Abstract
Colorectal cancer (CRC) mostly develops from a variety of polyps following mainly three different molecular pathways: chromosomal instability (CIN), microsatellite instability (MSI) and CpG island methylation (CIMP). Polyps are classified histologically as conventional adenomas, hyperplastic polyps, sessile serrated adenomas/polyps (SSA/P) and traditional serrated adenomas (TSA). However, the association of these polyps with the different types of CRCs and the underlying genetic and epigenetic aberrations has yet to be resolved. In order to address this question we analyzed 140 tumors and 20 matched mucosae by array comparative genomic hybridization, by sequence analysis of the oncogenes BRAF, KRAS, PI3K3CA, and by methylation arrays. MSI was tested indirectly by immunohistochemistry (IHC) and a loss of MLH1, MSH2, MSH6 or PMS2 was assigned as high microsatellite instability (MSI-H), while low microsatellite instability (MSI-L) was defined as MGMT IHC negativity only. CIN was detected in 78% of all MSI-H CRCs, most commonly as a gain of chromosome 8. Methylation data analyses allowed classification of samples into four groups and detected similar methylation profiles in SSA/P and MSI-H CRC. TSA also revealed aberrant methylation pattern, but clustered more heterogeneously and closer to microsatellite stable (MSS) CRCs. SSA/P, TSA and MSI-H CRCs had the highest degree of promotor methylation (CIMP pathway). Chromosomal instability, in contrast to the established doctrine, is a common phenomenon in MSI CRCs, yet to a lower extent and at later stages than in MSS CRCs. Methylation analyses suggest that SSA/P are precursors for MSI-H CRCs and follow the CIMP pathway.
Keywords: Serrated polyps, colorectal cancer, SSA/P, TSA, HP, MSI
Introduction
The world-wide incidence of colorectal cancer (CRC) amounts to approximately 1.2 million new cases per year.1 On the molecular level the development of CRC follows mainly three different molecular pathways: chromosomal instability (CIN), microsatellite instability (MSI) and CpG island methylation (CIMP).2, 3 Either one or a combination of these pathways can be involved in the stepwise transition from normal epithelium to polyps, and to CRC. Polyps are histologically classified as conventional adenomas, hyperplastic polyps (HP), traditional serrated adenomas (TSA), and sessile serrated adenoma/polyps (SSA/P).4 In addition to this classification, these premalignant lesions are grouped as either low-grade (LGD) or high-grade (HGD) dysplastic. HPs, by definition, do not harbor cellular or histological dysplasia.4 SSA/Ps also bear no cytological dysplasia but differ from HP by the presence of an abnormality of the histological architecture.4
In terms of tumor development, each of the above-mentioned precursor lesions might be initiated by a distinct molecular pathway (for review see:5-7).
Briefly, some 85% of CRCs and their conventional precursor adenomas are initiated by the CIN pathway.5 Epidemiologic data suggest a progression time from conventional adenoma to invasive CRC of five to 10 years.8
MSI, which is caused by mutations in DNA repair genes, is detectable in about 15% of CRCs and is associated with serrated polyps.5
The third pathway, CIMP, is observed in some 30% of CRCs and has recently gained attention because of the possibility that epigenetic changes may also result in genomic instability by changing chromosomal structure. This would explain why CIMP pathway overlaps with other pathways.9, 10
Polyps are precursor lesions for CRC. However, not all polyps harbor the same potential to develop into a carcinoma or require the same period of time before becoming an invasive disease. It is also not well established which type of polyps develops through which pathway into which form of CRC. Today's classification of polyps was introduced by Snover and colleagues in 2005. They established reproducible morphologic guidelines to distinguish polyps into conventional adenomas, HPs, TSAs and SSA/Ps.11 This discrimination is important since it is generally accepted that HPs are only very weakly, if at all, associated with cancer development.
While Snover established morphological criteria, no successful systematic molecular approach was undertaken to characterize the molecular driver behind the lesions or to solve the question of which precursor lesion leads to which carcinoma. The following study was conducted to explore whether the morphologically distinct pathways were associated with distinct patterns of genetic and epigenetic aberrations. We have therefore profiled 140 colorectal tumors and 20 matched normal mucosa samples by using array comparative genomic hybridization (aCGH), by sequence analysis of the oncogenes BRAF, KRAS, and PI3K3CA, by immunohistochemical detection of MLH1, MSH2, MSH6, PMS2, MGMT and by the GoldenGate™ Methylation Cancer Panel I Array. KRAS, PI3K3CA and BRAF were selected for mutation analyses since these genes are key players in the KRAS-BRAF and the PI3K3CA-PTEN-AKT pathways and somatic mutations tend to vary by different cancer types (MSI vs. MSS CRC).
Materials and Methods
Tumor Material
From 2008 to 2010 formalin-fixed, paraffin-embedded (FFPE) tumor samples were collected retrospectively from the archive of the Department of Pathology; Hospital of the Sozialstiftung Bamberg, Bamberg, Germany and from the Tumor Biobank of Hospital Clinic – IDIBAPS, Barcelona, Spain (Table 1). Normal mucosa was obtained in combination with the CRC resection. Normal mucosa was taken from the surgical specimen with a minimal distance of 3 cm from the tumor. In the majority, the distance was >10 cm, thereby reducing the chance for potential field effects.
Table 1.
Tumor samples according to study method.
| Histological type | Total sample numbers | Sample numbers for aCGH | Sample numbers for Methylation | Sample numbers for sequencing | Sample numbers for IHC |
|---|---|---|---|---|---|
| Normal mucosa | 20 | - | 20 | - | - |
| HP | 11 | 11 | 9 | 9 | 11 |
| Adenoma (LGD) | 27 | 27 | 21 | 21 | 27 |
| Adenoma (HGD) | 8 | 8 | 8 | 9 | 8 |
| TSA | 12 | 12 | 10 | 7 | 12 |
| SSA/P | 26 | 26 | 19 | 23 | 26 |
| MSI-H CRC | 18 | 18 | 17 | 9 | 18 |
| MSI-L CRC | 18 | 10 | 6 | 18 | 18 |
| MSS CRC | 20 | 13 | 20 | - | 20 |
| SUM | 160 | 125 | 130 | 96 | 140 |
Abbreviations: HP, hyperplastic polyp; LGD, low grade dysplasia; HGD, high grade dysplasia; TSA, traditional serrated adenoma; SSA/P, sessile serrated adenoma/polyp; CRC, colorectal cancer; MSI-H, high microsatellite instable; MSI-L, low microsatellite instable; MSS, microsatellite stable.
For each lesion a single FFPE block was included in this study. The study and sample acquisition was conducted in accordance with the regulation of the local ethics committees.
Morphology and clinical appearance
All lesions were diagnosed according to the current WHO-classification from 2010 (Fig. 1).4 Dysplasia of polyps was graded as either LGD or HGD based on the criteria established by the national polyp group and according to the WHO-classification.4, 12 Adenoma with LGD were grouped into small adenomas (diameter<10 mm) and large adenomas (diameter≥10 mm) in this study.
