Abstract
Objectives
There is an incomplete understanding of the host humoral immune response to severe acute respiratory syndrome (SARS)‐coronavirus (CoV)‐2, which underlies COVID‐19, during acute infection. Host factors such as age and sex as well as the kinetics and functionality of antibody responses are important factors to consider as vaccine development proceeds. The receptor‐binding domain of the CoV spike (RBD‐S) protein mediates host cell binding and infection and is a major target for vaccine design to elicit neutralising antibodies.
Methods
We assessed serum anti‐SARS‐CoV‐2 RBD‐S IgG, IgM and IgA antibodies by a two‐step ELISA and neutralising antibodies in a cross‐sectional study of hospitalised COVID‐19 patients of varying disease severities. Anti‐RBD‐S IgG levels were also determined in asymptomatic seropositives.
Results
We found equivalent levels of anti‐RBD‐S antibodies in male and female patients and no age‐related deficiencies even out to 93 years of age. The anti‐RBD‐S response was evident as little as 6 days after onset of symptoms and for at least 5 weeks after symptom onset. Anti‐RBD‐S IgG, IgM and IgA responses were simultaneously induced within 10 days after onset, with anti‐RBD‐S IgG sustained over a 5‐week period. Anti‐RBD‐S antibodies strongly correlated with neutralising activity. Lastly, anti‐RBD‐S IgG responses were higher in symptomatic COVID‐19 patients during acute infection compared with asymptomatic seropositive donors.
Conclusion
Our results suggest that anti‐RBD‐S IgG reflect functional immune responses to SARS‐CoV‐2, but do not completely explain age‐ and sex‐related disparities in COVID‐19 fatalities.
Keywords: COVID‐19, humoral immune response, isotypes, neutralising antibody, SARS‐CoV‐2, spike protein
Antibodies to severe acute respiratory syndrome‐coronaviruses‐2 spike arise within 1 week after COVID‐19 symptoms and are not affected by patient age or biological sex. Neutralising activity correlates with receptor‐binding domain reactivity and may reflect viral replication rather than acute protection.
![]()
Introduction
Human pathogenic coronaviruses (CoV) such as severe acute respiratory syndrome (SARS)‐CoV‐1, Middle East respiratory syndrome (MERS)‐CoV and SARS‐CoV‐2 (all β‐CoVs) have resulted from zoonoses and utilise cellular receptors to bind and access host cells for productive infection. 1 , 2 , 3 CoV spike (S) proteins are large (> 200 kDa) glycosylated trimeric structures that protrude from viral particles and enable binding of CoV to cellular receptors. SARS‐CoV‐2 interacts with angiotensin‐converting enzyme‐2 via a flexible receptor‐binding domain (RBD) located on the distal tip of the S protein. 4 , 5 , 6 , 7 After binding, several proteases act upon S, priming it to adopt large conformational shifts that facilitate entry into host cells. 8 First, the S1 domain (which contains RBD) is cleaved from the C‐terminal S2 domain. For SARS‐CoV‐2, this process may involve furin in the host cell membrane due to a novel furin‐recognition site in the S1/S2 region. 9 , 10 , 11 The S2 domain is further processed by other serine and cysteine proteases such as trypsin, cathepsin and TMPRSS2 to facilitate viral entry into the host cell. 4 , 12
Neutralising antibodies to SARS‐CoV‐1 have been isolated and were found to target spike glycoprotein RBD (RBD‐S). 13 One of these mAbs CR3022 was also found to bind SARS‐CoV‐2 RBD‐S. 14 At the polyclonal level, the quantity of anti‐RBD S IgG antibodies against SARS‐CoV‐2 correlates well with neutralising activity. 15 , 16 , 17 , 18 Cross‐neutralisation among SARS viruses by RBD‐S‐targeting antibodies can occur. 17 , 19 , 20 , 21 However, sequence homology for RBD‐S is low for non‐SARS β‐CoVs such as MERS, OC43 and HKU‐1 and for α‐CoVs such as NL63 and 229E. 16 , 18 For these reasons, serology for SARS‐CoV‐2 RBD‐S is being used to help identify recovered COVID‐19 patients as plasma donors for passive immunotherapy. 22
There are several risk factors for COVID‐19 mortality but the relationship between two of these – age and biological sex – with the SARS‐CoV‐2 RBD‐S response is not completely clear. Most serology studies have been done in the setting of severe COVID‐19 disease and, save for one, 18 without the benefit of detailed kinetics. Vermont in contrast to most of the United States and many nations has had a low number of cases (under 1700 cases total to date), and here, we present an examination of RBD‐S responses during acute COVID‐19 and during asymptomatic exposure in a setting of a low force of infection.
Results
We chose a two‐step ELISA‐based RBD‐S‐focused approach to serology in our study population. Reagents and pre‐print protocols were available in mid‐March 2020, which indicated that RBD‐S screening and full‐S confirmation could identify specific and functional antibodies and be quickly operationalised. Using the established protocol, 23 we confirmed the expected protein size of mammalian‐expressed RBD‐S (Figure 1a) and trimerised spike (Figure 1b) produced from DNA plasmids (gift from Florian Krammer, Mt Sinai School of Medicine). RBD‐S antibodies were specific and correlated with neutralisation, 15 findings that have been validated using similar RBD‐S‐focused assays. 16 , 18 We confirmed RBD‐S and S protein conformation by binding of CR3022 human IgG1 (Figure 1c and d). CR3022 was isolated as a SARS‐S1 domain‐binding single chain antibody fragment by phage display and is neutralising as an IgG1. 13 CR3022 binds adjacent to RBD‐S in trimeric S of SARS‐CoV‐2 in a glycosylation‐sensitive manner. 14 Mammalian expression of appropriate size proteins and recognition by CR3022 together confirm that our protein preparations exhibited the expected characteristics.
Figure 1.
