Abstract
Anthocyanidins are bioactive compounds found mostly in colored plants and fruits. Consumption of anthocyanidin-rich foods has been shown to reduce the risk of diabetes. However, limited information is available regarding the inhibitory effect and interactions of anthocyanidins on α-glucosidase, the key enzyme that controls diabetes through degrading carbohydrate. Therefore, we used computational docking analysis to investigate the degree and type of inhibition by α-glucosidase, and the structural interactions of enzyme-selected anthocyanidins. The results suggested that anthocyanidins exhibit half maximal inhibitory concentration of 4∼55 μM; the strongest and weakest α-glucosidase inhibitors were delphinidin and malvidin, respectively. Indeed, delphinidin inhibits α-glucosidase in a mixed type, close to non-competitive manner with an inhibitory constant of 78 nM. Addition of a glycoside (glucoside or galactoside) at C3 on the C ring of delphinidin significantly decreased inhibitory activity, and addition of glycosides at C3 on the C ring and C5 on the A ring of delphinidin prevented all inhibitory activity. Molecular docking and free binding energy accurately confirmed the mode of inhibition determined by enzyme kinetics. These data will inform the use of alternative sources of anthocyanidins in functional foods and dietary supplements for prevention of diabetes. The results provide useful information for evaluating possible molecular models using anthocyanins/anthocyanidins as templates and α-glucosidase as the key enzyme in management of diabetes.
Keywords: anthocyanidins, anthocyanins, anti-α-glucosidase activity, enzyme kinetics, molecular docking
INTRODUCTION
Type II diabetes mellitus (DM) is a metabolic disorder correlated with high blood glucose levels (hyperglycemia) due to insulin hormone resistance of β cells in the pancreas (Shrivastava et al., 2013). Numerous treatment options for type II DM are available, including non-pharmacological therapy (e.g. appropriate diet, exercise, weight loss, and lifestyle modification) and pharmacological interventions. The latter focuses on inhibition of enzymes that control carbohydrate degradation, α-amylase, and α-glucosidase (Hui et al., 2009; Nyenwe et al., 2011). Inhibition of these enzymes retards carbohydrate digestion and glucose absorption, which, in turn, reduces the level of postprandial hyperglycemia following a meal, and decreases triglyceride synthesis in adipose tissue, liver, and intestinal walls (van de Laar, 2008; de la Garza et al., 2013). Several medicinal treatments have been proposed from a pharmacological perspective with an effective dose; however, synthetic drugs are expensive, and often have severe side effects with responses dependent on the individual. Therefore, bioactive compounds with potential antidiabetic properties from natural products have recently attracted increasing interest.
Bioactive compounds derived from plants are generally phenolics, and are defined as phenolic acids, coumarins, flavonoids, and tannins. Among the phenolics, flavonoids are effective anti-α-glucosidase agents with potentially higher half maximal inhibitory concentration (IC50) than phenolic acids, coumarins, and tannins (Tadera et al., 2006; Mohamed Sham Shihabudeen et al., 2011; Yin et al., 2014; Zhu et al., 2014). Many previous studies have suggested that consumption of foods containing high levels of flavonoids prevent diabetes through inhibiting α-glucosidase (Bahadoran et al., 2013; Testa et al., 2016). Therefore, flavonoid-rich foods have been predicted to represent a potential new method for management of diabetes with reduced adverse effects and increased economical effectiveness compared with commercial synthetic drugs. Interestingly, the inhibitory effectiveness of flavonoids against α-glucosidase was higher than for α-amylase, suggesting that the former enzyme may be a more practical target for controlling diabetes (Tadera et al., 2006; Mohamed Sham Shihabudeen et al., 2011; Yin et al., 2014; Zhu et al., 2014).
Anthocyanidins are a subclass of flavonoids. The most commonly consumed anthocyanidins are cyanidin, delphinidin, malvidin, pelargonidin, peonidin, and petunidin, found in foods such as berries and red fruits (Miguel, 2011). Compared with other flavonoids, cyanidin is an effective anti-α-glucosidase agent (Tadera et al., 2006). Glycosylation of anthocyanidins forms anthocyanins. Anthocyanins with 3-glucosides, such as cyanidin-3-glucosides, are the main anthocyanins found in berries (Chaves et al., 2018). However, the ability of anthocyanins to inhibit α-glucosidase is decreased by glycosylation, and by substitution of different types of sugar (Wang et al., 2010; Xu et al., 2018). Previous studies indicated that a high intake of anthocyanidins and anthocyanin-rich fruits (e.g. berries, grapes, blackcurrants, apples, and cowpeas) was associated with lower risk of type II DM (Jenkins et al., 2011; Wedick et al., 2012). Results from animal models also suggested that plant extracts rich in anthocyanidins reduce high blood glucose levels and decrease hemoglobin A1c by enhancing insulin secretion and improving insulin resistance (Zang et al., 2016). Specifically, cyanidin-3-glucoside was reported to reduce diabetes-associated hyperlipidemia in mouse fibroblast 3T3-L1 cells by suppressing hexosamine biosynthesis, FoxO1 and adipose triglyceride pathways (Guo et al., 2012). In addition, delphinidin was shown to decrease glucose absorption via the free fatty acid receptor-1 dependent pathway in human intestinal cell lines (Hidalgo et al., 2017). Thus, anthocyanins and anthocyanidins have potential to be used as nutritional bioactive compounds with therapeutic antidiabetic properties.
Limited information is available on the inhibitory activity of anthocyanidins on α-glucosidase, and on the inhibitory modes of anthocyanidins and their glycoside forms (anthocyanins). Thus, we investigated the inhibitory effects of anthocyanidins and their glycosides on α-glucosidase, including IC50, inhibitory constant (Ki), mode of inhibition, and structural interactions (molecular docking). Since high anthocyanidin contents are present in berries, strawberries, cherries, grapes, raspberries, red wine, tea, and fruit peel with dark pigments, the results of this research will be useful for development of functional foods with high anthocyanidin contents. Moreover, through using enzyme-inhibitor structural analysis, we described the molecular interactions between anthocyanidins as the templates and α-glucosidase as the key enzyme in type II DM prevention and treatment.
