Abstract
Background
The placenta is an essential organ for the normal development of mammalian fetuses. Most of our knowledge on the molecular mechanisms of placental development has come from the analyses of mice, especially histopathological examination of knockout mice. Choriocarcinoma and immortalized cell lines have also been used for basic research on the human placenta. However, these cells are quite different from normal trophoblast cells.
Methods
In this review, we first provide an overview of mouse and human placental development with particular focus on the differences in the anatomy, transcription factor networks, and epigenetic characteristics between these species. Next, we discuss pregnancy complications associated with abnormal placentation. Finally, we introduce emerging in vitro models to study the human placenta, including human trophoblast stem (TS) cells, trophoblast and endometrium organoids, and artificial embryos.
Main findings
The placental structure and development differ greatly between humans and mice. The recent establishment of human TS cells and trophoblast and endometrial organoids enhances our understanding of the mechanisms underlying human placental development.
Conclusion
These in vitro models will greatly advance our understanding of human placental development and potentially contribute to the elucidation of the causes of infertility and other pregnancy complications.
Keywords: DNA methylation, epigenetics, human placenta, organoid, trophoblast stem (TS) cells
1. INTRODUCTION
The placenta is a multifunctional organ that is essential for the survival and development of mammalian fetuses. The placenta contains both maternal and fetal cells and serves to provide nourishment and oxygen from the mother to the fetus. Besides, placental trophoblast cells are a major source of pregnancy‐related hormones and help protect the fetus from the immune system of the mother. 1 , 2 , 3 It is interesting that the fetus, which is a semi‐allograft to the mother, is not attacked by the maternal immune cells in the uterus. The uterus is not an immunologically isolated organ, and many lymphocytes reside there. Thus, after implantation, embryos are exposed to maternal immune cells but do not trigger immune rejection. Abnormal placentation is associated with pregnancy complications such as infertility, miscarriage, and hypertensive disorders of pregnancy (HDP). These disorders are collectively called the “great obstetrical syndromes”. 4 Although some pregnancy complications such as fetal growth restriction and HDP occur during the 2nd and 3rd trimester in humans, attention should be focused on the early stages of intrauterine development to understand the underlying mechanisms of these diseases. There is no doubt that the placenta plays crucial roles in the development and health of all offspring, but there is still little information on the human placenta.
Primary cultures, choriocarcinoma cell lines, and immortalized cell lines have traditionally been used in basic studies of the human placenta. However, it is been difficult to maintain primary trophoblast cells in culture, and various molecular mechanisms may be disrupted in choriocarcinoma and immortalized cell lines. Alternatively, mice have also been used to study the placenta because of their genetic uniformity and the ease of genome manipulation. Moreover, the availability of mouse trophoblast stem (TS) cells has greatly advanced research on the molecular mechanisms of trophoblast proliferation and differentiation. 5 However, placental anatomy and trophoblast cell types are significantly different between mice and humans. 6 Therefore, the establishment of human TS cells has long been awaited, but not achieved for about 20 years after the establishment of mouse TS cells. Recently, however, we succeeded in the establishment of human TS cells. Moreover, due to advances in bioengineering technologies and three‐dimensional culture systems, artificial embryos and trophoblast and endometrium organoids have become available.
In this review, we discuss the differences in the anatomy, transcription factor networks, and epigenetic properties between mice and humans during placental development and then describe the molecular pathology of perinatal complications. Finally, we introduce several recent technical breakthroughs in the field of human placental research.
2. PLACENTATION DURING EARLY PREGNANCY
Studies on the human placenta have been conducted using samples obtained from artificial abortion or preterm delivery. Animal models, such as the mouse, are also useful in elucidating the molecular mechanisms of placentation. However, it should be noted that although preimplantation development is similar between humans and mice, there are substantial differences in placental structure and trophoblast subtypes after implantation.
2.1. Placental development before and during implantation
The mature oocyte and sperm fertilize to form a totipotent zygote. After several cell divisions, a zygote gives rise to a blastocyst that is composed of inner cell mass (ICM) and surrounding trophectoderm (TE) (Figure 1). TE is classified into the polar and mural TE: The former is adjacent to ICM, and the latter is not in contact with ICM. Whereas the polar TE proliferates rapidly, the mural TE stops cell division and differentiates into primary trophoblast giant cells (TGCs) in mice. TGCs are multinucleated cells that replicate their genomic DNA without cell or nuclear division. ICM differentiates into epiblast and primitive endoderm, which are destined to form embryonic and yolk sac tissues, respectively. Mouse blastocysts implant in utero at around embryonic day 4.5 (E4.5). During implantation, endometrial stromal cells undergo a special process called decidualization. This response is driven by the sex steroid hormones, estrogen, and progesterone, in both mice and humans. 7 Decidualization begins after implantation in mice, whereas it is induced during the mid‐secretory to late secretory phase of the menstrual cycle irrespective of implantation in humans. 7 , 8 The interaction between trophoblasts and maternal decidua is particularly important for placental development. 9 Impaired decidualization, such as endometriosis, has been reported to increase the risk of infertility and perinatal complications. 7 , 10
FIGURE 1.
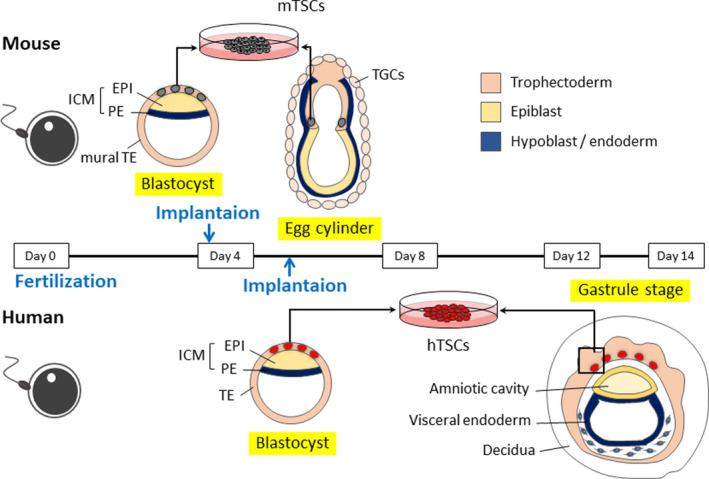
Comparison of peri‐implantation embryos in human and mouse. Epiblast and trophoblast directly interact in mice, but not in human. In mice, FGF4 from the epiblast is required for trophoblast proliferation. In humans, epiblasts and trophoblasts are separated by mesenchymal cells and cannot interact directly. Therefore, human trophoblast proliferation is likely to be epiblast (FGF4) EVT independent (maintenance of human TS cells requires EGF, not FGF). EPI: epiblast; PE: primitive endoderm; TE: trophectoderm; TSCs: trophoblast stem cells; TGCs: trophoblast giant cells
2.2. Placental development after implantation in mice
In mice, a longitudinally elongated structure, known as the egg cylinder, is formed at about E6.0 (Figure 1). The extraembryonic ectoderm and the ectoplacental cone protrude into the maternal decidua. At the periphery of the ectoplacental cone, secondary TGCs differentiate. FGF4 provided by ICM and epiblast is required for the proliferation of polar TE and extraembryonic ectoderm. 5 At E7.5, the ectoplacental cavity is formed. The base of the ectoplacental space is called the chorion. In the extraembryonic cavity, the extraembryonic mesoderm‐derived allantois extends from the fetal tail toward the chorion. At E9.0, the allantois fuses to the chorion and fetal blood vessels develop. At this stage, the chorionic ectoderm fuses with the roof of the ectoplacental space, and the ectoplacental space disappears. Failure of this process is the most frequent cause of mid‐pregnancy embryo lethality in mouse mutants. 11 , 12 By E10.0, the basic structure of the so‐called chorioallantoic placenta completes (Figure 2). The outermost layer (maternal side) of the placenta consists of secondary TGCs and serves to anchor the placenta to the uterus. A spongiotrophoblast layer is formed inside TGCs. After approximately E13.5, the spongiotrophoblast layer contains glycogen cells. The fetal side of the placenta, the labyrinthine layer, is invaded by fetal blood vessels from the allantois to form a capillary network.