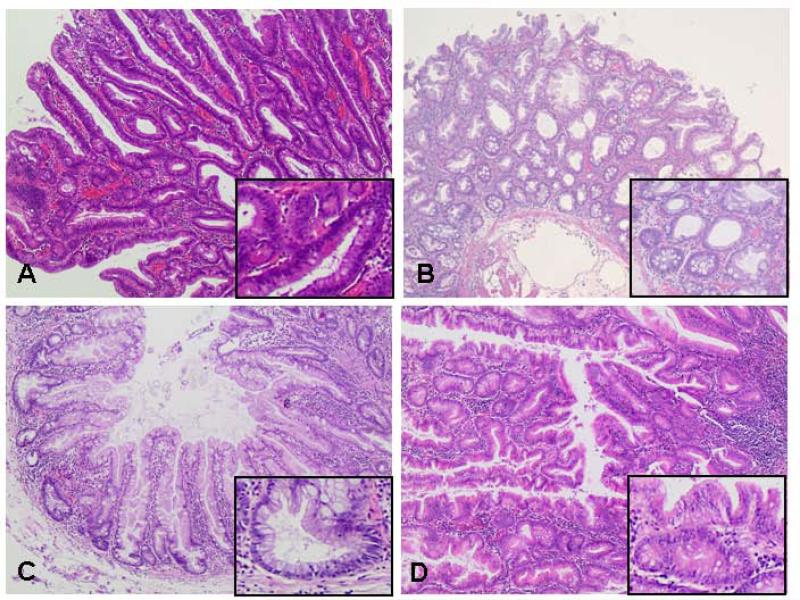
Figure 1.
Histological images of conventional adenoma and serrated lesions. (a) Conventional tubular adenoma with low-grade dysplasia. Inlay: hyperchromatic cells with multi-layered irregular nuclei with a high nucleus-to-cytoplasm ratio and a discrete loss of mucin. (b) Hyperplastic polyp with elongated but relatively straight crypts. Serration predominate visible near the luminal end of the crypts. Inlay: lower part of the polyp with small, basally placed nuclei without atypia in cytology or architecture. (c) Sessile serrated adenoma/polyp with distorted and dilated, L-shaped, inverted T-shaped or anchor-shaped crypts. Inlay: lower part of the polyp with L-shaped crypt. (d) Traditional serrated adenoma with complex and prominent “villiform” configuration. Inlay: cells lining the villi are tall and contain eosinophilic cytoplasm.
DNA preparation from histology slides
After histological evaluation of each HE slide by two board-certified pathologist (GS, TG) consecutive 20 μm thick sections were prepared from the corresponding paraffin block and mounted on standard charged glass slides. Based on the HE section, regions with >75% of tumor cells were marked on each of the 20 μm slides. Macrodissection was performed with a scalpel and DNA was prepared as described previously.13
Array Comparative Genomic Hybridization (aCGH)
The genome wide analysis of DNA copy number changes (aCGH) was performed using the SurePrint G3 Unrestricted CGH ISCA v2, 8x60K microarrays (Agilent, Santa Clara, CA). Labeling and hybridization were performed according to the manufacturer's protocol version 6.0 (Agilent). Briefly, 500 ng DNA from each of the patient samples and from sex matched reference genomic DNA (Promega, Madison, WI) were digested, purified, labeled and hybridized at 65°C for 40h at 20 rpm. Slides were then washed and scanned with a microarray scanner G2505B (Agilent) and analyzed using CGH Analytics software 4.0.76 (Agilent). The quality of the slides was assessed using quality control metrics provided by CGH Analytics. Aberration calls were based on the Aberration Detection Method 2 algorithm included in CGH Analytics. Overall, we applied a rather conservative threshold of 10.
BRAF, KRAS, PI3K3CA sequencing
Primer extension method was used to detect KRAS (exons 2-4), BRAF (exon 15) and PI3K3CA (exons 9 and 20) mutations. These regions (Supplementary Table 1 and 2) were amplified by multiplex PCR, and the obtained fragments were subjected to direct sequencing in both directions on an ABI-3100 Sequencer (Applied Biosystems, Foster City, CA), performed as previously reported.14
Microsatellite instability detection
Microsatellite instability phenotype was detected indirectly by loss of mismatch repair protein expression. Immunohistochemical staining was performed using monoclonal antibodies against MLH1, MSH2, MSH6, PMS2 and MGMT as described previously.15 A loss of MLH1, MSH2, MSH6 or PMS2 was assigned as MSI-H while MSI-L was defined only by MGMT IHC negativity. All MSI CRC cases had a negative CRC family history and MSH2 protein loss, the IHC marker for Lynch syndrome, was not detectable, which suggests the sporadic form of MSI CRC.
Methylation Analysis
A total of 1μg purified DNA for each sample was bisulfite converted using the Zymo Methylation Gold Kit (Zymo Research). Subsequently, methylation status of 1505 CpG sites were evaluated by using the GoldenGate Methylation Cancer Panel I Array (Illumina, San Diego, CA) as described previously.16 Image processing and data analyses were performed with Illumina-supplied equipment. The combination of five on the array presented genes, so called “CIMP markers”, CDKN2A, IGF2, MLH1, BDNF and CALCA were used to build a CIMP score. This procedure and the selection of the specific genes were based on findings by Weisenberger and colleagues.17 Any CIMP marker with a methylation intensity of more than 0.4 (range 0 - 1.0) above average of normal tissue adds one to the CIMP score. The groups were defined as follow: CIMP-negative: 0 (none of the five marker 0.4 above normal average); CIMP-Low= 1 (one of the five marker 0.4 above normal average); CIMP-Middle=2 (two of the five marker 0.4 above normal average); CIMP-High= 3,4,5 (3-5 markers 0.4 above normal average).
Statistical analysis
Differences in mutation rates were checked by using Fisher's exact test. A p-value of <0.05 was considered significant. P-values of significant aCGH results were evaluated with permutations with fixed number of total mutations as well as fixed number of mutations for each individual sample. The permutations for chromosome gain and loss were performed independently. The null hypothesis is that chromosomes are randomly gained or lost. Differential methylation was computed using t-tested p-values between normal and the sample (p<0.05 chosen as a cut-off). A Bonferroni correction was applied for multiple comparisons. Clustering analysis was done with Euclidean distance and Ward's method. For unsupervised clustering all data on the array except probes on X chromosome and probes with a p-value>0.05 in more than 10% of the samples were analyzed (total of 1,222 probes). Supervised clustering used 368 probes which are differentially methylated and have Bonferroni corrected two side Anova p-value <0.05 and a standard deviation above 0.1.
Results
Chromosomal gains and losses by aCGH
We successfully hybridized and analyzed 125 (11 HPs, 27 adenoma with LGD, 8 adenoma with HGD, 12 TSAs, 26 SSA/Ps, 10 MSI-L CRCs, 18 MSI-H CRCs and 13 MSS CRCs) colorectal tumor samples by aCGH (Table 2). In the group of the conventional adenomas chromosomal aberrations were found in eight out of 35 cases (23%). Among these samples, three of eight (38%) adenomas with HGD showed chromosomal aberrations, while only five of 27 (19%) adenomas with LGD had gains and losses. One out of 26 (4%) SSA/Ps and one out of 12 TSAs (8%) showed chromosomal aberrations. Considering the size as a distinguishing parameter within the low-grade adenomas, five out of 11 (46%) large adenomas (size ≥ 10 mm) showed chromosomal aberrations, mainly gains of chromosome arm 20q and chromosome 7, while none of the 16 small adenoma (size < 10 mm) showed aberrations, and chromosomal aberrations in the adenomas with HGD were more variable and frequent than the ones observed in the large adenomas.
