Key Points
Question
What is the yield and utility of germline genetic testing following tumor DNA sequencing in patients with cancer?
Findings
In this cohort study of 2023 patients, 30.5% harbored a pathogenic germline variant, most of which were potentially actionable, and these variants were prevalent across diverse cancer types, genes, and patient ages. However, 8.1% of these pathogenic variants were missed by tumor tests, and approximately 20% of germline-positive patients did not meet criteria for germline follow-up testing.
Meaning
The results of this study support the utility of germline analysis both following tumor sequencing and independent of tumor sequencing in appropriate patients.
Abstract
Importance
Both germline genetic testing and tumor DNA sequencing are increasingly used in cancer care. The indications for testing and utility of these 2 tests differ, and guidelines recommend that germline analysis follow tumor sequencing in certain patients to determine whether particular variants are of somatic or germline origin. Broad clinical experience with such follow-up testing has not yet been thoroughly described.
Objective
To examine the yield and utility of germline testing following tumor DNA sequencing in a large, diverse patient population.
Design, Setting, and Participants
A retrospective cohort study examined germline testing through a laboratory supporting multiple academic and community clinics. Participants included 2023 patients with cancer who received germline testing and previously underwent tumor DNA sequencing. These patients received germline testing between January 5, 2015, and January 31, 2020, although most (81% of patients) received testing between January 2, 2018, and January 31, 2020.
Main Outcomes and Measures
The prevalence of pathogenic germline variants (PGVs) was calculated by gene, cancer type, and age at diagnosis. Potential actionability of these findings was determined based on current management guidelines, precision therapy labels, and clinical trial eligibility criteria. Patient records were reviewed to determine whether germline follow-up testing would have been recommended by current guidelines.
Results
Among 2023 eligible patients, 1085 were female (53.6%), and the median age at cancer diagnosis was 56 (range, 0-92) years. Pathogenic germline variants were detected in 617 patients (30.5%; 95% CI, 28.5%-32.6%) and were prevalent across patient ages (1-85 years) and cancer types, including cancers known to be strongly associated with germline variance (eg, breast, colorectal) as well as others (eg, renal, lung, and bladder). Many patients (78%-82%) with PGVs met criteria for germline follow-up testing, and 8.1% of PGVs were missed by tumor sequencing. Among those with germline-positive findings, 69 patients (11.2%) had PGVs identified only after presenting with a second primary cancer that possibly could have been detected earlier or prevented given current gene-specific surveillance and risk-reduction recommendations.
Conclusions and Relevance
The findings of this study suggest that germline analysis following tumor sequencing often produces findings that may impact patient care by influencing systemic therapy choices, surgical decisions, additional cancer screening, and genetic counseling in families. Current guidelines and tumor testing approaches appear to capture many, but not all, of these germline findings, reinforcing the utility of both expanded germline follow-up testing as well as germline analysis independent of tumor sequencing in appropriate patients.
This cohort study examines the prevalence of pathogenic germline variants in patients with cancer who had received DNA sequencing.
Introduction
Genetic counseling and germline testing are recommended for patients with cancer who have suspected hereditary disease based on each patient’s presentation and family history.1,2 Separately, tumor DNA sequencing is increasingly used, most often in patients with advanced disease.3,4,5 The detection of either inherited germline variants or acquired somatic variants (ie, mutations in the tumor) can inform patient care, including systemic therapy selection.6,7 However, the 2 biomarker types are complementary, and different recommendations can follow from each.
In principle, tumor sequencing detects both somatic and germline variation, and studies have shown that pathogenic germline variants (PGVs) are indeed encountered in tumor testing.8,9 However, many of these data come from academic laboratories using paired tumor/normal sequencing, while the most commonly ordered tests sequence tumor tissue alone, a technique that cannot reliably distinguish between somatic and germline changes.8,10 Thus, 2019 updates to certain oncology practice guidelines recommend that germline testing follow tumor sequencing in patients meeting various criteria to determine whether particular variants are of germline or somatic origin.1,2,11,12,13 Other practice guidelines do not yet make such recommendations, even in cancers in which tumor sequencing is common (eg, lung cancer14).
Broad clinical experience with germline analysis following tumor sequencing has not yet been thoroughly described. We examined the yield and utility of germline findings in more than 2000 patients, representing diverse cancer types, patient ages, test indications, and clinical settings.
