaneous injection of tumor cells into the lower right flank, mice were maintained in a pathogen-free environment with free access to food. Mice were sacrificed when tumor size reached ~2000 mm3. Tumors were fixed overnight in buffered zinc formalin fixative (Anatech), washed twice with PBS, and embedded in paraffin. Paraffin blocks were sectioned as 5 μm slices, and staining was performed using a BOND RX automated stainer (Leica) and the reagent BOND Polymer Refine Red Detection (Leica). SPANX antibody used at a dilution of 1:500. Slides were scanned using an Aperio AT2 slide scanner (Leica).
Enzyme-linked immunosorbent assay (ELISA)
Conditioned medium used for ELISA was generated as follows. 1×105 cells were seeded in 12-well plates . Media were collected 64 h later and stored at −80° C. ELISA was performed using a kit (Abcam). Optical density was measured using an Infinite 2000 Pro reader (Tecan). Optical density was normalized to the cell number estimated by measurement of the protein quantity of the cell lysates.
Statistical analysis
All analyses were performed using GraphPad Prism software (GraphPad, La Jolla, CA, USA). Values are represented as means ± standard error of the mean. The statistical significance of differences between the means (of at least three independent assays) was evaluated using Student’s t-tests, if 2 groups were compared, one-way ANOVA, if more than 2 groups were compared, or two-way ANOVA for growth assays over time. The post hoc test used is indicated in figure legends and was determined by verifying normality of samples using a Kolmogorov–Smirnov test. P values below 0.05 (*), <0.01 (**) and <0.001 (***)were considered significant.
A list of shRNA sequences is found in the Supplementary Material and Methods.
RESULTS
SPANX proteins control tumor cell growth
To determine the proportion of SPANX-positive melanoma specimens, we monitored its transcript levels in a panel of melanoma patient-derived xenografts (PDX) (Fig 1A, Table S1). Consistent with previous reports (22), SPANX mRNA expression was detectable at various levels in most samples. To determine whether SPANX expression contributes to melanoma growth, we assessed potential changes in viability in melanoma lines subjected to SPANX depletion using short hairpin (sh) RNAs (Fig S1A) or small interfering (si) RNAs (Fig S1B). Melanoma cell lines chosen for this analysis exhibit notable levels of SPANX expression (Fig S1B–D). SPANX knockdown inhibited cell viability of A375 (Fig 1B and 1C, Fig S1A and S1B), WM1366 and UACC612 melanoma lines (Fig S1C and S1D). Furthermore, SPANX depletion reduced colony formation in both anchorage-dependent and -independent focus formation assays (Fig 1D and S1E and S1F). SPANX depletion also suppressed growth of primary melanoma cultures (Fig 1E). Conversely, ectopic expression of either SPANXA or SPANXC increased the size of melanoma spheroids and the number of colonies in soft agar relative to empty vector-transduced cells (Fig 1F and 1G and Fig S1G). Notably, ectopic expression of SPANX affected 3D but not 2D proliferation phenotypes (Fig S1H), pointing to the possibility that basal level of SPANX expression may be sufficient for 2D proliferation. SPANX expression may be subject to post translational modification, reflected in a doublet seen in western blots (Fig 1G). These observations suggest that SPANX is required for melanoma growth proliferation and survival, potentially by altering cellular organization, given that SPANXA or SPANXC promotes 3D growth.
Figure 1: SPANX expression promotes tumor cell growth.

A. RT-qPCR analysis of SPANX expression in melanoma patient-derived xenografts (PDX).
B. RT-qPCR and western blot analysis of SPANX expression in A375 cells transduced with empty vector (EV) or SPANX-targeting shRNA (n=3). C. A375 cells transduced with EV or SPANX-targeting shRNA were monitored for altered viability using a Cell Titer Glo luminescence-based kit (n=4). Statistical significance was assessed by two-way ANOVA with post hoc Tukey’s test, ***p-value < 0.001. D. Colony formation assay (left) and quantification (right) of A375 colony number following transduction with SPANX shRNA or EV control (n=3). E. Viability assay based on crystal violet staining of MM150922 cells transfected with control (ctrl) or SPANX-targeting siRNA (left) and quantification (right, upper) (n=3). Statistical significance was assessed by Student’s t-test, *p-value < 0.05. (right, lower) Western blot analysis of SPANX expression in MM150922 cells transfected with ctrl or SPANX-targeting si-RNA. F. Western blot analysis of A375 cells transduced with EV (pLVX) or vector expressing SPANXA (A) or SPANXC (C). SPANX expression was evaluated 4 days after induction with doxycycline. G. Sphere formation assay of A375 cells transduced as in (F) and pre-treated for 3 days with doxycycline. (left) Representative pictures of the obtained spheroids. Scale bar: 100μm. (right) Calculation of sphere area 8 days after cell seeding (n=3, 6 spheres per independent experiment). Error bars represent means ± SEM. Statistical significance was assessed by one-way ANOVA with post hoc Tukey’s test, **p-value < 0.01, ***p-value < 0.001 in panels B, D and G.
SPANX depletion induces G1/S arrest
To investigate mechanisms underlying SPANX knockdown phenotypes, we performed RNAseq to map changes in gene expression using SPANX shRNA in A375 cell line, relative to empty vector-transduced cells. Of 609 genes deregulated in both shRNA clones, 454 were upregulated and 123 were downregulated (Fig 2A). Gene signatures associated with cell division were downregulated by SPANX KD, while those associated with 3-phosphoinositide biosynthesis were upregulated (Fig 2B). Notably, Ingenuity Pathway Analysis (IPA) identified the cell cycle as the primary pathway affected upon SPANX depletion (Fig 2B). Further analysis using the Reactome database (30) revealed that most of those cell cycle-related genes were associated with the G1/S transition (Fig 2C). RT-qPCR validation confirmed down-regulation of genes functioning in cell division and progression following SPANX KD in A375 and WM1366 cells (Fig 2D and Fig S2A). FACS analysis confirmed G1 arrest of A375 cells subjected to SPANX depletion (Fig 2E and Fig S2B). Coupled with G1 arrest, SPANX downregulation increased expression of the cell cycle inhibitor CDKN1A/p21 (Fig 2F, 2G and Fig S2C), which is implicated in G1/S arrest and senescence (31). Conversely, SPANXA overexpression in A375 cells grown in 3D culture decreased CDKN1A/p21 expression relative to vector control cells (Fig 2H). We also observed increased senescence-associated β-galactosidase staining in SPANX KD A375 cells (Fig 2I), suggesting that sustained G1/S arrest promotes senescence in these cells. Overall, these observations suggest that SPANX is required for cell cycle progression of melanoma cells and thus its loss promotes cell cycle arrest and likely senescence.
Figure 2: SPANX depletion induces G1/S arrest.
