Abstract
Background and objective:
Topoisomerase TOP-IIA (TTOP-IIA) is widely used as a significant target for cancer therapeutics because of its involvement in cell proliferation. Steroidal drugs have been suggested for breast cancer treatment as aromatase enzymes inhibitors . TTOP-IIA inhibitors can be used as a target for the development of new cancer therapeutics.
Materials and Methods:
In this study, we conducted a docking study on steroidal drugs Anastrozole (ANA), Letrozole (LET), and exemestane (EXE) with TTOP-IIA to explore the therapeutic area of these drugs.
Results:
The binding interaction of EXE drug had significant docking interaction which is followed by ANA and LET. Thus, all these drugs could be used to inhibit the TTOP-IIA mediated cell proliferation and could be a hope to treat the other types of cancers. Among all three tested steroidal drugs, EXE showed binding energy -7.05 kcal/mol, hydrogen bond length1.78289 Å and amino acid involved in an interaction was A: LYS723:HZ3 -: UNK1:O6.
Conclusion:
The obtained data showed the most significant binding interaction analyzed with the tested enzyme. Thus, in vitro laboratory experimentation and in vivo research are necessary to put forward therapeutic repositioning of these drugs to establish them as a broad spectrum potential anticancer drugs.
Key Words: Topoisomerase, steroidal drugs, anticancer, docking, binding energy
Introduction
The topology of DNA is maintained by nuclear enzyme topoisomerases. They catalyze the provisional cutting and ligation processes of DNA, and classified as topoisomerases TOP-I (TTOP-I), TTOP-IIA, and topoisomerase TOP-IIB (TTOP-IIB) (Gupta et al., 1995). The TTOP-I cuts single-stranded DNA without the expenditure of ATP (Lin et al., 2014), while topoisomerase-II cuts DNA from double-strand with the use of ATP. TTOP-IIA catalyzes a cut on the 3′ end of DNA, while TTOP-IIB catalyzes by covalently linked to the 5’ end of the DNA (Pommier et al., 1994). The topoisomerase IA is associated with dividing the nature of the cells, while Topo B is found in neuron and not associated with dividing the nature of the cells. According to previously published research, TTOP-I is associated with cancer, and TTOP-II plays a significant role in cancer development. Thus, these enzymes contribute much more for cancer detection and treatment (Pommier et al., 1998; Andoh and Ishida 1998; Nitiss 2009).
TTOP-IIA is widely used as a significant cancer therapeutic because of its involvement with cell proliferation. In many cancer cells, the divergent expression of TTOP-IIA has been reported. Due to this characteristic, it can be utilized as a biomarker for the diagnosis of different types of cancer (Pommier et al., 1994; Yousaf et al., 2015). This study presented the topoisomerase integration and analysis to explore the understanding of TTOP-IIA in various carcinomas. Therefore, many scientific reports show contradictions within a similar area of study, the outline of this study that may help to accelerate the research for additional exploration of cancer therapeutics . A previous topoisomerase inhibitor analysis depicted the Aromatase inhibitor (AI) as a sweet overtreatment for various types of carcinomas particularly in liver carcinoma (Bromberg and Osheroff, 2003; Wang, 1996; Thakur, 2011). Wang (1996) a biochemist from America, for the first time revealed that topology of Escherichia coli DNA was regulated by a special nuclear enzyme. The telomerase activity was also reported in other prokaryotes and eukaryotes including, humans (Lin et al., 2014). Many compounds that work as poisons for topoisomerase work as anticancer drugs that cleave complex of DNA and enzyme. Topoisomerase can be linked to breast cancer and its overexpression has been seen in many cancers. Thus, analysis of implication, overexpression, and co-amplification of many genes can contribute to understanding of the stages of cancer. Under urinary bladder cancer, ovarian carcinoma, prostate cancer, soft tissue sarcoma, brain-related cancers, non-small cell lung cancer and hepatic cancers (Bromberg and Osheroff, 2003).
AIs block estrogen synthesis and thus have been tested well as an alternative treatment for estrogen-dependent breast cancer. They act through inhibiting the function of the aromatase enzyme complex, which catalyzes this conversion (Brodie et al., 1977) and incapable of binding on its actual site of androgen substrate. Aromatase forms estrogen in ovaries and in several tissues of the body (Reva et al., 2011). AIs are divided into type I, steroidal and type II, non-steroidal. The steroidal ones are formestane and EXE which are structurally similar to natural substrates, testosterone ,and androstenedione (Miller et al., 2008).