Table 2.
Chromosomal gains and losses based on aCGH.
| Entity (number of cases) | Chromosomal gains (number of cases, if > 1) | Chromosomal losses (number of cases, if > 1) | % of samples with CIN |
|---|---|---|---|
| MSS CRC (13) | 2q32.2-33.1, 3p11.1-14.2, 3q11.2-13.11, 4q12-13.1, 5, 8, 8q (2), 7 (4), 12, 12p11.1-13.33, 12q12-24.11, 13 (5), 13q21.1-34, 14q24.1, 16, 16p, 16p11.2, 16q12.1-13, 17p11.2, 17q11.2-12, 17q21.1-21.2, 17q21.31-24.3, 19, 20 (3), 20p11.1-12, 20p11.21-11.23, 20p12.1-13, 20q (2) 20q11.21-11.23, X (2) | 1p11.1-22.3, 3p14.1-25.3, 3p14.2-26.3, 4 (4), 5q14.1, 5q14.2-23.3, 5q21.3-23.1, 8p (3), 8p12-23.3 (2), 8q11.21-11.22, 10q22.3-24.2, 10q23.31-25.3, 11, 11q22.1-23.3, 14 (2), 14q23.1-32.33, 15 (2), 15q11.2-13.3, 15q22.31-26.3, 16, 16p12.1-13.3, 16p12.3-13.3, 16q13-24.3, 16q21-24.3, 17p (4), 17p11.2-13.3, 17q12, 17q21.2-21.31, 18 (4), 18q12.1-23, 18q12.3-23, 19, 20p (2), 20p11.23-13, 20p12.1, 21, X | 100% |
| MSI-H CRC (18) | 1q (2), 6p, 7, 8 (4), 8q, 9 (2), 12, 13 (3), 17 (2), 17q, 19, 20, 22 | 3p, 3q, 4q (2), 5q12.2-14.3, 5q 14.3-22.2, 6q, 8p (2), 9p, 9q22.32-34.2, 10p15.3-13, 12, 12q, 15q11.2-13.1, 15q15.32-13.2, 15q25.1-26.3, 16q, 17, 18q, 20p, 22 | 78 % |
| MSI-L CRC (10) | 2 (2), 8q, 12 (2), 13 (2), 20, 20p, 20q | 1p, 4, 5q (2), 7, 8p (2), 10p, 10q, 14q, 17, 17p (2), 17q, 18 (2), 19p, 20p, 21 | 60% |
| HP (11) | - | - | 0 % |
| Small adenoma (LGD) (16) | - | - | 0 % |
| Large adenoma (LGD) (11) | 7 (2), 13, 20q (2) | 1p, 3p, 8q, 11q, 18q, 20p, 5p21.1-22.2 | 46 % |
| Adenoma (HGD (8) | 6p, 7, 8, 14, 16, 17, 17q, 20q (2), 22 | 1q, 2q, 2p, 4, 5q (2), 9p, 7, 18, 20p, 22 | 38 % |
| TSA (12) | 7, 13 | - | 8 % |
| SSA/P (26) | 15q12-13.2 | - | 4 % |
| SUM 125 | |||
Abbreviations: CRC colorectal cancer; MSS, microsatellite stable; MSI-H, high microsatellite instable; MSI-L, low microsatellite instable; HP, hyperplastic polyp; LGD, low grade dysplasia; HGD, high grade dysplasia; TSA, traditional serrated adenoma; SSA/P, sessile serrated adenoma/polyp.
In the carcinoma group, 14 out of 18 (78%) MSI-H CRCs revealed chromosomal aberrations in contrast to six out of 10 (60%) MSI-L CRCs. Conventional MSS CRCs contained chromosomal aberrations in all 13 cases (100%). Most striking and statistically significant were a loss of 18q and a gain of 20q in this group, which are typical findings in CRC.18, 19
No chromosomal aberrations were detected in hyperplastic polyps and small adenomas (Table 2). In large adenomas, adenomas with HGD, TSAs and SSA/Ps chromosomal aberrations were present, but with low frequency. Gains of 20q were the most frequent aberrations in large adenomas and adenomas with HGD (18% and 25%). One out of 12 TSAs (8.3%) showed gains of chromosomes 7 and 13. The only aberration present in the SSA/Ps was a small loss on 15q. Regarding the MSI-H CRCs, we mapped gains of chromosomes 1q (11%), 8 (28%), 9 (11%), 13 (17%) and 17 (17%) and losses of chromosomes 4q, 5q, 8p, 15q and 21 (all 11%). In MSI-L CRCs we identified gains of chromosomes 2, 12, 13 (all 20%) and 20 (30%) and losses of chromosomes 5q, 8p (both 20%) and 17 (30%). The average number of chromosomal copy number alterations (calculated by dividing the total number of gains and losses by the number of cases in the respective groups) in MSI-H CRCs amounted to three and in MSI-L CRCs to five. Both MSI groups (MSI-L CRCs and MSI-H CRCs) showed far fewer chromosomal aberrations compared to conventional MSS CRCs, for which the average value reached seven. This is consistent with previous reports.20, 21
In summary, conventional adenoma showed significantly more chromosomal aberrations than TSAs together with SSA/Ps. The number of chromosomal aberrations in MSI CRCs compared to serrated adenoma was considerably higher. This implies that (i) chromosomal imbalances are also present in MSI CRCs and (ii) that they appear late. Chromosomal aberrations in MSI CRCs are more frequently intrachromosomal, and less common compared to MSS CRCs.
Mutation analysis of the BRAF, KRAS, and PI3K3CA oncogenes
Activating mutations in the oncogenes BRAF, KRAS, and PI3K3CA are common events in colorectal tumorigenesis. In order to establish the frequency and chronology of occurrence of such mutations, we sequenced 96 cases of seven different subgroups. In total, 20 out of 96 (21%) cases showed mutations in either BRAF, KRAS, or PI3K3CA. Only one large adenoma showed two mutated genes (PI3K3CA and KRAS).
BRAF mutations were rare. Only one SSA/P (1781A>T), one TSA (1799T>A) and one large adenoma (1781A>T) revealed a BRAF mutation (Table 3A). No significant differences for BRAF mutation status were observed between the groups (Table 3B). KRAS mutations were found in higher frequencies. While only one out of 12 (14%) TSAs showed a KRAS mutation (35G>C), three out of 11 (27%) large adenomas and two out of 10 (20%) small adenomas were mutated (Table 3A). No significant differences for KRAS mutation status were observed between the groups (Table 3B). The highest mutation frequencies were observed for PI3K3CA. Four out of nine (44%) high-grade adenomas showed mutations (Table 3A). Serrated adenomas (12 TSAs + 26 SSA/Ps) showed no mutations while MSI-H CRC presented mutations in three out of seventeen cases (18%), therefore resulting in a significant difference between the groups (p=0.047; Table 3B).
Table 3.