FIGURE 2.
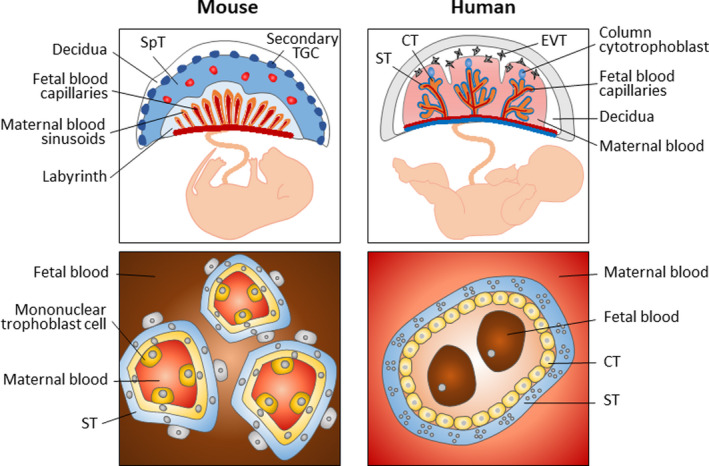
Structure of human and mouse mature placenta. Structure of the mouse placenta. The inset details the fetal‐maternal interface in the labyrinth. Structure of the human placenta. The inset image shows a cross section of a chorionic villus; trophoblast‐derived structures (blue) and mesoderm‐derived tissues (orange). The inset images illustrate the number and type of cell layers between the maternal and fetal blood. CT, cytotrophoblast; EVT, extravillous cytotrophoblast; SpT, spongiotrophoblast; ST, syncytiotrophoblast; TGC, trophoblast giant cell
2.3. Placental development after implantation in humans
In humans, implantation occurs at E6.0‐8.0. Trophoblasts adhere to endometrial epithelial cells and penetrate decidualized endometrial stroma. In mice, this invasion is triggered by TGCs, but in humans, primitive syncytiotrophoblast cells appear around undifferentiated cytotrophoblast (CT) cells and invade the decidua. 13 , 14 Primitive syncytiotrophoblast cells secrete enzymes that digest the decidua and enlarge the space for the embryo. In contrast to the mouse epiblast that acquires a columnar morphology, the human epiblast forms a disk‐like structure. Although the gross morphology is different, both mouse and human epiblasts form pseudostratified columnar epithelium. In the human embryo, epiblast cells adjacent to the trophoblast form squamous epithelium known as amnion and primitive endoderm‐derived yolk sac is formed. By E14.0, primitive streaks appear, gastrulation starts, and primordial germ cells (PGCs) are specified.
The basic structure of the human placenta is formed by about week 4 of gestation, and the maternal blood supply to the placenta is established by about 10 to 12 weeks (Figure 2). 15 The mature placenta consists of three types of trophoblast cells: cytotrophoblast (CT), extravillous cytotrophoblast (EVT), and syncytiotrophoblast (ST). CT cells are highly proliferative epithelial cells that can differentiate into EVT and ST cells. EVT cells differentiate from CT cells at the tips of villi, invade the endometrium, and remodel the spiral arteries to control the maternal blood flow. These cells infiltrate to one‐third of the thickness of the uterine wall. ST cells are multinucleated cells formed by the fusion of CT cells and mediate nutrient and gas exchange between the fetal and maternal blood circulation. ST cells are also involved in hormone production. In addition to these trophoblasts, placental villi also contain ICM‐derived fibroblasts, vascular endothelial cells, and macrophages called Hofbauer cells. These cells are collectively called stromal cells.
Although the mouse and human placentas are morphologically different, they are both classified as the hemochorial placenta. 16 , 17 In humans, the major sites of maternal nutrient and gas uptake are lined by a single layer of ST cells. In mice, there are two layers of syncytium. The feto‐maternal interface is highly variable among mammalian species and believed to be optimized by species. 18 At this interface, placental development proceeds through complex and sophisticated interactions between the trophoblast and the endometrium. Indeed, the maternal endometrium is known to play important roles in placental development. A study using human placental explant culture showed decidual stroma cell (DSC)‐derived Neuregulin‐1 (NRG‐1) promotes EVT differentiation. 19 Moreover, DSCs secretes a variety of growth factors and cytokines. They alter the expression of MMPs and integrins in trophoblast, which promotes trophoblast invasion. 20 At the same time, DSCs also secretes inhibitors of MMPs which suppress excessive trophoblast invasion. 21 Thus, the trophoblast differentiation and invasion into maternal tissues are thought to be exquisitely controlled in part by the paracrine factors released from DSCs. 22 The endometrial epithelium also plays an important role for placentation. As described above, the maternal arterial circulation to the placenta is not fully established until 10‐12 weeks of pregnancy. Endometrial glands are thought to be necessary to provide nutrients for the conceptus before the maternal blood supply. Indeed, the intervillous space before the entry of the maternal spiral artery is filled with secretions from the endometrium. 23 Endometrial glands secrete amino acids, lipids, proteins, and sugars that are nutrients of the conceptus. 24 Endometrial glands also supply growth factors such as VEGF, LIF, and EGF. 25 Studies using human villus explants showed that EGF stimulates CT cell proliferation at 4‐5 weeks of gestation and the secretion of hCG and human placental lactogen (hPL) from ST cells at 6‐10 weeks of gestation. 26 Thus, secretion from both of stromal cells and epithelial cells is important at each stage of trophoblast growth and differentiation.
3. TRANSCRIPTION FACTOR NETWORKS REGULATING TROPHOBLAST DEVELOPMENT
In mouse preimplantation embryos, Oct4 (Pou5f1) and Cdx2 antagonize each other and play an important role in the segregation of the embryonic and extraembryonic lineages. 27 , 28 Oct4 protein is expressed in all cells of the morula but gradually restricted to ICM. Cdx2 is expressed in the outer cells of the morula and completely restricted to TE at the blastocyst stage. 29 The Hippo signal transcriptional coactivator Yap preferentially induces preferential differentiation into TE. Yap translocates to the nucleus of the cells on the outer cells of the morula and activates the expression of the transcription factors Tead4 and Cdx2 in mice. 30 , 31 CDX2 and OCT4 exhibit similar expression patterns in human blastocysts, 32 but YAP is localized to the nucleus in both ICM and TE at the late blastocyst stage. TEAD4 expression, on the other hand, seems to show well‐conserved expression patterns between mice and humans. The role of Hippo signaling during human preimplantation development has not been well studied. 33 Whereas Cdx2 expression persists in the extraembryonic ectoderm after implantation in mice, it is controversial whether CDX2 is expressed in trophoblast lineages after implantation in humans. 34 , 35 , 36
During mouse placental development, Tead4 and Cdx2 act in concert with multiple transcription factors, including Eomes, Elf5, Ets2, Esrrb, Gata3, Sox2, and Tfap2c. 37 , 38 Dysfunction of these genes leads to abnormal placentation and embryonic lethality shortly after implantation. 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 Mouse TS cells have been used to analyze the interactions and regulation of these transcription factors. For example, Esrrb and Sox2 are found to be targets of the fibroblast growth factor (FGF) signaling mediated by Fgfr2c. Removal of FGF or addition of FGF signaling inhibitors rapidly reduces TS cell‐specific gene expression. Interestingly, the expression of ESRRB and SOX2 is negligible in TE and CT cells in humans. This observation is consistent with the absence of FGFR2C in human TE and CT cells. 44 , 45 , 51 Therefore, ESRRB and SOX2 might be dispensable for human trophoblast development. Additionally, human TE and CT cells do not express EOMES. 32 , 35 , 52 In contrast, GATA3, TFAP2C, ELF5, and ETS2 are expressed in TE and/or CT cells in humans. 32 , 35 , 36 , 53 , 54 These differences and similarities of the expression patterns of transcription factors in human and mouse trophoblast cells are summarized in Figure 3.
FIGURE 3.