MATERIALS AND METHODS
Chemicals and reagents
The enzyme [Saccharomyces cerevisiae (S. cerevisiae) α-glucosidase (type 1, ≥10 unit/mg)] and substrate [p-nitrophenyla-D-glucopyranoside (pNPG)] were sourced from Sigma-Aldrich Co. (St. Louis, MO, USA). Standards, including cyanidin chloride (≥95%), malvidin chloride (≥ 95%), pelargonidin chloride (≥98%), peonidin chloride (≥95%), delphinidin chloride (≥95%), and petunidin chloride (≥98%), were purchased from Sigma-Aldrich Co.. Delphin chloride (≥97%), delphinidin-3-galactoside chloride (≥95%), and delphinidin-3-glucoside chloride (≥95%) were purchased from Extrasynthese (Genay, Lyon, France).
Enzyme assays for α-glucosidase inhibitory activity
Inhibition of α-glucosidase was assessed in assays containing 5 μL of anthocyanins/anthocyanidins (various concentrations), 10 μL of S. cerevisiae α-glucosidase (0.01 U/mL), 25 μL of pNPG (1.25 mM) in 160 μL of 50 mM KPB (pH 6). The reaction was monitored at a wavelength of 405 nm at 37°C for 30 min using a SynergyTM HT 96-well UV-visible microplate reader with a Gen5 data analysis software program (BioTek Instruments, Inc., Winooski, VT, USA). Result was calculated as a percentage of inhibitory activity using the following equation:
where VoA is the initial velocity of the control reaction with enzyme (control), Voa is the initial velocity of the control reaction without enzyme (control blank), VoB is the initial velocity of the enzyme reaction with sample, and Vob is the initial velocity of the reaction with sample but without enzyme (sample blank).
The efficiency of anthocyanins and anthocyanidins for inhibiting α-glucosidase was determined by the IC50, analyzed using a dose-response plot of anthocyanins/anthocyanidins versus percentage of inhibition using a Graph-Pad Prism (version 5.00) for Windows (GraphPad Software, San Diego, CA, USA, www.graphpad.com).
Enzyme kinetics of α-glucosidase inhibitory activity
The Ki and mode of inhibition for inhibition of S. cerevisiae α-glucosidase (0.01 U/mL) by anthocyanins/anthocyanidins were determined using with 10 different concentrations of pNPG (0.1∼1.2 mM) in 50 mM KPB (pH 6). Assays were performed using the 96-well microplate reader as mentioned previously. Investigation of the kinetic behaviours of anthocyanins/anthocyanidins on α-glucosidase was performed using the Michaelis-Menten equation with a least squares fit parameter using the GraphPad Prism (version 5.00) for Windows (GraphPad Software). The Michaelis-Menten equation presents a relationship between substrate concentrations and reaction velocity and is expressed as:
where V is the initial velocity of the reaction, Vmax is the maximum velocity of reaction rate obtained with excess substrate and saturated enzyme, Km is the Michaelis-Menten constant indicating a binding affinity of enzyme and substrate at 50% of the reaction rate, and S is the concentration of substrate.
The Lineweaver-Burk plot was generated using Graph-Pad Prism (version 5.00) for Windows by plotting the inverse of substrate concentration and reaction velocity to determine Vmax and Km. The intercept on the y-axis represents 1/Vmax, the intercept on the X-axis indicates −1/Km, and the slope is Km/Vmax. The Lineweaver-Burk plot is expressed as:
Conversely, a Lineweaver-Burk double reciprocal plot was transformed from the Michaelis-Menten equation to determine the mode of inhibition. Ki was determined by the slope of the Lineweaver-Burk double reciprocal plot and the concentrations of anthocyanins and anthocyanidins.
Molecular docking
The X-ray crystallographic structure of α-glucosidase was retrieved from the Protein Data Bank (http://www.rcsb.org/pdb). Several human α-glucosidases, including the intestinal N-terminal maltase glucoamylase (PDB ID: 2QMJ), the C-terminal maltase glucoamylase (PDB ID: 3TOP), and the N-terminal sucrose isomaltase (PDB ID: 3LPP), have been submitted to and are available on the protein database. These proteins have been used for predicting binding interactions using molecular docking analysis in previous studies (Shang et al., 2013; Priscilla et al., 2014; Flores-Bocanegra et al., 2015; Zhang et al., 2016). The results showed that the N-terminal maltase gluco-amylase shared high structural similarity with the C-terminal maltase glucoamylase and the N-terminal sucrose isomaltase. Interestingly, the N-terminal maltase gluco-amylase was the most selective molecular model for studying α-glucosidase inhibition (Naumov, 2007; Quezada-Calvillo et al., 2007). Thus, the human intestinal N-terminal maltase glucoamylase was chosen as the template in this study.