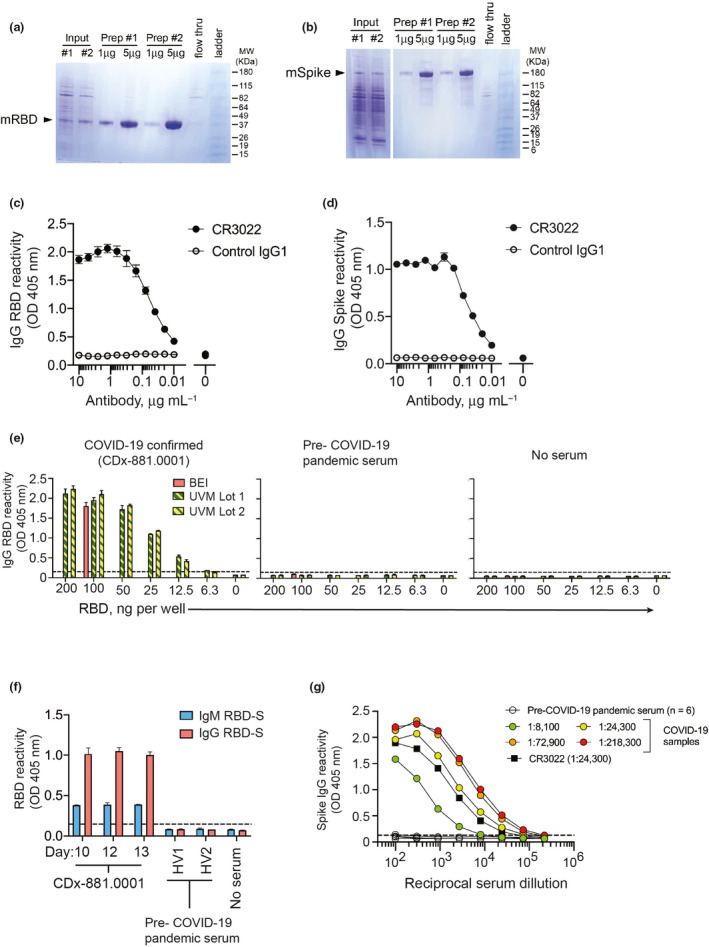
Validation of SARS‐CoV‐2 RBD‐S and spike antigens in COVID‐19 samples. Reducing SDS‐PAGE analysis of (a) RBD‐S and (b) trimeric spike purified from transiently transfected mammalian HEK293 cells. (c) Binding of CR3022 IgG1 mAb to SARS‐CoV‐2 RBD‐S and (d) trimerised spike. The anti‐dengue virus 1M7 IgG1 mAb 42 was used as a control. (e) Detection of serum IgG from a COVID‐19 patient (left), but not from pre‐2020 serum (centre) or no serum control (right). (f) Detection of IgM and IgG to RBD‐S in serial time the course serum samples from a COVID‐19 patient and not in pre‐COVID‐19 pandemic healthy volunteer sera (HV1,HV2). All sera diluted 1:50, and for the COVID‐19 patient (CD×‐881.0001), day after symptoms onset is shown on the x‐axis. (g) Anti‐spike IgG reactivity by ELISA for pre‐COVID‐19 pandemic sera (n = 6), the anti‐SARS‐CoV‐1/2 mAb CR3022, and COVID‐19 samples of varying titres (indicated in parentheses).
We first piloted our antigen preps for the RBD‐S IgG screening assay using serum samples from a PCR‐confirmed severe COVID‐19 patient (defined as admission to the intensive care unit, ICU) who was admitted to the hospital 10 days following symptom onset and based on an early report suggesting that SARS‐CoV‐2 could trigger antibody responses in this timeframe. 24 We compared IgG reactivity in this sample to decreasing amounts of our RBD‐S antigen preparations against a fixed, recommended amount of commercially produced RBD‐S protein derived from the protocol we used. 23 We found that a wide range of locally produced RBD‐S antigen yielded IgG reactivity equivalent to 100 ng of commercial antigen in an acute serum sample from this COVID‐19‐positive patient, but not in healthy pre‐COVID‐19 pandemic serum or in the absence of serum (Figure 1e). Using the standard 100 ng amount of RBD‐S hereafter, we found that RBD‐S‐binding IgM and IgG were present at 10–13 days after symptom onset. We did not detect any RBD‐S binding in healthy pre‐pandemic sera (Figure 1f) in agreement with previous results. 15 There was negligible day‐to‐day assay variability (Supplementary figure 1). Due to different secondary antibodies for IgM and IgG detection, we cannot conclude whether absolute levels of RBD‐S IgG were higher than RBD‐S IgM. A range of anti‐spike IgG titres was evident in different COVID‐19 patients, but not in healthy pre‐COVID‐19 pandemic samples (Figure 1g).
For a cross‐sectional COVID‐19 serological survey, we collected serum samples from 32 patients that tested COVID‐19 positive by nasopharyngeal swab RT‐qPCR testing. All patients had been admitted to the hospital, and 13/32 (40%) were admitted to the ICU. Twenty‐five patients were subsequently discharged and seven died. One to five serum samples were collected from each patient with the first sample being taken within approximately 9 days after diagnosis, in which diagnosis occurred about 5 days after symptom onset (Table 1). There was a 53%:47% male: female distribution, and patients were on average 68 ± 14 years of age (range 30−93 years; Table 1).
Table 1.
Hospitalised COVID‐19 patient characteristics
| COVID‐19 subjects | Male/female |
AGE ± SD [Range] |
Days from symptoms to Dx | Days between Dx and 1st serum |
|---|---|---|---|---|
| Swab PCR+ (n = 32) | 17/15 | 68 ± 14 [30–93] | 5.4 ± 4.7 [0–14] | 8.6 ± 7.5 [0–35] |
A male bias in COVID‐19 mortality was reported early during the pandemic 25 , 26 , 27 and has been confirmed worldwide in a recent meta‐analysis. 28 One of the hypotheses to explain this is differences in adaptive immunity between males and females. Although the mean serum RBD‐S IgG reactivity level appeared higher in male samples (O.D. = 1.8, n = 17) than in female samples (O.D. = 1.0, n = 15), this difference was not significant and the same maximum reactivity values were found in males and females (Figure 2a). Similarly, anti‐spike IgG endpoint titres trended higher in male patients compared to female patients, but this difference was not significant (Figure 2b).