Mutation analyses for BRAF, KRAS, PI3K3CA within different subgroups. (a) Mutation frequency for BRAF, KRAS, PI3K3CA. (b) P-values (Fisher exact test) for group comparisons.
| A) | |||
|---|---|---|---|
| Entity | BRAF | KRAS | PI3K3CA |
| HP (9) | 0% | 0% | 11% |
| Small adenoma (LGD) (10) | 0% | 20% | 10% |
| Large adenoma (LGD) (11) | 9% | 27% | 18% |
| Adenoma with HGD (9) | 0% | 11% | 44% |
| SSA/P (23) | 4% | 0% | 0% |
| TSA (7) | 14% | 14% | 0% |
| MSI-L CRC(9) | 0% | 0% | 0% |
| MSI-H CRC (18) | 0% | 6% | 18% |
| SUM (96) | |||
| B) | |||
|---|---|---|---|
| Entity | p-value | p-value | p-value |
| BRAF | PI3K3CA | KRAS | |
| SSA/P vs. TSA | 0.42 | 1.00 | 0.24 |
| Large adenoma vs. small adenoma | 1.00 | 1.00 | 1.00 |
| Adenoma (LGD) vs. adenoma (HGD) | 1.00 | 0.15 | 0.64 |
| SSA/P + TSA vs. small adenoma | 1.00 | 0.25 | 0.15 |
| SSA/P + TSA vs. MSI-H CRC | 0.52 | 0.047 | 1.00 |
| SSA/P vs. MSI-H CRC | 1.00 | 0.07 | 0.44 |
| TSA vs. MSI-H CRC | 0.28 | 0.53 | 0.49 |
Abbreviations: HP, hyperplastic polyps; LGD, low grade dysplasia; HGD, high grade dysplasia; SSA/P, sessile serrated adenoma/polyp; TSA, traditional serrated adenoma; CRC colorectal cancer; MSI-H, high microsatellite instable; MSI-L, low microsatellite instable.
Immunohistochemical detection of the mismatch repair proteins MLH1, MSH2, MSH6, PMS2 and MGMT
Polyps were only tested for MLH1, since it is already reported in the literature that mismatch repair (MMR) protein are only rarely lost in polyps.22 None of the 84 polyps showed a loss of MLH1, with all cases demonstrating a moderate to strong nuclear staining pattern. All 18 MSI-H carcinomas showed an immunohistochemical loss of at least two of the MMR proteins (MLH1, MSH2, MSH6, PMS2). Since MLH1/PMS2 and MSH2/MSH6 act as co-functional dimers, the loss of one MMR protein was connected with the loss of the corresponding partner. The MSI-L CRCs were selected as proposed by Esteller and colleagues by a complete loss of MGMT while MLH1, MSH2, MSH6 and PMS2 protein expression was preserved.23 No loss of MSH2 was detected within the MSI CRCs, therefore excluding Lynch Syndrome cases.
Methylation
We next explored to which extent aberrant methylation patterns of promotor regions occurred in our samples. Therefore, 136 samples were hybridized to a methylation array. One hundred thirty samples passed our quality control criteria with at least 75% loci with detection p-value <0.05 and at least 50% non-zero loci.
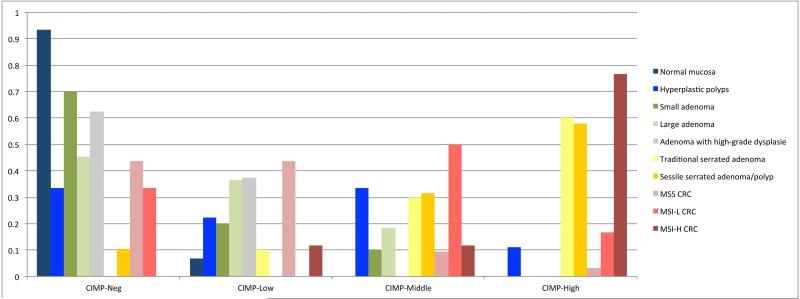
Weisenberger and colleagues identified a robust new marker panel to classify CIMP positive tumors based on 19 genes.17 Of these, six genes (CDKN2A, IGF2, MLH1, BDNF, CALCA, RUNX3) were present on the GoldenGate Methylation Cancer Panel I array. The gene RUNX3 had to be excluded due to an insufficient dynamic range among the samples. Evaluating CIMP-scores for the different groups showed that SSA/P, TSA and MSI-H CRCs had the highest degree of promotor methylation, followed by MSI-L CRCs and HP. Large adenoma and MSS CRCs demonstrated very low methylation levels (median CIMP score 1 and 0.5) (Fig. 2 and Supplementary Fig. 1). In general, the majority of TSAs (6/10), SSA/Ps (11/19) and MSI-H CRCs (13/17) were CIMP-High. 50% of the MSI-L CRCs (3/6) were CIMP-intermediate, while the remaining MSI-L CRCs samples were scored CIMP-negative. Strikingly, normal mucosa, MSS CRC, small- and large adenoma and adenoma with HGD were nearly completely CIMP-low or CIMP-negative. HP samples showed the highest degree of heterogeneity and were distributed among all four CIMP-scores (Fig. 2 and Supplementary Fig. 1). Analysis of the methylation status of specific genes revealed 174 differentially methylated CpGs sites in 127 genes when comparing normal mucosae and tumor samples, 159 of the 174 showed hypermethylation and 15 hypomethylation (Table 4). The most differentially methylated CpG were observed for TSAs (130 differential CpGs), SSA/Ps (110 differential CpGs) and MSI-H CRCs (106 differential CpGs). Of the 174 differentially methylated sites, 116 had the same methylation status in SSA/Ps compared to MSI-H CRCs. TSAs had 94 CpG sites in common with MSI-H CRCs (Table 4). This possibly suggests that SSA/P is more closely related to MSI-H CRC than to any other group.
Figure 2.
Distribution of the pathological subgroups into four different CIMP groups. The groups are defined as followed: CIMP-negative: 0 = none of the five genes CDKN2A, IGF2, MLH1, BDNF and CALCA is hypermethylated. CIMP-Low= one of the five marker is hypermethylated; CIMP-Middle= two of the five markers are hypermethylated); CIMP-High=3-5 of the five markers are hypermethylated. The majority of TSAs, SSA/Ps and MSI-H CRCs are CIMP-High. 50% of the MSI-L-CRCs are CIMP-middle, while the other ~50% are CIMP-negative (n=6!). Normal mucosa, MSS CRC, small- and large-adenoma and high grade adenoma were nearly completely CIMP-negative or CIMP-low. Hyperplastic polyps were distributed among all four CIMP-scores. Abbreviations: CIMP: CpG island methylation; CRC: colorectal cancer; MSS: microsatellite stable; MSI-L: low microsatellite instable; MSI-H: high microsatellite instable.
Table 4.
Number of significant CpG target sites being hyper- or hypo-methylated in the different subgroups (aligned to normal mucosa).
| Number of significant CpG targets hypermethylation | Number of significant CpG targets hypomethylation | Sum | |
|---|---|---|---|
| HP | 48 | 2 | 50 |
| Small adenoma (LGD) | 3 | 2 | 5 |
| Large adenoma (LGD) | 73 | 4 | 77 |
| Adenoma (HGD) | 20 | 0 | 20 |
| TSA | 125 | 5 | 130 |
| SSA/P | 110 | 0 | 110 |
| MSS CRC | 35 | 5 | 40 |
| MSI-L CRC | 40 | 2 | 42 |
| MSI-H CRC | 99 | 7 | 106 |
| SUM | 159 | 15 | 174 |
Abbreviations: HP, hyperplastic polyps; LGD, low grade dysplasia; HGD, high grade dysplasia; TSA, traditional serrated adenoma; SSA/P, sessile serrated adenoma/polyp; CRC, colorectal cancer; MSS, microsatellite stable; MSI-L, low microsatellite instable; MSI-H, high microsatellite instable.