They are more beneficial and safe in clinical studies than tamoxifen. Aromatase is a cytochrome P450 enzyme complex that expresses CYP19 gene at chromosome number 15q21.2. Its expression is found in ovaries, extragonadal tissues, fat, brain, liver, bones, endothelium, and mesenchymal cells of adipose tissue in the breast. Aromatase converts androgens (androstenedione and testosterone) to estrogen and estradiol (Zhao et al., 2016).
First, aromatase inhibitor, formestane, was used to compare it with tamoxifen effect in clinical trials and similar effect was reported. Other AIs, including ANA (Bonneterre et al.,2000), LET (Mouridsen et al., 2003), and EXE (Mokbel et al., 2002; Paridaens et al., 2003; Rocío et al., 2013) had higher efficiency when used in combination compared to use tamoxifen alone. AIs ANA and LET are given as a first therapy to post-menopausal hormone-sensitive women having advanced breast cancer (Mouridsen et al., 2003; Cuzick et al., 2010). However, AIs are not given to premenopausal women since they are not able to inhibit the enzyme as it is formed more in ovaries of these women (Mauri et al., 2006). These drugs are effective in these women when given in combination with other drugs inhibiting ovarian function (National Cancer Institute, 2012). Few side effects are reported for AIs, such as blood clots, stroke, bone loss, and heart problems (Breast Cancer Organization, 2014). Aromatase inhibitor is used for the prevention and treatment of breast cancer in patients whose ovaries stop functioning. Inhibitors have been used to activate ovulation in anovulatory infertility. They are used in combination with tamoxifen (Usluogullari et al., 2015). AIs reduce the recurrence and side effects, hence increasing survival of breast cancer patients. AIs inhibit estrogen synthesis by binding to the estrogen receptors and modulate cell proliferation. Topoisomerases are also involved in cell proliferation, AIs can be considered as an alternative treatment to inhibit topoisomerases activity. Computational methods like molecular docking analysis help us to identify the interaction and binding pattern of ligand molecules with receptor biomolecules and using 3 Dimensional (3D) visualization techniques it could be well depicted in the form of graphics for better understanding. Through implementing docking techniques, many studies have been already done to find out better TTOP-IIA inhibitors. Drwal et al., (2014) tested TTOP-IIA inhibitors through computational virtual screening methods and identified 8 potential compounds. Another docking study suggested camptothecin as TTOP-IIA inhibitor (Laco et al., 2002). AIs are well trusted against breast cancer. To explore the therapeutic application of AIs and their interaction, in silico studies are conducted using topoisomerase. Therefore, in this study, TTOP-IIA inhibitions were tested with steroidal drugs.
Materials and Methods
Drug molecule preparation
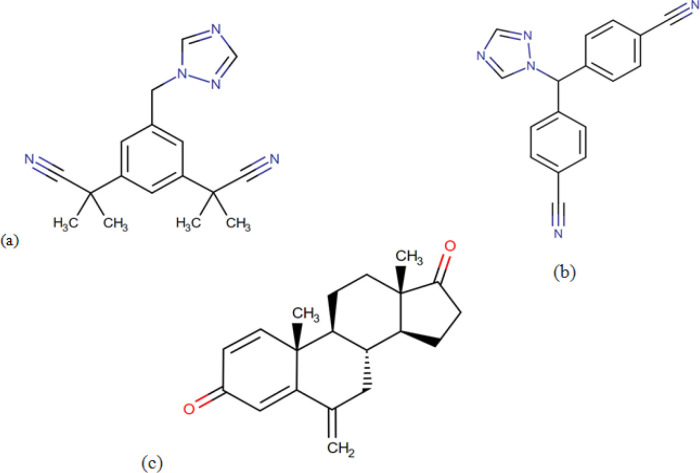
Two-dimensional (2D) structures of drugs molecule ANA (DB01217) (Figure 1(a) ), EXE (DB00990) (Figure 1(b)) and LET (DB01006) (Figure 1 (c)) (Table 1) were downloaded from DrugBank Database (www.drugbank.ca) (Wishart et al., 2018). The 2D drugs compound structure cannot be used directly to run docking studies and it should be further converted to .pdb files and minimized using CHARMm forcefield (Brooks et al., 2009) energy minimization procedure available in Discovery Studio visualizer 2019v (Dassault Systèmes BIOVIA, 2019).