All water molecules and miscellaneous molecules were removed from the receptor model using the Discovery Studio Client (version 2.5) program from Dassault Systèmes BIOVIA (San Diego, CA, USA) (Mikulic-Petkovsek et al., 2017) to avoid an unclear binding pocket and distorted structural model. Hydrogen atoms and Gasteiger-Huckel charges were then added to the model to generate a stabilized chemical structure. The 3D structures of all flavonoids were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov) and computed using Sybyl Molecular Modeling Package (Tripos, Inc., St. Louis, MO, USA) (Vujović et al., 2017). Binding energy was calculated using AutoDockTools (version 4.2, AutoDock, La Jolla, CA, USA) from The Scripps Research Institute (TSRI) (La Jolla, CA, USA) (Morris et al., 2009) to predict the interaction between the α-glucosidase model and anthocyanins/anthocyanidins. The grid box size was 84× 83×81 Å in the x, y, and z dimensions, respectively, with a spacing of 0.375 Å. During this procedure, the genetic algorithm (GA) was set at 30 runs, and the Lamarckian GA was chosen for docking calculations. Analyses were performed using the data collection tool Cygwin Terminal (version 2.8.0) from CygwinTM API library (https://cygwin.com) (Racine, 2000). Text on molecular docking was edited using a text editor program, EditPlus (version 4.3, ES-Computing, Jinju, Korea, https://www.editplus.com). Structural molecular analyses and molecular images were visualized using the UCSF Chimera package (version 1.11.2) from the Resource for Biocomputing, Visualization and Informatics, University of California, San Francisco (supported by NIH P41 RR-01081) (Pettersen et al., 2004), and the PyMOL Molecular Graphics System (version 1.8) from Schrödinger, LLC. (New York, NY, USA) (https://www.pymol.org) (Salam et al., 2014). The optimal docked model was selected based on the lowest binding free energy being the most favorable binding on α-glucosidase.
Statistical analysis
All experiments were carried out in triplicate with data expressed as mean±standard deviation (SD). All statistical analyses were evaluated using SPSS version 19 from IBM Corp. (New York, NY, USA). Unpaired sample t-test was performed to determine the significant differences between IC50 values of anthocyanins/anthocyanidins and that of acarbose. Significant differences was defined at P <0.05.
RESULTS AND DISCUSSION
Inhibition of α-glucosidase
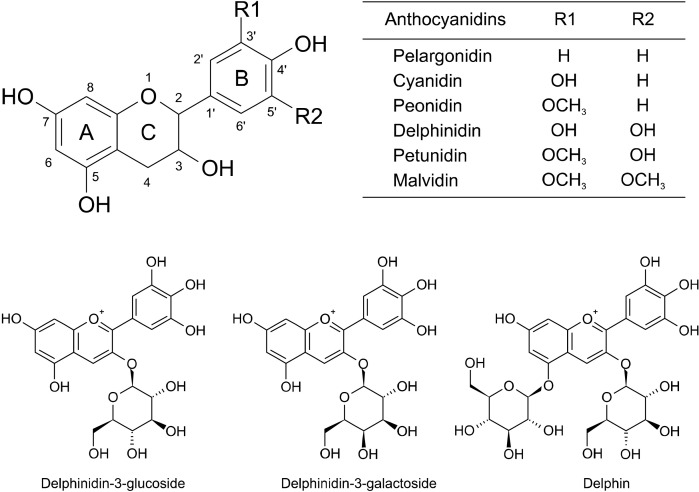
Anthocyanidins found in nature include cyanidin, delphinidin, malvidin, pelargonidin, peonidin, and petunidin (Miguel, 2011). These anthocyanidins exhibited IC50 values against α-glucosidase ranging from 4.11 to 54.69 μM, which are higher than the diabetic drug, acarbose (IC50 of 0.53±0.06 μM) (Table 1). The inhibitory actions of the anthocyanidins were in the following order of magnitude: delphinidin> cyanidin> petunidin> peonidin> pelargonidin> malvidin. The chemical structure of delphinidin (three hydroxyl moieties on the B ring) caused delphinidin (IC50 of 4.11±0.49 μM) to exhibit greater inhibitory activity than cyanidin and petunidin (IC50 of 17.01±1.35 and 30.78±1.17 μM, respectively). Cyanidin and petunidin both have two hydroxyl moieties, which suggests that the presence of more hydroxyl moieties at C-3’, C-4’, and C-5’ of the B ring promotes inhibitory activity (Table 1 and Fig. 1). The hydroxyl moieties at C-3’ and C-4’ of the B ring of cyanidin exhibited higher inhibitory activity than hydroxyl moieties at the C-4’ and C-5’ of the B ring of petunidin (Table 1 and Fig. 1), suggesting a more significant role of hydroxyl moieties at C-3’ than at C-5’ of the B ring. The significance of hydroxyl moieties was also revealed in the chemical structures of peonidin and petunidin (IC50 of 36.11±2.72 and 30.78±1.17 μM, respectively). Petunidin (hydroxyl moiety at C-5’ of the B ring) exhibited higher inhibitory activity than peonidin (no hydroxyl moiety at this position) (Table 1 and Fig. 1). Replacement of hydroxyl moieties with meth-oxyl groups decreased inhibitory activity, as observed in malvidin (IC50 of 54.69±2.14 μM) (Table 1). Malvidin is composed of two methoxyl moieties at C-3’ and C-5’ of the B ring, and exhibited lower inhibitory activity than petunidin and peonidin (methoxyl moiety at C-3’) (Table 1 and Fig. 1). Pelargonidin, which only has one hydroxyl moiety at C-4’ of the B ring, exhibited lower inhibitory activity than delphinidin, cyaniding, and petunidin, which all have two or three hydroxyl moieties in the B ring (Table 1 and Fig. 1). However, peonidin (methoxyl moiety at C-3’) exhibited higher inhibitory activity than pelargonidin (IC50 of 50.35±4.00 μM) with hydrogen at C-3’. However, compared with malvidin (two methoxyl moieties at C-3’ and C-5’), pelargonidin exhibited greater inhibitory activity, suggesting that the presence of a methoxyl moiety at C-5’ decreased the inhibitory activity to a greater extent than when the methoxyl moiety is at the C-3’ position. The presence of methoxylation and hydroxylation also increased the inhibition of α-glucosidase. Previous studies have indicated that hydroxylation of the B ring of the flavonoid structure increases the inhibitory potential (IC50) on α-glucosidase (Kim et al., 2006; Xiao and Högger, 2015). Indeed, the presence of more hydroxyl groups enhanced inhibition of α-glucosidase (Gao and Kawabata, 2004). However, the presence of methoxyl groups reduced inhibition of α-glucosidase (Xiao et al., 2013).