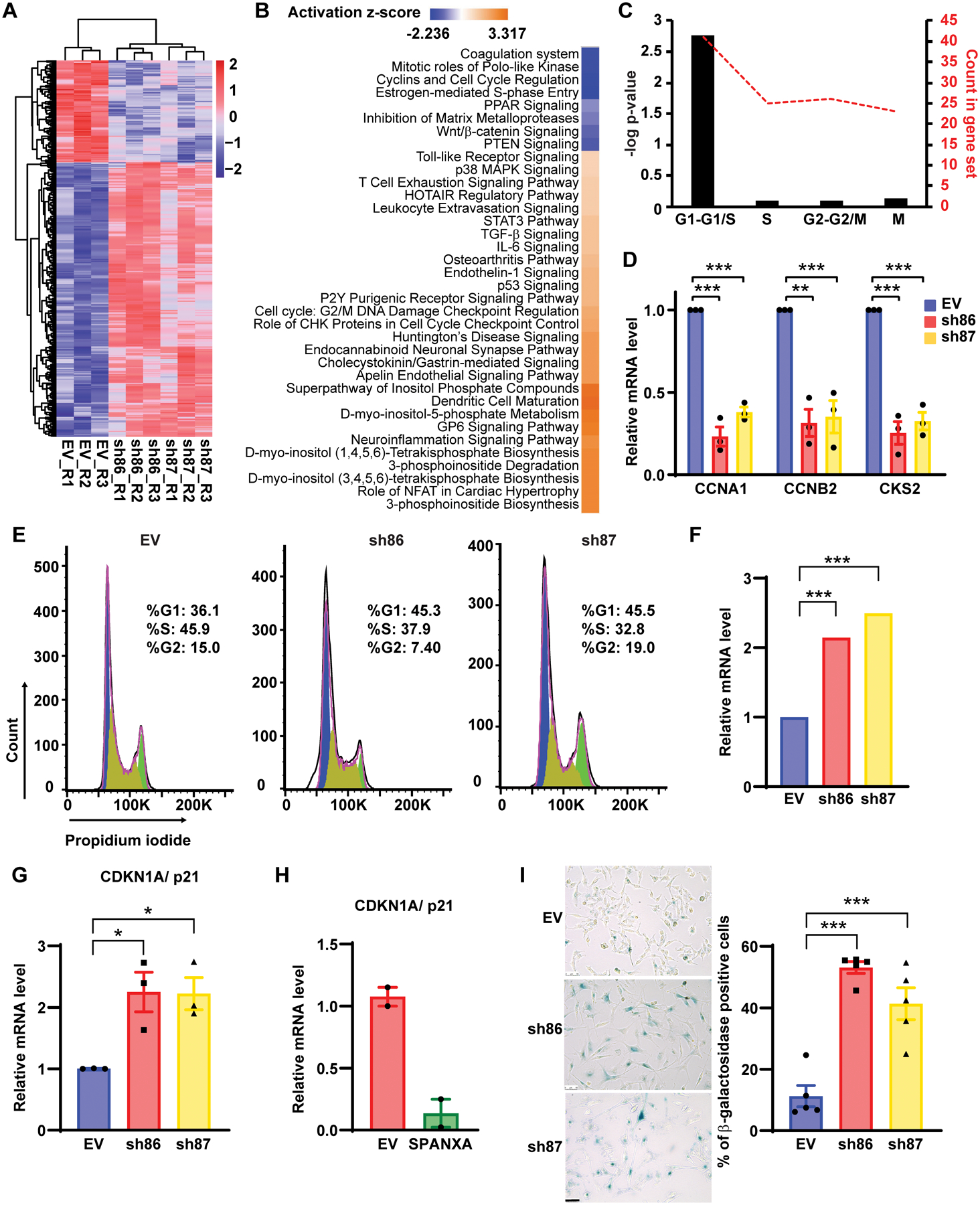
A. Heat map representing RNAseq data in A375 cells transduced with EV or SPANX-targeting shRNA (log2 fold-change >0.4 and p-value <0.01false discovery rate [FDR] adjusted p-value < 0.05 by Benjamini-Hochberg method. R: replicate. B. Ingenuity Pathway Analysis of canonical pathways associated wih deregulated genes identified by RNAseq in A375 cells subjected to SPANX KD. C. Analysis using the Reactome database of cell cycle-related genes identified as downregulated by RNAseq in SPANX KD A375 cells. D. RT-qPCR confirmation of selected genes identified by RNAseq in A375 cells transduced with EV or SPANX-targeting shRNAs. E. Cell cycle analysis of A375 cells transduced with EV or SPANX-targeting shRNAs. F. Relative CDKN1A/p21 expression as determined by RNAseq in A375 cells transduced with EV or SPANX-targeting shRNAs. Statistical significance was assessed by Benjamini-Hochberg method, ***p-value < 0.001. G. RT-qPCR analysis of CDKN1A/p21 expression in A375 cells transduced with EV or SPANX-targeting shRNAs. H. RT-qPCR analysis of CDKN1A/p21 expression in A375 cells transduced with EV or an inducible vector encoding SPANXA grown in 3D, 3 days after doxycycline treatment (n=2). I. (left) β-galactosidase staining in A375 cells transduced with EV or SPANX-targeting shRNAs. Scale bars: 50μm. (right) Quantification of β-galactosidase-positive cells (n=5). Error bars represent means ± SEM. Statistical significance was assessed by one-way ANOVA with post hoc Tukey’s test, *p-value < 0.05, ***p-value < 0.001 in panels D, G and I.
SPANX interacts with A-type lamins
To further define SPANX function in melanoma cells, we searched for SPANX-interacting proteins using mass spectrometry analysis of immunoprecipitated SPANXA. One of the most prominent SPANXA interacting partners was LMNA (lamin A/C), which is encoded by LMNA, a gene whose deregulation is implicated in cell division and senescent phenotypes (6) (Table S2 and Fig 3A). We confirmed interaction of SPANX with lamin A by immunoprecipitation (IP) using SPANXA-FLAG (Fig 3B, Fig S3A and S3B) or by reciprocal IP using hemagglutinin-tagged lamin A (HA-lamin A) as a bait (Fig 3C and Fig S3C). Since lamins serve as a hub for several regulatory proteins (32), we next mapped the lamin A domain required for SPANX interaction. Analysis of nested deletions of key lamin domains identified the immunoglobulin fold-like domain located within the lamin A tail as the SPANX binding region (Fig 3D–E and Fig S3D–E). Immunostaining and microscopic analysis of SPANX identified perinuclear staining overlapping with that of LMNA (Fig 3F, Fig S3F). Indeed, SPANX staining was seen in nucleoplasmic foci that co-localized with LMNA (Fig 3F and Movie S1 and S2), suggesting their proximity and possible interaction. Noteworthy is the heterogenous nature of SPANX expression (Fig 3F), which points to intrinsic cellular signaling which impacts its expression, or that SPANX expression is seen in a select melanoma cells (see Discussion). To further analyze SPANX subcellular localization we generated an antibody suitable for immunohistochemistry (Fig S3G). Immunostaining of a xenograft of a human melanoma tumor or of human melanoma specimens confirmed SPANX perinuclear staining (Fig 3G and Fig S3H), as well as the heterogenous nature of SPANX staining in human melanoma specimens (Fig 3G). Since the antibodies were not proven useful for IP of endogenous SPANX, we carried out super resolution microscopy (SRRF), which revealed SPANX-positive filamentous structures (Fig 3H) resembling LMNA. The latter may be attributed to SPANX polymerization, as suggested by SPANXA self-interaction and interaction with other SPANX proteins (Fig 3A and Fig S3I). Overall, these observations suggest that SPANX is a nuclear lamin A-interacting protein in melanoma cells.