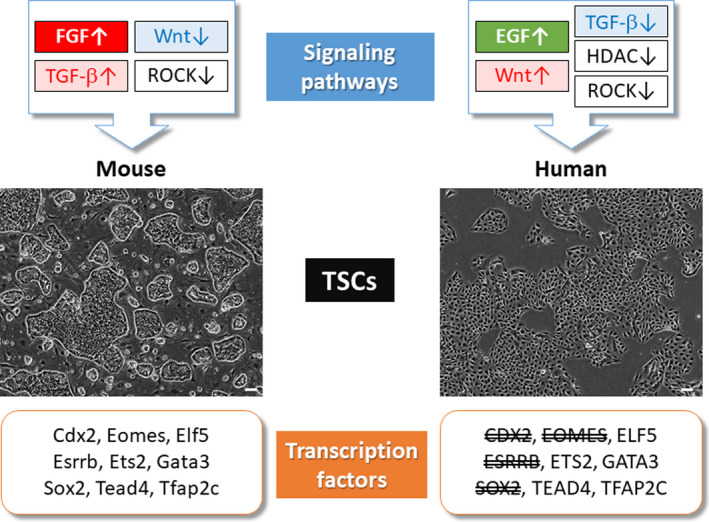
Comparison of signaling pathway and transcription factors between human and mouse TSCs. Wnt and TGF‐β signals are inversely involved in mTSCs and hTSCs. Some of the transcription factors required to maintain mTSCs are not expressed in hTSCs. In mTSCs, Esrrb and Sox2 are known to be activated downstream of FGF signals. ESRRB and SOX2 are not expressed in hTSCs, which is consistent with the FGF‐independent nature of hTSCs. The specific transcription factors for hTSCs are currently unknown
Most of the transcription factors that regulate trophoblast development do not show trophoblast‐specific expression patterns. In mice, all transcription factors described above are also expressed in non‐trophoblast cells. It is been recognized that transcription factors can change their partners in a cell type‐dependent manner. For example, Sox2 and Esrrb are essential for both mouse embryonic stem (ES) and TS cells. Sox2 forms a complex with Tfap2c in TS cells, 44 but Sox2 interacts with Oct4 in ES cells. Esrrb interacts with histone demethylase Kdm1a and RNA polymerase II‐associated integrator complexes in TS cells but not in ES cells. 45 Another important aspect is the relative ratio of interacting transcription factors. Additionally, the EOMES‐ELF5 protein complex acts to inhibit differentiation of mouse TS cells, 55 and the TFAP2C‐ELF5 protein promotes differentiation. 56 Collectively, the transcription factor networks regulating mouse TS cells have gradually been understood. However, little is known in humans.
4. EPIGENETIC REGULATION IN THE PLACENTA
Epigenetics is important not only for cell fate determination but also for stabilization of lineage‐specific gene expression patterns. Genetic information must be properly regulated by epigenetic modifications. 57 Epigenetic modifications such as DNA methylation and histone modifications play crucial roles in placental development.
4.1. DNA methylation reprogramming after fertilization
Among epigenetic modifications, methylation of cytosine residues in CpG dinucleotides has been well studied. DNA methylation plays important roles not only in cell lineage specification but also in the control of the genome imprinting and the X chromosome inactivation. DNA methylation goes through two dramatic reprogramming during mammalian development. One occurs during gametogenesis, and the other does after fertilization. 58 These two waves of reprogramming are observed in both humans and mice. Global DNA demethylation during gametogenesis allows expression of genes required for meiosis and also resets the parent‐of‐origin‐dependent DNA methylation at the imprinted loci. Some retrotransposons are resistant to demethylation and mediate the inheritance of epigenetic information across generations. 59 Global DNA demethylation also occurs after fertilization, but the imprinted loci are protected at this stage. In mice, the sperm‐derived genome is demethylated by the ten‐eleven translocation (Tet) 3 demethylase, which is known as active demethylation. On the other hand, the oocyte‐derived genome is protected from demethylation by Tet3. However, due to the weak activity of the maintenance DNA methyltransferase Dnmt1, methylated DNA is lost at each cell division, which is known as passive demethylation. DNA methylation patterns of sperm and oocytes are similar between humans and mice, 60 , 61 , 62 and the sperm‐derived genome also go through active demethylation in human embryos. However, passive demethylation seems to be incomplete in humans, and the oocyte‐derived genome shows higher methylation levels in humans than in mice (Figure 4).
FIGURE 4.
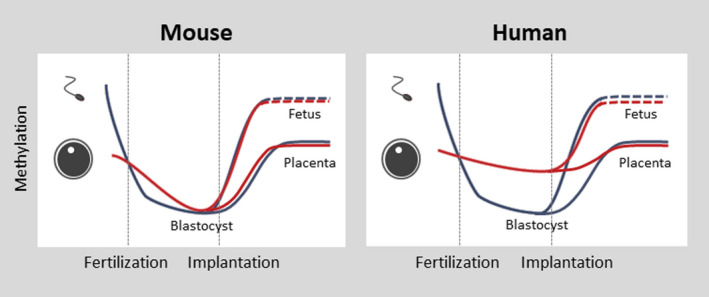
Dynamic DNA methylation change of early embryo in human and mouse. The maternal genome was demethylated to a much lesser extent in human blastocysts than in mouse blastocysts, which increase the number of imprinted DNA methylation
After implantation, DNA methylation is regained in both embryonic and extraembryonic cells, which is mediated by the de novo DNA methyltransferases (DNMT) 3A and DNMT3B 63 , 64 , 65 . However, whereas ~80% of CpG sites are methylated in epiblast cells, only 40‐50% of CpG sites are methylated in trophoblast cells. In both embryonic and extraembryonic tissues, the promoters are largely unmethylated and the gene body regions of actively transcribed genes are highly methylated. However, compared to epiblast cells, trophoblast cells have less DNA methylation at the gene body regions of weakly expressed genes and intergenic regions. The placenta is thought to have the lowest DNA methylation levels among tissues, 66 , 67 , 68 , 69 and the methylome of trophoblast cells are characterized by partial methylation domains (PMDs). 68 , 70 , 71 These PMDs are the same in mice and humans, but their functional significance is unknown.
4.2. Regulation of placenta‐specific gene expression by DNA methylation
Many genes have been identified to show placenta‐specific expression, 72 and some of them are regulated by DNA methylation. For example, the promoter of ELF5 is hypomethylated in trophoblast cells but hypermethylated in most embryonic lineage cells in both humans and mice. 36 The promoters of INSL4 and DSCR4 show similar methylation patterns. 73 , 74 We also recently identified 55 promoters that exhibit placenta‐specific hypomethylation in humans. 52
The DNA‐binding capacity of many transcription factors is influenced by DNA methylation. In vitro studies using synthetic methylated and unmethylated DNA‐binding domains have shown that the binding affinity of about 60% of human transcription factors is affected by DNA methylation. 75 , 76 Transcription factors that are important in trophoblasts, such as ETS2, ELF5, and TFAP2C, bind unmethylated DNA.
Long terminal repeat (LTR) sequences of endogenous retroviruses are associated with placental evolution in mammals. LTRs are rich in transcription factor binding motifs and exhibit unique functions in extraembryonic tissues. 77 , 78 DNA methylation is responsible for the stable silencing of foreign DNA. 79 , 80 , 81 In mouse and human trophoblast cells, many LTRs are hypomethylated. 82 , 83 Recently, it has been reported that LTRs act to stimulate the secretion of corticotropin‐releasing hormone, a regulator of gestational age. 84 It has also been reported that LTRs upstream of the nitric oxide synthase gene NOS3 drive placenta‐specific transcriptional isoforms in the human placenta. Moreover, syncytins, envelope genes derived from retroviruses, play an important role in both mouse and human ST cells. 85 , 86 Thus, retrotransposons and their DNA methylation patterns may contribute to the evolution of the placenta.