Table 1.
Effect of anthocyanidins, anthocyanins and acarbose on Saccharomyces cerevisiae α-glucosidase half maximal inhibitory concentrations (IC50), inhibition constants (Ki), and modes of inhibition
| Compounds | IC50 (μM) | Ki (μM) | Mode of inhibition |
|---|---|---|---|
| Anthocyanidins | |||
| Pelargonidin | 50.35±4.00 | 31.36 | Mixed type, close to non-competitive |
| Cyanidin | 17.01±1.35 | 2.43 | Mixed type, close to non-competitive |
| Peonidin | 36.11±2.72 | 1.23 | Non-competitive |
| Delphinidin | 4.11±0.49 | 0.078 | Mixed type, close to non-competitive |
| Petunidin | 30.78±1.17 | 1.52 | Competitive |
| Malvidin | 54.69±2.14 | 35.89 | Mixed type, close to non-competitive |
| Anthocyanins | |||
| Delphinidin-3-glucoside | 364.32±15.19 | 59.83 | Mixed type, close to non-competitive |
| Delphinidin-3-galactoside | 368.87±13.81 | 98.69 | Mixed type, close to non-competitive |
| Delphin | ND | − | − |
| Synthetic drug | |||
| Acarbose | 0.53±0.06 | 0.069 | Competitive |
Values are presented as mean±SD of three replications (n=3).
Significant differences between the IC50 values of anthocyanins/anthocyanidins and that of acarbose were reported using unpaired sample t-test (P<0.05).
ND, not detected.
Fig. 1.
Chemical structures of anthocyanins and anthocyanidins used in this experiment.
Since delphinidin exhibited the highest inhibitory activity with the lowest IC50 value, we further investigated its glycosides. Results suggested that all the glycosides (delphinidin-3-glucoside, delphinidin-3-galactoside, and delphin) were poorer inhibitors of α-glucosidase than delphinidin (Table 1). Substitution of sugar moieties (glucose and galactose) at C3 on the C ring of delphinidin weakened the inhibitory effect on α-glucosidase. This was indicated by increased IC50 values (delphinidin-3-glucoside, 364.32±15.19 μM; delphinidin-3-galactoside, 368.87 ±13.81 μM). Furthermore, delphinidin-3,5-diglucoside (delphin) was the weakest inhibitor of α-glucosidase since no inhibitory action was observed (Table 1). These results imply that replacement of the hydroxyl group at C3 of the C ring by either glucose or galactose moieties lowered its potential for α-glucosidase inhibition. Moreover, addition of glucose at C3 of the C ring and C5 of the A ring of the delphinidin skeleton drastically decreased the inhibition of α-glucosidase, suggesting that steric hindrance of bulky structures negatively effects binding interactions between delphinidin glycosides and α-glucosidase.
A previous study showed that glycosylation of the flavonoid skeleton decreased inhibition of α-glucosidase (Wang et al., 2010). The inhibitory activity was decreased to a greater extent with a larger number of glycoside substitutions on flavonoids (i.e. polyglycoside vs. monoglycoside residues) (Kim et al., 2000). The increase in bulkiness and polarity of the chemical structure causes a steric effect, which tends to possess a non-planar structure, thus reducing the favorable binding interactions between the flavonoids and α-glucosidase (Xiao et al., 2010). Other studies have indicated that the inhibitory activity of flavonoid glycosylation on α-glucosidase depended on the type, position, molecular size, and polarity of the sugar moiety, which is consistent with the results of our present study on anthocyanins and anthocyanidins (Kawabata et al., 2003; Shibano et al., 2008; Fan et al., 2010; Xiao et al., 2013).
Enzyme kinetics and mode of inhibition
Enzyme kinetics (Table 1) revealed that acarbose and petunidin behaved as competitive inhibitors of α-glucosidase, suggesting that the inhibitors competed with the substrate for the active site of the enzyme. Therefore, the inhibitor blocked the substrate from reaching the active site, thereby interfering with enzymatic activity. This finding was consistent with a previous report that indicated that acarbose inhibits in a competitive manner (Hansawasdi and Kawabata, 2006). However, peonidin inhibited α-glucosidase in a non-competitive manner (Table 1), indicating binding to either the free enzyme or enzyme-substrate complex. Interactions occurring outside of the active site alter the structure of the enzyme and prevent the substrate from entering the active site. The other anthocyanidins and anthocyanins (with the exception of delphin) inhibited α-glucosidase in mixed-type, close to non-competitive inhibitors manner, suggesting that these compounds bind the free enzymes and enzyme-substrate complexes (Table 1). The type of inhibition could not be determined for delphin (two molecules of glucose attached to C3 of the C ring and C5 of the A ring) due to low inhibitory activity.
The determination of Ki for the inhibition of α-glucosidase showed that anthocyanidins and anthocyanins had Ki value of the following order of magnitude: delphinidin< peonidin< petunidin< cyanidin< pelargonidin< malvidin< delphinidin-3-glucoside< delphinidin-3-galactoside (Table 1). A smaller Ki implies stronger inhibition or a higher affinity for binding α-glucosidase (tight binding). The Ki value also indicates the effectiveness of ligand binding to the enzyme. Based on these data, delphinidin had the strongest binding to α-glucosidase, whereas delphinidin-3-galactoside had the poorest (Ki of 0.078 μM and 98.69 μM, respectively). This finding was in good agreement with the IC50 values (delphinidin had the lowest IC50, whereas delphinidin-3-galactoside had the highest IC50). Interestingly, delphinidin exhibited a similar Ki value as acarbose (Ki=0.069 μM), suggesting delphinidin is an effective anti-α-glucosidase agent.