Figure 1.
2D Structures of Drug Compounds (a), ANA; (b), EXE; (c) LET extracted from the DrugBank database
Table 1.
Showing Detailed Information of Selected Drugs Molecule
| S.No. | Drugs Compound | IUPAC Name | Chemical Formula | Molecular Weight | SMILES ID |
|---|---|---|---|---|---|
| 1 | Anastrozole | 2-[3-(1-cyano-1-methylethyl)-5-[(1H-1,2,4-triazol-1-yl)methyl]phenyl]-2-methylpropanenitrile | C17H19N5 | 293.4 g/mol | CC(C)(C#N)C1=CC(=CC(=C1)CN2C=NC=N2)C(C)(C)C#N |
| 2 | Letrozole | 4-[(4-cyanophenyl)(1H-1,2,4-triazol-1-yl)methyl]benzonitrile | C17H11N5 | 285.3 g/mol | C1=CC(=CC=C1C#N)C(C2=CC=C(C=C2)C#N)N3C=NC=N3 |
| 3 | Exemestane | (1S,2R,10R,11S,15S)-2,15-dimethyl-8-methylidenetetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadeca-3,6-diene-5,14-dione | C20H24O2 | 296.4 g/mol | CC12CCC3C(C1CCC2=O)CC(=C)C4=CC(=O)C=CC34C |
Receptor molecule preparation
The well-known protein structure database Protein Data Bank (PDB) governed by Research Collaboratory of Structural Bioinformatics (RCSB) (www.rcsb.org) (Berman et al., 2000) was used to download the 3D structure of human topoisomerase II alpha (HTIIa) (PDB ID: 5GWK) (Figure 2). The structure was selected on the basis of available experimental data like the X-ray diffraction method used to develop the 3D structure with the obtained resolution of 3.152 Å, R-value Free 0.244, and R-value work 0.203 (Wang et al., 2017). The water molecules and HETATM were removed from the 5GWK.pdb file, and CHARMm force field was applied for energy minimization (Brooks et al., 2009) using Discovery Studio visualizer 2019v (Dassault Systèmes BIOVIA, 2019).
Figure 2.
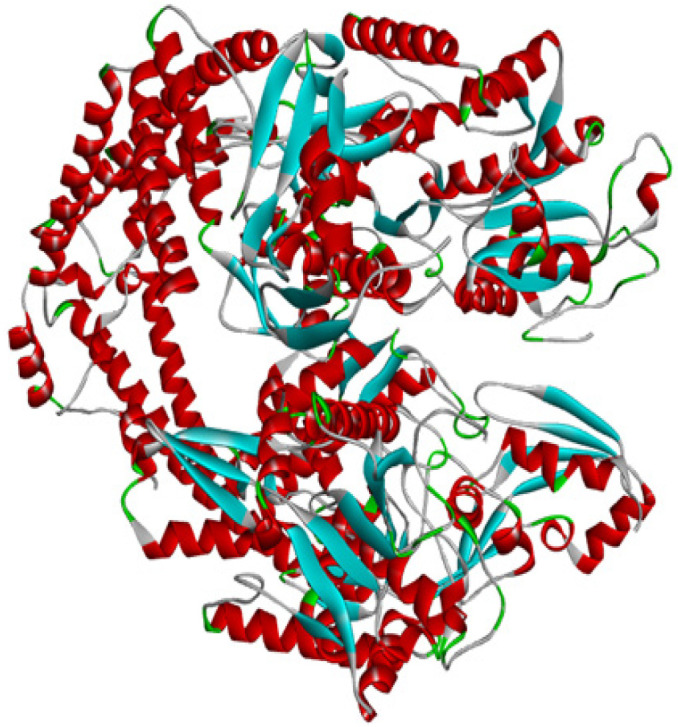
3D Crystallography Structure of HTIIa (PDB ID: 5GWK)
In silico interaction analysis
MGL tool Autodock was used to predict the binding affinity between drug compounds and topoisomerase enzymes. Interaction analysis was carried out using the Lamarckian genetic algorithm (LGA). Molecular docking methods followed by probing the best conformation of enzyme and drugs compound-complex on the calculation and obtained binding energy (∆G).
