Abstract
Background
The histone H3K36me3 mark regulates transcription elongation, pre-mRNA splicing, DNA methylation, and DNA damage repair. However, knowledge of the regulation of the enzyme SETD2, which deposits this functionally important mark, is very limited.
Results
Here, we show that the poorly characterized N-terminal region of SETD2 plays a determining role in regulating the stability of SETD2. This stretch of 1–1403 amino acids contributes to the robust degradation of SETD2 by the proteasome. Besides, the SETD2 protein is aggregate prone and forms insoluble bodies in nuclei especially upon proteasome inhibition. Removal of the N-terminal segment results in the stabilization of SETD2 and leads to a marked increase in global H3K36me3 which, uncharacteristically, happens in a Pol II-independent manner.
Conclusion
The functionally uncharacterized N-terminal segment of SETD2 regulates its half-life to maintain the requisite cellular amount of the protein. The absence of SETD2 proteolysis results in a Pol II-independent H3K36me3 deposition and protein aggregation.
Keywords: Chromatin, Proteasome, Histone, Methylation, Aggregation
Background
The N-terminal tails of histones protrude from the nucleosome and are hotspots for the occurrence of a variety of post-translational modifications (PTMs) that play key roles in regulating epigenetic processes. H3K36me3 is one such important functionally characterized PTM. In yeast, this mark suppresses cryptic transcription from within the coding region of genes by preventing histone exchange [1]. In mammalian cells, it is involved in the recruitment of DNA repair machinery, in splicing and also, in establishing DNA methylation patterns by acting as a binding site for the enzyme DNMT3a [2–6]. Recent reports have emphasized the tumor-suppressive role of H3K36me3 in renal cancer especially, where the gene coding for the SETD2 histone methyltransferase is often deleted or mutated [7–9].
In yeast, the SET domain-containing protein Set2 (ySet2) is the sole H3K36 methyltransferase [10]. ySet2 interacts with the large subunit of the RNA polymerase II, Rpb1, through its SRI (Set2–Rpb1 Interaction) domain, and co-transcriptionally deposits H3K36me3 [11]. The deletion of the SRI domain from ySet2 abolishes both the Set2–RNA Pol II interaction and H3K36me3 methylation in yeast [12]. H3K36 methylation is a highly conserved histone mark and Set2 homologs are found in more complex eukaryotes [13]. These homologs share the conserved features like the AWS (associated with SET), SET [Su(var)3–9, Enhancer-of-zeste and Trithorax] and Post-SET domains that are required for the catalytic activity of the enzyme, and also, the protein–protein interaction domains such as the WW and the SRI. Notably, the mammalian homolog, SETD2, has a long N-terminal segment that is not present in ySet2. The function of this region has remained obscure [13, 14].
Here, we show that SETD2 is an inherently aggregate-prone protein and its N-terminal region regulates its half-life. This, in turn, is important for the fidelity of the deposition of the functionally important H3K36me3 mark. Our findings reveal that the previously uncharacterized N-terminal region of SETD2 is important in governing appropriate SETD2 function and activity.
Results
SETD2 is robustly degraded by the ubiquitin–proteasome pathway
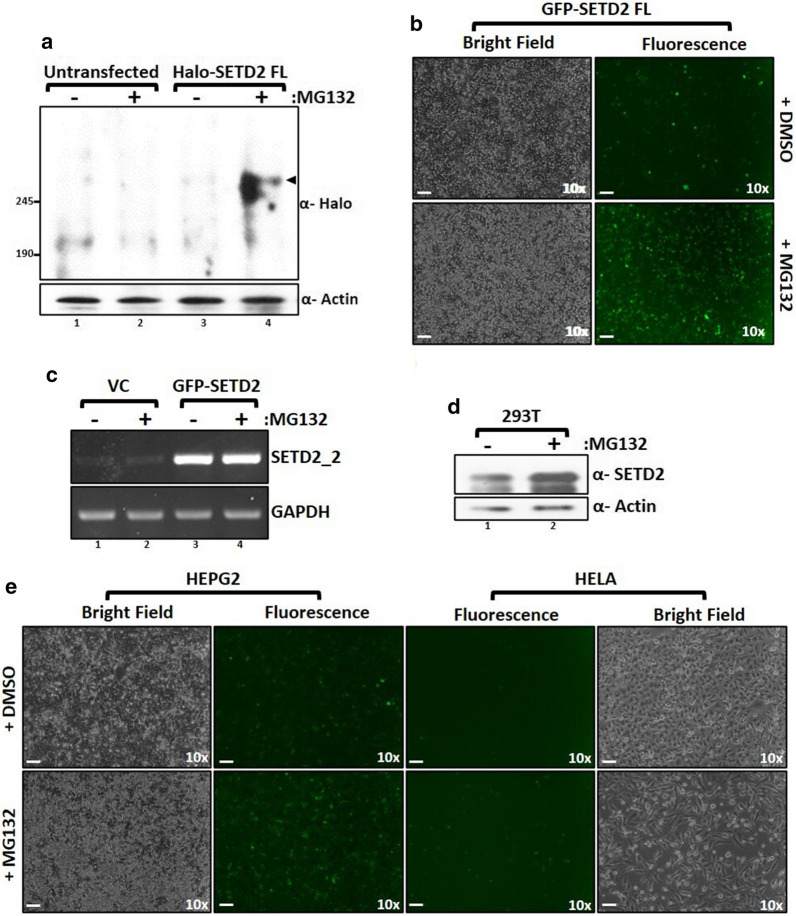
To understand the regulation of the SETD2 enzyme in human cells, we introduced a construct to express Halo- or GFP-tagged SETD2 FL (full length) under the control of the CMV promoter in 293T cells. Strikingly, the expression of GFP-SETD2 FL was barely detected post-72 h of transfection although the expression of GFP in vector control transfected cells was robust (Additional file 1: Figure S1a). Also, RT-PCR revealed a marked increase in the transcript level of SETD2 suggesting a robust transcription from the constructs introduced (Additional file 1: Figure S1b). Similar results were obtained with Halo-SETD2 FL transfected cells in which the expression was not detected with the western blotting of whole-cell lysates with an anti-Halo antibody (Fig. 1a, lane 3).
Fig. 1.
SETD2 is robustly degraded by the proteasome. a Western blot of whole-cell lysates probed with the depicted antibodies. Lysates of wild type 293T (untransfected) cells or expressing SETD2 full-length (FL) were prepared after 12 h of MG132 (10 µM) treatment. The expected band for the target protein is indicated by an arrow. b Microscopy images showing the effect of MG132 treatment on expression of GFP-SETD2 FL in 293T cells. The scale bar is 1 mm. c RNA was isolated from GFP-SETD2 FL transfected cells and RT-PCR was performed to check the transcript levels. GAPDH was used as a normalization control. VC- empty vector control. d Whole-cell lysates of 293T cells were prepared after 12 h of MG132 (10 µM) treatment and probed with the antibodies depicted. e Microscopy images showing the effect of MG132 treatment on the expression of GFP-SETD2 FL in the cell lines described. The scale bar is 1 mm
Autophagy and the ubiquitin–proteasome system (UPS) are the major pathways for protein degradation in mammalian cells [15]. To investigate whether autophagy plays a role in SETD2 turn-over, 293T cells expressing GFP-SETD2 FL were treated with increasing concentration of the lysosome inhibitor, chloroquine. Chloroquine treatment did not have an apparent effect on the GFP-SETD2 FL protein level (Additional file 1: Figure S1c).