Figure 2.
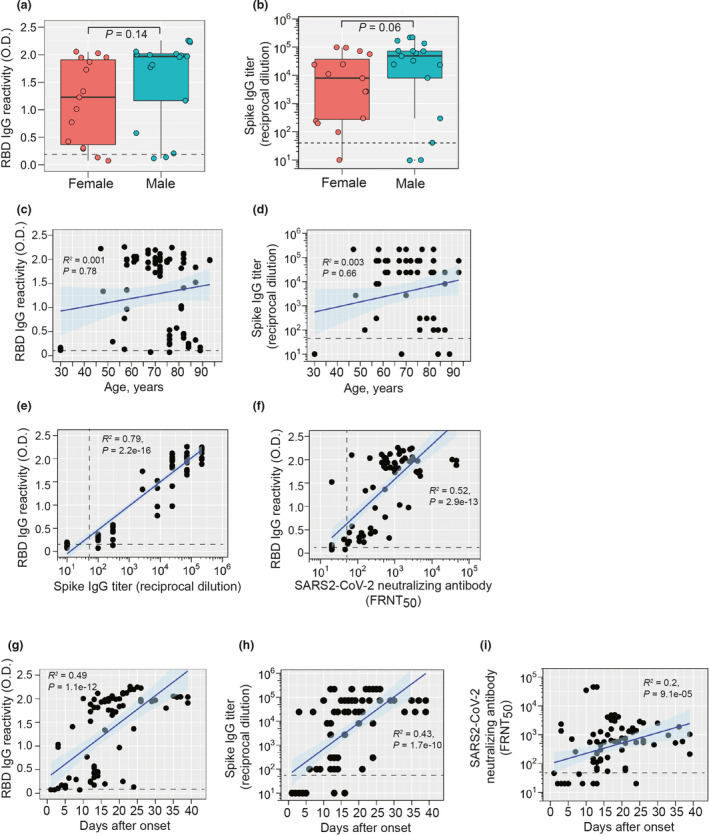
IgG responses to SARS‐CoV‐2 RBD‐S and spike. (a) Comparison of RBD‐S IgG reactivity (OD 405 nm) or (b) anti‐spike IgG endpoint titres levels in male (n = 17) or female (n = 15) patients. Some patients had multiple samples that were averaged for this analysis. Boxplots show the 25–75th percentiles, with median as horizontal line and whiskers as 95% confidence level with subjects as symbols and groups were analysed by Mann–Whitney U‐tests. All patient samples were plotted individually. (c) Anti‐RBD‐S IgG and (d) anti‐spike IgG are expressed as a function of age with all individual samples from a patient plotted (n = 77 total). (e) Anti‐RBD‐S IgG reactivity is plotted against anti‐spike IgG endpoint titres. (f) SARS‐CoV‐2 microneutralisation titres [50% focus reduction neutralisation titres (FRNT50)] are plotted against RBD‐S IgG reactivity (n = 77). (g) RBD‐S IgG reactivity, (h) anti‐spike IgG endpoint titres, and (i) SARS‐CoV‐2 neutralising antibody titres are plotted against days after symptoms onset. (Cut‐off values for each assay are shown by dashed lines. Spearman's Rho coefficient (R 2), 95% confidence interval (shading), and P‐values are shown for panels c–i).
Although not absolute, it appears that irrespective of comorbidities, there is a higher risk of COVID‐19 mortality and morbidity in older individuals (60 years of age and over). 29 , 30 , 31 We therefore assessed RBD‐S IgG antibodies by age. There was a broad range of RBD‐S IgG and anti‐spike IgG endpoint titre responses that did not differ as a function of age as assessed by correlation analysis (R 2 < 0.01, Figure 2c and d). Notably, one of the highest anti‐RBD‐S/spike IgG responses was from a 93‐year‐old patient. A serum sample from a 30‐year‐old COVID‐19 patient was negative for RBD‐S IgG, but this sample was taken just 3 days after symptom onset, which may be too early for induction of robust IgG responses. Taken together, we did not find evidence of biological sex‐ or age‐related deficiencies in RBD‐S IgG responses in COVID‐19 patients.
To further validate our approach, we confirmed each sample (whether RBD‐positive or not) with an endpoint titration for reactivity against the full spike ectodomain trimer. 15 Samples that were RBD‐S‐negative were also low for spike titre (Figures 2e). Furthermore, we found a very strong correlation between RBD‐S and spike IgG levels (Figure 2e). We also tested for neutralising antibodies against SARS‐CoV‐2 with a focus microneutralisation assay (Supplementary figure 2). We observed a strong correlation between RDB‐S IgG and microneutralisation titres (Figure 2f), confirming the utility of a two‐step RBD‐S serology approach for estimation of functional neutralising antibodies in agreement with other studies. 15 , 16 , 18 We next assessed the kinetics of the anti‐spike RBD‐S response in confirmed COVID‐19 infection. RBD‐S‐reactive serum IgG was detected in five of 12 (42%) samples that were taken within 10 days of symptom onset (Figure 2g). After day 10 of symptoms, > 98% of samples were positive for RBD IgG (Figure 2g). Anti‐spike IgG paralleled the kinetics of the anti‐RBD‐S response and correlated with days of onset (Figure 2h). We found a weaker, although still significant positive correlation of SARS‐CoV‐2 neutralising antibodies and days post‐onset, with the majority strong responses occurring at least 10 days after symptoms onset (Figure 2i).