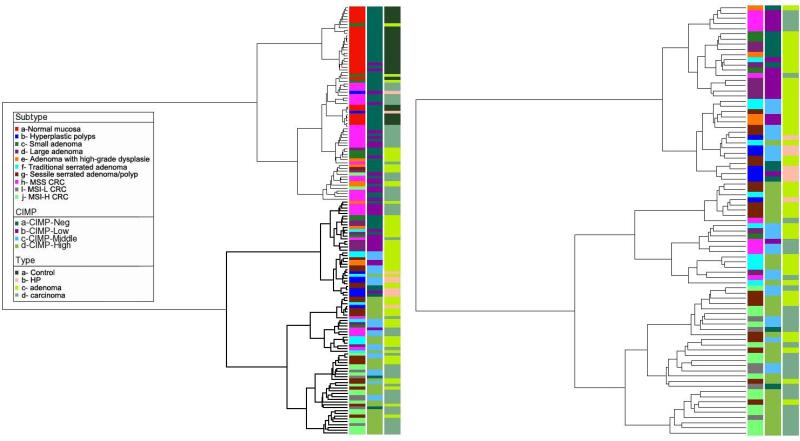
Supervised and unsupervised clustering revealed very similar results. Supervised hierarchical clustering of methylation data suggested the existence of four groups: Group 1, majority of normal samples; group 2, majority of conventional carcinomas and small adenomas; group 3, majority of HPs, large adenomas, TSAs and half of SSA/Ps, group 4, majority of MSI-H, MSI-L carcinomas and about 50% of the SSA/Ps (Fig. 3). Additional evidence for the validity of tumor segregation was also acquired through a Principal Component Analysis (PCA). Normal mucosa was grouped perfectly together and clearly segregated from the rest of the tumor samples, and only three small adenoma samples were close to this group. MSI-H CRCs were located in close proximity to MSI-L CRCs and SSA/P, but separated from TSAs (Supplementary Fig. 2A,B).
Figure 3.
Supervised hierarchical clustering of methylation data suggests the existence of four groups: Branch 1, majority of normal samples; Branch 2, majority of MSS CRCs and small adenomas; Branch 3, majority of HPs, large adenomas, TSAs and half of SSA/Ps, Branch 4, majority of MSI-H CRC, MSI-L CRC and half of SSA/Ps. SSA/Ps located in close proximity to MSI-H CRCs, TSAs cluster closer to MSS CRCs. MSI-L CRCs are located close to MSI-H CRCs. Abbreviations: CIMP: CpG island methylation; HP: hyperplastic polyp; CRC: colorectal cancer; MSS: microsatellite stable; MSI-L: low microsatellite instable; MSI-H: high microsatellite instable.
On the single gene level, MLH1, the most important DNA mismatch repair gene, was highly methylated in MSI CRCs compared to normal mucosae. Furthermore, DCC, IGFBP3, BMP6 and DBC1 were hypermethylated in SSA/Ps and MSI CRCs.
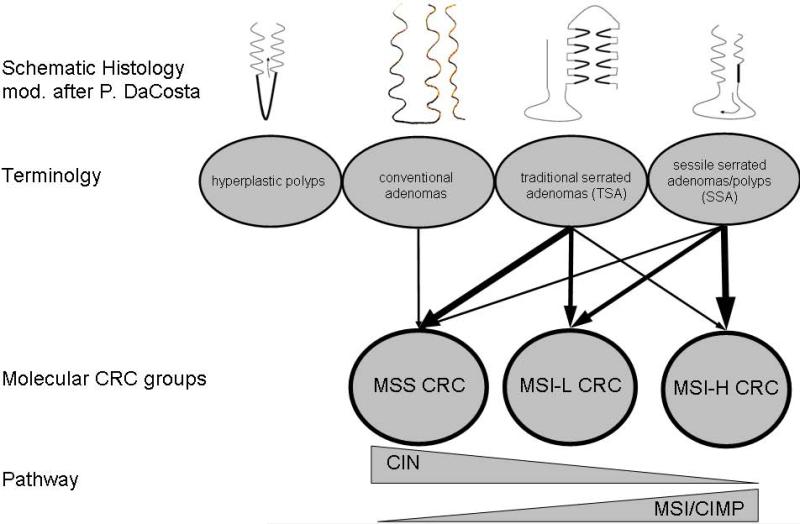
In conclusion, the methylation profile is consistent with the hypothesis that SSA/Ps could be the precursors of MSI-H CRCs. The similarity of the general methylation pattern between SSA/Ps and MSI-H CRC suggests that the MSI-H CRC methylation could have been established in the early adenoma stage and carried through later stages. Following the methylation cluster it appears that TSAs are just a special growth pattern of conventional adenoma leading also to MSS CRC. The scheme shown in Fig.4 summarizes the relation of the different polyps and the molecular CRC groups.
Figure 4.
Hypothetical relationship between different polyps and molecular colorectal cancer groups. While hyperplastic polyps do not harbor a noteworthy malignant potential, conventional adenomas develop through the chromosomal instability pathway into MSS CRC. TSA like SSA/P are supposed to follow the microsatellite instability and/or CpG island methylation pathway and develop mainly into MSI-L and MSI-H CRC. Abbreviations: CRC, colorectal cancer; MSS, microsatellite stable; MSI-L, low microsatellite instable; MSI-H, high microsatellite instable.
Discussion
In 1988 Fearon and Vogelstein proposed a genetic model for CRC development by demonstrating that a stepwise acquisition of mutations causes the progression from a small adenoma to an invasive carcinoma.24 In addition to specific mutations, sporadic CRCs were also defined by the non-random distribution of chromosomal gains and losses.21, 25 However, some CRCs seem to be unaware of that theorem and showed a normal or almost normal karyotype.26 These tumors were assigned as MSI, and defective DNA MMR genes were regarded as their molecular basis. Further studies revealed a third mechanism for developing CRC, i.e., the methylation of CpG islands.3 This pathway is called CIMP. Initially thought to be mutually exclusive, it became obvious that the CIMP pathway overlaps with the majority of MSI positive cancers, but also to a low percentage with MSS CRCs. Since polyps are the precursor lesion for CRC, we conducted the present study to better understand the connection between specific polyps and CRC defined by either the CIN, MSI or CIMP pathway. BRAF, KRAS and PI3K3CA are the most frequently mutated genes in polyps and CRC. In the literature, 60-100% of SSA/Ps showed BRAF mutations, whereas KRAS mutations were observed in 70% of conventional adenomas. Mutations in both genes were mutually exclusive. 27-29 In the present study the mutation frequencies for BRAF and KRAS were in general lower than reported,30 making it difficult to observe differences between subgroups. The reasons for that could not be clarified. So far, activating BRAF mutations were speculated to act as an anti-apoptotic stimulus, therefore causing the serrated morphology.31 Nevertheless, our data suggest that BRAF mutations are not the main initial proliferation impetus for serrated adenoma. Regarding PI3K3CA mutations which induce uncontrolled proliferation and cell migration,32 we identified only significant difference between serrated adenomas (TSA and SSA/Ps) and MSI-H CRC. Our data therefore indicate that PI3K3CA mutations occur in parallel with tumor progression but are not specific for any form of CRCs. In agreement with published results,33, 34 we did not find an association of specific mutations with a certain polyp or a certain cancer subtype.