Molecular docking
To understand the interactions between anthocyanidins, especially delphinidin, and α-glucosidase, a molecular docking technique was used to investigate docking of enzyme-anthocyanidin interactions, both on the centered macromolecule and the enzyme active site. We used the X-ray crystallographic structure of human intestinal N-terminal maltase glucoamylase as the macromolecule receptor to determine an optimally conformational position of anthocyanidins on α-glucosidase. Molecular docking was predicted by the scores of the lowest binding free energy to indicate potential interactions between anthocyanidins and α-glucosidase.
The docking results indicated that enzyme-anthocyanidins interactions throughout the macromolecule were not the same as interactions at the active site. A previous report suggested that docking analysis of the competitive inhibitor, acarbose, on the N-terminal subunit of maltase glucoamylase occurred between the catalytic triad (Asp327, Asp443, and Asp542) and active site residues (Thr205 and Arg526) (Sim et al., 2008). Similar results were observed in our experiment (Table 2). However, the interaction between acarbose and the enzyme active site differed from that between acarbose and the entire enzyme, during which acarbose preferred forming hydrogen bonds with Gln272, Gln275, Asp649, His657, Glu658, Asp697, and Arg730 of the enzyme. Considering the chemical structure of acarbose on human α-glucosidase, the interaction mostly occurred through the hydroxyl moieties of acarbose (Xiao et al., 2013; Proença et al., 2017). The mode of inhibition of acarbose, the competitive inhibitor, corresponded with the free binding energy. Indeed, the free binding energy of docking to the whole enzyme (−0.91 kcal/mol) was higher than that of docking into the enzyme active site (−2.98 kcal/mol) (Table 2). Therefore, acarbose likely accessed the catalytic pocket of α-glucosidase and formed interactions with the active-site residues rather than interacting away from the active site.
Table 2.
Structural interactions between different anthocyanidins/anthocyanins and Saccharomyces cerevisiae α-glucosidase
| Compounds | Interactions centered on macromolecules | Interactions centered on active sites | |||
|---|---|---|---|---|---|
| EFBE (kcal/mol) | Interactions with enzyme | EFBE (kcal/mol) | Interactions with enzyme | ||
| Anthocyanidins | |||||
| Pelargonidin | −7.14 | Phe535, Asp777, Thr778, Val779 | −4.69 | Asp327, Asp366, Asp542 | |
| Cyanidin | −8.60 | His645, Val779 | −8.18 | Asp443, Asp542, Tyr605 | |
| Peonidin | −6.02 | Ile523, Asp777, Thr778 | −6.40 | Asp203, Asp327, Ser448 | |
| Delphinidin | −9.00 | Arg520, Ser521, Ile523, Phe535, Asp777, Thr778 | −8.38 | Asp443, Arg526, Asp542, Gln603, Tyr605 | |
| Petunidin | −8.12 | Lys513, Arg520, Lys534, Phe535 | −7.98 | Asp327, Asp443, Asp542, Tyr605 | |
| Malvidin | −6.53 | Ser288, Ile523, Lys776, Asp777 | −3.97 | Arg202, Asp203, Arg526 | |
| Anthocyanins | |||||
| Delphinidin-3-glucoside | −4.71 | Pro17, Gln19, Ser40, Val41, Arg471 | −5.91 | Asp203, Asp327, Asp443, Arg526, Asp542, Gln603 | |
| Delphinidin-3-galactoside | −4.35 | Pro17, Gln19, Ser40, Val41, Arg471 | −5.32 | Asp203, Asp327, Asp443, Asp542, Tyr605 | |
| Delphin | −3.75 | Ser40, Val41, Thr196, Arg254, Arg471 | −2.60 | Asp203, Thr205, Asp327, Asp542, Gln603 | |
| Synthetic drug | |||||
| Acarbose | −0.91 | Gln272, Gln275, Asp649, His657, Glu658, Asp697, Arg730 | −2.98 | Thr205, Asp327, Asp443, Asp542, Arg526 | |
Estimated free binding energy (EFBE) and interactions with enzyme ligands are presented for interactions centered on whole macromolecules and the enzyme active sites.
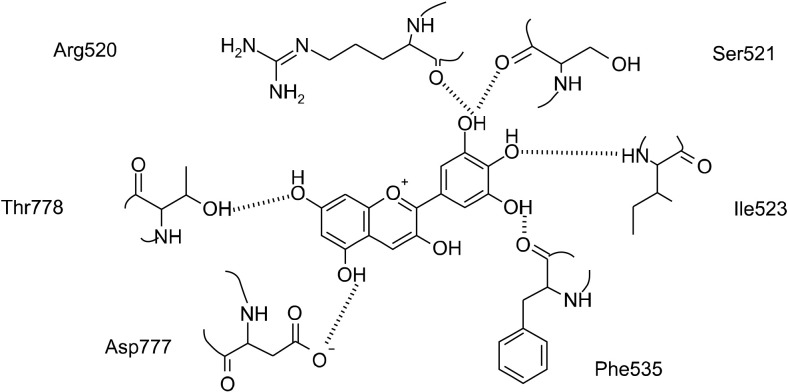
Structural prediction of molecular docking in the centered macromolecule of the enzyme to create a flexible binding mode indicated free binding energy scores ranging from −9.00 kcal/mol to −6.02 kcal/mol (Table 2). Molecular docking analysis showed that delphinidin exhibited the lowest estimated free binding energy (−9.00 kcal/mol), suggesting that this anthocyanidin has potential to form strong interactions with α-glucosidase. The results from our molecular docking analysis, especially the estimated free binding energy, correspond with results from enzyme kinetics, suggesting delphinidin with the lowest Ki and IC50 values is a strong α-glucosidase inhibitor. Our structural predictions indicated that delphinidin forms six hydrogen bonds with enzyme residues located outside of the active site (Fig. 2). Indeed, the hydrogen atom on the hydroxyl moiety at C-3’ of the B ring interacts with oxygen atoms of the backbone carboxyl moiety of Phe535, while the oxygen atom at the C-4’ hydroxyl moiety directly interacts with hydrogen atoms on the backbone amine moiety of Ile523. In addition, the hydrogen atom of the C-5’ hydroxyl moiety of the B ring interacts with oxygen atoms of the backbone carboxyl moieties of Arg520 and Ser521, while oxygen atoms on the hydroxyl moieties at C-5 and C-7 of the A ring interact with hydrogen atoms of the backbone amine moieties of Asp777 and Thr778, respectively.