4.3. Development of trophoblast cells and histone modifications
Histone modifications are essential for trophoblast development in mice. Embryos deficient in EHMT2 (also known as G9A), which mediates H3K9 dimethylation, fail to fuse the chorioallantoic membranes, which is lethal in mid‐pregnancy. 87 Polycomb group proteins responsible for H3K27me3 modification are essential for chorionic villus formation, and their defects result in abnormalities from early‐ to mid‐gestation. 88 , 89 H3K9 dimethylation, trimethylation (H3K9me 2/3), and H3K27me3 are inhibitory marks and often function in overlap with DNA methylation. Mouse ES cells deficient in H3K9 trimethylation‐mediated histone methyltransferase (SETDB1) express Cdx2 and differentiate into trophoblast cells. 90 , 91 , 92 Lysine demethylase (KDM1A)‐deficient mouse ES cells exhibit similar characteristics. H3K27me3 and H3K9me2 are also involved in the regulation of imprinted genes in the mouse placenta. 93 , 94 , 95 Despite these important findings in mice, the functional roles of histone modifications are poorly understood in human trophoblast cells.
4.4. Placenta‐specific genomic imprinting:
In mice, the development of both female‐developing embryos (embryos composed of the oocyte‐derived nuclei) and androgenetic embryos (embryos composed of sperm‐derived nuclei) is lethal. So, the development of these causes the death of the embryo. However, their phenotypes are very different. Parthenogenetic embryos rarely form the placenta, whereas androgenetic embryos show placental hyperplasia. Similarly, in humans, androgenesis is associated with complete hydatidiform mole, a disease characterized by trophoblast overgrowth. Thus, genomic imprinting is important for placental development.
Some imprinted genes show uniparental expression in a tissue‐specific manner, and many tissue‐specific imprinted genes have been found in mice, especially in the placenta and brain. We performed a comprehensive analysis of the imprinted genes in the mouse placenta, 96 which revealed that many placenta‐specific imprinted genes reported by that time were due to contamination of maternal tissues, and only 12 genes actually underwent imprinting. We also demonstrated that uniparental expression of these placenta‐specific imprinted genes was disrupted in the placentas of somatic cell nuclear transfer (SCNT) mouse embryos, which are typically characterized by placental hypertrophy. These findings reveal the importance of placenta‐specific imprinted genes in the placental development. 97 , 98 However, human homologs of mouse placenta‐specific imprinted genes do not show uniparental expression in human placentas. 96 There are about 110 human placenta‐specific imprinted genes, far more than the mouse imprinted genes. 64 This is likely due to incomplete demethylation of the oocyte‐derived genome after fertilization in humans. In addition to protein‐coding genes and long non‐coding RNAs, some miRNAs are imprinted in the placenta. There are about 2000 miRNAs in the human and mouse genomes. MiRNAs bind to target mRNAs and inhibit translation. Some miRNAs form clusters, and the miRNA clusters on chromosomes 14 and 19 are regulated by genomic imprinting in the human placenta. 99 , 100 , 101 The reason why humans and mice have acquired almost completely different sets of placenta‐specific imprinted genes is currently unknown, but it is possible that they contribute to differences in placental structure, gestational period, or litter size between these species.
4.5. Incomplete X chromosome inactivation in human placenta:
One of two X chromosomes is inactivated in female mammalian cells. In marsupials, both embryonic and extraembryonic tissues show imprinted X chromosome inactivation, in which the paternal X chromosome is selectively inactivated. In mice, inactivation of the X chromosome in embryonic tissues is random, whereas the paternal X chromosome is selectively inactivated in the placenta. It has been reported that X chromosome inactivation is skewed in the human placenta. The paternal X chromosome is preferentially inactivated in the human placenta but the degree varies among individuals. 64 , 102
5. EPIGENOMIC ABNORMALITIES AND PLACENTAL DISORDERS
Large‐scale knockout studies in mice indicate that about 25‐30% of all genes are essential for survival. However, with few exceptions, such analyses have focused on embryos and little attention has been paid to placentas. 103 A recent study revealed that morphological abnormalities of the placenta are observed in approximately two‐thirds of embryonic lethal mutant mice. 12 It is also known that placental abnormalities tend to be associated with abnormalities of the heart, brain, and vascular embryos. In humans, infants with congenital heart disease have also been reported to exhibit a significantly higher frequency of placental abnormalities than healthy controls. 104 , 105 , 106
Abnormalities in genomic imprinting may be associated with the development of small for gestational age (SGA), intrauterine growth restriction (IUGR), and HDP in humans. For example, in the placenta of IUGR, there is a trend toward increased expression of the paternally imprinted gene PHLDA2 and decreased expression of the maternally imprinted genes MEST and PLAGL1. Decreased expression of paternally imprinted genes MEG3 and GNAS has also been reported. 121
HDP is also thought to be associated with epigenetic abnormalities. The expression of several miRNAs is reported to be dysregulated in preeclamptic placentas compared to normal placentas. 122 , 123 The human placenta‐specific imprinted gene CUL7 is also reported to be hypomethylated and shows increased expression in human fetal growth restriction placentas. 124 Deficiency of CUL7 also causes fetal growth restriction, severe post‐natal growth restriction, and 3M syndrome type I, a congenital anomaly syndrome characterized by characteristic facies. 125 Furthermore, it has been reported that Cul7‐deficient mice cause vascular abnormalities in the decidua and exhibit phenotypes such as impaired placental development and fetal growth restriction. 124 CYP2J2, which is a placenta‐specific imprinted gene 64 and encodes one of the cytochrome P 450 enzymes known as drug‐metabolizing enzymes, 126 is reported to be highly expressed in HDP patients. 126 In addition, the metabolite EET of CYP2J2 is also increased in a rat model of HDP. These results support the hypothesis that aberrant expression of imprinted genes may be associated with IUGR and HDP.
A complete hydatidiform mole (CHM) is caused by androgenesis and is characterized by abnormal proliferation of placental trophoblasts. CHM is also associated with a high rate of secondary tumors and cancers, making this disease one of the most important pregnancy complications for clinical diagnosis and management. Abnormalities in the expression of imprinted genes have been thought to play a major role in the pathogenesis of CHM, but it is not clear which imprinted genes are involved in the pathogenesis. Recently, we established cell lines from CHM samples and demonstrated that silencing of the imprinted gene P57KIP2 confers resistance to cell cycle arrest by contact inhibition in these cell lines. 127
6. DEVELOPMENT OF USEFUL TOOLS TO STUDY HUMAN PLACENTAL DEVELOPMENT
The establishment of mouse TS cells was first reported in 1998. 5 It was only recently that human TS cells were established. 52 Furthermore, in recent years, various useful tools have been developed to study placental development, such as artificial mouse embryos and human trophoblast and endometrial organoids (Figure 5).
FIGURE 5.
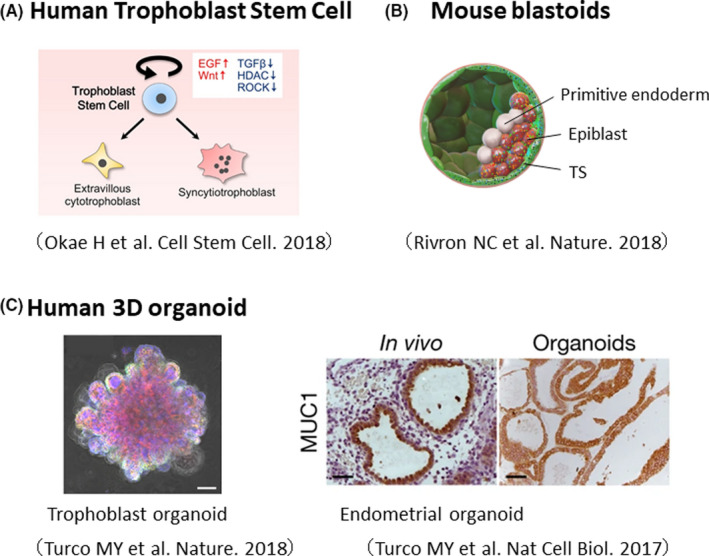
Development of useful tools to study human placental development
6.1. Human trophoblast stem (TS) cells:
In 1998, mouse TS cells were first derived from blastocysts or extraembryonic ectoderm of post‐implantation embryos using FGF4, heparin, and mouse embryonic fibroblasts (MEF). 5 However, the culture conditions of mouse TS cells cannot be applied to human TS cells, probably due to the differences in the molecular mechanisms regulating trophoblast proliferation and differentiation. For example, FGFR2c, a receptor for FGF4, is expressed in mouse blastocysts but not in human blastocysts. 51 Although various models have been generated and used to study human trophoblast cells (Table 1), the establishment of human TS cells has not been achieved for a long time.