To test whether SETD2 is targeted for degradation by the UPS, SETD2 FL expression was checked after treating the cells with the proteasome inhibitor MG132. Proteasome inhibition led to an increase in the accumulation of SETD2. This was confirmed by western blotting of whole-cell lysate of Halo-SETD2 FL-expressing cells as well as microscopy visualization of GFP-SETD2 FL expression (Fig. 1a, b). The addition of MG132 did not have a prominent effect on Halo or GFP expression (Additional file 1: Figure S1d, e). Also, RT-PCR did not reveal any change in the transcript abundance of both endogenous and exogenous SETD2 upon MG132 treatment, suggesting that the increased SETD2 protein abundance observed is due to protein stabilization (Fig. 1c). Importantly, an increase in the accumulation of the endogenous SETD2 was also observed upon MG132 treatment of WT 293T cells, implying that SETD2 degradation by the proteasome is not limited to the recombinant form of the protein (Fig. 1d). Degradation of SETD2 leads to the appearance of lower molecular weight bands on a denaturing gel. Hence, the specificity of the SETD2 antibody was validated (Additional file 1: Figure S2). Next, the expression of GFP-SETD2 FL was tested in HEPG2 and HELA cells. SETD2 behaved similarly in these cell lines. Very weak expression of GFP-SETD2 FL was observed which increased upon proteasome inhibition by MG132 treatment (Fig. 1e).
Our results suggest that the short half-life of SETD2 is due to UPS-mediated decay which is consistent with previous findings [16]. Also, this mechanism for SETD2 proteolysis is not cell line specific.
The removal of N-terminus region stabilizes SETD2
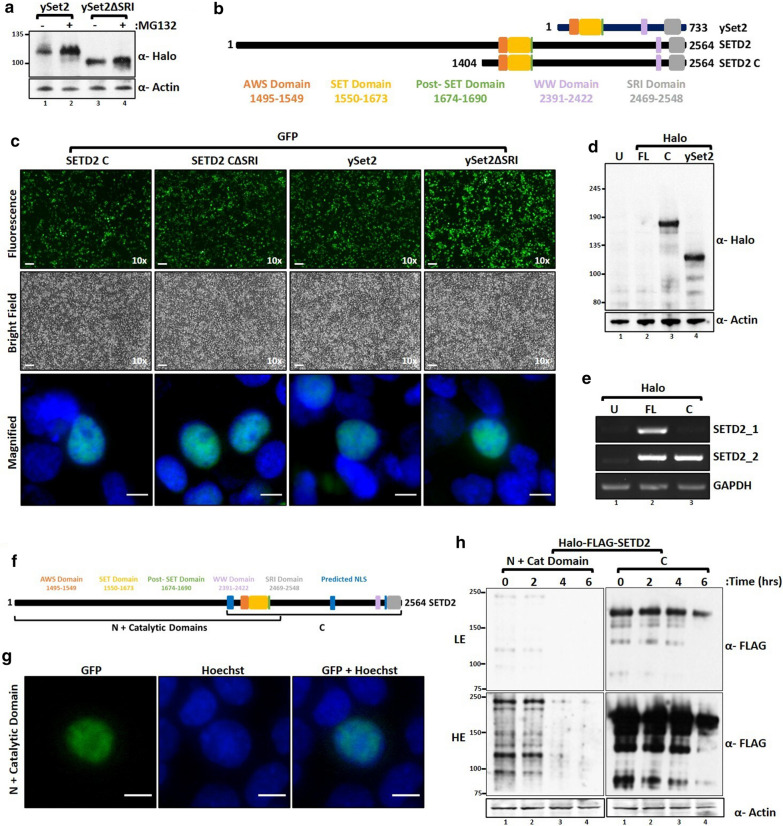
Previously, the expression of yeast Set2 (ySet2) has been used in human cells to rescue H3K36me3 [17]. Hence, we next investigated the expression of Halo-ySet2 in 293T cells. Western blotting of whole-cell lysate with an anti-Halo antibody revealed a robust expression of ySet2 (Fig. 2a). ySet2 is a well-characterized protein that is degraded by the proteasome in yeast [18]. The UPS and the architecture of the proteasome itself are conserved from yeast to mammals [19, 20]. Consistent with that, our data show that ySet2 responds to MG132 treatment in 293T cells (Fig. 2a). This suggests that ySet2 is targeted for degradation through the UPS in human cells too, although, its expression level is much higher than that of SETD2 FL. SETD2 has an N-terminal region which is absent in ySet2 (Fig. 2b). We speculated that the disparity in the expression between ySet2 and SETD2 FL could be due to the presence of this segment in SETD2.
Fig. 2.
Removal of the N-terminal region of SETD2 leads to its stabilization. a, d Western blot of whole-cell lysates probed with the depicted antibodies. Lysates of 293T cells expressing Halo-ySet2 constructs were prepared after 12 h of MG132 (10 µM) treatment. U-untransfected. b Cartoon illustrating the known domains of yeast Set2 (ySet2), SETD2, and the SETD2 C-terminus region that shares the conserved domains with ySet2. c Microscopy images showing the expression and localization of GFP-SETD2 C and ySet2. The scale bar is 1 mm for 10 × images and 10 µm for the magnified ones. e RNA was isolated from transfected cells described in d and RT-PCR was performed to check the SETD2 transcript levels. GAPDH was used as a normalization control. SETD2_1 primer pair binds to the N-terminal segment of SETD2 transcripts, whereas, SETD2_2 binds to the C-terminal region which is common in both SETD2 FL and C. f Cartoon illustrating the SETD2 segments used in the cycloheximide-chase experiment. g Microscopy images showing the expression and localization of the GFP-SETD2 N + Catalytic Domain of SETD2. The scale bar is 10 µm. h Western blot of whole-cell lysates probed with the depicted antibodies. Lysates of 293T cells expressing Halo-FLAG-SETD2 constructs were prepared after cycloheximide (10 µg/ml) treatment. The duration of the treatment is shown in the figure. HE- High Exposure, LE-Low Exposure
For this, we tested the expression level of a truncated mutant of SETD2, SETD2 C. Consistent with our hypothesis, microscopy of cells expressing the GFP-tagged proteins revealed that the expression level of SETD2 C was comparable to that of ySet2 (Fig. 2c). Analysis of whole-cell lysate with anti-Halo western blotting revealed that the expression of SETD2 C was markedly higher than SETD2 FL and was comparable to ySet2 (Fig. 2d). RT-PCR using primers specific for SETD2 FL and SETD2 C transcripts confirmed that the transcripts were produced robustly (Fig. 2e). Also, the transcript levels of SETD2 FL and SETD2 C were comparable.
To ascertain that the differences in protein levels observed were indeed due to the differences in protein half-lives, we performed a cycloheximide-chase experiment. For this, two overlapping segments of SETD2 were used (Fig. 2f). The catalytic domain of SETD2 was included in both the segments in case the SETD2 methyltransferase activity is important for its half-life. Microscopy with the GFP-tagged version of the protein revealed that the N-terminal + Catalytic domain of SETD2 was nuclear (Fig. 2g). Next, 293T cells expressing the Halo-FLAG-tagged N-terminal + Catalytic domain of SETD2 or SETD2 C were treated with cycloheximide to inhibit protein translation. Whole-cell extracts were prepared at different time-points and the expression level of SETD2 was scored by probing with an anti-FLAG antibody. The protein level of the N + Catalytic Domain segment was barely detectable 4-h post-treatment though SETD2 C still remains abundant (Fig. 2h). This suggests that the difference in protein half-lives that arises upon the removal of the N-terminal region of SETD2 is a significant contributor towards the greater accumulation of SETD2 C.
Collectively, the results suggest that the proteolytic instability of the full-length SETD2 might be a key contributor to the differences observed in protein abundance between SETD2 FL and SETD2 C.