∆G binding = ∆Ggauss + ∆Grepulsion + ∆Ghbond + ∆Ghydrophobic + ∆Gtors
In the above mentioned formula, ∆G gauss is dispersion of two gaussian functions, ∆Grepulsion is square of the distance if closer than a threshold value, ∆Ghbond is ramp function, it is also used for interactions with metal ions, ∆Ghydrophobic is ramp function, and ∆Gtors is proportional to the number of rotatable bonds (Morris et al., 1998; Morris et al., 2009).
Further, water molecules were removed from the 3D structure of enzyme (i.e. PDB ID: 5GWK). Before docking and hydrogen atoms, Kollman united charges and salvation parameters were added to the 5GWK. Gasteiger charge was added to the drug compounds. The Grid box was set to cover the maximum part of 5GWK and drug compounds. The values were set to 60×60×60° in X, Y, and Z-axis of a grid point. The default grid points, spacing was 0.375 Å. Lamarckian Genetic Algorithm (LGA) (Goodshell et al., 1996 and Tsai et al., 2012) was used for 5GWK-drug compounds flexible docking calculations. The default LGA parameters like population size (ga_pop_size), energy evaluations (ga_num_generation), mutation rate, crossover rate, and step size were set to 150, 2500,000, 27,000, 0.02, 0.8 and 0.2 Å, respectively. The LGA runs were set at 10 runs. After successful execution of docking steps, obtained conformations of 5GWK and drugs-compound complexes were analyzed for the interactions and binding energy using Discovery Studio 2019v molecular visualization software (Dassault Systèmes BIOVIA, 2019).
Results
The topological properties of DNA are regulated by enzyme topoisomerase (Wang et al., 2002). It also has been involved in DNA repair, transcription, recombination, and replication (Marinello et al., 2013). One of the major ways to kill cancer cells through anticancer drugs is inhibition of TTOP-II (Azarova et al., 2007). Thus, topoisomerase is the target of many drugs, including fluroquinones, mitoxantrone, etoposide, doxorubicin, and amascrine. Its ligand-protein interaction has been analyzed by in silico docking studies (Shen et al., 1989). Previous computational analysis revealed that fluoroquinolines could target TTOP-IIA inhibitors during cancer treatment (Jadhav et al., 2017). A recent study used molecular docking techniques and identified rubidazone and other ligands, such as daunorubicin, m-amsacrine, bisantrene, and mitoxantrone, as the best TTOP-II inhibitors (Arthur, 2019). Thus, steroidal drugs could be considered as an alternative drugs to inhibit the targeted enzymes.
Therefore, in this study, we analyzed the binding interaction affinities of ANA, EXE, and LET with topoisomerase II by implementing in silico docking studies. All three tested steroidal drugs had significant binding affinities with HTIIa in this study. These drugs have already been used as breast cancer drugs. In this study, an interaction analysis was performed to explore the therapeutic area of these drugs given that HTIIa may involve in maintaining the topology of DNA and any default in this mechanism may result in a different cancer. All drugs used in this study significantly interacted with enzyme, which may loss of interacting native structure and result in inhibition of the topoisomerase activity.