Figure 3: SPANX interacts with A type lamins.
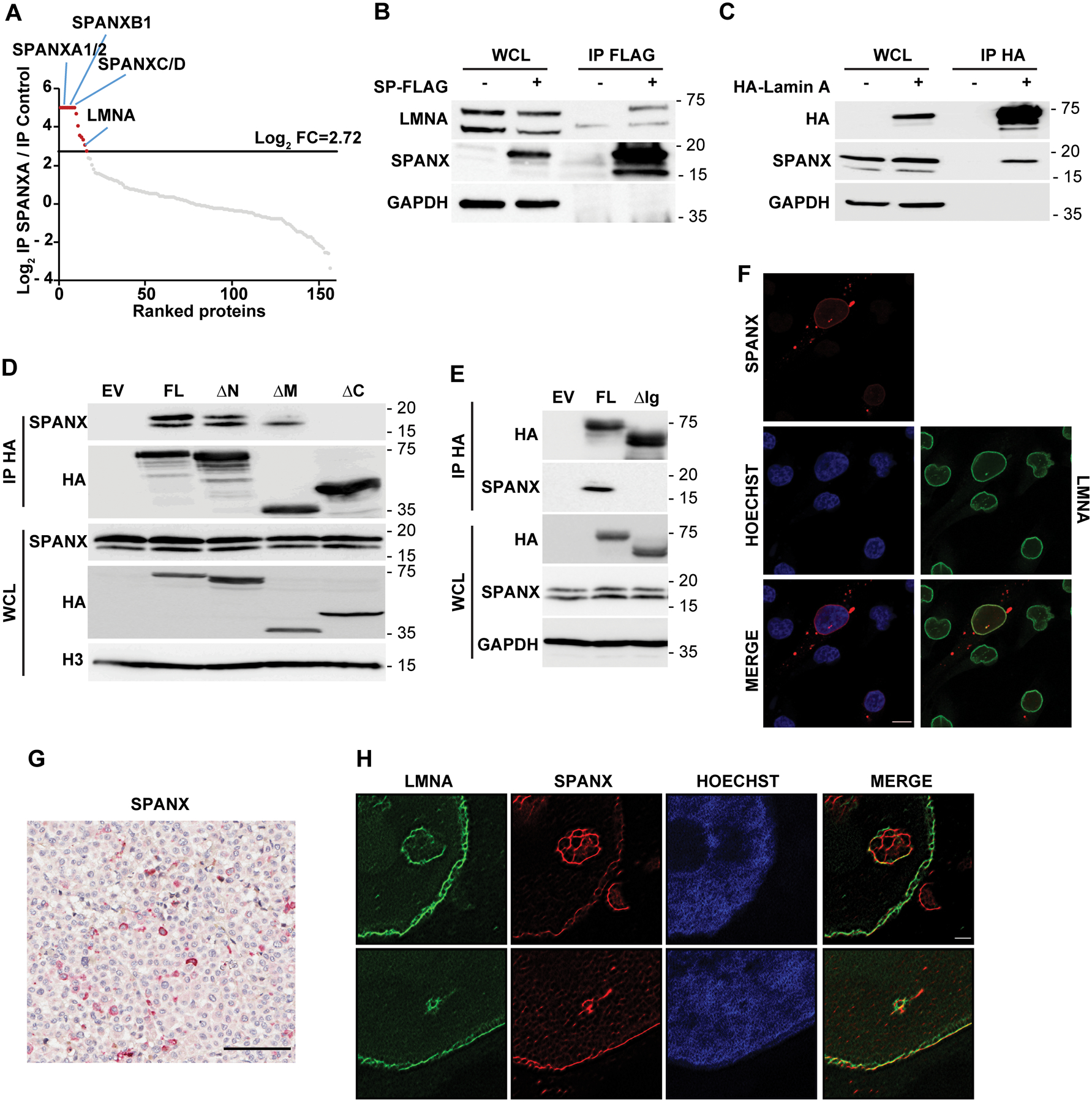
A. A375 cells transduced with EV (pLVX) or vector expressing SPANXA-FLAG were treated for 3 days with doxycycline before seeding, lysis, immunoprecipitation (IP) using anti-FLAG beads. Precipitates were analyzed by nano LC-MS/MS. Graph shows log2 ratio of SPANXA-FLAG IP over IP control as measured by nano LC-MS/MS. Proteins are ranked from highest to lowest ratio. The log2 fold-change (FC) threshold of 2.72 was determined as described in Materials and Methods.
B. IP with anti-FLAG beads of A375 cells expressing FLAG-tagged SPANXA (SP) followed by immunoblot with indicated antibodies. WCL, whole cell lysate. C. IP with anti-HA beads of A375 cells expressing HA-tagged lamin A followed by immunoblot with indicated antibodies. D. IP of lamin A mutants (described in Fig S3D) followed by immunoblot for indicated proteins. WCL, whole cell lysate; FL, full length; ΔN, deletion of lamin A head; ΔM, deletion of lamin A central rod domain; ΔC, deletion of lamin A tail. E. IP of either FL lamin A or lamin A deleted of the Ig-like domain (ΔIg), as described in Fig S3E, followed by immunoblot for indicated proteins.
F. Immunofluorescence staining of endogenous SPANX and LMNA in A375 cells, as detected by confocal microscopy. Scale bar: 10μm. G. Immunohistochemical staining of SPANX in a tumor derived from a patient with melanoma. Sample is counterstained with hematoxylin. Scale bar: 100μm. H. Super-Resolution Radial Fluctuations (SRRF) microscopy analysis of endogenous SPANX and LMNA in A375 cells. Scale bar: 1μm.
SPANX effects on cell proliferation are, in part, lamin A-dependent.
To determine the contribution of lamin A interaction to SPANX activity we assessed cell growth phenotypes following ectopic expression of SPANXA constructs lacking the capacity to interact with lamin A. To do so, we first mapped SPANXA amino acid residues required for lamin A interaction. IP analysis in HEK293T cells transduced with FLAG-tagged SPANXA constructs and HA-tagged lamin A revealed that mutations within a C-terminal acidic domain (Fig S4A), which is conserved among all SPANX proteins (Fig S4B), largely disrupted SPANXA interaction with lamin A (Fig 4A). Intriguingly, ectopic expression of that SPANXA mutant form was detectable in HEK293T cells (Fig 4A–B), but required the presence of the proteasome inhibitor, MG132, in melanoma cells (Fig S4C), implying that this mutant is subjected to a tighter regulation in melanoma cells. Overexpression of WT SPANXA alone increased colony size (Fig 4B–D) but not number (Fig S4D) relative to empty vector-transduced cells. Colong size was further increased upon co-expression of SPANXA with lamin A (Fig 4C–D). Notably, effects on colony size were blocked when cells were transfected with the SPANXA C-terminal mutant, either with or without lamin A (Fig 4C–D). These observations suggest that SPANX effects on cell growth depend, at least in part, on its ability to interact with lamin A.
Figure 4: SPANX effects on cell proliferation are, in part, lamin A-dependent.