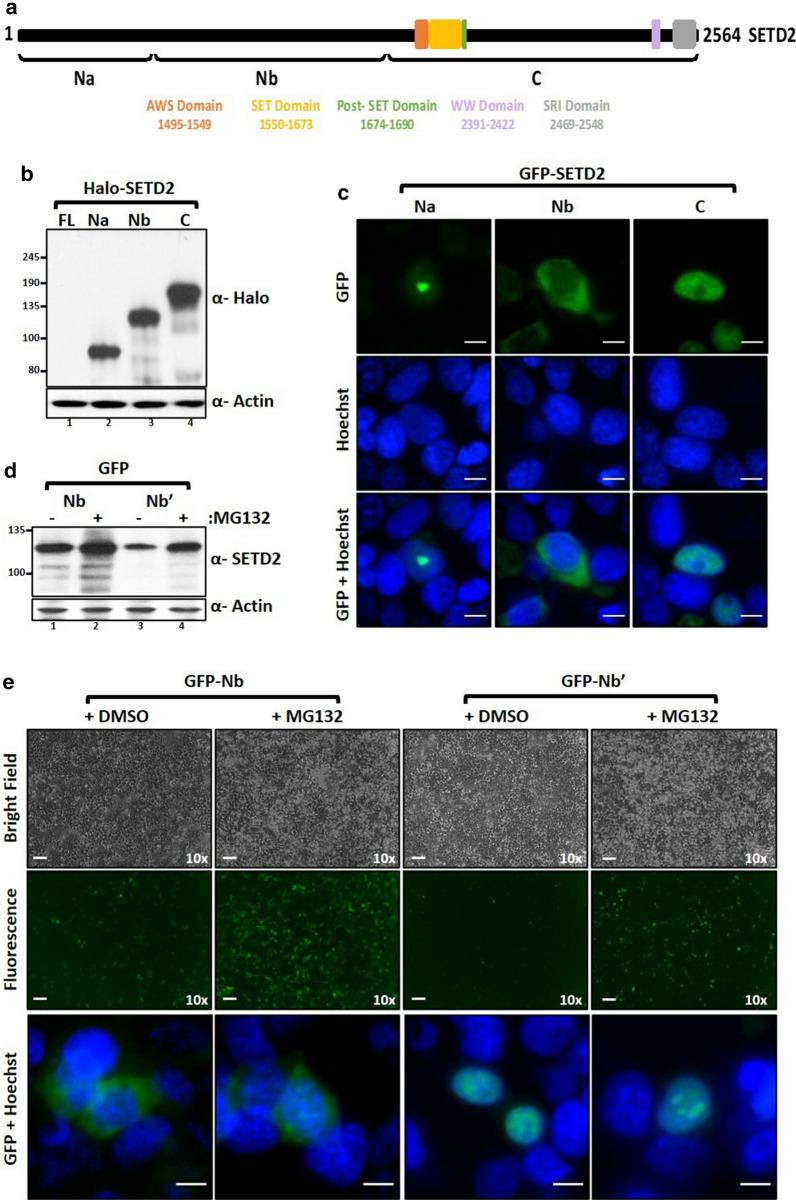
Unlike the full-length protein, SETD2 fragments express robustly
We wanted to test whether a specific stretch of the N-terminal region of SETD2 causes its robust degradation. To test that, we made sub-fragments of the N-terminal region, 1–503 (Na) and 504–1403 (Nb), and tested their expression (Fig. 3a). Strikingly, western blotting of whole-cell lysates with an anti-Halo antibody revealed that both the fragments expressed robustly in 293T cells with their expression level similar to SETD2 C; whereas, SETD2 FL could not be detected (Fig. 3b). Notably, microscopy analyses revealed that the fragment Nb was cytoplasmic, unlike Na and C which were nuclear (Fig. 3c). To test whether the localization of this fragment alters its stability, the c-myc nuclear localization signal (NLS) was added to GFP-Nb (Nb’) and the expression was checked. Western blotting of the whole-cell lysates revealed a reduced expression level of Nb’ as compared to Nb with continued sensitivity to MG132 treatment (Fig. 3d). Microscopy confirmed that the addition of NLS resulted in the nuclear translocation of Nb and also, reduced its expression level (Fig. 3e). Nevertheless, the expression of all the SETD2 fragments was very robust compared to SETD2 FL and continued to display sensitivity to MG132.
Fig. 3.
SETD2 fragments express robustly. a Cartoon illustrating the fragments of SETD2 along with its known domains. b Western blot of whole-cell lysates of 293T cells expressing Halo-SETD2 constructs probed with the depicted antibodies. c, e Microscopy images showing the localization of GFP-SETD2 fragments. The scale bar is 1 mm for 10 × images and 10 µm for the magnified ones. See the text for more details. d Western blot of whole-cell lysates of cells shown in e probed with the depicted antibodies
From these experiments, no specific region emerged in the SETD2 protein that is particularly targeted for UPS-mediated decay.
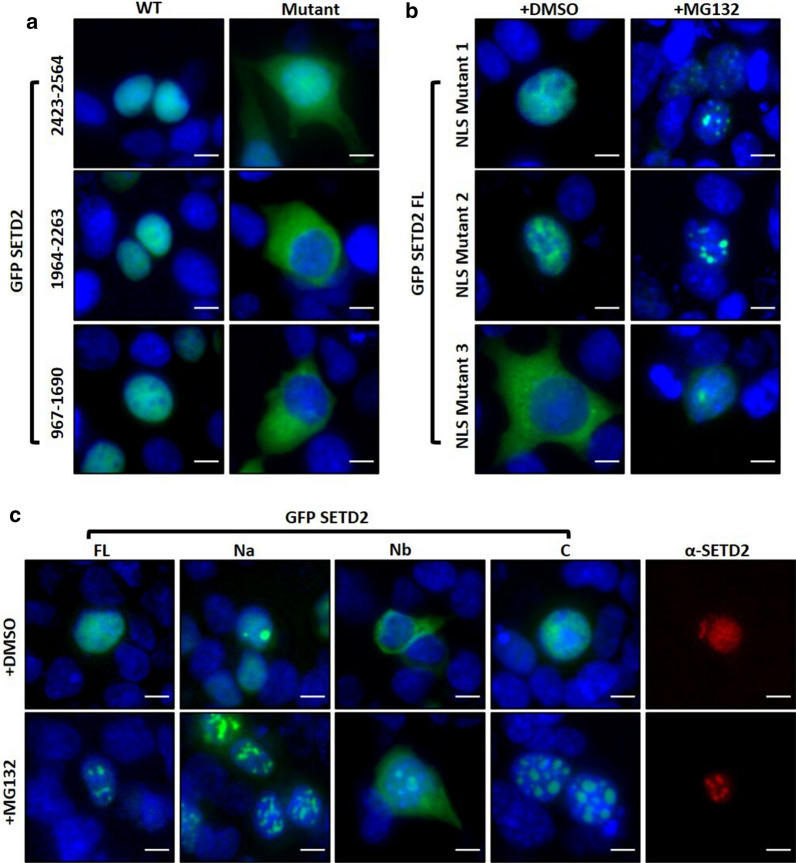
SETD2 forms nuclear puncta
Microscopy revealed that GFP-SETD2 Na is nuclear and formed puncta (Fig. 3c). This was surprising because this segment lacks a putative NLS with a significant score as per NLS mapper prediction (https://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) [21]. We decided to characterize the NLS of SETD2 to better understand the unexpected localization of the SETD2 fragments. To this end, the localization of a series of GFP-SETD2 fragments was checked using fluorescence microscopy (Additional file 1: Figure S3). Of all the SETD2 fragments tested, the data revealed the presence of putative NLS in three fragments of SETD2: 967–1690, 1964–2263, and 2423–2564 (Fig. 4a). This was consistent with the NLS mapper prediction that revealed the presence of NLS in each of these segments (Additional file 1: Figure S3). To validate these NLS, site-directed mutagenesis was performed to mutate the lysine (K) and arginine (R) residues to alanine (A) and the disruption of the nuclear localization of the mutated GFP-SETD2 fragments was confirmed by microscopy (Fig. 4a, Additional file 1: Figure S3b). Further, to validate that the full-length SETD2 has only these three NLSs, site-directed mutagenesis was performed to mutate these NLS one by one (Fig. 4b). The cytoplasmic localization of the GFP-SETD2 FL mutant, in which all the three NLS were disrupted, confirmed that SETD2 has three NLS (Fig. 4b). Importantly, this also shows that the SETD2 fragment Na forms nuclear puncta without an NLS.