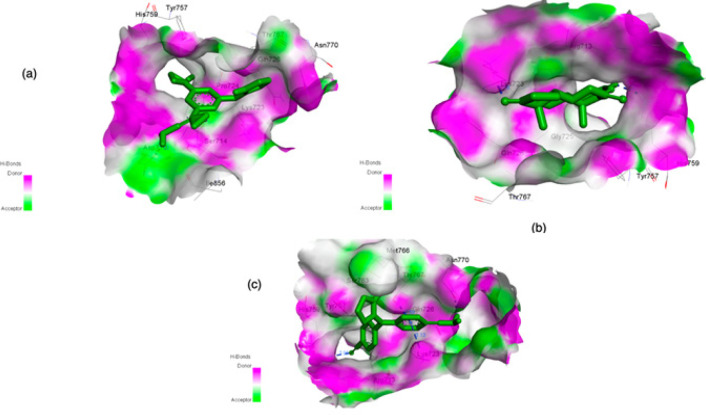
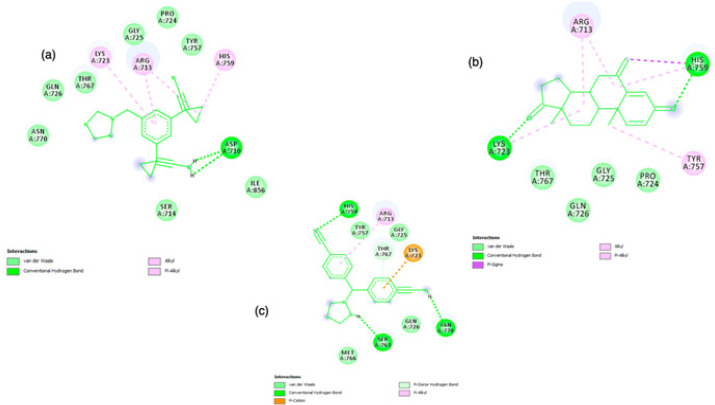
The docking interaction of ANA with HTIIa showed -6.04 kcal/mol binding energy. The negative value of interacting binding energy indicated significance of binding interaction. The analyzed inhibition constant was 277.73 uM, signifing the binding interaction between ANA and HTIIa. The involved hydrogen bond-forming amino acids were UNK0:H16 - A: ASP710:OD2 and UNK0:H17 - A: ASP710:OD2 with a hydrogen bond length 2.33347 Å and 1.99519 Å, respectively (Figure 3 (a), Table 1). Beside hydrogen bond formation, the hydrophobic interactions were observed with the involvement of amino acid Asp710, Arg713, Ser714, Lys723, Pro724, Gly725, Gln726, Tyr757, His759, Thr767, Asn770, and Ile856. Different types of interaction were also found to provide more stability to the complex structure (e.g. Lys723, Arg713 and His759 involved in alkyl and pi-alkyl interaction) (Figure 4 (a), Table 1).
Figure 3.
3D Visualization of the Drugs Molecule (a) ANA (b) EXE and (c) LET settlement in the binding pocket of HTIIa (PDB ID:3GWK). Blue dotted lines shows hydrogen bonds. Graphics generated by Discovery Studio Visualizer 2019v
Figure 4.
2D Graphics of Drugs Compound (a) ANA (b) EXE and (c) LET binding interactions with HTIIa (PDB ID: 3GWK) and residues involved in the hydrophobic interaction. Graphics generated by Discovery Studio Visualizer 2019v
In addition, analyzing docking interaction of EXE with HTIIa showed -7.05 kcal/mol binding energy. The negative value of interacting binding energy indicated the significance of binding interaction. The analyzed inhibition constant was 6.76 uM, demonstrating the binding interaction between EXE and HTIIa. The involved hydrogen bond-forming amino acids were A: LYS723:HZ3 -: UNK1:O6 and A: HIS759:HE2 -: UNK1:O21 with a hydrogen bond length 1.78289 Å and 2.26904 Å, respectively (Figure 3 (b), Table 1). Beside the hydrogen bond formation, the hydrophobic interactions were also observed with the involvement of amino acid Arg713, Lys723, Pro724, Gly725, Gln726, Tyr757, His759, and Thr767. It was also observed that different types of interaction provided more stability to the complex structure (e.g. Arg713 and Tyr757 involved in alkyl and pi-alkyl interaction and one pi-Sigma interaction found with His759) (Figure 4 (b), Table 1).