TABLE 1.
Previous in vitro models for human trophoblast cell research
| Name | Origin | Established methods | Marker | Section references |
|---|---|---|---|---|
| Trophoblast cell line | First‐trimester placenta | Explants | CK, hCG | 107 |
| Long‐term cytotrophoblast culture | First‐trimester placenta | Explants | CK, hCG, Trop‐1, Trop‐2 | 108 |
| HTR‐8/SVneo | Early placenta | SV40 large T antigen (immortalization) | CK, hCG | 109 |
| HT | Full‐term placenta | Primary culture (transformed trophoblasts) | CK, hCG, Placental alkaline phosphatase, Trop‐2 | 110 |
| TCL‐1 | Chorionic membrane of placenta | SV40 large T antigen(immortalization) | hCG | 111 |
| NPC | First‐trimester placenta | EGF, Insulin, Dexamethasone | CK18, GnRH, Neuropeptide Y, Neurotensin, Leucine‐enkephalin, Dopamine, 5‐HT, Progesteron, hCG | 112 |
| IST‐1 | First‐trimester placenta | Retrovirus encoding HPV16 E6 and E7 proteins(immortalization) | CK7, CK18, hPL, Mel‐CAM (CD146) | 113 |
| BMP4‐hESC | ES cell | BMP (FGF2, MEF‐CM) | TFAP2, MSX2, SSI3, GATA2, GATA3, HEY1, CG‐α, CG‐β | 114 |
| CTBS | ES cell | Embyoid body formation, FGF4, heparin, MEF‐CM | CDX2, HLA‐G, CD9,CK7 | 115 |
| HPT‐8, HPT‐8‐HBV | First‐trimester placenta | Primary culure, singel cell cloning, HBV (immortalization) | CK7, CK18, Vimentin, CD9, EGFR, SDF1, Prolactin, E2, Progesterone, hCG, HLA‐G | 116 |
| TBPCs | Chorion membrane of first‐trimester placenta | FGF2, SB431542 (TGF‐β inhibitor), gelatin substrates | OCT4, ZO‐1, GATA4, Nestin, CK7 | 117 |
| iTP | Human fetal fibroblast (IMR90) | Lentivirus(CDX2, ELF5, C‐MYC, KLF4, EOMES) | CDX2, EOMES, ELF5, CK7, GATA3, TEAD4 | 118 |
| BAP‐hESC | ES cell | BMP4, A83‐01 (ALK4/5/7 inhibitor), PD173074 (FGF2‐signaling inhibitor), MEF‐CM | CK, T, HLA‐G | 119 |
| TSCs from UCSFB | ES cell lines from blastomeres | Embryoid body formation, FGF2, SB431542 | CDX2, TEAD4, GATA3, ELF5, GDF15, β‐catenin | 120 |
We recently succeeded in establishing human TS cells from blastocysts and CT cells of first‐trimester placental villi. Human TS cells can be maintained in an undifferentiated state for a long period (80 passages or more) and meet following four criteria for trophoblast cells. 52 , 53 (1) expression of trophoblast markers such as GATA3 and TFAP2C; (2) decreased expression of HLA class I molecules; (3) hypomethylation of the ELF5 gene promoter; and (4) expression of the placenta‐specific miRNA cluster C19MC. We found that activation of Wnt and EGF signaling and inhibition of TGF‐β signaling, HDAC, and ROCK are important for human TS cell derivation. When treated with the adenylate cyclase activator forskolin, human TS cells fuse to differentiate into multinucleated ST cells. Human TS cells can also differentiate into spindle‐shaped extravillous cytotrophoblast (EVT) cells when they are treated with neuregulin (NRG1) and a TGF‐β inhibitor. The methylation patterns of human TS cells are highly correlated with those of primary trophoblast cells. Furthermore, when human TS cells were implanted subcutaneously in immunocompromised mice, they infiltrated the dermis and subcutaneous tissue of the mice and differentiated into EVT‐ and ST‐like cells. Interestingly, some of the ST‐like cells were found to be vacuolated and have an influx of mouse blood flow. This structure closely resembles the primordial syncytial cells, which are specialized cells produced when human blastocysts implant in the uterus. In summary, human TS cells retain unique characteristics of trophoblast cells and therefore are very useful to analyze human placental development and function.
6.2. Artificial embryos generated using TS and ES cells:
TS cells have also been used for modeling of early embryos. Mouse TS cells, when combined with ES cells, self‐organize to form post‐implantation embryo‐like structures that mimic early embryo development. 128 Blastocyst‐like structures were also generated using mouse ES and TS cells. These “blastoids” have the potential to implant into the mouse uterus and induce decidualization. Although ethical issues must be addressed, the development of human artificial embryos using TS and ES cells will open up great potential for the study of human embryogenesis.
6.3. 3D trophoblast organoids
Soon after human TS cell establishment was reported, three‐dimensional trophoblast organoids have been generated using CT cells purified from first‐trimester placental tissue. 129 , 130 These organoids contain CT‐ and ST‐like cells and can be maintained for a long time. They also give rise to EVT‐like cells as human TS cells do. Such trophoblast organoids are useful for studying placental development and function under more physiological conditions.
6.4. New models and data resources to study the feto‐maternal interface
The generation of organoid models has also been reported for the maternal endometrium. Three‐dimensional culture of endometrial glands leads to self‐organized cyst‐like structures, which respond to sex hormones such as estrogen and progesterone. Endometrial organoids can also replicate the phenotype of endometriosis and endometrial cancer. 131 Endometrial receptivity is essential for successful implantation and pregnancy. Indeed, two‐thirds of implantation failure is caused by endometrial receptivity. 132 The combination of organoid technologies that mimic the development of embryos, trophoblast cells, and endometrium has the potential to recapitulate implantation and placentation more precisely than the conventional trophoblast‐endometrium coculture assay. 133 , 134 This would expand our understanding of the human early developmental process, which has been a “black box” due to ethical constraints.
Recently, single‐cell RNA‐seq analysis has provided a comprehensive picture of the cell populations present in the human placenta and decidua. This approach also enables the identification of previously unknown cell populations. Moreover, the analysis of ligand‐receptor interactions based on the expression data makes it possible to estimate the dynamic interactions between fetal and maternal cells at the feto‐maternal interface. 135 , 136
7. CONCLUSIONS AND FUTURE PERSPECTIVES
Maternal nutrition, physical activity, and psychological stress during pregnancy can affect not only fetal and placental growth but also the risk of cancer and lifestyle‐related diseases such as cardiovascular disease and diabetes in adulthood. It is known as the “Developmental Origins of Health and Disease (DOHaD)” theory. 137 , 138 The placenta is sensitive to environmental changes in utero and may be involved in the lifelong health of humans. Studies of the placenta have made significant progress, led by studies on mouse models. However, as we have seen, the placental structure and development differ greatly between humans and mice. The recent establishment of human TS cells and trophoblast and endometrial organoids enhances our understanding of the mechanisms underlying human placental development. Moreover, further advancement of these techniques will lead to a better understanding of embryogenesis and implantation and the treatment of diseases such as infertility and pregnancy‐induced hypertension.
Disclosures
Our derivation of human TS cells was approved by the Institutional Review Board (IRB) approval at Tohoku university (2014‐1‐879).
Conflict of interest: Authors have no conflict of interest to be declared.
Acknowledgments
We would like to thank all the member of our laboratory for their support and valuable suggestions. This work was supported by Japan Society for the promotion of science Grants‐in‐Aid for Scientific Research (JSPS KAKENHI) Grants 18K09216 (H.O) and 19H05757 (H.O), Japan Agency for Medical Research and Development (AMED) Grants JP20gm1310001h0002 (T.A) and JP20bm0704021h0003 (H.O).