In the patient‐specific RBD IgG data (Supplementary figure 3a), we found several patterns: initial seroconversion (e.g. patients 0003, and 0017), rapid increases (e.g. patients 0005, 0006, 0009, 0011, 0020, occurring between days 10 and 20), and plateaued responses (e.g. patients 0012 and 0021, occurring mainly after day 20). These responses were concordant with temporal patient‐specific S IgG titres (Supplementary figure 3b). Anti‐S titres in patients with a negative RBD‐S test were generally low and in RBD‐positive samples, followed the same trends as RBD reactivity, providing further confirmation of robust serological responses to SARS‐CoV‐2 during acute COVID‐19. At the patient level, neutralising activity was observed after as few as 5 days after symptom onset and throughout the study period and was predominantly found in those samples with positive RBD‐S IgG (Supplementary figure 4).
To assess antibody isotype dynamics during acute SARS‐CoV‐2, we followed RBD‐S and full spike‐specific IgM and IgA levels in the same samples for which RBD‐S and spike IgG were determined. At the patient level, we found robust co‐occurrence of IgM, IgG and IgA antibodies reactive to RBD‐S in most samples, particularly in post‐day 10 samples (Supplementary figure 5). Pooling all the data revealed that all pre‐day 10 RBD‐S responses for all isotypes were low. At about day 10, IgM targeting RBD‐S as well as the switched isotypes IgG and IgA simultaneously rose. While RBD‐reactive IgM and IgA responses tapered after 3 weeks post‐onset (although they remained higher than baseline), those for IgG continued to rise to a plateau that was sustained up to 5 weeks after symptoms onset (the most protracted timepoint measured, Figure 3a). Similar patterns were obtained for full spike‐reactive antibodies (Figure 3b). These results showed during acute infection COVID‐19 patients undergo rapid seroconversion across isotypes to SARS‐CoV‐2. Neither anti‐RBD‐S IgM nor IgA responses were different by sex (P = 0.78 for IgM and P = 0.08 for IgA; Supplementary figure 6a and b).
Figure 3.
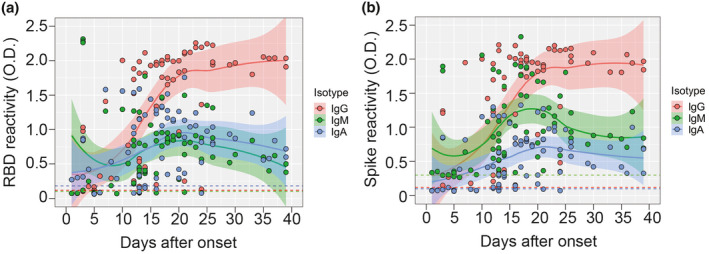
Antibody isotype usage during the response to SARS‐CoV‐2 RBD‐S and spike. (a) RBD‐S IgM, IgG and IgA in serum (diluted 1:50) were determined by ELISA and plotted against days post‐onset of symptoms (n = 77 samples). LOESS‐smoothed lines and 95% confidence intervals are shown for each isotype. (b) Spike‐reactive IgM, IgG and IgA in serum (diluted 1:100) were determined by ELISA and plotted against days post‐onset of symptoms. LOESS‐smoothed lines and 95% confidence intervals are shown for each isotype.
Lastly, we assessed anti‐RBD‐IgG responses by clinical severity. All the patients in this study were hospitalised, and 40% of were admitted to the intensive care unit. When we stratified by ICU admission and compared RBS‐S IgG levels, we found a trend towards higher levels in those requiring ICU‐level care (P = 0.09; Figure 4a). Additionally, we observed a significant association between RBD‐S IgG and duration of ICU admission (Figure 4b). Anti‐RBD‐IgM, but not IgA, responses were higher in ICU patients compared to non‐ICU patients (Supplementary figure 6c and d). Lastly, seven of 32 (22%) patients succumbed to COVID‐19. While a significant difference in the median RBD‐S IgG was not observed between survivors and decedents, a smaller range trending towards higher RBD‐S reactivity was observed in those patients that died (Figure 4c). To further test the relationship of anti‐RBD antibodies to clinical disease severity, we compared levels in symptomatic patients with 17 seropositive asymptomatic subjects. RBD‐S IgG levels (Figure 4d) and anti‐spike IgG titres (Figure 4e) were significantly higher in symptomatic patients compared to asymptomatic convalescent donors.
Figure 4.
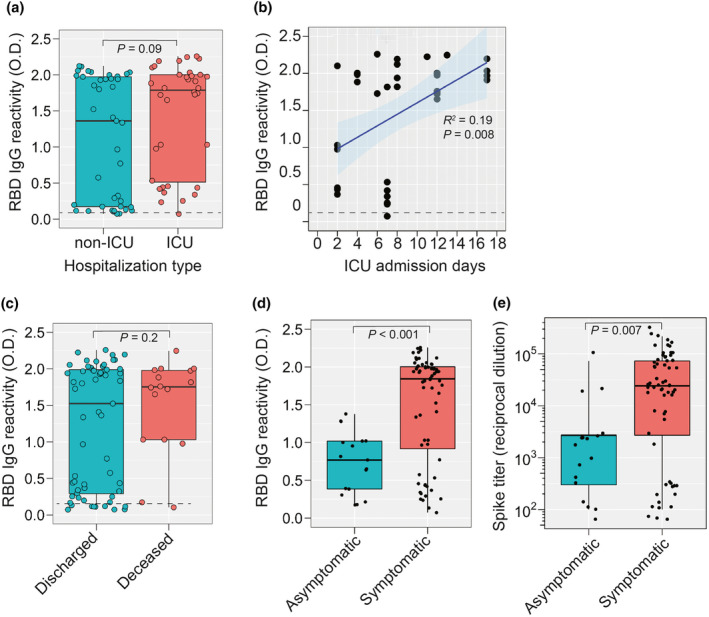
SARS‐CoV‐2 RBD‐S IgG responses during hospitalisation. (a) RBD‐S IgG in patients that were hospitalised in the ICU or not. (b) For ICU‐hospitalised patients, all RBD‐S IgG values are presented as a function of ICU admission days. Spearman's Rho coefficient (R 2), 95% confidence interval, and P‐value are shown. (c) RBD‐S IgG in patients that were deceased (n = 16 samples from seven patients) or discharged (n = 61 samples from 25 patients). (d) RBD‐S IgG reactivity and (e) anti‐S IgG titres in symptomatic COVID‐19 patients (n = 77 samples) versus convalescent seropositive COVID‐19‐exposed volunteers (n = 17). Boxplots in a, c–e show the median, 95% confidence level and all individual samples. The Student's t‐test P‐value is shown.