Further, the docking interaction of LET with HTIIa showed -6.22 kcal/mol binding energy. The negative value of interacting binding energy of this studied drug also indicated the significance of the binding interaction between LET and HTIIa. The analyzed inhibition constant was 140.71 uM, revealing the binding interaction between ANA and topoisomerase, but this value was less as compared to that of ANA. The involved hydrogen bond-forming amino acids were A: HIS759: HE2 - : UNK0:N3 with a hydrogen bond length 2.96051 Å,: UNK0:H1 - A:SER763:O. Hydrogen bond length of this group was 2.04926 Å,: UNK0:H3 - A: ASN770: OD1 with a hydrogen bond length 2.2549 Å. The other amino acid involved in hydrogen bonding were A: LYS723: HZ3 -: UNK0 and A: THR767: HG1- :UNK0 and with the bond length of 3.12925 Å and 2.59146 Å, respectively (Figure 3 (c), Table 1). In addition to hydrogen bond formation, the hydrophobic interactions were observed with the involvement of amino acid Arg713, Lys723, Gly725, Gln726, Tyr757, His759, Ser763, Met766, Thr767, and Asn770. Same to other interaction analyses in the complex also different interacting bonds found e.g. Lys723 formed pication bond, Arg713 formed pi-alkyl bond to provide more stability to ligand-receptor complex (Figure 4 (b), Table 1). Moreover, it was found that the binding energy of EXE with HTIIa was minimum among the studied drugs. This ANA could be bound more efficiently to inhibit the enzyme more efficiently.
Table 2.
Data Obtained through Molecular Interaction Analysis between Drugs Compound and PDB ID:5GWK Receptor Using Auto Dock Tool. The table reflects that EXE has the lowest binding energy -7.05 kcal/mol with 6.76 uM inhibition constant
| S.No. | Drugs Compound | Receptor Molecule PDB ID:5GWK |
Hydrogen bonds | Length of Hydrogen bond ( Å ) |
Final Intermolecular Energy (vdW + Hbond + desolv Energy) | Inhibition Constant | Residues involved in hydrophobic interaction |
|---|---|---|---|---|---|---|---|
| 1 | ANA | Human topoisomerase II alpha |
:UNK0:H16 - A:ASP710:OD2 | 2.33347 | -6.04 kcal/mol | 277.73 uM | Asp710,Arg713,Ser714,Lys723,Pro724,Gly725,Gln726,Tyr757,His759,Thr767,Asn770,Ile856 |
| :UNK0:H17 - A:ASP710:OD2 | 1.99519 | ||||||
| 2 | EXE | Human topoisomerase II alpha |
A:LYS723:HZ3 - :UNK1:O6 | 1.78289 | -7.05 kcal/mol | 6.76 uM | Arg713,Lys723,Pro724,Gly725,Gln726,Tyr757,His759,Thr767 |
| A:HIS759:HE2 - :UNK1:O21 | 2.26904 | ||||||
| 3 | LET | Human topoisomerase II alpha |
A:HIS759:HE2 - :UNK0:N3 | 2.96051 | -6.22 kcal/mol | 140.71 uM | Arg713,Lys723,Gly725,Gln726,Tyr757,His759,Ser763,Met766,Thr767,Asn770 |
| :UNK0:H1 - A:SER763:O | 2.04926 | ||||||
| :UNK0:H3 - A:ASN770:OD1 | 2.2549 | ||||||
| A:LYS723:HZ3 - :UNK0 | 3.12925 | ||||||
| A:THR767:HG1 - :UNK0 | 2.59146 |
Discussion
This analysis explored the promising potential anticancer activity of AI, and it was observed it could be used as an effective drug against many cancers. Thus, in vitro laboratory experimentation and in vivo research are necessary to put forward therapeutic repositioning of AI to establish it as a potential anticancer drug against cancers other than breast cancer.
In conclusion, topoisomerases catalyze an important cellular process to maintain the topology of DNA. Their role has also been described in cell growth and development which can be abnormal and resultes in the development of cancers. Many inhibitors have been developed to control cancers originated from the malfunctioning of topoisomerase, including breast, which could be due to the estrogen synthesis . Thus, aromatase inhibitors block estrogen synthesis and can be considred as alternative treatment for estrogen-dependent breast cancer. The in silico interaction studies on steroidal drugs signified their therapeutic potentialities against topoisomerases. Among all three tested steroidal drugs, EXE has shown binding energy -7.05 kcal/mol, hydrogen bond length 1.78289 Å and amino acid involved in an interaction was A: LYS723:HZ3 -: UNK1:O6. It was the most significant binding interaction analyzed by a tested enzyme. Thus, in conclusion, in vitro laboratory experimentation and in vivo research are needed to put forward therapeutic repositioning of these drugs to establish them as a broad spectrum anticancer drugs.