Shibata S, Kobayashi EH, Kobayashi N, Oike A, Okae H, Arima T. Unique features and emerging in vitro models of human placental development. Reprod Med Biol. 2020;19:301–313. 10.1002/rmb2.12347
Funding information
Japan Society for the promotion of science Grants‐in‐Aid for Scientific Research (JSPS KAKENHI) Grants 18K09216 (H.O) and 19H05757 (H.O), Japan Agency for Medical Research and Development (AMED) Grants JP20gm1310001h0002 (T.A) and JP20bm0704021h0003 (H.O).
References
- 1. Napso T, Yong HEJ, Lopez‐Tello J, Sferruzzi‐Perri AN. The role of placental hormones in mediating maternal adaptations to support pregnancy and lactation. Front Physiol. 2018;9:1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. PrabhuDas M, Bonney E, Caron K, et al. Immune mechanisms at the maternal‐fetal interface: perspectives and challenges. Nat Immunol. 2015;16(4):328‐334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6(8):584‐594. [DOI] [PubMed] [Google Scholar]
- 4. Brosens I, Pijnenborg R, Vercruysse L, Romero R. The, “Great obstetrical syndromes” are associated with disorders of deep placentation. Am J Obstet Gynecol. 2011;204(3):193‐201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282(5396):2072‐2075. [DOI] [PubMed] [Google Scholar]
- 6. Hemberger M, Hanna CW, Dean W. Mechanisms of early placental development in mouse and humans. Nat Rev Genet. 2020;21(1):27‐43. [DOI] [PubMed] [Google Scholar]
- 7. Ramathal CY, Bagchi IC, Taylor RN, Bagchi MK. Endometrial decidualization: of mice and men. Semin Reprod Med. 2010;28(1):17‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shuya LL, Menkhorst EM, Yap J, Li P, Lane N, Dimitriadis E. Leukemia inhibitory factor enhances endometrial stromal cell decidualization in humans and mice. PLoS ONE. 2011;6(9):e25288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woods L, Perez‐Garcia V, Kieckbusch J, et al. Decidualisation and placentation defects are a major cause of age‐related reproductive decline. Nat Commun. 2017;8(1):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klemmt PA, Carver JG, Kennedy SH, Koninckx PR, Mardon HJ. Stromal cells from endometriotic lesions and endometrium from women with endometriosis have reduced decidualization capacity. Fertil Steril. 2006;85(3):564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Copp AJ. Death before birth: clues from gene knockouts and mutations. Trends Genet: TIG. 1995;11(3):87‐93. [DOI] [PubMed] [Google Scholar]
- 12. Perez‐Garcia V, Fineberg E, Wilson R, et al. Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature. 2018;555(7697):463‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. James JL, Carter AM, Chamley LW. Human placentation from nidation to 5 weeks of gestation. Part I: what do we know about formative placental development following implantation? Placenta. 2012;33(5):327‐334. [DOI] [PubMed] [Google Scholar]
- 14. Knofler M, Haider S, Saleh L, Pollheimer J, Gamage T, James J. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol Life Sci: CMLS. 2019;76(18):3479‐3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boyd JD, Hamilton WJ. The Human Placenta. Cambridge: Heffer & Sons; 1970. [Google Scholar]
- 16. Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2(7):538‐548. [DOI] [PubMed] [Google Scholar]
- 17. Woods L, Perez‐Garcia V, Hemberger M. Regulation of placental development and its impact on fetal growth‐new insights from mouse models. Front Endocrinol. 2018;9:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adamson SL, Lu Y, Whiteley KJ, et al. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev Biol. 2002;250(2):358‐373. [DOI] [PubMed] [Google Scholar]
- 19. Fock V, Plessl K, Draxler P, et al. Neuregulin‐1‐mediated ErbB2‐ErbB3 signalling protects human trophoblasts against apoptosis to preserve differentiation. J Cell Sci. 2015;128(23):4306‐4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Godbole G, Suman P, Gupta SK, Modi D. Decidualized endometrial stromal cell derived factors promote trophoblast invasion. Fertil Steril. 2011;95(4):1278‐1283. [DOI] [PubMed] [Google Scholar]
- 21. Roth I, Fisher SJ. IL‐10 is an autocrine inhibitor of human placental cytotrophoblast MMP‐9 production and invasion. Dev Biol. 1999;205(1):194‐204. [DOI] [PubMed] [Google Scholar]
- 22. Sharma S, Godbole G, Modi D. Decidual control of trophoblast invasion. Am J Reprod Immunol. 2016;75(3):341‐350. [DOI] [PubMed] [Google Scholar]
- 23. Burton GJ, Scioscia M, Rademacher TW. Endometrial secretions: creating a stimulatory microenvironment within the human early placenta and implications for the aetiopathogenesis of preeclampsia. J Reprod Immunol. 2011;89(2):118‐125. [DOI] [PubMed] [Google Scholar]
- 24. Hempstock J, Cindrova‐Davies T, Jauniaux E, Burton GJ. Endometrial glands as a source of nutrients, growth factors and cytokines during the first trimester of human pregnancy: a morphological and immunohistochemical study. Reprod Biol Endocrinol. 2004;2:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burton GJ, Jauniaux E, Charnock‐Jones DS. Human early placental development: potential roles of the endometrial glands. Placenta. 2007;28(Suppl A):S64‐S69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maruo T, Matsuo H, Murata K, Mochizuki M. Gestational age‐dependent dual action of epidermal growth factor on human placenta early in gestation. J Clin Endocrinol Metab. 1992;75(5):1362‐1367. [DOI] [PubMed] [Google Scholar]
- 27. Niwa H, Toyooka Y, Shimosato D, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123(5):917‐929. [DOI] [PubMed] [Google Scholar]
- 28. Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10(2):91‐103. [DOI] [PubMed] [Google Scholar]
- 29. Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre‐implantation embryo. Development. 2007;134(23):4219‐4231. [DOI] [PubMed] [Google Scholar]
- 30. Nishioka N, Yamamoto S, Kiyonari H, et al. Tead4 is required for specification of trophectoderm in pre‐implantation mouse embryos. Mech Dev. 2008;125(3‐4):270‐283. [DOI] [PubMed] [Google Scholar]
- 31. Nishioka N, Inoue K, Adachi K, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16(3):398‐410. [DOI] [PubMed] [Google Scholar]
- 32. Blakeley P, Fogarty NM, Del Valle I, et al. Defining the three cell lineages of the human blastocyst by single‐cell RNA‐seq. Development. 2015;142(20):3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qin H, Hejna M, Liu Y, et al. YAP induces human naive pluripotency. Cell Rep. 2016;14(10):2301‐2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Horii M, Li Y, Wakeland AK, et al. Human pluripotent stem cells as a model of trophoblast differentiation in both normal development and disease. Proc Natl Acad Sci USA. 2016;113(27):E3882‐E3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soncin F, Khater M, To C, et al. Comparative analysis of mouse and human placentae across gestation reveals species‐specific regulators of placental development. Development. 2018;145(2): dev156273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hemberger M, Udayashankar R, Tesar P, Moore H, Burton GJ. ELF5‐enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet. 2010;19(12):2456‐2467. [DOI] [PubMed] [Google Scholar]
- 37. Kidder BL, Palmer S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 2010;20(4):458‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Latos PA, Hemberger M. Review: the transcriptional and signalling networks of mouse trophoblast stem cells. Placenta. 2014;35(Suppl):S81‐S85. [DOI] [PubMed] [Google Scholar]
- 39. Russ AP, Wattler S, Colledge WH, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404(6773):95‐99. [DOI] [PubMed] [Google Scholar]
- 40. Donnison M, Beaton A, Davey HW, Broadhurst R, L'Huillier P, Pfeffer PL. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132(10):2299‐2308. [DOI] [PubMed] [Google Scholar]
- 41. Yamamoto H, Flannery ML, Kupriyanov S, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12(9):1315‐1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wen F, Tynan JA, Cecena G, et al. Ets2 is required for trophoblast stem cell self‐renewal. Dev Biol. 2007;312(1):284‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luo J, Sladek R, Bader JA, Matthyssen A, Rossant J, Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR‐beta. Nature. 1997;388(6644):778‐782. [DOI] [PubMed] [Google Scholar]
- 44. Adachi K, Nikaido I, Ohta H, et al. Context‐dependent wiring of Sox2 regulatory networks for self‐renewal of embryonic and trophoblast stem cells. Mol Cell. 2013;52(3):380‐392. [DOI] [PubMed] [Google Scholar]