Discussion
Here, we comprehensively surveyed SARS‐CoV‐2 spike RBD antibodies in confirmed COVID‐19 patients in the context of risk factors for COVID‐19, clinical severity, and relation to immune responses in asymptomatic donors. Neither RBD‐S nor S antibodies were significantly different as a function of biological sex in symptomatic COVID‐19 patients. Anti‐RBD‐S and spike IgG responses were induced across six decades of age with robust responses found in several samples from patients ≥ 80 years old.
Our results suggest a paucity of anti‐SARS‐CoV‐2 anti‐spike responses across serotypes in very early blood samples taken prior to day 10 after symptoms onset in agreement with others. 18 , 24 , 32 We also assessed anti‐spike RBD responses as a function of level of hospital care and disease severity and found that duration of ICU‐level care was associated with higher responses, possibly due to an extended period of SARS‐CoV‐2 replication during severe disease. Interestingly, anti‐RBD‐S IgM responses were higher in ICU patients compared to non‐ICU patients, which could suggest a delay in class‐switching in some patients, although we have not examined this at the B‐cell level. However, anti‐SARS‐CoV‐2 spike responses can also be found after mild COVID‐19. 33 , 34 There are several limitations of our study. Our study population of COVID‐19 patients admitted to hospital was relatively small. The reason for this is that Vermont has to date (September 11, 2020) had the lowest transmission rate (< 300 cases per 100 000 residents) in the United States and compared to many nations. This, in turn, however allowed for serological analysis in the absence of a high force of infection which could confound accurate diagnoses. It is possible that day of sampling (which was random and taken as available with no pre‐selection) could be a potential confounder for the age and sex analyses. Another limitation is that we did not continuously monitor viral load in these patients during hospitalisations. Thus, we infer that the correlation between RBD‐S IgG levels and length of ICU stay could reflect ongoing viral replication during more severe disease. This was supported by the lower anti‐spike levels in asymptomatic subjects taken within 2 months of our acute patients, where substantial antibody decay is not yet evident. 35
Wang et al. 36 found lower neutralising antibodies in inpatients than in convalescent volunteers. Our data differ on this point through multiple lines of evidence. Anti‐RBD‐S levels trended higher in ICU‐hospitalised subjects than in non‐ICU patients, and in deceased than in discharged patients. In our study, the length of ICU stay correlated positively with RBD‐S IgG, and symptomatic patients exhibited substantially higher anti‐RBD‐S responses than contemporaneous non‐hospitalised COVID‐19 seropositive volunteers. It is possible that differences in methodology or community force of infection underlie these discrepant results. We performed our measurements in three independent tests: screening with RBD‐S, then confirmation with full‐length spike, and finally assessment of functionality with direct neutralisation assays that correlate strongly with RBD‐S reactivity. Regarding age, Wang et al. also found higher neutralising antibody responses in patients > 30 years of age than in those 16–30, but not among those > 30 years of age. We did not have any subjects < 30 years of age in our cohort. In agreement with Wang et al., we found no clear correlation between RBD‐S IgG levels and age. Regarding possible sex differences in COVID‐19 immune responses, our results are consistent with Zeng et al. who found no difference with male and female COVID‐19 patients. 37
We did not directly assess whether the RBD‐specific antibodies we studied were neutralising at the clonal level. We did observe a strong association with polyclonal RBD‐S IgG responses and SARS‐CoV‐2 neutralising activity. This is in agreement with other reports which confirm that RBD‐S IgG levels correlate with neutralising activity and that the RBD of SARS‐CoV‐2 is a potent target for neutralising antibodies. 16 , 17 , 18 , 20 , 21 , 38 All RBD‐S‐positive samples were also spike‐reactive, reinforcing the utility of this two‐step approach. There was a low level of spike reactivity in RBD‐negative samples could indicate a baseline cross‐reactivity against other human coronaviruses. 39 S cross‐reactivity would presumably occur in regions outside the RBD given the low conservation of SARS‐CoV‐2 RBD compared to other human CoVs with the exception of SARS‐CoV‐1. 16
Lastly, we found robust induction of IgM, IgG and IgA responses in a contemporaneous fashion within 10 days after onset of symptoms. Indeed, a recent report at the cellular level showed isotype‐switching and the appearance of near‐germline SARS‐CoV‐2 neutralising antibodies early after infection, suggesting that neutralising antibodies may not require extensive diversification. 40 It will be important to determine whether anti‐RBD IgA or even IgM antibodies contribute to blocking activity. In sum, natural SARS‐CoV‐2 infection induces a robust RBD‐S antibody response equally in men and women irrespective of age and may reflect viral burden and acute COVID‐19 disease severity.