Fig. 2.
Schematic diagram of delphinidin docking to the centered macromolecule of human intestinal N-terminal maltase glucoamylase (PDB: 2QMJ). Potential interactions with enzyme residues are shown with dashed lines.
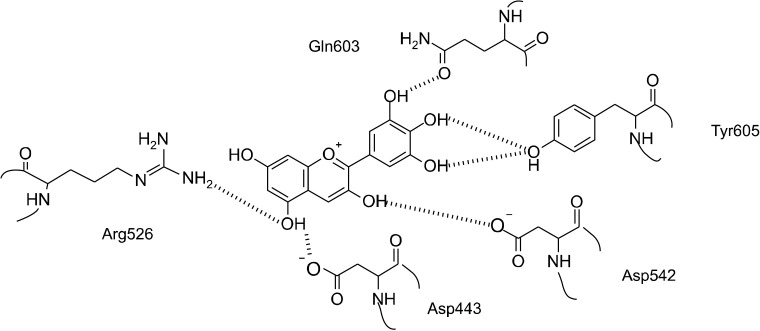
Structural prediction of molecular docking into the catalytic pocket of the enzyme indicated free binding energy scores ranging from −8.38 kcal/mol to −3.97 kcal /mol (Table 2). Again, delphinidin had the lowest estimated free binding energy, and was predicted to form hydrogen bonds with enzyme residues inside the active site via Asp443, Arg526, Asp542, Gln603, and Tyr605 (Fig. 3). Two catalytic residues from the catalytic triad of human intestinal N-terminal maltase glucoamylase were present as Asp443 and Asp542, whereas one catalytic residue, Asp327, was missing from the structural interactions. The distance of this ligand from delphinidin was greater than 3 Å, thus we predicted no interaction was formed. This result indicates that the hydrogen atom of C-3’ hydroxyl moiety of the B ring directly interacts with the side chain carbonyl of Gln603, while hydrogen atoms of the C-4’ and C-5’ hydroxyl moieties interact with the η-hydroxyl side chain of Tyr605. The hydrogen atom of the C-3 hydroxyl moiety of the C ring interacts with the oxygen atom on the γ-acidic side chain of Asp542. Moreover, the hydrogen atom of the C-5 hydroxyl moiety of the A ring interacts with the γ-acidic side chain of Asp443, whereas its oxygen atom interacts with the side chain η1-amine of Arg526.
Fig. 3.
Schematic diagram of delphinidin docking into the catalytic pocket of human intestinal N-terminal maltase glucoamylase (PDB: 2QMJ). Potential interactions with enzyme residues are shown with dashed lines.
Through determining the free binding energy of delphinidin docking into the centered macromolecule and the catalytic pocket of the enzyme, we showed that delphinidin preferred to bind at a site far away from the active site of α-glucosidase (−9.00 kcal/mol) rather than within the active site (−8.38 kcal/mol). Interestingly, the free binding energy was in agreement with our kinetic results that indicated delphinidin is a mixed-type, close to non-competitive α-glucosidase inhibitor. Based on these results, we concluded that delphinidin prefers to interact with either the free enzyme or the enzyme-substrate complex, thus interfering with the enzyme-substrate intermediate. In addition, molecular docking analysis confirmed that delphinidin acts as a mixed-type inhibition as it interacts with enzyme residues in the region of the enzyme located far away from the active sites. The structure-activity relationship suggests that 3’, 4’, and 5’ trihydroxyl moieties on the B ring play important roles in the inhibitory activity of delphinidin as these trihydroxyl moieties form hydrogen bonds with α-glucosidase. Moreover, the presence of the hydroxyl group may be essential to maintain interactions between delphinidin and α-glucosidase, which retard the entrance of substrates into the enzyme active site.
These data suggest that delphinidin is a potent α-glucosidase mixed-type, close to non-competitive inhibitor. Therefore, delphinidin shows potential as an effective anti-α-glucosidase agent and future diabetic drug. However, its bioaccessibility, toxicity and ability to inhibit α-glucosidase activity in vitro and in vivo are currently unknown.
ACKNOWLEDGEMENTS
Financial support throughout this graduate study for a Master Degree of Food and Nutritional Toxicology was received from the Institute of Nutrition, Mahidol University, Nakhon Pathom, Thailand.