- 45. Latos PA, Goncalves A, Oxley D, Mohammed H, Turro E, Hemberger M. Fgf and Esrrb integrate epigenetic and transcriptional networks that regulate self‐renewal of trophoblast stem cells. Nat Commun. 2015;6:7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knott JG, Paul S. Transcriptional regulators of the trophoblast lineage in mammals with hemochorial placentation. Reproduction. 2014;148(6):R121‐R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ralston A, Cox BJ, Nishioka N, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137(3):395‐403. [DOI] [PubMed] [Google Scholar]
- 48. Home P, Ray S, Dutta D, Bronshteyn I, Larson M, Paul S. GATA3 is selectively expressed in the trophectoderm of peri‐implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284(42):28729‐28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell‐Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126‐140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kuckenberg P, Buhl S, Woynecki T, et al. The transcription factor TCFAP2C/AP‐2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol. 2010;30(13):3310‐3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kunath T, Yamanaka Y, Detmar J, et al. Developmental differences in the expression of FGF receptors between human and mouse embryos. Placenta. 2014;35(12):1079‐1088. [DOI] [PubMed] [Google Scholar]
- 52. Okae H, Toh H, Sato T, et al. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22(1):50‐63 e56. [DOI] [PubMed] [Google Scholar]
- 53. Lee CQ, Gardner L, Turco M, et al. What is trophoblast? a combination of criteria define human first‐trimester trophoblast. Stem Cell Reports. 2016;6(2):257‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee Y, Kim KR, McKeon F, et al. A unifying concept of trophoblastic differentiation and malignancy defined by biomarker expression. Hum Pathol. 2007;38(7):1003‐1013. [DOI] [PubMed] [Google Scholar]
- 55. Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct‐3/4 defines differentiation, dedifferentiation or self‐renewal of ES cells. Nat Genet. 2000;24(4):372‐376. [DOI] [PubMed] [Google Scholar]
- 56. Latos PA, Sienerth AR, Murray A, et al. Elf5‐centered transcription factor hub controls trophoblast stem cell self‐renewal and differentiation through stoichiometry‐sensitive shifts in target gene networks. Genes Dev. 2015;29(23):2435‐2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487‐500. [DOI] [PubMed] [Google Scholar]
- 58. Dean W, Santos F, Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin Cell Dev Biol. 2003;14(1):93‐100. [DOI] [PubMed] [Google Scholar]
- 59. Seisenberger S, Andrews S, Krueger F, et al. The dynamics of genome‐wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48(6):849‐862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okae H, Chiba H, Hiura H, et al. Genome‐wide analysis of DNA methylation dynamics during early human development. PLoS Genet. 2014;10(12):e1004868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606‐610. [DOI] [PubMed] [Google Scholar]
- 62. Smith ZD, Chan MM, Humm KC, et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511(7511):611‐615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y, Xiang Y, Yin Q, et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat Genet. 2018;50(1):96‐105. [DOI] [PubMed] [Google Scholar]
- 64. Hamada H, Okae H, Toh H, et al. Allele‐specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am J Hum Genet. 2016;99(5):1045‐1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hanna CW, Penaherrera MS, Saadeh H, et al. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016;26(6):756‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gamage T, Schierding W, Tsai P, et al. Human trophoblasts are primarily distinguished from somatic cells by differences in the pattern rather than the degree of global CpG methylation. Biol Open. 2018;7(8), bio034884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Robinson WP, Price EM. The human placental methylome. Cold Spring Harb Perspect Med. 2015;5(5):a023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schroeder DI, Blair JD, Lott P, et al. The human placenta methylome. Proc Natl Acad Sci USA. 2013;110(15):6037‐6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bianco‐Miotto T, Mayne BT, Buckberry S, Breen J, Rodriguez Lopez CM, Roberts CT. Recent progress towards understanding the role of DNA methylation in human placental development. Reproduction. 2016;152(1):R23‐R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Smith ZD, Shi J, Gu H, et al. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature. 2017;549(7673):543‐547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Luo C, Hajkova P, Ecker JR. Dynamic DNA methylation: in the right place at the right time. Science. 2018;361(6409):1336‐1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rawn SM, Cross JC. The evolution, regulation, and function of placenta‐specific genes. Annu Rev Cell Dev Biol. 2008;24:159‐181. [DOI] [PubMed] [Google Scholar]
- 73. Du Y, Zhang J, Wang H, et al. Hypomethylated DSCR4 is a placenta‐derived epigenetic marker for trisomy 21. Prenat Diagn. 2011;31(2):207‐214. [DOI] [PubMed] [Google Scholar]
- 74. Macaulay EC, Weeks RJ, Andrews S, Morison IM. Hypomethylation of functional retrotransposon‐derived genes in the human placenta. Mamm Genome. 2011;22(11‐12):722‐735. [DOI] [PubMed] [Google Scholar]
- 75. Hu S, Wan J, Su Y, et al. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013;2:e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yin Y, Morgunova E, Jolma A, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356(6337):eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bourque G, Leong B, Vega VB, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18(11):1752‐1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pavlicev M, Hiratsuka K, Swaggart KA, Dunn C, Muglia L. Detecting endogenous retrovirus‐driven tissue‐specific gene transcription. Genome Biol Evol. 2015;7(4):1082‐1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20(2):116‐117. [DOI] [PubMed] [Google Scholar]
- 80. Bestor TH, Bourc'his D. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb Symp Quant Biol. 2004;69:381‐387. [DOI] [PubMed] [Google Scholar]
- 81. Lavie L, Kitova M, Maldener E, Meese E, Mayer J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV‐K(HML‐2). J Virol. 2005;79(2):876‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Macfarlan TS, Gifford WD, Agarwal S, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25(6):594‐607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cohen CJ, Rebollo R, Babovic S, Dai EL, Robinson WP, Mager DL. Placenta‐specific expression of the interleukin‐2 (IL‐2) receptor beta subunit from an endogenous retroviral promoter. J Biol Chem. 2011;286(41):35543‐35552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dunn‐Fletcher CE, Muglia LM, Pavlicev M, et al. Anthropoid primate‐specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS Biol. 2018;16(9):e2006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mi S, Lee X, Li X, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785‐789. [DOI] [PubMed] [Google Scholar]
- 86. Dupressoir A, Vernochet C, Harper F, et al. A pair of co‐opted retroviral envelope syncytin genes is required for formation of the two‐layered murine placental syncytiotrophoblast. Proc Natl Acad Sci USA. 2011;108(46):E1164‐E1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tachibana M, Sugimoto K, Nozaki M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779‐1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb‐group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330‐4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23(20):4061‐4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3‐K9 methyltransferase ESET is essential for early development. Mol Cell Biol. 2004;24(6):2478‐2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yeap LS, Hayashi K, Surani MA. ERG‐associated protein with SET domain (ESET)‐Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin. 2009;2(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yuan P, Han J, Guo G, et al. Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 2009;23(21):2507‐2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lewis A, Mitsuya K, Umlauf D, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36(12):1291‐1295. [DOI] [PubMed] [Google Scholar]
- 94. Umlauf D, Goto Y, Cao R, et al. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36(12):1296‐1300. [DOI] [PubMed] [Google Scholar]
- 95. Wagschal A, Feil R. Genomic imprinting in the placenta. Cytogenetic and genome research. 2006;113(1‐4):90‐98. [DOI] [PubMed] [Google Scholar]