Fig. 4.
SETD2 forms nuclear puncta. a, b Microscopy images showing localization of GFP-SETD2 fragments that have putative NLS and the NLS mutants. See the text for more details. c Microscopy images showing the localization of SETD2 and its fragments. The scale bar is 10 µm
To test whether the full-length SETD2 protein also behaves similarly, the localization of NLS mutants of SETD2 was tested upon MG132 treatment. Strikingly, all the mutants exhibited the formation of nuclear puncta (Fig. 4b). To characterize the aggregate-prone tendency of SETD2 further, 293T cells expressing GFP-SETD2 truncations (described in Fig. 2) were observed under the microscope with or without MG132 treatment. Remarkably, all the SETD2 fragments responded to the treatment with MG132 and demonstrated a marked increase in the formation of puncta (Fig. 4c). Notably, the completely cytoplasmic fragment Nb showed a pan-cellular distribution with a tendency to form puncta upon MG132 treatment similar to SETD2 FL NLS Mutant 3. Furthermore, puncta were also formed by WT GFP-SETD2 FL protein suggesting that it might not be an artifact caused due to protein misfolding resulting from the truncations or mutations (Fig. 4c). To test whether the endogenous SETD2 behaves similarly, immunofluorescence of 293T cells was performed with an anti-SETD2 antibody. Importantly, proteasomal inhibition caused speckle-like staining, revealing that endogenous SETD2 also forms puncta (Fig. 4c). This observation, together with the fact that despite very weak expression, SETD2 FL is aggregate prone suggests that aggregation is an intrinsic property of the SETD2 protein that is exacerbated by an increased protein abundance.
SETD2 forms ubiquitinated insoluble aggregates
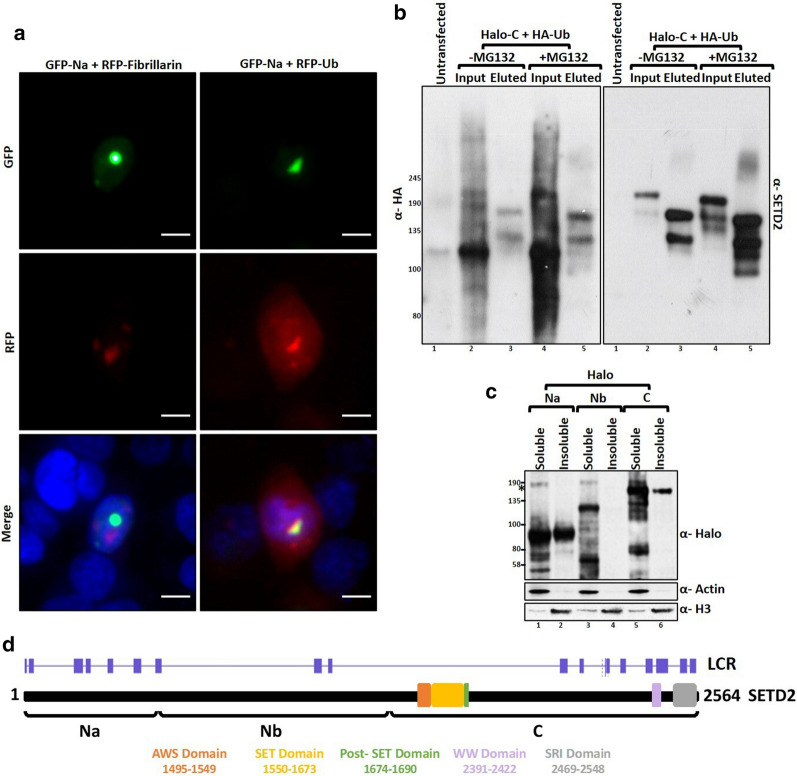
Exogenously expressed as well as the endogenous SETD2 formed puncta, especially upon MG132 treatment, that are reminiscent of inclusion bodies formed by aggregated proteins. The most striking pattern was observed with the fragment SETD2 Na that formed distinct puncta even in the absence of proteasome inhibition. As aggregates are often ubiquitinated, to confirm that the puncta formed by SETD2 are aggregated structures, the colocalization of RFP-ubiquitin with GFP- SETD2 Na was tested. Clear colocalization was observed suggesting that SETD2 Na puncta are indeed ubiquitinated aggregate structures (Fig. 5a). GFP-SETD2 Na did not colocalize with RFP-Fibrillarin indicating that the puncta were not nucleolar (Fig. 5a). To biochemically substantiate that SETD2 is ubiquitinated, Halo-C was affinity purified from HEK293T cells co-expressing HA-ubiquitin with or without MG132 treatment. The purified proteins were then resolved on a gel and analyzed for the presence of ubiquitination. Western blotting with an anti-HA antibody revealed that SETD2 C was indeed ubiquitinated (Fig. 5b).
Fig. 5.
SETD2 forms insoluble ubiquitinated aggregates. a Microscopy images showing the colocalization of GFP-SETD2 Na fragment with RFP-Fibrillarin or RFP-Ub. The scale bar is 10 µm. b Halo purification was performed from extracts of 293T cells co-expressing Halo-C and HA-Ub. Eluted and 0.5% of Input samples were resolved on a gel and probed with the depicted antibodies. c Soluble and insoluble fractions were separated from 293T cells expressing Halo-SETD2 fragments, resolved on a gel, and probed with the depicted antibodies. *Non-specific. d Cartoon depicting the predicted low-complexity regions (LCRs) in SETD2. The prediction was performed using the webserver at https://repeat.biol.ucy.ac.cy/fgb2/gbrowse/swissprot/
To confirm further that SETD2 forms aggregates, we checked the solubility of Halo-SETD2 fragments Na, Nb, and C. 293T cells expressing the Halo-SETD2 fragments were lysed, their soluble and insoluble fractions separated and analyzed by western blotting with an anti-Halo antibody. Correlating with the microscopy observations, SETD2 Na that formed spontaneous puncta was highly insoluble; C was insoluble to a lesser extent (Fig. 5c). The segment Nb was soluble (Fig. 5c). Interestingly, the data show that the different regions of the same protein have different aggregation propensity under similar experimental conditions and are not merely a consequence of expression from a strong CMV promoter. Furthermore, the presence of low-complexity regions (LCR) in a protein is linked to its aggregation propensity [22–24]. Hence, we analyzed the SETD2 protein sequence for the presence of LCR. Strikingly, the solubility profile observed for the SETD2 fragments correlated very well with the LCR distribution of the protein (Fig. 5d).
Collectively, our data show that SETD2 forms aggregated ubiquitinated puncta (discussed in the Discussion section).