In our study the selection of MSI CRCs was based on IHC. In addition, all polyps were tested for MLH1 loss but in accordance with previous findings, MMR proteins were not lost in early adenoma stage.22
Concerning the CIN pathway, initial studies revealed none or only few chromosomal aberrations in MSI-H CRC.35 However, these studies used conventional CGH, which has a much lower resolution than aCGH. Therefore, the possibility that serrated lesions and MSI-H CRCs also follow a multistep carcinogenesis pathway has not been ruled out. In our study, the first observable chromosomal copy number change occurred at the stage of large adenoma (size ≥10 mm). This was also reported by Leslie and colleagues who described early, small adenomas as not necessarily aneuploid.36 Some authors stated in this context that single gene mutations are initial steps leading to adenoma formation while CINs accumulate later.37 Others hypothesized that chromosomal gains and losses start as clonal events which need time to expand.38 Both concepts can explain why in our study chromosomal gains and losses were only detectable in large adenomas. Although more chromosomal copy number changes could be observed in adenomas with HGD vs. adenomas with LGD, the difference failed to reach statistical significance, probably due to the small numbers of adenomas with HGD. In serrated adenoma compared with MSI-H CRCs, the number of chromosomal aberration was significantly higher in the carcinoma group. MSI-H CRCs showed copy number changes in 78%, some with whole chromosome arm or whole chromosome gains and losses. This is clearly different from the general perception that MSI-H CRCs are chromosomally stable and it is in agreement with previous studies which have shown a similar pattern of chromosomal imbalances in MSI-H CRC.39, 40 The distribution of genomic alterations in MSI CRC is similar to the one observed in MSS CRCs, however, 18q was lost in only one of 18 (6%) MSI-H CRCs but in six of 13 (46%) MSS CRCs. Gain of whole chromosome 8 was observed in four of 18 (22%) MSI-H CRCs but in only one of 13 (8%) MSS CRCs. This finding is in alignment with data from Trautmann and colleagues reporting gains of chromosome 8 also as the most frequent imbalance in MSI-H CRCs.41 While the gain of certain regions of chromosome 8 (8q23–24) were associated with lymph node and distant metastases it is not clear whether other genes beside MYC on that chromosome arm are drivers of tumorigenesis.42
Concerning the average number of chromosomal copy number alterations (calculated by dividing the total number of gains and losses by the number of cases in the respective groups) MSI CRCs showed a significantly lower value. While MSS CRC showed the expected chromosomal gains and losses18, 43 with an average value of seven, MSI-H CRCs showed only three alterations per case (MSI-L showed five imbalances per sample). We submit the conclusion that CIN is a common event in MSI CRCs, but occurs later and to a lower degree compared to MSS CRCs. A number of studies showed that MSI-H CRCs follow a less aggressive clinical course and patients survive longer after surgery.44-47 In particular, the study by Malesci with 893 patients showed MSI status as a stage-dependent predictor of survival in patients with CRC. 48 In our data, measurement of methylation status allowed the most precise classification of the different tumor subgroups. It became obvious that MSI-H CRCs as well as SSA/Ps follow the CIMP pathway because they were associated with a high CIMP score; with respect to MSI-H CRCs this was already demonstrated by Cheng and colleagues.49 Clustering analyses revealed that SSA/Ps are potential precursors for MSI-H CRCs. MSI-L CRCs clustered between MSI-H and MSS CRCs and showed a mainly intermediate CIMP level. TSAs were located closer to MSS CRCs, but were defined by a more heterogeneous pattern, with some cases similar to MSI-L CRCs. This suggests that TSAs can be considered as precursors for MSS and MSI CRCs.
On the single gene level, numerous relevant tumor suppressor genes were highly methylated in SSA/Ps and MSI CRCs compared to normal mucosae, including DBC1, DCC and IGFBP3. MLH-1 was highly methylated in MSI CRCs but not in SSA/Ps. This confirms the rationale for the IHC based selection of the MSI-H CRCs cases and the already mentioned observation that MSI is a relatively late event in MSI cancer development. DBC1 (deleted in bladder cancer), located on chromosome band 9q32-33 is frequently lost in CRCs and bladder cancer. Hypermethylation of this gene was so far only reported in bladder cancer. Our data suggest a role of DBC1 in MSI CRC as well, which will be further investigated. DCC (deleted in colon cancer) located on chromosome band 18q21.3, a region also frequently lost in advanced CRCs, is supposed to prevent cell growth. Hypermethylation presumably will result in a lower expression and therefore in a growth stimulating effect. IGFBP3 (insulin-like growth factor binding protein 3) acts as a positive regulator of apoptosis in CRC cells and protein expression decreases during the progression of prostate cancer to its metastatic form.
In summary, the data presented here clearly demonstrate that the three pathways, CIN, CIMP and MSI, are not mutually exclusive and CRCs are often defective in more than one of these pathways. Methylation profiling of SSA/Ps and MSI-H CRCs support the general hypothesis that SSA/Ps are the progenitors of MSI-H CRCs and need to be monitored properly when detected by endoscopy. TSA together with conventional adenoma mainly progress to MSS CRCs, but can also give rise to MSI-L CRCs. Chromosomal instability is a common phenomenon in MSS CRCs, but also, albeit to a lower extent and at later stages, in MSI CRCs.
Supplementary Material
Figure S1: Comparison of CIMP-scores for all subgroups listed case by case. CIMP-scores ranged from 0-5 based on the methylation level of CDKN2A, IGF2, MLH1, BDNF and CALCA. SSA/Ps, TSAs and MSI-H CRCs were highly methylated, followed by MSI-L CRCs and HPs. Large adenomas and MSS CRCs demonstrated lower levels but were higher methylated than small adenomas and high grade adenoma samples. Normal mucosa did not show a relevant methylation level. Abbreviations: N, normal mucosa; HP, hyperplastic polyps, SA, small adenoma with low grade dysplasia; BA, large adenoma with low grade dysplasia; HGA, adenoma with high-grade dysplasia; TSA, traditional serrated adenoma; SSA/P, sessile serrated adenoma/polyp; MSS CRC, microsatellite stable colorectal cancer; MSI-L CRC, colorectal cancer with low microsatellite instability; MSI-H CRC; colorectal cancer with high microsatellite instability.
Figure S2: Principal component analysis (PCA) of gene methylation within the subgroups. (a) The PCA showed a clear separation of normal mucosa from the rest of the samples. Only a few outliers of MSS CRC were located close to the majority of normal samples. (b) MSI CRCs were separated from MSS CRC but no clear separation for MSI-L CRCs from MSI-H CRCs was achieved. TSAs were in general located closer to MSS CRCs than MSI CRCs but some were connected closer to MSI-L CRCs. SSA/Ps showed close relation to MSI-H CRCs. Conventional adenomas clustered nearby MSS CRCs. Abbreviations: HP, hyperplastic polyps, MSS CRC, microsatellite stable colorectal cancer; MSI-L CRC, colorectal cancer with low microsatellite instability; MSI-H CRC; colorectal cancer with high microsatellite instability; CIMP: CpG island methylation.