Footnotes
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
References
- Bahadoran Z, Mirmiran P, Azizi F. Dietary polyphenols as potential nutraceuticals in management of diabetes: a review. J Diabetes Metab Disord. 2013;12:43. doi: 10.1186/2251-6581-12-43. https://doi.org/10.1186/2251-6581-12-43 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves VC, Boff L, Vizzotto M, Calvete E, Reginatto FH, Simões CM. Berries grown in Brazil: anthocyanin profiles and biological properties. J Sci Food Agric. 2018;98:4331–4338. doi: 10.1002/jsfa.8959. [DOI] [PubMed] [Google Scholar]
- de la Garza AL, Etxeberria U, Lostao MP, San Román B, Barrenetxe J, Martínez JA, et al. Helichrysum and grapefruit extracts inhibit carbohydrate digestion and absorption, improving postprandial glucose levels and hyperinsulinemia in rats. J Agric Food Chem. 2013;61:12012–12019. doi: 10.1021/jf4021569. [DOI] [PubMed] [Google Scholar]
- Fan P, Terrier L, Hay AE, Marston A, Hostettmann K. Antioxidant and enzyme inhibition activities and chemical profiles of Polygonum sachalinensis F. Schmidt ex Maxim (Polygonaceae) Fitoterapia. 2010;81:124–131. doi: 10.1016/j.fitote.2009.08.019. [DOI] [PubMed] [Google Scholar]
- Flores-Bocanegra L, Pérez-Vásquez A, Torres-Piedra M, Bye R, Linares E, Mata R. α-Glucosidase inhibitors from Vauquelinia corymbosa. Molecules. 2015;20:15330–15342. doi: 10.3390/molecules200815330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Kawabata J. Importance of the B ring and its substitution on the alpha-glucosidase inhibitory activity of baicalein, 5,6,7-trihydroxyflavone. Biosci Biotechnol Biochem. 2004;68:1858– 1864. doi: 10.1271/bbb.68.1858. [DOI] [PubMed] [Google Scholar]
- Guo H, Guo J, Jiang X, Li Z, Ling W. Cyanidin-3-Ob-glucoside, a typical anthocyanin, exhibits antilipolytic effects in 3T3-L1 adipocytes during hyperglycemia: involvement of FoxO1-mediated transcription of adipose triglyceride lipase. Food Chem Toxicol. 2012;50:3040–3047. doi: 10.1016/j.fct.2012.06.015. [DOI] [PubMed] [Google Scholar]
- Hansawasdi C, Kawabata J. α-Glucosidase inhibitory effect of mulberry (Morus alba) leaves on Caco-2. Fitoterapia. 77:568–573. doi: 10.1016/j.fitote.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Hidalgo J, Teuber S, Morera FJ, Ojeda C, Flores CA, Hidalgo MA, et al. Delphinidin reduces glucose uptake in mice jejunal tissue and human intestinal cells lines through FFA1/GPR40. Int J Mol Sci. 2017;18:750. doi: 10.3390/ijms18040750. https://doi.org/10.3390/ijms18040750 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui H, Tang G, Go VL. Hypoglycemic herbs and their action mechanisms. Chin Med. 2009;4:11. doi: 10.1186/1749-8546-4-11. https://doi.org/10.1186/1749-8546-4-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins DJ, Srichaikul K, Kendall CW, Sievenpiper JL, Abdulnour S, Mirrahimi A, et al. The relation of low glycaemic index fruit consumption to glycaemic control and risk factors for coronary heart disease in type 2 diabetes. Diabetologia. 2011;54:271– 279. doi: 10.1007/s00125-010-1927-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata J, Mizuhata K, Sato E, Nishioka T, Aoyama Y, Kasai T. 6-Hydroxyflavonoids as α-glucosidase inhibitors from marjoram (Origanum majorana) leaves. Biosci Biotechnol Biochem. 2003;67:445–447. doi: 10.1271/bbb.67.445. [DOI] [PubMed] [Google Scholar]
- Kim JH, Ryu YB, Kang NS, Lee BW, Heo JS, Jeong IY, et al. Glycosidase inhibitory flavonoids from Sophora flavescens. Biol Pharm Bull. 2006;29:302–305. doi: 10.1248/bpb.29.302. [DOI] [PubMed] [Google Scholar]
- Kim JS, Kwon CS, Son KH. Inhibition of alpha-glucosidase and amylase by luteolin, a flavonoid. Biosci Biotechnol Biochem. 2000;64:2458–2461. doi: 10.1271/bbb.64.2458. [DOI] [PubMed] [Google Scholar]
- Miguel MG. Anthocyanins: antioxidant and/or anti-inflammatory activities. J Appl Pharm Sci. 2011;1:7–15. [Google Scholar]
- Mikulic-Petkovsek M, Koron D, Zorenc Z, Veberic R. Do optimally ripe blackberries contain the highest levels of metabolites? Food Chem. 2017;215:41–49. doi: 10.1016/j.foodchem.2016.07.144. [DOI] [PubMed] [Google Scholar]
- Mohamed Sham Shihabudeen H, Hansi Priscilla D, Thirumurugan K. Cinnamon extract inhibits α-glucosidase activity and dampens postprandial glucose excursion in diabetic rats. Nutr Metab. 2011;8:46. doi: 10.1186/1743-7075-8-46. https://doi.org/10.1186/1743-7075-8-46 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov DG. Structure and evolution of mammalian maltase-glucoamylase and sucrase-isomaltase genes. Mol Biol. 2007;41:1056–1068. doi: 10.1134/S0026893307060131. [DOI] [PubMed] [Google Scholar]
- Nyenwe EA, Jerkins TW, Umpierrez GE, Kitabchi AE. Management of type 2 diabetes: evolving strategies for the treatment of patients with type 2 diabetes. Metabolism. 2011;60:1–23. doi: 10.1016/j.metabol.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Priscilla DH, Roy D, Suresh A, Kumar V, Thirumurugan K. Naringenin inhibits α-glucosidase activity: a promising strategy for the regulation of postprandial hyperglycemia in high fat diet fed streptozotocin induced diabetic rats. Chem Biol Interact. 2014;210:77–85. doi: 10.1016/j.cbi.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Proença C, Freitas M, Ribeiro D, Oliveira EFT, Sousa JLC, Tomé SM, et al. α-Glucosidase inhibition by flavonoids: an in vitro and in silico structure-activity relationship study. J Enzyme Inhib Med Chem. 2017;32:1216–1228. doi: 10.1080/14756366.2017.1368503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada-Calvillo R, Robayo-Torres CC, Opekun AR, Sen P, Ao Z, Hamaker BR, et al. Contribution of mucosal maltase-glucoamylase activities to mouse small intestinal starch alpha-glucogenesis. J Nutr. 2007;137:1725–1733. doi: 10.1093/jn/137.7.1725. [DOI] [PubMed] [Google Scholar]