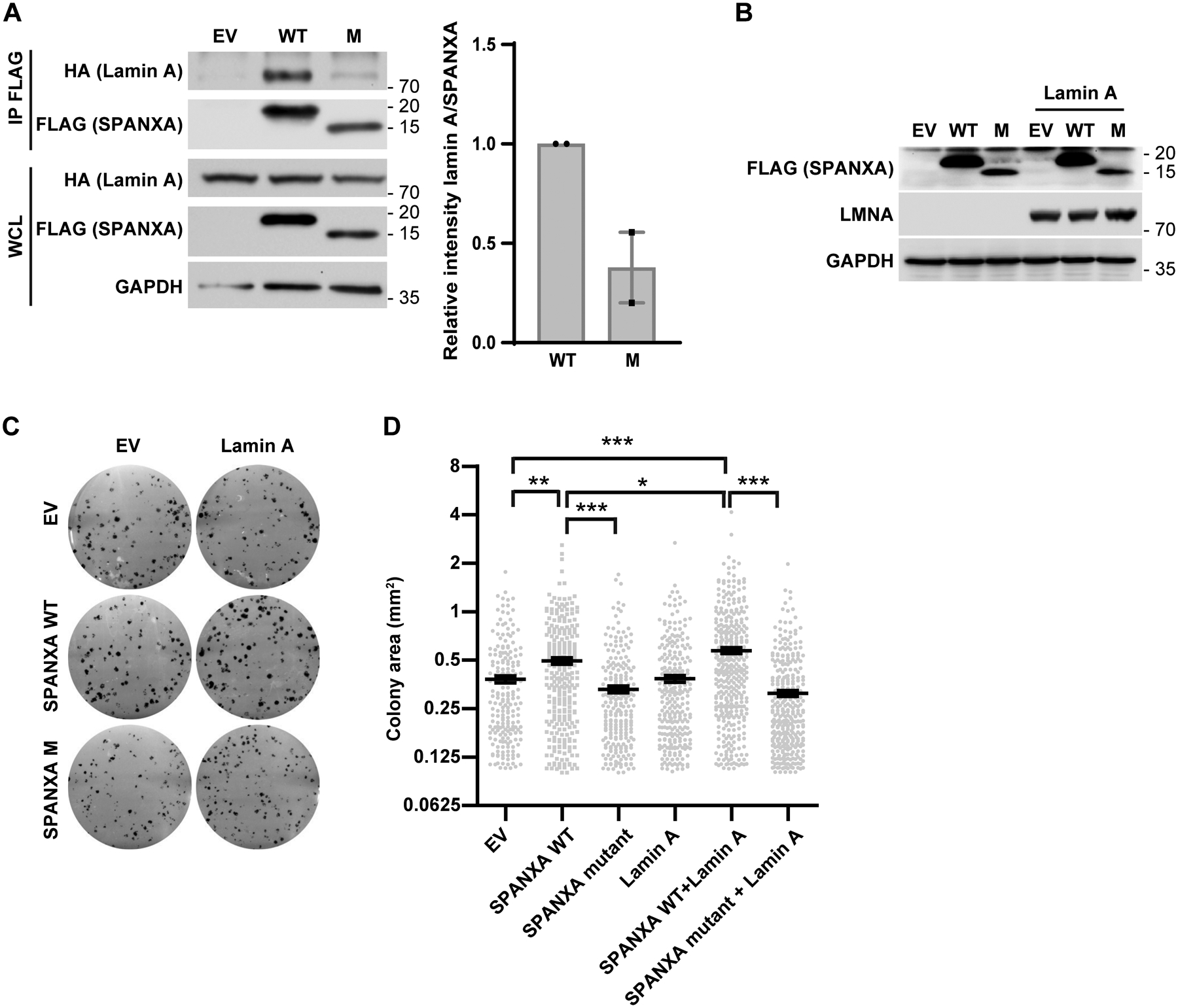
A. (left) HEK293T cells were transfected with wild type (WT) or mutant (M) FLAG-tagged SPANXA, plus or minus HA-lamin A. Immunoprecipitation (IP) of WT or M FLAG-tagged SPANXA was performed using anti-FLAG beads followed by immunoblot. WCL: whole cell lysate. (right) Quantification of results (n=2). B. Western blot analysis of indicated proteins in HEK293T cells transfected with WT or M FLAG-tagged SPANXA, plus or minus HA-lamin A.
C. Colony formation assay in HEK293T cells transfected with WT or M FLAG-tagged SPANXA, plus or minus HA-lamin A. D. Measurement of colony area in colony formation assays shown in C (n=3). Statistical significance was assessed by one-way ANOVA with post hoc Tukey’s test, *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001. Error bars represent means ± SEM.
SPANX loss alters LMNA organization and nuclear architecture
Given that SPANX interacts with LMNA and can exhibit a filamentous structure, we assessed effects of SPANX KD on LMNA structure. Super-resolution microscopy analysis of A375 cells revealed LMNA disorganization following SPANX knockdown (Fig 5A) but no change in lamin A expression, maturation (as determined by immunoprecipitation and immunoblot analysis) or phosphorylation (Fig S5A–D). SPANX knockdown also resulted in aberrantly shaped nuclei of melanoma cells, as revealed by fluorescence (Fig 5B and Fig S5E–F) and electron microscopy (Fig 5C). Similarly shaped nuclei have been reported in cells derived from specimens from patients harboring LMNA mutations (33). Moreover, nuclei in SPANX KD melanoma cells were larger and more elliptical (Fig 5D–E and Fig S5G–H), a phenotype also documented in laminopathies (5). SPANX KD A375 cells exhibited increase in nuclear size, compared with control cells (Fig 5F and Fig S5I). Increased nuclear size is among characteristic phenotypes of senescence, resembling changes observed upon SPANX KD (Fig 2). Taken together, these observations suggest that SPANX plays a role in LMNA organization and nuclear shape and that its loss in melanoma cells confers phenotypes seen in laminopathies.
Figure 5: SPANX loss alters LMNA organization and nuclear architecture.