Removal of the N-terminal region of SETD2 leads to a marked increase in global H3K36me3 levels
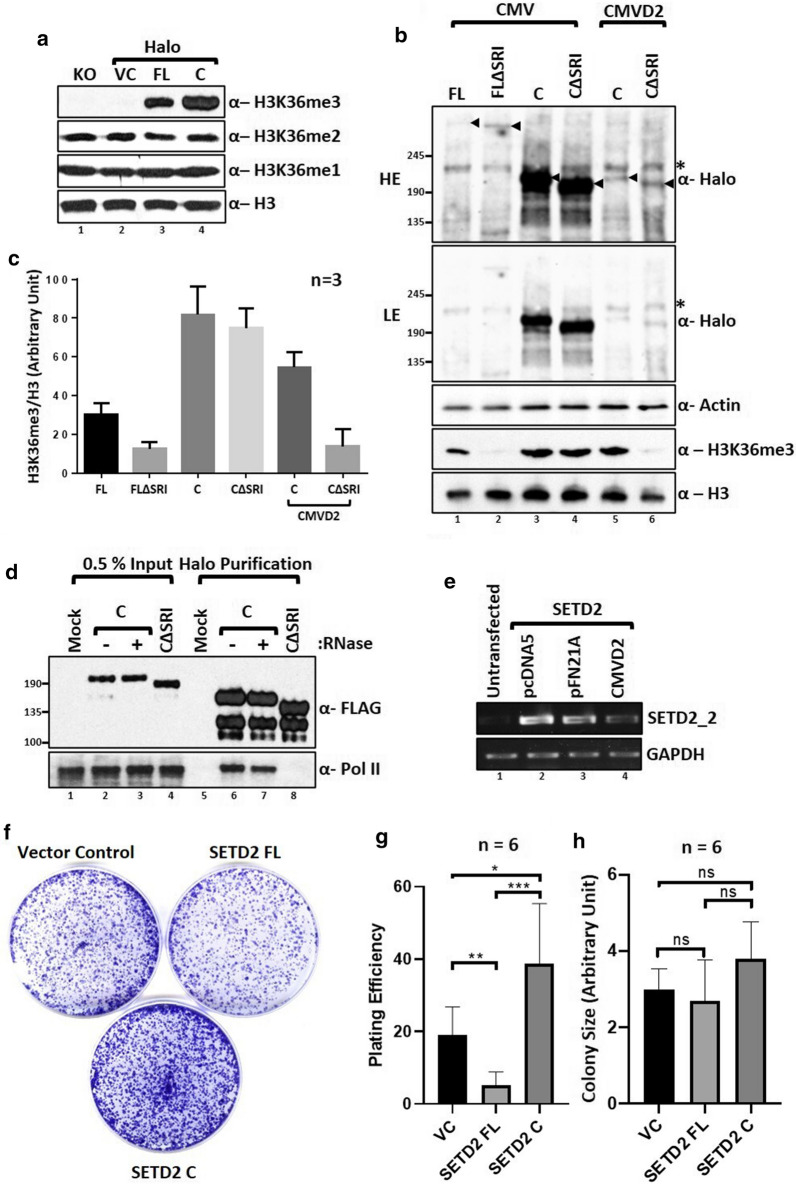
We found that the removal of the N-terminal region of SETD2 leads to the stabilization of the remaining portion that shares conserved domains with ySet2. Next, we investigated whether the removal of the N-terminal region affects the catalytic activity of SETD2. To check the activity of the exogenously introduced SETD2, setd2Δ (KO) 293T cells were used. Consistent with the role of SETD2 as the sole H3K36me3 depositor in humans, in the KO cells the H3K36me3 mark was not detected in the whole-cell lysates by immunoblotting (Fig. 6a). Next, constructs to express Halo-tagged SETD2 FL or SETD2 C were introduced in the KO cells by transfection. 72 h post-transfection, whole-cell lysates were prepared and analyzed by western blotting. The expression of the empty vector (VC) did not rescue H3K36me3 as expected (Fig. 6a). Strikingly, the expression of SETD2 C in KO cells led to a marked increase in the H3K36me3 level as compared to the rescue with SETD2 FL (Fig. 6a). The other two H3K36 methyl marks, H3K36me1, and H3K36me2, largely remained unchanged (Fig. 6a).
Fig. 6.
SETD2 has reduced Pol II dependency at high expression levels. a Western blot of whole-cell lysates probed with the depicted antibodies. setd2Δ 293T (KO) cells were transfected with Halo-vector control (VC), SETD2 full-length (FL) or SETD2 C (C), and the lysates were prepared 72 h post-transfection. b Western blot of whole-cell lysates of 293T cells expressing Halo-SETD2 constructs, under the control of either CMV or CMVD2, probed with the depicted antibodies. The expected bands are marked by arrows. *Non-specific. c Bar graph showing H3 normalized H3K36me3 signal intensity of data depicted in b. The data plotted are from three independent biological replicates. d Halo purification was performed from extracts of 293T cells expressing Halo-SETD2 C. Input and eluted samples were resolved on a gel and probed with the depicted antibodies. e RNA was isolated from 293T cells expressing SETD2 from different vectors and RT-PCR was performed to check transcript levels. GAPDH was used as a normalization control. f Plates showing the results of the colony formation assay to test the effect of introducing full-length SETD2 (FL) or SETD2 C under the control of a CMV promoter on the cell proliferation of setd2Δ 293T cells. g, h Quantification of the number and size of colonies obtained in the colony formation assay (n = 6). Unpaired t test was used for statistical analysis. P value is < 0.05
Thus, the removal of the N-terminal segment leads to a marked increase in the global H3K36me3 level. Also, these experiments demonstrate that in the absence of the N-terminal region, SETD2 retains its histone methyltransferase activity.
At high cellular levels, SETD2 has a reduced RNA pol II dependency for H3K36me3 deposition
Studies conducted in yeast have revealed that the deposition of the H3K36me3 mark is strictly dependent on the ySet2–Pol II association [12]. We were curious whether the marked increase in the global H3K36me3 level upon SETD2 C expression happens in an RNA Pol II-dependent manner.
To test this, Halo-SETD2 constructs without the SRI domain were introduced in setd2Δ 293T cells. Similar to the findings for ySet2 in yeast, removal of the SRI domain from full-length SETD2 protein (FLΔSRI) led to a marked decrease in H3K36me3 levels as compared to the FL (Fig. 6b, c). Strikingly though, removal of the SRI domain from SETD2 C (CΔSRI) had a very marginal effect on the H3K36me3 levels (Fig. 6b, c). To confirm that the removal of the SRI domain leads to the abolishment of SETD2–Pol II interaction, Halo-FLAG-SETD2 C and Halo-FLAG-SETD2 CΔSRI were affinity purified from 293T extracts using Halo ligand-conjugated magnetic resin. Elution of proteins purified using this technique involves cleaving the Halo tag with TEV protease, leaving the FLAG epitope which can be detected on the eluted bait by immunoblotting (Fig. 6d). Immunoblotting with an anti-Pol II antibody confirmed that the deletion of the SRI domain from SETD2 leads to the abolishment of SETD2–Pol II interaction (Fig. 6d).