- Racine J. The Cygwin tools: a GNU toolkit for Windows. J Appl Econ. 2000;15:331–341. doi: 10.1002/1099-1255(200005/06)15:3<331::AID-JAE558>3.0.CO;2-G. [DOI] [Google Scholar]
- Salam NK, Adzhigirey M, Sherman W, Pearlman DA. Structure-based approach to the prediction of disulfide bonds in proteins. Protein Eng Des Sel. 2014;27:365–374. doi: 10.1093/protein/gzu017. [DOI] [PubMed] [Google Scholar]
- Shang Q, Xiang J, Zhang H, Li Q, Tang Y. The effect of polyhydroxylated alkaloids on maltase-glucoamylase. PLoS One. 2013;8:e70841. doi: 10.1371/journal.pone.0070841. https://doi.org/10.1371/journal.pone.0070841 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibano M, Kakutani K, Taniguchi M, Yasuda M, Baba K. Antioxidant constituents in the dayflower (Commelina communis L.) and their alpha-glucosidase-inhibitory activity. J Nat Med. 2008;62:349–353. doi: 10.1007/s11418-008-0244-1. [DOI] [PubMed] [Google Scholar]
- Shrivastava SR, Shrivastava PS, Ramasamy J. Role of self-care in management of diabetes mellitus. J Diabetes Metab Disord. 2013;12:14. doi: 10.1186/2251-6581-12-14. https://doi.org/10.1186/2251-6581-12-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim L, Quezada-Calvillo R, Sterchi EE, Nichols BL, Rose DR. Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J Mol Biol. 2008;375:782–792. doi: 10.1016/j.jmb.2007.10.069. [DOI] [PubMed] [Google Scholar]
- Tadera K, Minami Y, Takamatsu K, Matsuoka T. Inhibition of alpha-glucosidase and alpha-amylase by flavonoids. J Nutr Sci Vitaminol. 2006;52:149–153. doi: 10.3177/jnsv.52.149. [DOI] [PubMed] [Google Scholar]
- Testa R, Bonfigli AR, Genovese S, De Nigris V, Ceriello A. The possible role of flavonoids in the prevention of diabetic complications. Nutrients. 2016;8:310. doi: 10.3390/nu8050310. https://doi.org/10.3390/nu8050310 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Laar FA. Alpha-glucosidase inhibitors in the early treatment of type 2 diabetes. Vasc Health Risk Manag. 2008;4:1189– 1195. doi: 10.2147/VHRM.S3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujović T, Ružić Đ, Cerović R, Leposavić A, Karaklajić-Stajić Ž, Mitrović O, et al. An assessment of the genetic integrity of micropropagated raspberry and blackberry plants. Sci Hortic. 2017;225:454–461. doi: 10.1016/j.scienta.2017.07.020. [DOI] [Google Scholar]
- Wang TTY, Schoene NW, Kim YS, Mizuno CS, Rimando AM. Differential effects of resveratrol and its naturally occurring methylether analogs on cell cycle and apoptosis in human androgen-responsive LNCaP cancer cells. Mol Nutr Food Res. 2010;54:335–344. doi: 10.1002/mnfr.200900143. [DOI] [PubMed] [Google Scholar]
- Wedick NM, Pan A, Cassidy A, Rimm EB, Sampson L, Rosner B, et al. Dietary flavonoid intakes and risk of type 2 diabetes in US men and women. Am J Clin Nutr. 2012;95:925–933. doi: 10.3945/ajcn.111.028894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Cao H, Wang Y, Yamamoto K, Wei X. Structure-affinity relationship of flavones on binding to serum albumins: effect of hydroxyl groups on ring A. Mol Nutr Food Res. 2010;54:S253–S260. doi: 10.1002/mnfr.200900454. [DOI] [PubMed] [Google Scholar]
- Xiao J, Kai G, Yamamoto K, Chen X. Advance in dietary polyphenols as α-glucosidases inhibitors: a review on structure-activity relationship aspect. Crit Rev Food Sci Nutr. 2013;53:818– 836. doi: 10.1080/10408398.2011.561379. [DOI] [PubMed] [Google Scholar]
- Xiao JB, Högger P. Dietary polyphenols and type 2 diabetes: current insights and future perspectives. Curr Med Chem. 2015;22:23–38. doi: 10.2174/0929867321666140706130807. [DOI] [PubMed] [Google Scholar]
- Xu Y, Xie L, Xie J, Liu Y, Chen W. Pelargonidin-3-O-rutinoside as a novel α-glucosidase inhibitor for improving postprandial hyperglycemia. Chem Commun. 2018;55:39–42. doi: 10.1039/C8CC07985D. [DOI] [PubMed] [Google Scholar]
- Yin Z, Zhang W, Feng F, Zhang Y, Kang W. α-Glucosidase inhibitors isolated from medicinal plants. Food Sci Hum Well. 2014;3:136–174. doi: 10.1016/j.fshw.2014.11.003. [DOI] [Google Scholar]
- Zang Y, Igarashi K, Li Y. Anti-diabetic effects of luteolin and luteolin-7-O-glucoside on KKAy mice. Biosci Biotechnol Biochem. 2016;80:1580–1586. doi: 10.1080/09168451.2015.1116928. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang N, Wang W, Wang J, Zhu Z, Li X. Molecular mechanisms of novel peptides from silkworm pupae that inhibit α-glucosidase. Peptides. 2016;76:45–50. doi: 10.1016/j.peptides.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Zhu F, Asada T, Sato A, Koi Y, Nishiwaki H, Tamura H. Rosmarinic acid extract for antioxidant, antiallergic, and α-glucosidase inhibitory activities, isolated by supramolecular technique and solvent extraction from Perilla leaves. J Agric Food Chem. 2014;62:885–892. doi: 10.1021/jf404318j. [DOI] [PubMed] [Google Scholar]