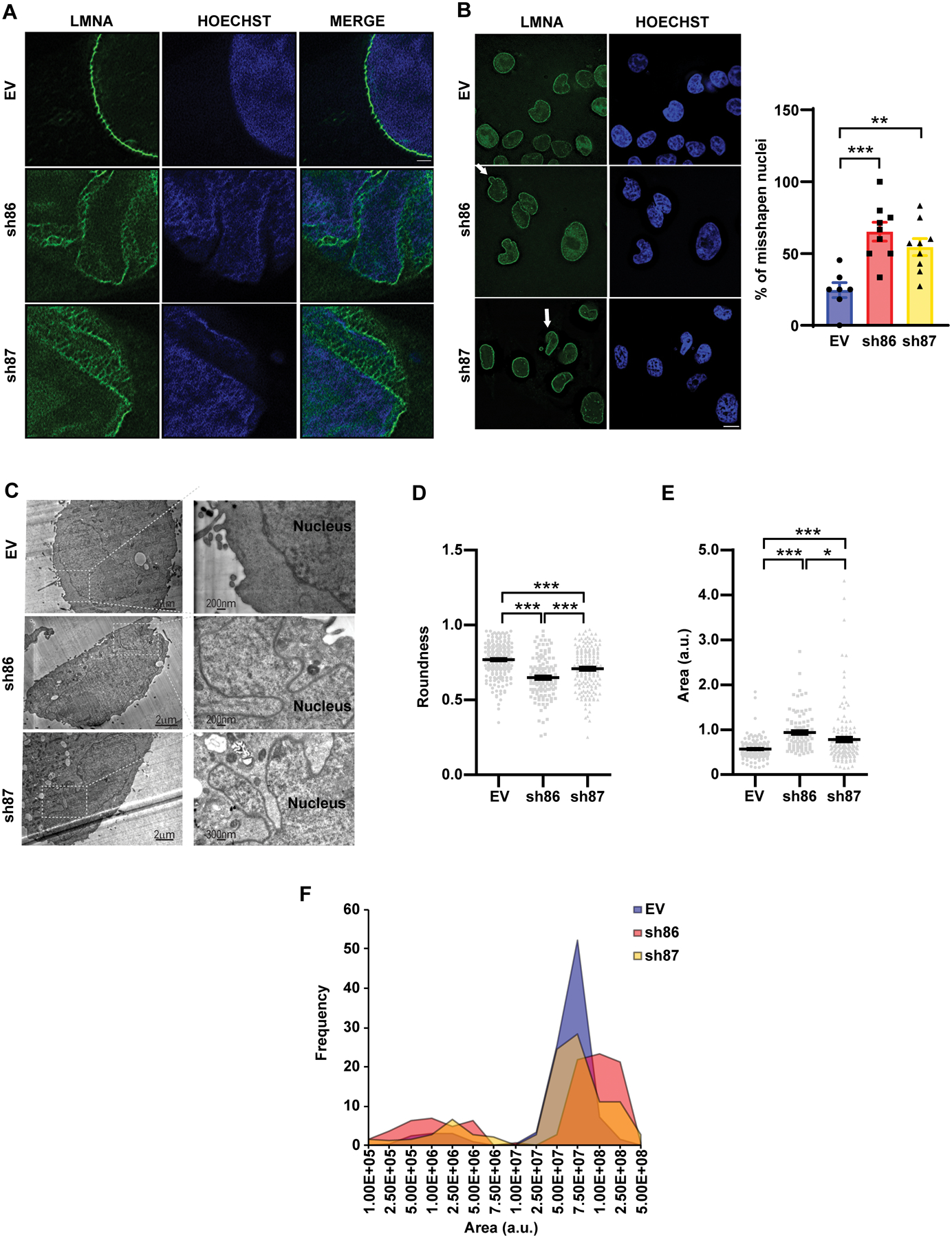
A. SRRF microscopy analysis of LMNA in A375 cells transduced with EV or shRNAs targeting SPANX. Scale bar: 1μm. B. (left) Immunostaining of LMNA followed by fluorescence microscopy of A375 cells transduced as in (A). Arrows indicate aberrant nuclear structures. Scale bar: 10μm. (right) Quantification of disorganized nuclei observed in these cells (n=3). C. Electron micrographs of A375 cells transduced as in (A). D. Analysis of degree of nuclear roundness using Image J software in A375 cells transduced as in (A) (n=3). E. Nuclear area measurement using Image J software in A375 cells transduced as in (A) (n=3). a.u., arbitrary units. F. Distribution of size of the nuclear area of A375 cells transduced as in (A). Error bars represent means ± SEM. Statistical significance was assessed by one-way ANOVA with post hoc Tukey’s test in panels B, D and E, *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001.
Growth arrest following SPANX loss is in part IRF3/IL1A-dependent
Among nuclear perturbations observed following SPANX KD in A375 and UACC612 cells was accumulation of cytosolic chromatin structures in the form of micronuclei (Fig 6A–C and Fig S6A), structures known to be unstable. Accordingly, conditions rendering the nuclear envelope unstable could expose chromatin to cytosolic factors (34) and potentially activate innate immune pathways such as IRF3 signaling (15). Consistent with the observed increase in cytosolic chromatin, SPANX knockdown in A375 cells showed a relatively higher percentage of cells exhibiting nuclear IRF3 and increased IRF3 phosphorylation on serine 386 (Fig 6D–E and Fig S6B–C), which marks active IRF3 (35). Accordingly, SPANX KD led to an increased expression of the IRF3 target gene IFNB1 (Fig S6D). Significantly, IRF3 depletion in SPANX KD A375 cells rescued proliferation to levels seen in SPANX non-depleted cells (Fig 6F and Fig S6E), suggesting that SPANX effects of on melanoma proliferation are mediated, at least in part, by IRF3.
Figure 6: SPANX loss-induced growth arrest is, in part, IRF3-dependent.

A. Immunostaining for LMNA followed by fluorescence microscopy analysis in A375 cells transduced with EV or shRNAs targeting SPANX. Arrows indicate micronuclei. Scale bar: 10μm.
B. Quantification of findings shown in (A) (n=4). C. Electron micrographs of A375 cells transduced as in (A). D. Immunostaining of IRF3 followed by fluorescence microscopy analysis in A375 cells transduced as in (A). Scale bar: 10μm. E. Quantification of findings shown in (D) (n=4).
F. Viability assay of A375 cell following transduction with EV or with SPANX- or IRF3-targeting shRNA, or a combination of both (n=3). Statistical significance was assessed by one-way ANOVA with post hoc Tukey’s test, *p-value < 0.05, ***p-value < 0.001. Error bars represent means ± SEM. Statistical significance was assessed by Student’s t-test, *p-value < 0.05 in panels B and E.
Given that IRF3 is implicated in cytokine expression (36), we asked whether SPANX expression non-autonomously regulates viability of melanoma cells via control of cytokine secretion. To do so, we collected conditioned media from SPANX-depleted or control A375 cells and assessed its possible effect on growth of naive melanoma cells. Media from SPANX-depleted cells reduced proliferation of naive A375 cells relative to control media (Fig 7A), suggesting that secreted factors function in SPANX-dependent effects on melanoma growth. Among cytokines identified as deregulated in our RNAseq analysis of SPANX KD cells was IL1A (Table S3). Notably, we observed significantly upregulated IL1A expression in SPANX-depleted A375 cells (Fig 7B). Interestingly, IL1A is implicated in senescence phenotypes (37) and IRF3 KO mice show lower than expected levels of circulating IL1A in response to infection (38), suggesting potential regulation of IL1A by IRF3. IL1A mRNA levels increased in both A375 and WM1366 melanoma cells following SPANX KD (Fig 7C and Fig S7A). Corresponding increases in IL1A secretion were seen in SPANX KD A375 cells (Fig 7D and Fig S7B). Notably, increased IL1A seen upon SPANX KD was attenuated following IRF3 KD (Fig 7E), supporting the idea that IL1A expression is dependent on IRF3. Likewise, we observed corresponding upregulation of expression of the IL1 receptor following SPANX KD (Fig S7C), supporting increases in IL1A signaling. Finally, inhibition of IL1A expression by shRNA in SPANX KD melanoma cells partially rescued cell viability (Fig 7F and Fig S7D), suggesting that the IRF3/IL1A axis mediates, at least in part, cell growth arrest induced by SPANX KD.
Figure 7: Growth arrest of A375 cells following SPANX loss is in part IRF3/IL1A-dependent.