Methods
Patient samples
Symptomatic COVID‐19 patients
Patients were admitted to the University of Vermont Medical Center (UVMMC), situated in a low‐density (26–112 persons km−2) catchment area with a COVID‐19 diagnosis from a PCR‐positive swab testing performed within a CLIA‐certified clinical laboratory. University of Vermont Institutional Review Board approval was granted under registration STUDY00881. Samples and patient data were obtained under Exemption 4, Waiver of Consent and UVM/UVMMC HIPAA Authorization under 46.116(f)(1)(3), 46.164.512(i)(1)(2). Patient IDs are coded here as ‘CDDx.001‐032’. Deidentified patient (age, sex) and clinical data (COVID‐19 diagnosis, dates of symptom onset, hospitalisation, intensive care unit admission) were obtained from the electronic health record. To identify asymptomatic COVID‐19‐exposed seropositives, we conducted two serosurveys. First, we surveyed anti‐SARS2 RBD‐S binding IgG in healthy UVMMC Intensive Care Unit staff between April and May 2020 (University of Vermont Institutional Review Board approval granted under registration STUDY00881). Seven of 174 (4.0%) of HCW were seropositive as described below. All HCW were defined as asymptomatic for COVID‐19 based on negative responses to an enrolment questionnaire to experiencing fever, cough, shortness of breath, or diarrhoea and a temperature < 100.4°F at the time of blood draw and in accordance with UVMMC policies. Second, we tested 454 healthy adults between 25 and 28 June 2020 in the Chittenden county catchment area for both presence of SARS‐CoV‐2 viraemia by PCR and for anti‐RBD‐S antibodies. None reported as having COVID‐19 symptoms (as above) at the time of blood draw. One subject was positive for PCR at the time of testing, but negative for serology. Ten were seropositive (2.2%), including all five subjects who previously (since March) had positive COVID‐19 testing or COVID‐19 symptoms per their primary care provider. University of Vermont Institutional Review Board approval for the population serology study was granted under registration STUDY00914.
RBD‐S and spike antigen preparations
pCAGGS plasmids containing hexahistidine‐tagged SARS‐CoV‐2 spike glycoprotein RBD (RBD‐S) and trimerised SARS‐CoV‐2 15 , 23 were obtained as Whatman spots from Florian Krammer (Mt. Sinai School of Medicine) and transformed into Escherichia coli to make plasmid stocks. We sequence verified these using pcaggs‐F (5′‐GTTCGGCTTCTGGCGTGT‐3′) and pcaggs‐R (5′‐TATGTCCTTCCGAGTGAGAG‐3′). Plasmids were then transfected into Expi293F cells (Thermo Fisher, Bedford, MA, USA, Cat. #A14527), and protein was purified by Ni‐NTA agarose resin (Qiagen, Germantown, MD, USA, Cat. #30230) as described. 23 Protein was quantified using bovine serum albumin as a standard (Sigma‐Aldrich, St. Louis, MO, USA, Cat. #A4505, Cohn Faction V) and Bradford reagent (Bio‐Rad, Hercules, CA, USA, Cat. #5000006). Protein was run on denaturing 4–20% recast protein gels (Bio‐Rad, Cat. #4561094) and visualised by Coomassie blue staining with a 10–190 kDa protein ladder (Thermo Fisher–Invitrogen Cat. #10748‐010).
Spike Glycoprotein RBD from SARS‐CoV‐2, Wuhan‐Hu‐1, was also used as a positive control during assay set up, and this reagent was produced in HEK293T cells under HHSN272201400008C and obtained through BEI Resources (NIAID, Bethesda, MD, USA): Spike Glycoprotein RBD from SARS‐Related Coronavirus 2, Wuhan‐Hu‐1, Recombinant form Cat. #NR‐52306.
Preparation of CR3022 monoclonal antibody
CR3022 is a SARS‐CoV S‐specific antibody originally isolated by single chain variable region phage display and then cloned as an IgG1/kappa monoclonal human IgG1/κ. 13 We received CR3022 heavy chain (HC) and light chain (LC) cloned into pFUSEss‐CHIg‐hG1 and pFUSE2ss‐CLIg‐hK, respectively (Invivogen, San Diego, CA, USA) from Florian Krammer spotted on filter paper. We resuspended spots in 100 µL TE and transformed 20 µL E. coli (NEB C2987H) with 1 µL followed by growth in the presence of Zeocin (25 µg mL−1, Invivogen, for CR3022‐HC) and blasticidin (100 µg mL−1, Invivogen for CR3022 LC). Midi‐preps were then sequenced confirmed CR3022HC (Genbank DQ168569) and LC (Genbank DQ168570) with primer HTLV‐5′UTR (forward) 5′‐GCTTGCTCAACTCTACGTC‐3′ and CR3022‐HC in the reverse direction by primer Fc (reverse): 5′CTCACGTCCACCACCACGCA‐3′. Recombinant CR3022 was expressed in 293A cells (Invitrogen) by polyethylenimine (Polysciences Inc., Warrington, PA, USA, Cat. #24765) transfection of 9 µg each of CR3022‐HC and LC, culture for 7 days, and protein A agarose bead purification as described. 41 IgG was quantified by sandwich ELISA with anti‐human IgG (Jackson Immunoresearch, Bar Harbor, ME, USA, Cat. #109‐005‐008) as capture and horseradish peroxidase‐conjugated anti‐human IgG (Jackson Immunoresearch, Cat. #109‐005‐008) as detection Ab with known human serum as a standard.
Clinical RBD‐S and S IgG ELISA testing
For IgG against RBD‐S from SARS‐CoV‐2, we followed Stadlbauer et al. 23 and the Emergency Use Authorization granted to MSSM by the Food and Drug Administration on 4/15/2020 (https://www.fda.gov/media/137029/download). Briefly, for RBD‐S IgG levels 96‐well plates were coated with 100 ng per well of purified RBD‐S and then blocked with 3% milk in phosphate‐buffered saline (PBS) containing 0.1% Tween‐20 (T). Heat‐inactivated (56°C for 1 h) serum samples were diluted 1:5 in PBS, and 20 µL of this was added to 180 µL of dilution buffer (PBS‐T + 1% milk) in each well for 1:50 final dilution of sample. One hundred microliter of sample is then added to each well, and after 1 hr incubation at room temperature and washing with PBS‐T using a Biotek ELx‐405 Select CW (Biotek, Winooski, VT, USA), IgG was detected with alkaline phosphatase‐conjugated cross‐adsorbed anti‐human IgG (Sigma‐Aldrich, Cat. #SAB3701277, diluted 1:2500 in blocking buffer), washing, and addition of p‐nitrophenylphosphate (Sigma‐Aldrich, Cat. #N2770) substrate. The colorimetric reaction (optical density at 405 nm) was detected with a Cytation 3 (Biotek). Two negative control samples of pre‐COVID‐19 pandemic (i.e. collected before 2019) serum were used on each plate, and the average + three standard deviations above the mean were used as the assay cut‐off for positivity.