Acknowledgments
The author is thankful to Qassim University, Saudi Arabia, for providing the necessary infrastructure facility to carry out the study.
Ethical statement
None.
Conflict of interest
The author declares no conflict of interest.
References
- Andoh T, Ishida R. Catalytic inhibitors of DNA topoisomerase II. Biochim Biophys Acta. 1998;1400:155–71. doi: 10.1016/s0167-4781(98)00133-x. [DOI] [PubMed] [Google Scholar]
- Arthur DE. Molecular docking studies of some topoisomerase II inhibitors: Implications in designing of novel anticancer drugs. Radiol of Infect Dis. 2019;6:68–79. [Google Scholar]
- Azarova AM, Lyu YL, Lin CP, et al. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc Natl Acad Sci U S A. 2007;104:11014–9. doi: 10.1073/pnas.0704002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–2. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneterre J, Thürlimann B, Robertson JR, et al. Anastrozole versus tamoxifen as first-line therapy for advanced breast cancer in 668 postmenopausal women: Results of the Tamoxifen or Arimidex Randomized Group Efficacy and Tolerability Study. J Clin Oncol. 2000;18:3748–7. doi: 10.1200/JCO.2000.18.22.3748. [DOI] [PubMed] [Google Scholar]
- Brodie AM, Schwarzel WC, Shaikh AA, et al. The effect of an Aromatase inhibitor, 4-Hydroxy-4-Androstene- 3,17-Dione, on Estrogen-Dependent Processes in reproduction and breast cancer. Endocrinology. 1977;100:1684–5. doi: 10.1210/endo-100-6-1684. [DOI] [PubMed] [Google Scholar]
- Bromberg KD, Osheroff N. Mechanism of action of topoisomerase II-targeted anticancer drugs. DNA Topoisomerases in Cancer Therapy. Boston, MA: Springer; 2003. pp. 53–78. [Google Scholar]
- Brooks BR, Brooks CL, Mackerell AD, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Sestak I, Baum M, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol. 2010;11:1135–41. doi: 10.1016/S1470-2045(10)70257-6. [DOI] [PubMed] [Google Scholar]
- Drwal MN, Marinello J, Manzo SG, et al. Novel DNA Topoisomerase IIα inhibitors from combined ligand- and structure-based virtual screening. PLoS One. 2014;9:e114904. doi: 10.1371/journal.pone.0114904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Becerra R, Santos N, Díaz L, Camacho J. Mechanisms of resistance to endocrine therapy in breast cancer: Focus on Signaling Pathways, miRNAs and Genetically Based Resistance. Int J Mol Sci. 2012;14:108–45. doi: 10.3390/ijms14010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: Applications of autodock. J Molecular Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Gupta M, Fujimori A, Pommier Y. Eukaryotic DNA topoisomerases I Biochim Biophys. Acta Gene Structure Expression. 1995;1262:1–14. doi: 10.1016/0167-4781(95)00029-g. [DOI] [PubMed] [Google Scholar]
- Higgins NP. A human TOP2A core DNA binding X-ray structure reveals Topoisomerase subunit dynamics and a potential mechanism for SUMO modulation of decatenation. J Mol Bio. 2012;424:105–18. doi: 10.1016/j.jmb.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Jadhav AK, Karuppayil SM. Molecular docking studies on thirteen fluoroquinolines with human topoisomerase II a and b. In Silico Pharmacol. 2017;5:1–12. doi: 10.1007/s40203-017-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laco GS, Collins JR, Luke BT, et al. Human Topoisomerase I inhibition: Docking camptothecin and derivatives into a structure-based active site model. Biochemistry. 2002;41:1428–35. doi: 10.1021/bi011774a. [DOI] [PubMed] [Google Scholar]
- Lin Y, Huang W, Chen M, Hsieh T. Toward discovering new anti-cancer agents targeting topoisomerase IIα: A Facile Screening Strategy Adaptable to High Throughput Platform. PLoS One. 2014;9:e97008. doi: 10.1371/journal.pone.0097008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinello J, Chillemi G, Bueno S, Manzo SG, Capranico G. Abstract 637: Antisense transcripts and R-loops caused by DNA topoisomerase I inhibition by camptothecin at human active CpG island promoters. Cancer Res. 2013;73 doi: 10.1093/nar/gkt778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauri D, Pavlidis N, Polyzos NP, Ioannidis JP. Survival with Aromatase inhibitors and inactivators versus standard hormonal therapy in advanced breast cancer: Meta-analysis. J Natl Cancer Inst. 2006;98:1285–91. doi: 10.1093/jnci/djj357. [DOI] [PubMed] [Google Scholar]