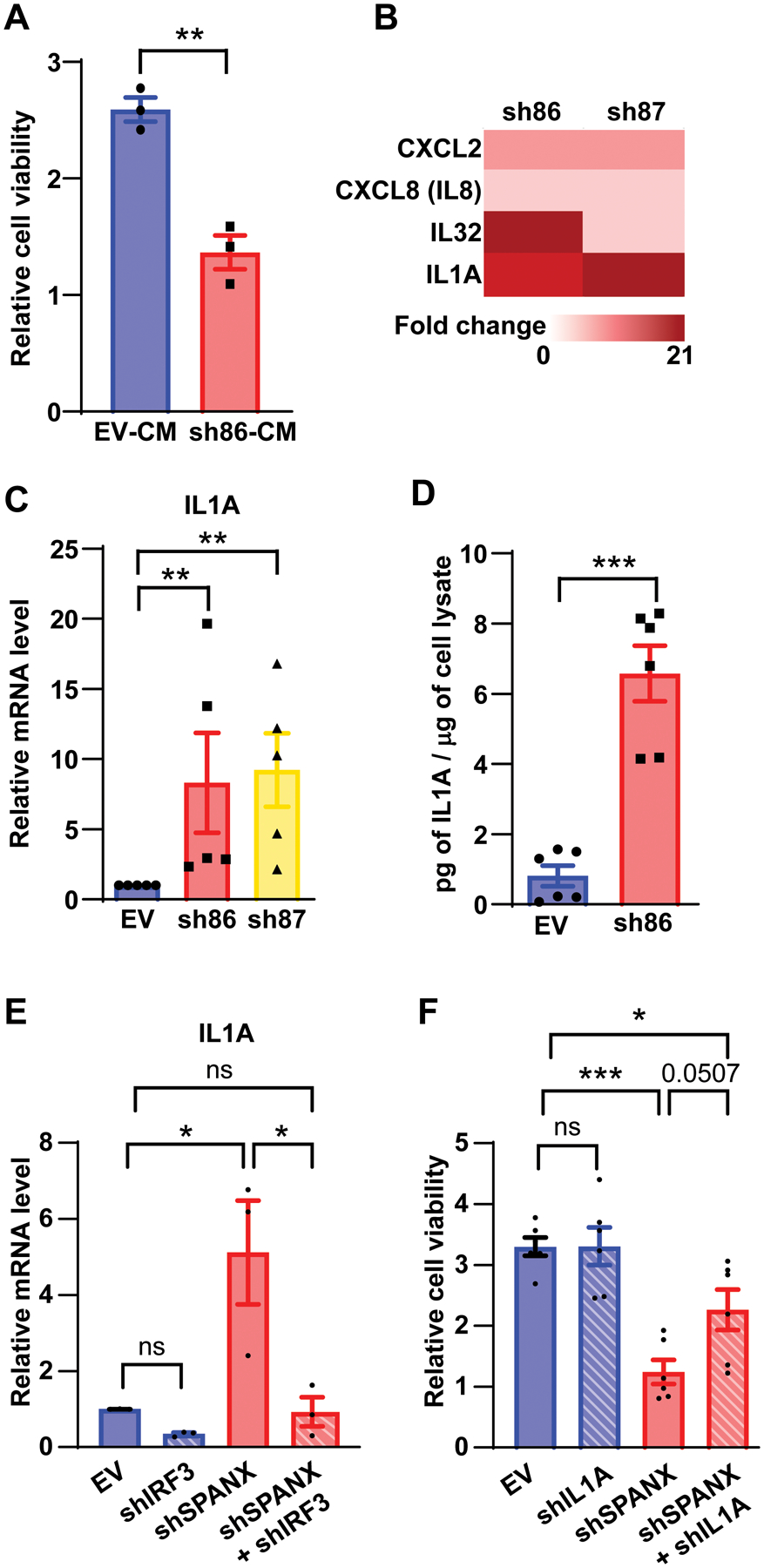
A. Viability assay of A375 cells incubated 48h with conditioned medium from A375 cells transduced with EV or SPANX-targeting shRNA (n=3), CM: conditioned media B. Heat map representing fold-change as determined from RNAseq data of indicated genes in A375 cells transduced with SPANX-targeting shRNA versus control cells, p-value < 0.01false discovery rate [FDR] adjusted p-value < 0.05 by Benjamini-Hochberg method. C. RT-qPCR analysis of IL1A expression in A375 cells transduced with EV or SPANX-targeting shRNA (n=3). D. Measurement by ELISA of IL1A protein levels in conditioned medium from A375 cells transduced with EV or SPANX-targeting shRNA (n=6). E. RT-qPCR analysis of IL1A expression in A375 cells transduced with the EV or with SPANX- or IRF3-targeting shRNA, or a combination of both (n=3).
F. Viability assay of A375 cells transduced with EV or with SPANX-targeting or IL1A-targeting shRNA, or a combination of both (n=3). Error bars represent means ± SEM. Statistical significance was assessed by Student’s t-test in panels A,C and D and by one-way ANOVA with post hoc Tukey’s test in panels E and F, ns: not significant, *p-value < 0.05, **p-value < 0.01, ***p-value < 0.001
DISCUSSION
The present study desmonstrates that SPANX interacts with and regulates LMNA structure, pointing to SPANX control of nuclear architecture. SPANX depletion effectively inhibited melanoma cell proliferation by promoting G1/S arrest and senescence-like phenotypes. Our study underscores the importance of SPANX in LMNA organization and function in melanoma cells and raises important questions relevant to LMNA control and dysregulation in cancer.
SPANXC/LMNA interaction was previously observed in a breast cancer model (25), where SPANX expression was linked with tumor invasiveness, and to a lesser degree with cell proliferation. Although the latter study demonstrated that manipulation of LMNA levels can alter tumor cell invasivity, a formal link between SPANX and LMNA had not been previously established (25). Here we show that decreased proliferation is the most striking phenotype induced upon SPANX depletion in melanoma cells. SPANX depletion also decreased melanoma cell migration, although this effect was seen in the NRAS mutant WM1366 line but not BRAF mutant A375 cells (Fig S8). These findings suggest that other cellular changes, associated with certain mutational landscape, are required for SPANX ability to affect cell migration.
Notably, the mechanism underlying the regulation of SPANX expression in non-testis tissues remains to be determined. Although analysis of the SPANX promoter showed an absence of CpG islands (21), SPANXB1 expression has been linked to cytosine methylation in a colorectal adenocarcinoma cell line (39). In addition to epigenetic mechanisms regulating SPANX expression, the SPANX promoter harbors binding sites for the SRY (Sex-determining region Y protein) transcription factor and a heat shock response element (21). The latter is largely involved in the cellular response to stress, including genotoxic stress (40). Moreover, SRY promotes acquisition of stemness-related properties in a hepatocellular carcinoma model, and SRY KD in these cells decreases self-renewal capacity and decreases chemoresistance (41). Accordingly, heterogeneous expression of SPANX seen in tumors raises the possibility that SPANX expression may be limited to select cell populations that may persist under harsh environmental conditions, based on their differentiation state, dormancy or stemness. Given that tumor heterogeneity has been implicated in tumor resistance (42) and metastasis (43), achiles hills of therapy effectiveness, studies of SPANX contribution to these phenotypes is needed.
Consistent with inhibition of proliferation, most genes downregulated following SPANX knockdown function in cell cycle progression, a phenotype consistent with the link between LMNA and control of cell division (44). Our mapping of the lamin A Ig-fold domain as the SPANX interaction domain, is consistent with an earlier report (45), and in accord with a previous study implicating this domain in cell cycle-related activity (8).