To determine anti‐spike titres, heat‐inactivated serum samples were diluted 1:5 in PBS, and then, 20 µL was added to 180 µL of dilution buffer in the starting well (for a final 1:100 starting dilution) and then serially diluted 1:3 to an endpoint dilution of 1:218 700 as needed. IgG detection was performed as described above with 100 µL of 1:100 sample. Endpoint titre was defined to be the last dilution at which the signal was above the cut‐off (defined as was done for RBD‐S above). Six pre‐COVID‐19 pandemic negative control samples did not exhibit reactivity above a no serum control.
Testing for RBD‐S and S IgM and IgA in clinical samples by ELISA
Samples were handled as above for IgG except that the detection steps used alkaline phosphatase‐conjugated anti‐human IgM (Sigma‐Aldrich, Cat. #A3437, diluted 1:1000 in blocking buffer) or IgA (Sigma‐Aldrich, Cat. #A3400, diluted 1:1000 in blocking buffer).
SARS‐CoV‐2 microneutralisation assay
All experiments featuring infectious SARS‐CoV‐2 were conducted at the UVM BSL‐3 facility under an approved Institutional Biosafety protocol. SARS‐CoV‐2 strain 2019‐nCoV/USA_USA‐WA1/2020 (WA1) was generously provided by Kenneth Plante and the World Reference Center for Emerging Viruses and Arboviruses at the University of Texas Medical Branch and propagated in African green monkey kidney cells (Vero E6) that were kindly provided by J.L Whitton. Vero E6 cells were maintained in complete Dulbecco's Modified Eagle Medium (cDMEM; Thermo Fisher, Cat. #11965–092) containing 10% foetal bovine serum (16140–071), 1% HEPES Buffer Solution (15630–130), and 1% penicillin–streptomycin (Thermo Fisher, Cat. #15140–122). Cells were grown in a humidified incubator at 37ºC with 5% CO2. To assess the neutralisation capacity of patient sera against authentic SARS‐CoV‐2, we conducted a focus reduction neutralisation test (FRNT). Each serum sample was heat inactivated via incubation at 56°C for 1 h. Samples were then diluted serially in 25 µL of cDMEM, mixed with an equal volume of cDMEM containing 175 focus forming units of SARS‐CoV‐2, and then incubated for 60 min at 37°C. Each serum sample was tested for neutralisation at an initial dilution of 1:50 and then serially at 1:2 dilutions until reaching an endpoint of 1:3200. The media from confluent Vero E6 cell monolayers in 96‐well white polystyrene microplates (Thermo Fisher, Cat. #07‐200‐628) were removed, and 50 µL of each antibody–virus mixture was inoculated onto the cells and incubated at 37°C in a 5% CO2 incubator for 60 min, after which the wells were overlaid with 1.2% methylcellulose in cDMEM and incubated at 37°C in a 5% CO2 incubator for 24 h. Infected cells were fixed in 25% formaldehyde in 3 × PBS. Cells were permeabilised with 0.1% 100X Triton in 1× PBS for 15 min and then incubated with a primary, cross‐reactive rabbit anti‐SARS‐CoV N monoclonal antibody (Sinobiological, distributed by Thermo Fisher, Cat. #40143‐R001 at a dilution of 1:20 000 followed by a peroxidase‐labelled goat anti‐rabbit antibody (SeraCare, Milford, MA, USA, Cat. #5220‐0336) diluted to 1:2000 and then the peroxidase substrate (SeraCare, Cat. #5510‐0030). Images of the wells were captured using a Zeiss AxioCam MRC Imager. M1 microscope and viral foci were quantified manually. Focus counts were normalised to virus only control wells. FRNT50 determinations were made using a non‐linear regression curve fit [log(inhibitor) versus normalised response − variable slope] in GraphPad Prism (GraphPad Software, San Diego, CA, USA).
Graphics and statistical testing
All statistics and graphics were performed using R version 3.6.1 (https://www.r‐project.org). Non‐parametric LOESS (LOcal regrESSion) was used for smoothing.
Author Contributions
Nancy R Graham: Data curation; Investigation; Methodology. Annalis N Whitaker: Data curation; Investigation; Methodology. Camilla A Strother: Investigation. Ashley K Miles: Data curation. Dore Grier: Data curation. Benjamin D McElvany: Investigation; Methodology. Emily A Bruce: Funding acquisition; Validation; Writing‐review & editing. Matthew E Poynter: Writing‐review & editing. Kristen K Pierce: Resources. Beth D Kirkpatrick: Funding acquisition; Resources. Renee D Stapleton: Resources; Writing‐review & editing. Eline van den Broek‐Altenburg: Investigation; Resources. Gary An: Funding acquisition; Resources. Jason W Botten: Conceptualization; Formal analysis; Funding acquisition; Supervision. Jessica W Crothers: Conceptualization; Funding acquisition; Supervision; Writing‐review & editing. Sean ADiehl: Conceptualization; Funding acquisition; Investigation; Project administration; Resources; Supervision; Writing‐original draft; Writing‐review & editing.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Acknowledgments
We thank all healthcare workers and laboratory personnel who contributed to treatment and diagnosis of these and other COVID‐19 patients. We thank the clinical research staff at the University of Vermont (UVM) Medical Center Pathology and Laboratory Medicine and the Vaccine Testing Center. We also thank the UVM Research Protections Office, Institutional Review Board, and Institutional Biosafety Committee for rapid turnaround of COVID‐19‐related projects. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This work was funded by a pilot grant to SAD and EB from the UVM Translational Global Infectious Disease Research Center (National Institute of Health grant P20GM125498). Additional funding was from NIH grant U01AI141997 to SAD, JWB, and BDK, the Office of the Vice President for Research at the University of Vermont to JWB, and the University of Vermont Larner College of Medicine Departments of Surgery and Radiology. Sequencing confirmation of reagents was performed in the Vermont Integrative Genomics Resource Sequencing Facility and was supported by the UVM Cancer Center, Lake Champlain Cancer Research Organization, UVM College of Agriculture and Life Sciences, and the UVM Larner College of Medicine.