We wondered whether the decreased dependency on Pol II interaction for the H3K36me3 activity of SETD2 C is due to the loss of a possible autoinhibition by the N-terminal region of SETD2 or is due to the increased expression of SETD2 C fragment as compared to the full-length protein. To address these possibilities, Halo-SETD2 C constructs under the control of the CMVD2 promoter were introduced in setd2Δ cells. CMVD2 promoter is a truncated form of CMV and exhibits a much weaker transcription activity. The weaker activity of the CMVD2 promoter was confirmed by RT-PCR (Fig. 6e). The reduced expression of SETD2 C under the regulation of CMVD2 promoter was verified by analyzing whole-cell lysate with an anti-Halo antibody (Fig. 6b). Notably, analysis of H3K36me3 revealed that the RNA Pol II dependency of SETD2 C was restored at a reduced expression level as SETD2 CΔSRI did not exhibit much activity when expressed using the CMVD2 promoter (Fig. 6b, c).
SETD2 is considered a tumor suppressor [25]. With this in mind, we tested the effect of SETD2 expression on cell proliferation by introducing full-length SETD2 (FL) or SETD2 C under the control of the CMV promoter in setd2Δ 293T cells. Consistent with its association with tumor-suppressive phenotype, the reintroduction of SETD2 FL led to a decrease in cell proliferation (Fig. 6f, g). Strikingly, expression of SETD2 C led to a significant increase in the proliferation of cells as judged by the results of the colony formation assay (Fig. 6f, g). An increase in the colony size was also observed; however, the differences were not statistically significant (Fig. 6h).
We conclude that high cellular levels of SETD2 result in an RNA Pol II-independent H3K36me3 deposition and an increase in cell proliferation.
Discussion
In recent years, many reports have highlighted the adverse effect of the loss of H3K36me3 that occurs upon SETD2 deletion. Here, we show that the other end of the spectrum can also be detrimental as an excess of SETD2 can lead to inadvertent consequences. Previously, SETD2 has been reported to be regulated by the proteasome [16]. We reveal that the absence of SETD2 proteolysis results in a Pol II-independent H3K36me3 deposition and protein aggregation. Our work illustrates the importance of the N-terminal segment of SETD2, which has been a mystery, in maintaining the requisite intracellular amount of the protein.
Regulation of SETD2 half-life is important for regulating its function
Altered protein half-life can lead to abnormal development and diseases such as cancer and neurodegeneration [26]. Considering the role of H3K36me3 in a variety of important cellular processes, it is reasonable that regulating the activity of the methyltransferase responsible for the deposition of this mark is important. The previously uncharacterized N-terminal region, which is absent in ySet2, plays an important role in SETD2 regulation. The gain or loss of protein segments may be an important contributor to the degradation rate of proteins during evolution [27]. The differences in half-lives between homologs might be needed to adjust for the differences in the mechanism of deposition of H3K36 methylation. For instance, unlike in yeast where ySet2 performs all three states of methylation of H3K36, SETD2 does not appear to be majorly responsible for me1 and me2 deposition. Therefore, even with a shorter half-life, SETD2 might be able to do the required H3K36me3 deposition. Some evidence for this is provided by studies on human cancers which show that the total H3K36me3 levels are not significantly impacted by a monoallelic loss of SETD2 [28, 29].
SETD2 over-abundance might have inadvertent consequences
We found that SETD2 is an inherently aggregate-prone protein and its aggregation tendency is positively correlated with the presence of LCRs. Aggregation induced by LCRs can be detrimental and there are more than 20 genetic disorders linked to the expansion of trinucleotide repeats within coding sequences that generates LCRs. The uncontrolled expansion of CAG triplets that leads to polyQ tracts is associated with Huntington disease and several ataxias [30]. Overaccumulation of SETD2 can lead to the formation of stable and insoluble protein aggregates impeding the normal function of other proteins. SETD2 aggregation might lead to the co-aggregation of other proteins leading to their inactivation and proteotoxic stress. Ubiquitinated aggregates like the ones formed by SETD2 can directly inhibit or clog proteasomes [31, 32]. As for aggregation of a protein a concentration threshold is required, possibly the robust degradation mechanism in cells ensures that SETD2 is kept at levels that maintain its solubility and activity. Interestingly, SETD2, aka, HYPB were initially identified in a screen to find interactors of the aggregate-prone protein Huntingtin (Htt) [33]. It is possible that the reported Huntingtin–SETD2 interaction was due to the aggregation propensities of these proteins. Interestingly, the N-terminus of SETD2 (SETD2 Na) behaves very similarly to the polyQ containing N-terminus of Huntingtin protein. Like mutant Htt, SETD2 Na localizes to the nucleus in the absence of an NLS and forms spontaneous puncta characteristic of aggregated proteins [34]. The nucleoplasm promotes the formation of such aberrant and insoluble protein aggregates due to the strong crowding forces from highly concentrated macromolecules (approximately 100 mg/ml) [35].
Instead of impeding cellular activities, aggregates can also play functionally important roles. Aggregates formed by SETD2 may indicate the formation of functional bodies such as the RNA granules. RNA granules are micron-sized membrane-less entities formed by phase separation [36]. RNA granules are complex structures that contain ribosomal subunits, translation factors, scaffold, and RNA-binding proteins. These factors control the stability, localization, and translation of RNA cargo [37]. Notably, RNA Pol II also forms granules [38]. Considering the established interaction between SETD2 and Pol II, there might be an interplay between the granules formed by SETD2 and RNA Pol II. Strikingly, the expression of SETD2 C led to an increase in the proliferation of cells. This is consistent with the previous report that SETD2 depletion caused decreased cell proliferation in 293T and HKC8 cells [9]. Increased accumulation of SETD2 might cause it to phase separate, which may lead to a stabilization of transcripts which, in turn, might induce signaling pathways leading to cell proliferation. This possibility along with the likelihood of SETD2 forming part of RNA granules to regulate transcription and translation will be interesting to investigate in the future.
Pol II association is required for enhancing SETD2 activity
Our data show that when the expression level of SETD2 is high, it has a reduced dependency on RNA Pol II association for H3K36me3 deposition. Possibly, the interaction with Pol II is required for the activation of SETD2 enzymatic activity. When present in its normal cellular amounts, SETD2 is not active enough to lead to a robust H3K36me3 deposition without the Pol II association. Pol II association enhances SETD2 activity and hence, although SETD2 FL protein is barely detectable, the rescue of H3K36me3 can be readily seen in the setd2Δ cells. At high abundance, despite its low activity, a robust H3K36me3 deposition is seen likely due to the increased copy number of the SETD2 protein in cells. Recent studies in yeast have also challenged the notion that the Pol II association is required for chromatin recruitment of Set2 [39]. The study found that the engagement of Set2 and Pol II through the SRI domain is instead required for the activation of Set2. Recent findings highlight the importance of the regulation of SETD2 activity in cells. This report shows that H3.3S31ph can enhance H3K36me3 deposition by augmenting SETD2 activity, leading to rapid, high-level expression of stimulation-induced genes [40]. At a high cellular level, SETD2′s dependency on such factors might be reduced which can lead to unintended transcriptional changes. By keeping the SETD2 expression level low and thus, maintaining its requirement for activation by factors such as H3.3S31ph and Pol II, the cells can regulate H3K36me3 deposition and avoid unintended transcriptional changes. Additionally, the Pol II-independent deposition of H3K36me3 can result in the redistribution of the H3K36me3 mark resulting in the mistargeting of important epigenetic regulators. The PWWP domain-containing proteins DNMT3a, MutSα, and MORF depend on H3K36me3 for proper recruitment [4, 5, 17, 41]. It is important to note that besides histone H3, SETD2 also has non-histone targets like tubulin, the methylation of which is important for the metaphase transition [42]. Hence, the consequences of an increase in SETD2 abundance might not be limited to changes in histone methylation.