Why would tumor cells benefit from SPANX expression, which is normally restricted to the testis? SPANX expression might provide an advantage to tumor cells by competing with other proteins functioning in normal nuclear organization and cell division or promoting plasticity required for tumor cells to adapt to harsh environmental conditions. The link between SPANX expression and cell growth arrest might also be attributed to LMNA interaction with mitotic chromosomes (46) or regulation of mitotic spindle assembly and positioning (9). Lamin A deregulation as observed in laminopathic or some cancer cells leads to chromosome missegregation, accumulation of micronuclei and aneuploidy (47). In as much, SPANX expression may serve to prevent tumor cells from being killed by mechanisms that under normal circumstances would promote cell death or senescence.
Our findings also identify IRF3 as a possible mediator of SPANX-dependent phenotypes, given that IRF3 depletion partially rescues melanoma growth inhibition seen upon SPANX knockdown. IRF3 activation in this context may be attributable to an increase in cytosolic chromatin structures, which we observed following SPANX depletion and which are commonly seen in LMNA mutant samples (48). SPANX KD also results in abnormally-shaped nuclei; thus, IRF3 activation may result from nuclear envelope breaks at the primary nucleus site. Given that phosphorylated IRF3 accumulates in a cGAS- (Cyclic GMP-AMP synthase) dependent manner (49) and that IRF3 activation engages innate immune responses (11), our finding that IRF3 is activated upon SPANX loss suggests that SPANX loss may also affect anti-tumor immunity. Our RNAseq data indicated increased expression of HLA genes following SPANX knockdown (Table S3), a change also seen following IRF3 activation (11,50). SPANX expression by tumor cells may thus promote tumor immune evasion. Paradoxically, however, SPANX is a cancer-testis antigen (21), and its overexpression may thus impact tumor cell antigen presentation, thereby altering immune recognition. Notably, overexpression of SPANXB1 is sufficient to induce killing of tumor cells in vitro by peripheral blood CD4+ T cells stimulated with autologous dendritic cells (51).
In all, SPANX modulates melanoma cell growth via intrinsic and extrinsic mechanisms. Our findings provide the justification for further analysis of SPANX in cancer, which is expected to shed new light on tumor intrinsic mechanisms that govern development, progression and possibly anti-tumor immunity. That phenotypes seen in SPANX knockdown melanoma cells resemble those seen in laminopathies, in which LMNA is mutated, points to the possible role IRF3 and IL1A signaling may play in these patients.
Supplementary Material
IMPLICATIONS.
SPANX, a testis protein, interacts with LAMNA and controls nuclear architecture and melanoma growth.
ACKNOWLEDGEMENTS
We thank SBP and Technion Core facilities for help in different phases of this study including the Smoler Protein Research Center (Technion), The Biomedical Core Facilities (Electron Microscopy, Imaging and Flow Cytometry Units; Faculty of Medicine, Technion); the Histology Core (SBP) and The Genomics Core (SBP). We thank Dr Tom Misteli (NIH) for useful discussions, and Vitto Rebeca for help with PDX samples. Z.A.R. gratefully acknowledges support by a National Cancer Institute grant (R01CA202021) and a Melanoma Research Alliance grant (MRA 509524). I.L. was supported by a Rubenstein Family Post-Doctorate fellowship at the Technion. Sanford Burnham Prebys Shared Resources are supported by the NCI Cancer Center Support Grant (P30 CA030199). M.H. is supported by NIH grants 5P01CA114046, 5P50CA174523, and 1U54CA224070.
Footnotes
Competing Interests: ZR is a co-founder and serves as scientific advisor to Pangea Therapeutics. All other authors declare no competing interests.
REFERENCES
- 1.Miller AJ, Mihm MC Jr. Melanoma. N Engl J Med 2006;355(1):51–65 doi 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Jain RK, Martin JD, Stylianopoulos T. The role of mechanical forces in tumor growth and therapy. Annu Rev Biomed Eng 2014;16:321–46 doi 10.1146/annurev-bioeng-071813-105259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. Nuclear lamins. Cold Spring Harb Perspect Biol 2010;2(11):a000547 doi 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 2015;84:131–64 doi 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 5.Davidson PM, Lammerding J. Broken nuclei--lamins, nuclear mechanics, and disease. Trends Cell Biol 2014;24(4):247–56 doi 10.1016/j.tcb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature 2005;437(7058):564–8 doi 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy BK, Pennypacker JK. RB and lamins in cell cycle regulation and aging. Adv Exp Med Biol 2014;773:127–42 doi 10.1007/978-1-4899-8032-8_6. [DOI] [PubMed] [Google Scholar]
- 8.Shumaker DK, Solimando L, Sengupta K, Shimi T, Adam SA, Grunwald A, et al. The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J Cell Biol 2008;181(2):269–80 doi 10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi R, Xu N, Wang G, Ren H, Li S, Lei J, et al. The lamin-A/C-LAP2alpha-BAF1 protein complex regulates mitotic spindle assembly and positioning. J Cell Sci 2015;128(15):2830–41 doi 10.1242/jcs.164566. [DOI] [PubMed] [Google Scholar]
- 10.Wolf C, Rapp A, Berndt N, Staroske W, Schuster M, Dobrick-Mattheuer M, et al. RPA and Rad51 constitute a cell intrinsic mechanism to protect the cytosol from self DNA. Nat Commun 2016;7:11752 doi 10.1038/ncomms11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 2016;17(10):1142–9 doi 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 12.Kreienkamp R, Graziano S, Coll-Bonfill N, Bedia-Diaz G, Cybulla E, Vindigni A, et al. A Cell-Intrinsic Interferon-like Response Links Replication Stress to Cellular Aging Caused by Progerin. Cell Rep 2018;22(8):2006–15 doi 10.1016/j.celrep.2018.01.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalsbeek D, Golsteyn RM. G2/M-Phase Checkpoint Adaptation and Micronuclei Formation as Mechanisms That Contribute to Genomic Instability in Human Cells. Int J Mol Sci 2017;18(11) doi 10.3390/ijms18112344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017;550(7676):402–6 doi 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017;548(7668):466–70 doi 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553(7689):467–72 doi 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouprina N, Mullokandov M, Rogozin IB, Collins NK, Solomon G, Otstot J, et al. The SPANX gene family of cancer/testis-specific antigens: rapid evolution and amplification in African great apes and hominids. Proc Natl Acad Sci U S A 2004;101(9):3077–82 doi 10.1073/pnas.0308532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Zhang Y, Liu H, Salati E, Chiriva-Internati M, Lim SH. Gene expression and immunologic consequence of SPAN-Xb in myeloma and other hematologic malignancies. Blood 2003;101(3):955–60 doi 10.1182/blood-2002-06-1930. [DOI] [PubMed] [Google Scholar]