References
- 1. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019; 17: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J et al Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou P, Yang XL, Wang XG et al A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoffmann M, Kleine‐Weber H, Schroeder S et al SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade‐long structural studies of SARS coronavirus. J Virol 2020; 94: e00127‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science 2020; 369: 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wrapp D, Wang N, Corbett KS et al Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science 2020; 367: 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res 2016; 96: 29–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann M, Kleine‐Weber H, Pohlmann S. A multibasic cleavage site in the spike protein of SARS‐CoV‐2 is essential for infection of human lung cells. Mol Cell 2020; 78: 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jaimes JA, Andre NM, Chappie JS, Millet JK, Whittaker GR. Phylogenetic analysis and structural modeling of SARS‐CoV‐2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 2020; 432: 3309–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS‐CoV‐2 spike protein and the role of the novel S1/S2 Site. iScience 2020; 23: 101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ou X, Liu Y, Lei X et al Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun 2020; 11: 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. ter Meulen J, van den Brink EN, Poon LL et al Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med 2006; 3: e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan M, Wu NC, Zhu X et al A highly conserved cryptic epitope in the receptor binding domains of SARS‐CoV‐2 and SARS‐CoV. Science 2020; 368: 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Amanat F, Stadlbauer D, Strohmeier S et al A serological assay to detect SARS‐CoV‐2 seroconversion in humans. Nat Med 2020; 26: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Premkumar L, Segovia‐Chumbez B, Jadi R et al The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS‐CoV‐2 patients. Sci Immunol 2020; 5: eabc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ju B, Zhang Q, Ge J et al Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature 2020; 584: 115–119. [DOI] [PubMed] [Google Scholar]
- 18. Suthar MS, Zimmerman MG, Kauffman RC et al Rapid generation of neutralizing antibody responses in COVID‐19 patients. Cell Rep Med 2020; 1: 100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell 2020; 181: 281–292.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pinto D, Park YJ, Beltramello M et al Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature 2020; 583: 290–295. [DOI] [PubMed] [Google Scholar]
- 21. Wec AZ, Wrapp D, Herbert AS et al Broad neutralization of SARS‐related viruses by human monoclonal antibodies. Science 2020; 369: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stadlbauer D, Baine I, Amanat F et al Anti‐SARS‐CoV‐2 spike antibodies are stable in convalescent plasma when stored at 4 degrees celsius for at least 6 weeks. Transfusion 2020: trf.16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stadlbauer D, Amanat F, Chromikova V et al SARS‐CoV‐2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 2020; 57: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. To KK, Tsang OT, Leung WS et al Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis 2020; 20: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the chinese center for disease control and prevention. JAMA 2020; 323: 1239–1242. [DOI] [PubMed] [Google Scholar]
- 26. Chen N, Zhou M, Dong X et al Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395: 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guan WJ, Ni ZY, Hu Y et al Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL. Considering how biological sex impacts immune responses and COVID‐19 outcomes. Nat Rev Immunol 2020; 20: 442–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verity R, Okell LC, Dorigatti I et al Estimates of the severity of coronavirus disease 2019: a model‐based analysis. Lancet Infect Dis 2020; 20: 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iaccarino G, Grassi G, Borghi C et al Age and multimorbidity predict death among COVID‐19 patients: results of the SARS‐RAS Study of the Italian Society of Hypertension. Hypertension 2020; 76: 366–372. [DOI] [PubMed] [Google Scholar]
- 31. Miller IF, Becker AD, Grenfell BT, Metcalf CJE. Disease and healthcare burden of COVID‐19 in the United States. Nat Med 2020; 26: 1212–1217. [DOI] [PubMed] [Google Scholar]
- 32. Wolfel R, Corman VM, Guggemos W et al Virological assessment of hospitalized patients with COVID‐2019. Nature 2020; 581: 465–469. [DOI] [PubMed] [Google Scholar]
- 33. Fafi‐Kremer S, Bruel T, Madec Y et al Serologic responses to SARS‐CoV‐2 infection among hospital staff with mild disease in eastern France. EBioMedicine 2020; 59: e102915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu F, Liu M, Wang A et al Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID‐19 in Shanghai, China. JAMA Intern Med 2020; 4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gudbjartsson DF, Norddahl GL, Melsted P et al Humoral immune response to SARS‐CoV‐2 in Iceland. N Engl J Med 2020: NEJMoa2026116 10.1056/nejmoa2026116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X, Guo X, Xin Q et al Neutralizing antibodies responses to SARS‐CoV‐2 in COVID‐19 inpatients and convalescent patients. Clin Infect Dis 2020: ciaa721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeng F, Dai C, Cai P et al A comparison study of SARS‐CoV‐2 IgG antibody between male and female COVID‐19 patients: a possible reason underlying different outcome between sex. J Med Virol 2020:92:2050–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Q, Zhang Y, Wu L et al Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell 2020; 181: 894–904.e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meyer B, Drosten C, Muller MA. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res 2014; 194: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kreer C, Zehner M, Weber T et al Longitudinal isolation of potent near‐germline SARS‐CoV‐2‐neutralizing antibodies from COVID‐19 patients. Cell 2020; 182: 843–854.e812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith K, Garman L, Wrammert J et al Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 2009; 4: 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, Crowe JE Jr. Isolation of dengue virus‐specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol 2014; 88: 12233–12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