- 96. Okae H, Hiura H, Nishida Y, et al. Re‐investigation and RNA sequencing‐based identification of genes with placenta‐specific imprinted expression. Hum Mol Genet. 2012;21(3):548‐558. [DOI] [PubMed] [Google Scholar]
- 97. Okae H, Matoba S, Nagashima T, et al. RNA sequencing‐based identification of aberrant imprinting in cloned mice. Hum Mol Genet. 2014;23(4):992‐1001. [DOI] [PubMed] [Google Scholar]
- 98. Matoba S, Liu Y, Lu F, et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159(4):884‐895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Morales‐Prieto DM, Chaiwangyen W, Ospina‐Prieto S, et al. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33(9):725‐734. [DOI] [PubMed] [Google Scholar]
- 100. Doridot L, Miralles F, Barbaux S, Vaiman D. Trophoblasts, invasion, and microRNA. Front Genet. 2013;4:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Noguer‐Dance M, Abu‐Amero S, Al‐Khtib M, et al. The primate‐specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19(18):3566‐3582. [DOI] [PubMed] [Google Scholar]
- 102. Moreira de Mello JC, de Araujo ES, Stabellini R, et al. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele‐specific gene expression along the X chromosome. PLoS ONE. 2010;5(6):e10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Dickinson ME, Flenniken AM, Ji X, et al. High‐throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jones HN, Olbrych SK, Smith KL, et al. Hypoplastic left heart syndrome is associated with structural and vascular placental abnormalities and leptin dysregulation. Placenta. 2015;36(10):1078‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Matthiesen NB, Henriksen TB, Agergaard P, et al. Congenital heart defects and indices of placental and fetal growth in a nationwide study of 924 422 liveborn infants. Circulation. 2016;134(20):1546‐1556. [DOI] [PubMed] [Google Scholar]
- 106. Rychik J, Goff D, McKay E, et al. Characterization of the placenta in the newborn with congenital heart disease: distinctions based on type of cardiac malformation. Pediatr Cardiol. 2018;39(6):1165‐1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Goustin AS, Betsholtz C, Pfeifer‐Ohlsson S, et al. Coexpression of the sis and myc proto‐oncogenes in developing human placenta suggests autocrine control of trophoblast growth. Cell. 1985;41(1):301‐312. [DOI] [PubMed] [Google Scholar]
- 108. Yagel S, Casper RF, Powell W, Parhar RS, Lala PK. Characterization of pure human first‐trimester cytotrophoblast cells in long‐term culture: growth pattern, markers, and hormone production. Am J Obstet Gynecol. 1989;160(4):938‐945. [DOI] [PubMed] [Google Scholar]
- 109. Graham CH, Hawley TS, Hawley RG, et al. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res. 1993;206(2):204‐211. [DOI] [PubMed] [Google Scholar]
- 110. Ho CK, Li SY, Yu KJ, Wang CC, Chiang H, Wang SY. Characterization of a human tumorigenic, poorly differentiated trophoblast cell line . In Vitro Cell Develop Biol Animal.. 1994;30A(7):415‐417. [DOI] [PubMed] [Google Scholar]
- 111. Lewis MP, Clements M, Takeda S, et al. Partial characterization of an immortalized human trophoblast cell‐line, TCL‐1, which possesses a CSF‐1 autocrine loop. Placenta. 1996;17(2‐3):137‐146. [DOI] [PubMed] [Google Scholar]
- 112. Rong‐Hao L, Luo S, Zhuang LZ. Establishment and characterization of a cytotrophoblast cell line from normal placenta of human origin. Hum Reprod. 1996;11(6):1328‐1333. [DOI] [PubMed] [Google Scholar]
- 113. Shih I, Wang T, Wu T, Kurman RJ, Gearhart JD. Expression of Mel‐CAM in implantation site intermediate trophoblastic cell line, IST‐1, limits its migration on uterine smooth muscle cells. J Cell Sci. 1998;111(Pt 17):2655‐2664. [DOI] [PubMed] [Google Scholar]
- 114. Xu RH, Chen X, Li DS, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20(12):1261‐1264. [DOI] [PubMed] [Google Scholar]
- 115. Harun R, Ruban L, Matin M, et al. Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Hum Reprod. 2006;21(6):1349‐1358. [DOI] [PubMed] [Google Scholar]
- 116. Zhang L, Zhang W, Shao C, et al. Establishment and characterization of a spontaneously immortalized trophoblast cell line (HPT‐8) and its hepatitis B virus‐expressing clone. Hum Reprod. 2011;26(8):2146‐2156. [DOI] [PubMed] [Google Scholar]
- 117. Genbacev O, Donne M, Kapidzic M, et al. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells. 2011;29(9):1427‐1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chen Y, Wang K, Gong YG, Khoo SK, Leach R. Roles of CDX2 and EOMES in human induced trophoblast progenitor cells. Biochem Biophys Res Comm. 2013;431(2):197‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Amita M, Adachi K, Alexenko AP, et al. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci USA. 2013;110(13):E1212‐E1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zdravkovic T, Nazor KL, Larocque N, et al. Human stem cells from single blastomeres reveal pathways of embryonic or trophoblast fate specification. Development. 2015;142(23):4010‐4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. McMinn J, Wei M, Schupf N, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27(6‐7):540‐549. [DOI] [PubMed] [Google Scholar]
- 122. Pineles BL, Romero R, Montenegro D, et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196(3):261 e261‐266. [DOI] [PubMed] [Google Scholar]
- 123. Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200(6):661;e661‐667. [DOI] [PubMed] [Google Scholar]
- 124. Gascoin‐Lachambre G, Buffat C, Rebourcet R, et al. Cullins in human intra‐uterine growth restriction: expressional and epigenetic alterations. Placenta. 2010;31(2):151‐157. [DOI] [PubMed] [Google Scholar]
- 125. Arai T, Kasper JS, Skaar JR, Ali SH, Takahashi C, DeCaprio JA. Targeted disruption of p185/Cul7 gene results in abnormal vascular morphogenesis. Proc Natl Acad Sci USA. 2003;100(17):9855‐9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Herse F, Lamarca B, Hubel CA, et al. Cytochrome P450 subfamily 2J polypeptide 2 expression and circulating epoxyeicosatrienoic metabolites in preeclampsia. Circulation. 2012;126(25):2990‐2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Takahashi S, Okae H, Kobayashi N, et al. Loss of p57(KIP2) expression confers resistance to contact inhibition in human androgenetic trophoblast stem cells In: Proceedings of the National Academy of Sciences of the United States of America; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Harrison SE, Sozen B, Christodoulou N, Kyprianou C, Zernicka‐Goetz M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356(6334). [DOI] [PubMed] [Google Scholar]
- 129. Turco MY, Gardner L, Kay RG, et al. Trophoblast organoids as a model for maternal‐fetal interactions during human placentation. Nature. 2018;564(7735):263‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Haider S, Meinhardt G, Saleh L, et al. Self‐renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem Cell Reports. 2018;11(2):537‐551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Boretto M, Maenhoudt N, Luo X, et al. Patient‐derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat Cell Biol. 2019;21(8):1041‐1051. [DOI] [PubMed] [Google Scholar]
- 132. Achache H, Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update. 2006;12(6):731‐746. [DOI] [PubMed] [Google Scholar]
- 133. Wang H, Pilla F, Anderson S, et al. A novel model of human implantation: 3D endometrium‐like culture system to study attachment of human trophoblast (Jar) cell spheroids. Mol Hum Reprod. 2012;18(1):33‐43. [DOI] [PubMed] [Google Scholar]
- 134. Buck VU, Gellersen B, Leube RE, Classen‐Linke I. Interaction of human trophoblast cells with gland‐like endometrial spheroids: a model system for trophoblast invasion. Hum Reprod. 2015;30(4):906‐916. [DOI] [PubMed] [Google Scholar]
- 135. Suryawanshi H, Morozov P, Straus A, et al. A single‐cell survey of the human first‐trimester placenta and decidua. Sci Adv. 2018;4(10):eaau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Vento‐Tormo R, Efremova M, Botting RA, et al. Single‐cell reconstruction of the early maternal‐fetal interface in humans. Nature. 2018;563(7731):347‐353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Eichenwald EC, Stark AR. Management and outcomes of very low birth weight. N Eng J Med. 2008;358(16):1700‐1711. [DOI] [PubMed] [Google Scholar]
- 138. Murphy VE, Smith R, Giles WB, Clifton VL. Endocrine regulation of human fetal growth: the role of the mother, placenta, and fetus. Endocr Rev. 2006;27(2):141‐169. [DOI] [PubMed] [Google Scholar]