Conclusion
SETD2 is robustly degraded by the proteasome and the removal of its N-terminal region leads to its stabilization. We also discovered that SETD2 is an aggregate-prone protein. We characterized the nuclear localization signals (NLS) in SETD2 and found that strikingly, SETD2 can form nuclear puncta even without an intact NLS. This behavior is very similar to that exhibited by well-characterized aggregate-prone proteins like the mutant huntingtin. Importantly, it suggests that the robust degradation of SETD2 might be a mechanism to prevent inadvertent protein aggregation mediated by SETD2 and of SETD2 itself. This can also be a mechanism to regulate the possible liquid phase transition of SETD2 that needs to be investigated in future studies. Importantly, an increased accumulation of SETD2 also results in a reduced RNA Pol II dependency for H3K36me3 deposition, which is very uncharacteristic. SETD2 has a long N-terminal region that is missing in its yeast homolog Set2. Our work illustrates the importance of this N-terminal segment of SETD2 in maintaining the requisite intracellular amount of the protein.
Methods
Plasmids and oligos
SETD2-HaloTag® human ORF in pFN21A was procured from Promega. Deletion mutants of SETD2 were constructed by PCR (Phusion polymerase, NEB) using full-length SETD2 as a template and individual fragments were cloned. All constructs generated were confirmed by sequencing. SETD2-GFP, mRFP-Ub, pLenti puro HA-Ubiquitin, pTagRFP-C1-Fibrillarin and pCDNA3-ySet2 were procured from Addgene. siRNAs were purchased from Dharmacon and oligonucleotides were procured from Integrated DNA Technologies (IDT).
Cell line maintenance and drug treatment
The cell lines used in this study (HEK293T, HEPG2 and, HELA) were procured from ATCC. Cells were maintained in DMEM supplemented with 10% FBS and 2-mM l-glutamine at 37 °C with 5% CO2. MG132 (Sigma) was added at a final concentration of 10 μM for 12 h. Chloroquine (Sigma) treatment was done as indicated in the text. Cycloheximide (Sigma) was added at a final concentration of 10 μg/ml. Transfections were performed at cell confluency of 40% using Fugene HD (Promega) using a ratio of 1:4 of the plasmid (µg) to transfection reagent (µl).
Colony Formation Assay
The cells (n = 4000) were plated in triplicate in 60-mm tissue culture plates and were allowed to grow as a monolayer for 14 days. Cells were incubated in the complete culture medium, with media changes every 2–3 days. After 14 days, the cells were fixed with 4% paraformaldehyde for 1 h. The colonies were stained with 0.5% crystal violet (0.5% in 70% ethanol) for 1 h at room temperature, rinsed and air-dried. Images were captured and the quantification of the number and size of the colonies was done using ImageJ.
Isolation of total RNA and PCR
Total RNA was extracted from cells as per the manufacturer’s (Qiagen) instructions. It was further treated with DNaseI (NEB) for 30 min at 72 °C to degrade any possible DNA contamination. RNA (2 μg) was subjected to reverse transcription using the QScript cDNA synthesis mix according to the manufacturer’s instructions. cDNAs were then amplified with the corresponding gene-specific primer sets. For RT-PCR, PCR was conducted for 24 cycles using the condition of 30 s at 94 °C, 30 s at 60 °C and 30 s at 72 °C. The PCR products were analyzed on a 1% agarose gels containing 0.5 μg/ml ethidium bromide. The sequence of oligos is in Additional file 1: Figure S4.
Histone isolation and immunoblot analysis
Histones were isolated and analyzed as described previously [43]. For immunoblotting, histones were resolved on 15% SDS-polyacrylamide gel, transferred to PVDF membrane and probed with antibodies. Signals were detected using the ECL plus detection kit (ThermoFisher).
Antibodies
H3K36me3 (CST, 4909S), H3K36me2 (Active Motif, 39255), H3K36me1 (Abcam, ab9048), H3 (CST, 9715S), Halo (Promega, G9211), β-actin (Abcam, ab8224), SETD2 (Abclonal, A3194), FLAG (Sigma-Aldrich, A8592), Pol II (Abcam, ab5095).
Cell Fractionation
To prepare soluble and insoluble extracts, 293T cells were washed with 1xPBS, collected by centrifugation, and resuspended in lysis buffer (50-mM Tris, pH 7.5, 350-mM NaCl, 1% Triton-X 100, 0.1% Na-deoxycholate and a protease inhibitor mix). The lysed cells were centrifuged at 13,000 rpm for 20 min. The supernatant was collected as the soluble fraction. The pellet was washed with lysis buffer containing 600-mM NaCl). The remaining insoluble pellet following another centrifugation was resuspended in Laemmli buffer (Biorad) and solubilized by sonication on ice.
Affinity purification
293T cells expressing the protein of interest were harvested in 1xPBS and collected by centrifugation. The cells were lysed by resuspending in lysis buffer (50-mM Tris, pH 7.5, 150-mM NaCl, 1% Triton-X 100, 0.1% Na-deoxycholate, and a protease inhibitor cocktail). The lysed cells were centrifuged at 13,000 rpm for 20 min. The supernatant was collected and diluted 1:3 by adding dilution buffer (1× PBS, pH 7.5 with 1-mM DTT and 0.005% NP-40). The diluted lysate was added to 100 µl of pre-equilibrated Magne® HaloTag® Beads (Promega, G7282) and incubated overnight on a rotator at 4 °C. The beads were then washed with wash buffer (50-mM Tris–HCL, pH 7.5, 300-mM NaCl, 0.005% NP-40, and 1-mM DTT. 2 µl AcTEV (ThermoFisher, 12575015) protease was used per 100 µl of elution buffer.
Immunofluorescence
293T cells were plated onto glass coverslips in a 6-well plate. Cells were washed with 1× PBS and fixed in 4% paraformaldehyde for 20 min at 37 °C. Cells were then washed three times with cold 1 × PBS and permeabilized for 5 min with 1× PBS containing 0.2% Triton-X 100. Permeabilized cells were then blocked for 30 min with blocking buffer (3% BSA and 0.1% Triton-X in 1× PBS). Cells were stained with primary antibodies against SETD2 (1:1,000; Abclonal) for 1 h at room temperature. A secondary antibody conjugated with AlexaFluor 568 was applied for 1 h at room temperature. All the images depicted in the same panel were captured using the same settings.
Supplementary information
Acknowledgements
The authors are grateful to Dr. Kimryn Rathmell, Vanderbilt Institute for Infection, Immunology and Inflammation from providing SETD2 knock out 293T cells. The authors would like to thank the members of the Workman lab for their critical suggestions to improve the manuscript.
Abbreviations
- PTMs
Post-translational modifications
- SRI
Set2–Rpb1 Interaction
- AWS
Associated with SET
- SET
Su(var)3-9, Enhancer-of-zeste and Trithorax
- UPS
Ubiquitin–proteasome system
- NLS
Nuclear localization signal
- LCR
Low-complexity regions
Authors’ contributions
SB conceptualized the work, designed and performed the experiments. SB wrote the manuscript. JLW conceived the idea of the work, provided supervision, acquired funding, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by funding from the National Institute of General Medical Sciences (Grant No. R35GM118068) and the Stowers Institute for Medical Research to Jerry L Workman.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13072-020-00362-8.