- 19.Westbrook VA, Diekman AB, Naaby-Hansen S, Coonrod SA, Klotz KL, Thomas TS, et al. Differential nuclear localization of the cancer/testis-associated protein, SPAN-X/CTp11, in transfected cells and in 50% of human spermatozoa. Biol Reprod 2001;64(1):345–58 doi 10.1095/biolreprod64.1.345. [DOI] [PubMed] [Google Scholar]
- 20.Zendman AJ, Cornelissen IM, Weidle UH, Ruiter DJ, van Muijen GN. CTp11, a novel member of the family of human cancer/testis antigens. Cancer Res 1999;59(24):6223–9. [PubMed] [Google Scholar]
- 21.Zendman AJ, Zschocke J, van Kraats AA, de Wit NJ, Kurpisz M, Weidle UH, et al. The human SPANX multigene family: genomic organization, alignment and expression in male germ cells and tumor cell lines. Gene 2003;309(2):125–33 doi 10.1016/s0378-1119(03)00497-9. [DOI] [PubMed] [Google Scholar]
- 22.Westbrook VA, Schoppee PD, Diekman AB, Klotz KL, Allietta M, Hogan KT, et al. Genomic organization, incidence, and localization of the SPAN-x family of cancer-testis antigens in melanoma tumors and cell lines. Clin Cancer Res 2004;10(1 Pt 1):101–12 doi 10.1158/1078-0432.ccr-0647-3. [DOI] [PubMed] [Google Scholar]
- 23.Salemi M, Calogero AE, Vicari E, Migliore E, Zaccarello G, Cosentino A, et al. A high percentage of skin melanoma cells expresses SPANX proteins. Am J Dermatopathol 2009;31(2):182–6 doi 10.1097/DAD.0b013e3181978d6f. [DOI] [PubMed] [Google Scholar]
- 24.Hsiao YJ, Su KY, Hsu YC, Chang GC, Chen JS, Chen HY, et al. SPANXA suppresses EMT by inhibiting c-JUN/SNAI2 signaling in lung adenocarcinoma. Oncotarget 2016;7(28):44417–29 doi 10.18632/oncotarget.10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maine EA, Westcott JM, Prechtl AM, Dang TT, Whitehurst AW, Pearson GW. The cancer-testis antigens SPANX-A/C/D and CTAG2 promote breast cancer invasion. Oncotarget 2016;7(12):14708–26 doi 10.18632/oncotarget.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avitan-Hersh E, Feng Y, Vaisman AO, Ahmed YA, Zohar Y, Zhang T, et al. Regulation of eIF2alpha by RNF4 promotes melanoma tumorigenesis and therapy resistance. J Invest Dermatol 2020. doi 10.1016/j.jid.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raaijmakers MI, Widmer DS, Maudrich M, Koch T, Langer A, Flace A, et al. A new live-cell biobank workflow efficiently recovers heterogeneous melanoma cells from native biopsies. Exp Dermatol 2015;24(5):377–80 doi 10.1111/exd.12683. [DOI] [PubMed] [Google Scholar]
- 28.Krepler C, Xiao M, Sproesser K, Brafford PA, Shannan B, Beqiri M, et al. Personalized Preclinical Trials in BRAF Inhibitor-Resistant Patient-Derived Xenograft Models Identify Second-Line Combination Therapies. Clin Cancer Res 2016;22(7):1592–602 doi 10.1158/1078-0432.CCR-15-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabre B, Livneh I, Ziv T, Ciechanover A. Identification of proteins regulated by the proteasome following induction of endoplasmic reticulum stress. Biochem Biophys Res Commun 2019;517(2):188–92 doi 10.1016/j.bbrc.2019.07.040. [DOI] [PubMed] [Google Scholar]
- 30.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res 2018;46(D1):D649–D55 doi 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009;9(6):400–14 doi 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuela N, Bar DZ, Gruenbaum Y. Lamins in development, tissue maintenance and stress. EMBO Rep 2012;13(12):1070–8 doi 10.1038/embor.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, et al. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 2008;22(7):832–53 doi 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 2013;154(1):47–60 doi 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006;25(3):349–60 doi 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFkappaB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep 2013;4(5):959–73 doi 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol 2015;17(8):1049–61 doi 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Paolo NC, Doronin K, Baldwin LK, Papayannopoulou T, Shayakhmetov DM. The transcription factor IRF3 triggers “defensive suicide” necrosis in response to viral and bacterial pathogens. Cell Rep 2013;3(6):1840–6 doi 10.1016/j.celrep.2013.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yilmaz-Ozcan S, Sade A, Kucukkaraduman B, Kaygusuz Y, Senses KM, Banerjee S, et al. Epigenetic mechanisms underlying the dynamic expression of cancer-testis genes, PAGE2, −2B and SPANX-B, during mesenchymal-to-epithelial transition. PLoS One 2014;9(9):e107905 doi 10.1371/journal.pone.0107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubrez L, Causse S, Borges Bonan N, Dumetier B, Garrido C. Heat-shock proteins: chaperoning DNA repair. Oncogene 2019. doi 10.1038/s41388-019-1016-y. [DOI] [PubMed] [Google Scholar]
- 41.Murakami S, Ninomiya W, Sakamoto E, Shibata T, Akiyama H, Tashiro F. SRY and OCT4 Are Required for the Acquisition of Cancer Stem Cell-Like Properties and Are Potential Differentiation Therapy Targets. Stem Cells 2015;33(9):2652–63 doi 10.1002/stem.2059. [DOI] [PubMed] [Google Scholar]
- 42.Shannan B, Perego M, Somasundaram R, Herlyn M. Heterogeneity in Melanoma. Cancer Treat Res 2016;167:1–15 doi 10.1007/978-3-319-22539-5_1. [DOI] [PubMed] [Google Scholar]
- 43.Snyder D, Wang Y, Kaetzel DM. A rare subpopulation of melanoma cells with low expression of metastasis suppressor NME1 is highly metastatic in vivo. Sci Rep 2020;10(1):1971 doi 10.1038/s41598-020-58996-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moir RD, Spann TP, Lopez-Soler RI, Yoon M, Goldman AE, Khuon S, et al. Review: the dynamics of the nuclear lamins during the cell cycle-- relationship between structure and function. J Struct Biol 2000;129(2–3):324–34 doi 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- 45.Dittmer TA, Sahni N, Kubben N, Hill DE, Vidal M, Burgess RC, et al. Systematic identification of pathological lamin A interactors. Mol Biol Cell 2014;25(9):1493–510 doi 10.1091/mbc.E14-02-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glass JR, Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J Cell Biol 1990;111(3):1047–57 doi 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith ER, Capo-Chichi CD, Xu XX. Defective Nuclear Lamina in Aneuploidy and Carcinogenesis. Front Oncol 2018;8:529 doi 10.3389/fonc.2018.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meaburn KJ, Cabuy E, Bonne G, Levy N, Morris GE, Novelli G, et al. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell 2007;6(2):139–53 doi 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- 49.Zierhut C, Yamaguchi N, Paredes M, Luo JD, Carroll T, Funabiki H. The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 2019;178(2):302–15 e23 doi 10.1016/j.cell.2019.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 1998;95(26):15623–8 doi 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almanzar G, Olkhanud PB, Bodogai M, Dell’agnola C, Baatar D, Hewitt SM, et al. Sperm-derived SPANX-B is a clinically relevant tumor antigen that is expressed in human tumors and readily recognized by human CD4+ and CD8+ T cells. Clin Cancer Res 2009;15(6):1954–63 doi 10.1158/1078-0432.CCR-08-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