Novelty & Impact Statements.
We detected that chromosomal instability, in contrast to the established doctrine, was a common phenomenon in microsatellite instable colorectal cancers (CRC) yet to a lower extent and at later stages compared to microsatellite stable CRCs. Methylation analyses suggest that sessile serrated adenomas were precursors for CRCs with high microsatellite instability.
Acknowledgements
The authors thank Buddy Chen for excellent technical assistance with figures and Miia Margareta Suuriniemi for help with the aCGH protocol. Daniela Hirsch was supported by by the RISE program of the German Academic Exchange Service (DAAD).
The study was sponsored by the Intramural Research Program of the NIH, National Cancer Institute (USA); Ministerio de Ciencia e Innovacion (SAF2007-64167), Red Tematica de Investigacion Cooperativa en Cancer (RD06/0020/1020) and Generalitat de Catalunya (2009 SGR 1107) (Spain).
Abbreviations
- CRC
colorectal cancer
- CIN
chromosomal instability
- MSS
microsatellite stable
- MSI
microsatellite instable
- CIMP
CpG island methylation
- SSA/P
sessile serrated adenomas/polyps
- TSA
traditional serrated adenomas
- IHC
immunohistochemistry
- MSI-H
high microsatellite instable
- MSI-L
low microsatellite instable
- HP
hyperplastic polyps
- LGD
low-grade dysplasia
- HGD
high-grade dysplasia
- aCGH
array comparative genomic hybridization
- FFPE
formalin-fixed, paraffin-embedded tissue
- WHO
World Health Organization
- NM
normal mucosa
- PCA
principal component analysis
- SA
small adenomas with low grade dysplasia
- LA
large adenomas with low grade dysplasia
- HGA
adenomas with high-grade dysplasia
Footnotes
Conflict of interest statement: No potential “conflict of interest” has to be declared for any contributing author in this study.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Georgiades IB, Curtis LJ, Morris RM, Bird CC, Wyllie AH. Heterogeneity studies identify a subset of sporadic colorectal cancers without evidence for chromosomal or microsatellite instability. Oncogene. 1999;18:7933–40. doi: 10.1038/sj.onc.1203368. [DOI] [PubMed] [Google Scholar]
- 3.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the Digestive System, 2010/12/22 4th Edition ed. WHO PRESS; Lyon: 2010. [Google Scholar]
- 5.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–30. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 6.Issa JP. Colon cancer: it's CIN or CIMP. Clin Cancer Res. 2008;14:5939–40. doi: 10.1158/1078-0432.CCR-08-1596. [DOI] [PubMed] [Google Scholar]
- 7.Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- 8.Olsen HW, Lawrence WA, Snook CW, Mutch WM. Risk factors and screening techniques in 500 patients with benign and malignant colon polyps. An urban community experience. Dis Colon Rectum. 1988;31:216–21. doi: 10.1007/BF02552550. [DOI] [PubMed] [Google Scholar]
- 9.Makinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131–50. doi: 10.1111/j.1365-2559.2006.02548.x. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snover DC, Jass JR, Fenoglio-Preiser C, Batts KP. Serrated polyps of the large intestine: a morphologic and molecular review of an evolving concept. Am J Clin Pathol. 2005;124:380–91. doi: 10.1309/V2EP-TPLJ-RB3F-GHJL. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien MJ, Winawer SJ, Zauber AG, Gottlieb LS, Sternberg SS, Diaz B, Dickersin GR, Ewing S, Geller S, Kasimian D, et al. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology. 1990;98:371–9. [PubMed] [Google Scholar]
- 13.Gaiser T, Camps J, Meinhardt S, Wangsa D, Nguyen QT, Varma S, Dittfeld C, Kunz-Schughart LA, Kemmerling R, Becker MR, Heselmeyer-Haddad K, Ried T. Genome and transcriptome profiles of CD133-positive colorectal cancer cells. Am J Pathol. 2011;178:1478–88. doi: 10.1016/j.ajpath.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jo P, Jung K, Grade M, Conradi LC, Wolff HA, Kitz J, Becker H, Ruschoff J, Hartmann A, Beissbarth T, Muller-Dornieden A, Ghadimi M, et al. CpG island methylator phenotype infers a poor disease-free survival in locally advanced rectal cancer. Surgery. 2011 doi: 10.1016/j.surg.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Ruschoff J, Bocker T, Schlegel J, Stumm G, Hofstaedter F. Microsatellite instability: new aspects in the carcinogenesis of colorectal carcinoma. Virchows Arch. 1995;426:215–22. doi: 10.1007/BF00191357. [DOI] [PubMed] [Google Scholar]
- 16.Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149–63. doi: 10.1007/978-1-59745-522-0_12. [DOI] [PubMed] [Google Scholar]
- 17.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 18.Camps J, Grade M, Nguyen QT, Hormann P, Becker S, Hummon AB, Rodriguez V, Chandrasekharappa S, Chen Y, Difilippantonio MJ, Becker H, Ghadimi BM, et al. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–95. doi: 10.1158/0008-5472.CAN-07-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermsen M, Postma C, Baak J, Weiss M, Rapallo A, Sciutto A, Roemen G, Arends JW, Williams R, Giaretti W, De Goeij A, Meijer G. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology. 2002;123:1109–19. doi: 10.1053/gast.2002.36051. [DOI] [PubMed] [Google Scholar]
- 20.Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–90. [PMC free article] [PubMed] [Google Scholar]
- 21.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–45. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 22.Petko Z, Ghiassi M, Shuber A, Gorham J, Smalley W, Washington MK, Schultenover S, Gautam S, Markowitz SD, Grady WM. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res. 2005;11:1203–9. [PubMed] [Google Scholar]
- 23.Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JP, Sidransky D, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–71. [PubMed] [Google Scholar]
- 24.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 25.Meijer GA, Hermsen MA, Baak JP, van Diest PJ, Meuwissen SG, Belien JA, Hoovers JM, Joenje H, Snijders PJ, Walboomers JM. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol. 1998;51:901–9. doi: 10.1136/jcp.51.12.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bardi G, Fenger C, Johansson B, Mitelman F, Heim S. Tumor karyotype predicts clinical outcome in colorectal cancer patients. J Clin Oncol. 2004;22:2623–34. doi: 10.1200/JCO.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 27.Chan TL, Zhao W, Leung SY, Yuen ST. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878–81. [PubMed] [Google Scholar]
- 28.McLellan EA, Owen RA, Stepniewska KA, Sheffield JP, Lemoine NR. High frequency of K-ras mutations in sporadic colorectal adenomas. Gut. 1993;34:392–6. doi: 10.1136/gut.34.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S, Farraye FA, Mack C, Posnik O, O'Brien MJ. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol. 2004;28:1452–9. doi: 10.1097/01.pas.0000141404.56839.6a. [DOI] [PubMed] [Google Scholar]