References
- 1.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 2.Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, Liu Y, Qiu T, Jiang M, Pan Q, Han Y, Wang X, Li Q, Qin J. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest. 2017;127:3375–3391. doi: 10.1172/JCI94292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter AC, Humphrey TC. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7:2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang YC, Park IY, Terzo EA, Tripathi DN, Mason FM, Fahey CC, Karki M, Shuster CB, Sohn BH, Chowdhury P, Powell RT, Ohi R, Tsai YS, de Cubas AA, Khan A, Davis IJ, Strahl BD, Parker JS, Dere R, Walker CL, Rathmell WK. SETD2 haploinsufficiency for microtubule methylation is an early driver of genomic instability in renal cell carcinoma. Cancer Res. 2018;78:3135–3146. doi: 10.1158/0008-5472.CAN-17-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su X, Zhang J, Mouawad R, Comperat E, Roupret M, Allanic F, Parra J, Bitker MO, Thompson EJ, Gowrishankar B, Houldsworth J, Weinstein JN, Tost J, Broom BM, Khayat D, Spano JP, Tannir NM, Malouf GG. NSD1 inactivation and SETD2 mutation drive a convergence toward loss of function of H3K36 writers in clear cell renal cell carcinomas. Cancer Res. 2017;77:4835–4845. doi: 10.1158/0008-5472.CAN-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Kluiver J, Osinga J, Westers H, van Werkhoven MB, Seelen MA, Sijmons RH, van den Berg A, Kok K. Functional studies on primary tubular epithelial cells indicate a tumor suppressor role of SETD2 in Clear cell renal cell carcinoma. Neoplasia. 2016;18:339–346. doi: 10.1016/j.neo.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/MCB.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki S, Kato H, Suzuki Y, Chikashige Y, Hiraoka Y, Kimura H, Nagao K, Obuse C, Takahata S, Murakami Y. Histone H3K36 trimethylation is essential for multiple silencing mechanisms in fission yeast. Nucleic Acids Res. 2016;44:4147–4162. doi: 10.1093/nar/gkw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDaniel SL, Strahl BD. Shaping the cellular landscape with Set2/SETD2 methylation. Cell Mol Life Sci. 2017;74:3317–3334. doi: 10.1007/s00018-017-2517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker KE, Fahey CC, Shinsky SA, Chiang YJ, DiFiore JV, Jha DK, Vo AH, Shavit JA, Davis IJ, Strahl BD, Rathmell WK. Structure/function analysis of recurrent mutations in SETD2 protein reveals a critical and conserved role for a set domain residue in maintaining protein stability and histone H3 Lys-36 trimethylation. J Biol Chem. 2016;291:21283–21295. doi: 10.1074/jbc.M116.739375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper GM. The cell: a molecular approach. The development and causes of cancer. 2. Sunderland: Sinauer Associates; 2000. [Google Scholar]
- 16.Zhu K, Lei PJ, Ju LG, Wang X, Huang K, Yang B, Shao C, Zhu Y, Wei G, Fu XD, Li L, Wu M. SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. Nucleic Acids Res. 2017;45:92–105. doi: 10.1093/nar/gkw814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. J Biol Chem. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Fonseca PC, He J, Morris EP. Molecular model of the human 26S proteasome. Mol Cell. 2012;46:54–66. doi: 10.1016/j.molcel.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 20.Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci USA. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cascarina SM, Elder MR, Ross ED. Atypical structural tendencies among low-complexity domains in the Protein Data Bank proteome. PLoS Comput Biol. 2020;16:e1007487. doi: 10.1371/journal.pcbi.1007487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grignaschi E, Cereghetti G, Grigolato F, Kopp MRG, Caimi S, Faltova L, Saad S, Peter M, Arosio P. A hydrophobic low-complexity region regulates aggregation of the yeast pyruvate kinase Cdc19 into amyloid-like aggregates in vitro. J Biol Chem. 2018;293:11424–11432. doi: 10.1074/jbc.RA117.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murthy AC, Dignon GL, Kan Y, Zerze GH, Parekh SH, Mittal J, Fawzi NL. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat Struct Mol Biol. 2019;26:637–648. doi: 10.1038/s41594-019-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget. 2016;7:50719–50734. doi: 10.18632/oncotarget.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Cell Death Differ. 2005;12:1178–1190. doi: 10.1038/sj.cdd.4401692. [DOI] [PubMed] [Google Scholar]
- 27.Lee R, Lang B, Kruse K, Gsponer J, Sánchez de Groot N, Huynen MA, Matouschek A, Fuxreiter M, Babu MM. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell reports. 2014;8:1832–1844. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho TH, Park IY, Zhao H, Tong P, Champion MD, Yan H, Monzon FA, Hoang A, Tamboli P, Parker AS, Joseph RW, Qiao W, Dykema K, Tannir NM, Castle EP, Nunez-Nateras R, Teh BT, Wang J, Walker CL, Hung MC, Jonasch E. High-resolution profiling of histone h3 lysine 36 trimethylation in metastatic renal cell carcinoma. Oncogene. 2016;35:1565–1574. doi: 10.1038/onc.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberti A, Dobay MP, Bisig B, Vallois D, Boéchat C, Lanitis E, Bouchindhomme B, Parrens M-C, Bossard C, Quintanilla-Martinez L, Missiaglia E, Gaulard P, de Leval L. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat Commun. 2016;7:12602–12602. doi: 10.1038/ncomms12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 31.Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito RR. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J Cell Biol. 2012;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nonaka T, Hasegawa M. A Cellular Model To Monitor Proteasome Dysfunction by α-synuclein. Biochemistry. 2009;48:8014–8022. doi: 10.1021/bi900619j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 34.Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, Dawson VL, Dawson TM, Ross CA. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 35.Mikecz A. Pathology and function of nuclear amyloid. Protein homeostasis matters. Nucleus. 2014;5:311–317. doi: 10.4161/nucl.29404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidibé H, Vande VC. RNA granules and their role in neurodegenerative diseases. The biology of mRNA: structure and function. In: Oeffinger M, Zenklusen D, editors. The biology of mRNA: structure and function. Cham: Springer International Publishing; 2019. pp. 195–245. [Google Scholar]
- 37.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson VF, Victor RA, Morera AA, Moinpour M, Liu MN, Kisiel CC, Pickrel K, Springhower CE, Schwartz JC. Transcription-dependent formation of nuclear granules containing FUS and RNA Pol II. Biochemistry. 2018;57:7021–7032. doi: 10.1021/acs.biochem.8b01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Niu Y, Li B. Balancing acts of SRI and an auto-inhibitory domain specify Set2 function at transcribed chromatin. Nucleic Acids Res. 2015;43:4881–4892. doi: 10.1093/nar/gkv393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armache A, Yang S, MartínezdePaz A, Robbins LE, Durmaz C, Cheong JQ, Ravishankar A, Daman AW, Ahimovic DJ, Klevorn T, Yue Y, Arslan T, Lin S, Panchenko T, Hrit J, Wang M, Thudium S, Garcia BA, Korb E, Armache KJ, Rothbart SB, Hake SB, Allis CD, Li H, Josefowicz SZ. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature. 2020;583:852–857. doi: 10.1038/s41586-020-2533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Gu L, Li G-M. H3K36me3-mediated mismatch repair preferentially protects actively transcribed genes from mutation. J Biol Chem. 2018;293:7811–7823. doi: 10.1074/jbc.RA118.002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park IY, Powell RT, Tripathi DN, Dere R, Ho TH, Blasius TL, Chiang Y-C, Davis IJ, Fahey CC, Hacker KE, Verhey KJ, Bedford MT, Jonasch E, Rathmell WK, Walker CL. Dual chromatin and cytoskeletal remodeling by SETD2. Cell. 2016;166:950–962. doi: 10.1016/j.cell.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharya S, Reddy D, Jani V, Gadewal N, Shah S, Reddy R, Bose K, Sonavane U, Joshi R, Smoot D, Ashktorab H, Gupta S. Histone isoform H2A1H promotes attainment of distinct physiological states by altering chromatin dynamics. Epigenet Chrom. 2017;10:48. doi: 10.1186/s13072-017-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.