- Miller WR, Bartlett J, Brodie AM, et al. Aromatase inhibitors: Are there differences between steroidal and nonsteroidal aromatase inhibitors and do they matter? Oncologist. 2008;13:829–37. doi: 10.1634/theoncologist.2008-0055. [DOI] [PubMed] [Google Scholar]
- Mokbel K. The evolving role of AI in breast cancer. Int J Clin Oncol. 2002;7:279–83. doi: 10.1007/s101470200040. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–62. [Google Scholar]
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouridsen H, Gershanovich M, Sun Y, et al. Phase III study of Letrozole versus Tamoxifen as first-line therapy of advanced breast cancer in postmenopausal women: Analysis of Survival and Update of Efficacy From the International Letrozole Breast Cancer Group. J Clin Oncol. 2003;21:2101–9. doi: 10.1200/JCO.2003.04.194. [DOI] [PubMed] [Google Scholar]
- Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–37. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaens R, Dirix L, Lohrisch C, et al. Mature results of a randomized phase II multicenter study of exemestane versus tamoxifen as first-line hormone therapy for postmenopausal women with metastatic breast cancer. Ann Oncol. 2003;14:1391–8. doi: 10.1093/annonc/mdg362. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Tanizawa A, Kohn KW. Mechanisms of topoisomerase I inhibition by anticancer drugs. Adv Pharmacol. 1994;29:73–92. doi: 10.1016/s1054-3589(08)61132-1. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Leo E, Zhang H, Marchand C. DNA Topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–33. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Barakat A, Pippen JE, Osborne C. Aromatase inhibitors in the treatment of breast cancer in post-menopausal female patients: an update. Breast Cancer (Dove Med Press) 2011;3:113–25. doi: 10.2147/BCTT.S22905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LL, Baranowski J, Pernet AG. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: specificity and cooperativity of drug binding to DNA. Biochemistry. 1989;28:3879–85. doi: 10.1021/bi00435a038. [DOI] [PubMed] [Google Scholar]
- Thakur DS. Topoisomerase II, inhibitors in cancer treatment. Int J Pharm Sci Nanotechnol. 2011;3:1173–81. [Google Scholar]
- Tsai CW, Chen JL, Yang CS. An improved LGA for protein ligand docking prediction. Proceedings of IEEE congress on evolutionary computation. 2012:1–6. [Google Scholar]
- Usluogullari B, Duvan C, Usluogullari C. Use of aromatase inhibitors in practice of gynecology. J Ovarian Res. 2015;8:1–7. doi: 10.1186/s13048-015-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Miller W, Milton J, et al. Structure–activity relationships for analogues of the phenazine-based dual topoisomerase I/II inhibitor XR11576. Bioorg Med Chem Lett. 2002;12:415–18. doi: 10.1016/s0960-894x(01)00770-3. [DOI] [PubMed] [Google Scholar]
- Wang YR, Chen SF, Wu CC, et al. Producing irreversible topoisomerase II-mediated DNA breaks by site-specific Pt(II)-methionine coordination chemistry. Nucleic Acids Res. 2017;45:10861–871. doi: 10.1093/nar/gkx742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC. DNA Topoisomerases. Annu Rev Biochem. 1996;65:635–92. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Guo AC, et al. DrugBank 50: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2017;46:1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousaf A, Hamid SA, Bunnori NM, Ishola AA. Applications of calixarenes in cancer chemotherapy: facts and perspectives. Drug Des Devel Ther. 2015;9:2831–8. doi: 10.2147/DDDT.S83213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhou L, Shangguan AJ, Bulun SE. Aromatase expression and regulation in breast and endometrial cancer. J Mol Endocrinol. 2016;57:19–33. doi: 10.1530/JME-15-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]