- 30.Rosty C, Buchanan DD, Walsh MD, Pearson SA, Pavluk E, Walters RJ, Clendenning M, Spring KJ, Jenkins MA, Win AK, Hopper JL, Sweet K, et al. Phenotype and polyp landscape in serrated polyposis syndrome: a series of 100 patients from genetics clinics. Am J Surg Pathol. 2012;36:876–82. doi: 10.1097/PAS.0b013e31824e133f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–87. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 32.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 33.Velho S, Oliveira C, Ferreira A, Ferreira AC, Suriano G, Schwartz S, Jr., Duval A, Carneiro F, Machado JC, Hamelin R, Seruca R. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur J Cancer. 2005;41:1649–54. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 34.Middeldorp A, van Eijk R, Oosting J, Forte GI, van Puijenbroek M, van Nieuwenhuizen M, Corver WE, Ruano D, Caldes T, Wijnen J, Morreau H, van Wezel T. Increased frequency of 20q gain and copy-neutral loss of heterozygosity in mismatch repair proficient familial colorectal carcinomas. Int J Cancer. 2012;130:837–46. doi: 10.1002/ijc.26093. [DOI] [PubMed] [Google Scholar]
- 35.Ortega P, Moran A, de Juan C, Frias C, Hernandez S, Lopez-Asenjo JA, Sanchez-Pernaute A, Torres A, Iniesta P, Benito M. Differential Wnt pathway gene expression and E-cadherin truncation in sporadic colorectal cancers with and without microsatellite instability. Clin Cancer Res. 2008;14:995–1001. doi: 10.1158/1078-0432.CCR-07-1588. [DOI] [PubMed] [Google Scholar]
- 36.Leslie A, Stewart A, Baty DU, Mechan D, McGreavey L, Smith G, Wolf CR, Sales M, Pratt NR, Steele RJ, Carey FA. Chromosomal changes in colorectal adenomas: relationship to gene mutations and potential for clinical utility. Genes Chromosomes Cancer. 2006;45:126–35. doi: 10.1002/gcc.20271. [DOI] [PubMed] [Google Scholar]
- 37.Smith G, Carey FA, Beattie J, Wilkie MJ, Lightfoot TJ, Coxhead J, Garner RC, Steele RJ, Wolf CR. Mutations in APC, Kirsten-ras, and p53--alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci U S A. 2002;99:9433–8. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–17. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camps J, Armengol G, del Rey J, Lozano JJ, Vauhkonen H, Prat E, Egozcue J, Sumoy L, Knuutila S, Miro R. Genome-wide differences between microsatellite stable and unstable colorectal tumors. Carcinogenesis. 2006;27:419–28. doi: 10.1093/carcin/bgi244. [DOI] [PubMed] [Google Scholar]
- 40.Douglas EJ, Fiegler H, Rowan A, Halford S, Bicknell DC, Bodmer W, Tomlinson IP, Carter NP. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res. 2004;64:4817–25. doi: 10.1158/0008-5472.CAN-04-0328. [DOI] [PubMed] [Google Scholar]
- 41.Trautmann K, Terdiman JP, French AJ, Roydasgupta R, Sein N, Kakar S, Fridlyand J, Snijders AM, Albertson DG, Thibodeau SN, Waldman FM. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin Cancer Res. 2006;12:6379–85. doi: 10.1158/1078-0432.CCR-06-1248. [DOI] [PubMed] [Google Scholar]
- 42.Ghadimi BM, Grade M, Liersch T, Langer C, Siemer A, Fuzesi L, Becker H. Gain of chromosome 8q23-24 is a predictive marker for lymph node positivity in colorectal cancer. Clin Cancer Res. 2003;9:1808–14. [PubMed] [Google Scholar]
- 43.Sayagues JM, Fontanillo C, Abad Mdel M, Gonzalez-Gonzalez M, Sarasquete ME, Chillon Mdel C, Garcia E, Bengoechea O, Fonseca E, Gonzalez-Diaz M, De las Rivas J, Munoz-Bellvis L, et al. Mapping of genetic abnormalities of primary tumours from metastatic CRC by high-resolution SNP arrays. PLoS One. 2010;5:e13752. doi: 10.1371/journal.pone.0013752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elsaleh H, Powell B, McCaul K, Grieu F, Grant R, Joseph D, Iacopetta B. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res. 2001;7:1343–9. [PubMed] [Google Scholar]
- 45.Halling KC, French AJ, McDonnell SK, Burgart LJ, Schaid DJ, Peterson BJ, Moon-Tasson L, Mahoney MR, Sargent DJ, O'Connell MJ, Witzig TE, Farr GH, Jr., et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 46.Hemminki A, Mecklin JP, Jarvinen H, Aaltonen LA, Joensuu H. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–8. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 47.Wright CM, Dent OF, Barker M, Newland RC, Chapuis PH, Bokey EL, Young JP, Leggett BA, Jass JR, Macdonald GA. Prognostic significance of extensive microsatellite instability in sporadic clinicopathological stage C colorectal cancer. Br J Surg. 2000;87:1197–202. doi: 10.1046/j.1365-2168.2000.01508.x. [DOI] [PubMed] [Google Scholar]
- 48.Malesci A, Laghi L, Bianchi P, Delconte G, Randolph A, Torri V, Carnaghi C, Doci R, Rosati R, Montorsi M, Roncalli M, Gennari L, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res. 2007;13:3831–9. doi: 10.1158/1078-0432.CCR-07-0366. [DOI] [PubMed] [Google Scholar]
- 49.Cheng YW, Pincas H, Bacolod MD, Schemmann G, Giardina SF, Huang J, Barral S, Idrees K, Khan SA, Zeng Z, Rosenberg S, Notterman DA, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–13. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Comparison of CIMP-scores for all subgroups listed case by case. CIMP-scores ranged from 0-5 based on the methylation level of CDKN2A, IGF2, MLH1, BDNF and CALCA. SSA/Ps, TSAs and MSI-H CRCs were highly methylated, followed by MSI-L CRCs and HPs. Large adenomas and MSS CRCs demonstrated lower levels but were higher methylated than small adenomas and high grade adenoma samples. Normal mucosa did not show a relevant methylation level. Abbreviations: N, normal mucosa; HP, hyperplastic polyps, SA, small adenoma with low grade dysplasia; BA, large adenoma with low grade dysplasia; HGA, adenoma with high-grade dysplasia; TSA, traditional serrated adenoma; SSA/P, sessile serrated adenoma/polyp; MSS CRC, microsatellite stable colorectal cancer; MSI-L CRC, colorectal cancer with low microsatellite instability; MSI-H CRC; colorectal cancer with high microsatellite instability.
Figure S2: Principal component analysis (PCA) of gene methylation within the subgroups. (a) The PCA showed a clear separation of normal mucosa from the rest of the samples. Only a few outliers of MSS CRC were located close to the majority of normal samples. (b) MSI CRCs were separated from MSS CRC but no clear separation for MSI-L CRCs from MSI-H CRCs was achieved. TSAs were in general located closer to MSS CRCs than MSI CRCs but some were connected closer to MSI-L CRCs. SSA/Ps showed close relation to MSI-H CRCs. Conventional adenomas clustered nearby MSS CRCs. Abbreviations: HP, hyperplastic polyps, MSS CRC, microsatellite stable colorectal cancer; MSI-L CRC, colorectal cancer with low microsatellite instability; MSI-H CRC; colorectal cancer with high microsatellite instability; CIMP: CpG island methylation.