Methods
We retrospectively analyzed a cohort of patients receiving germline testing of hereditary cancer predisposition genes who had a current diagnosis or personal history of cancer and previously had received tumor DNA sequencing. Germline testing occurred through a laboratory supporting multiple academic and community clinics. Indications for germline testing were diverse and included tumor findings of potential germline origin, treatment guidance or surgical planning, personal or family history, and patient concern. These patients received germline testing between January 5, 2015, and January 31, 2020, although most (81% of patients) received testing between January 2, 2018, and January 31, 2020. Patient records were reviewed to determine whether germline follow-up testing would have been recommended by current guidelines. The specific germline and tumor tests varied at clinician discretion and were performed by different laboratories. Pathogenic (including likely pathogenic) germline variants conferring a high or moderate cancer risk were analyzed in this study; variants of uncertain significance and low-penetrance variants were excluded (eMethods in the Supplement). This study was performed under protocol 20161796 approved by the Western Institutional Review Board; data were deidentified and informed consent was waived. This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline for cohort studies.
Statistical Analysis
Statistics were computed using Python, version 3.7 (Python Software Foundation). The Wilson method was used to calculate 95% CIs. The significance of differences between proportions was calculated using 2-sided z scores.
Results
In total, 2023 patients met study criteria (eMethods in the Supplement) representing a mix of sexes (1085 [53.6%] female), ages at cancer diagnosis (median [range], 56 [0-92] years), and race/ethnicity (Table 1). Of these patients, 617 (30.5%; 95% CI, 28.5%-32.6%) were found to harbor 1 or more PGVs in known cancer predisposition genes. As expected,12 clinically important variants in certain genes were more likely to be of germline origin compared with others (Figure 1). For example, 39% of pathogenic variants in BRCA1 and BRCA2 and 28% in MLH1, MSH2, MSH6, and PMS2 (the Lynch syndrome mismatch repair genes) were determined to be of germline origin. In contrast, only 4% of pathogenic TP53 variants were germline, although most (64%) of the germline TP53 carriers did not meet the Chompret criteria for germline TP53 testing.1 Pathogenic germline variants were prevalent among all cancer types studied (Figure 2), including cancers known to be strongly gene associated (eg, breast, colorectal) as well as others (eg, renal, lung, and bladder). In germline-positive cases, PGVs were often detected in patients with cancers that were not strongly associated with their particular germline findings (eg, BRCA1 and BRCA2 PGVs were uncovered in patients with colorectal, thyroid, endometrial, and almost every other cancer type studied). Pathogenic germline variants were modestly enriched in patients with first cancer diagnosis between age 30 and 49 years but were prevalent (ie, observed in 25%-38% of patients) across all age groups (Figure 3).
Table 1. Patient Demographic and Clinical Characteristics (N = 2023).
| Characteristic | No. (%) |
|---|---|
| Sex | |
| Female | 1085 (56.3) |
| Male | 938 (46.4) |
| Self-reported race/ethnicitya | |
| White/Caucasian | 1318 (65.2) |
| Ashkenazi Jewish | 143 (7.1) |
| Black/African American | 121 (6.0) |
| Asian | 107 (5.3) |
| Hispanic | 97 (4.8) |
| Other/mixed | 176 (8.7) |
| Unknown | 61 (3.0) |
| Cancer diagnoses | |
| Single primary | 1779 (87.9) |
| Multiple primaries | 244 (12.1) |
| Age at first cancer diagnosis, ya,b | |
| 0-19 | 87 (5.8) |
| 20-29 | 53 (3.5) |
| 30-39 | 143 (9.6) |
| 40-49 | 233 (15.6) |
| 50-59 | 378 (25.3) |
| 60-69 | 376 (25.2) |
| 70-79 | 184 (12.3) |
| ≥80 | 39 (2.6) |
| Unknown | 530 (26.2) |
| Age at germline test, ya | |
| 0-19 | 117 (5.8) |
| 20-29 | 47 (2.3) |
| 30-39 | 132 (6.5) |
| 40-49 | 244 (12.1) |
| 50-59 | 414 (20.5) |
| 60-69 | 527 (26.1) |
| 70-79 | 420 (20.8) |
| ≥80 | 122 (6.0) |
| No. of genes tested in the germline | |
| 1-5 | 404 (20.0) |
| 6-15 | 138 (6.8) |
| 16-50 | 864 (42.7) |
| ≥51 | 617 (30.5) |
Percent does not equal 100% due to rounding.
Percent calculated excluding unknowns (n = 1493 as denominator).
Figure 1. Germline and Somatic Findings, by Gene.
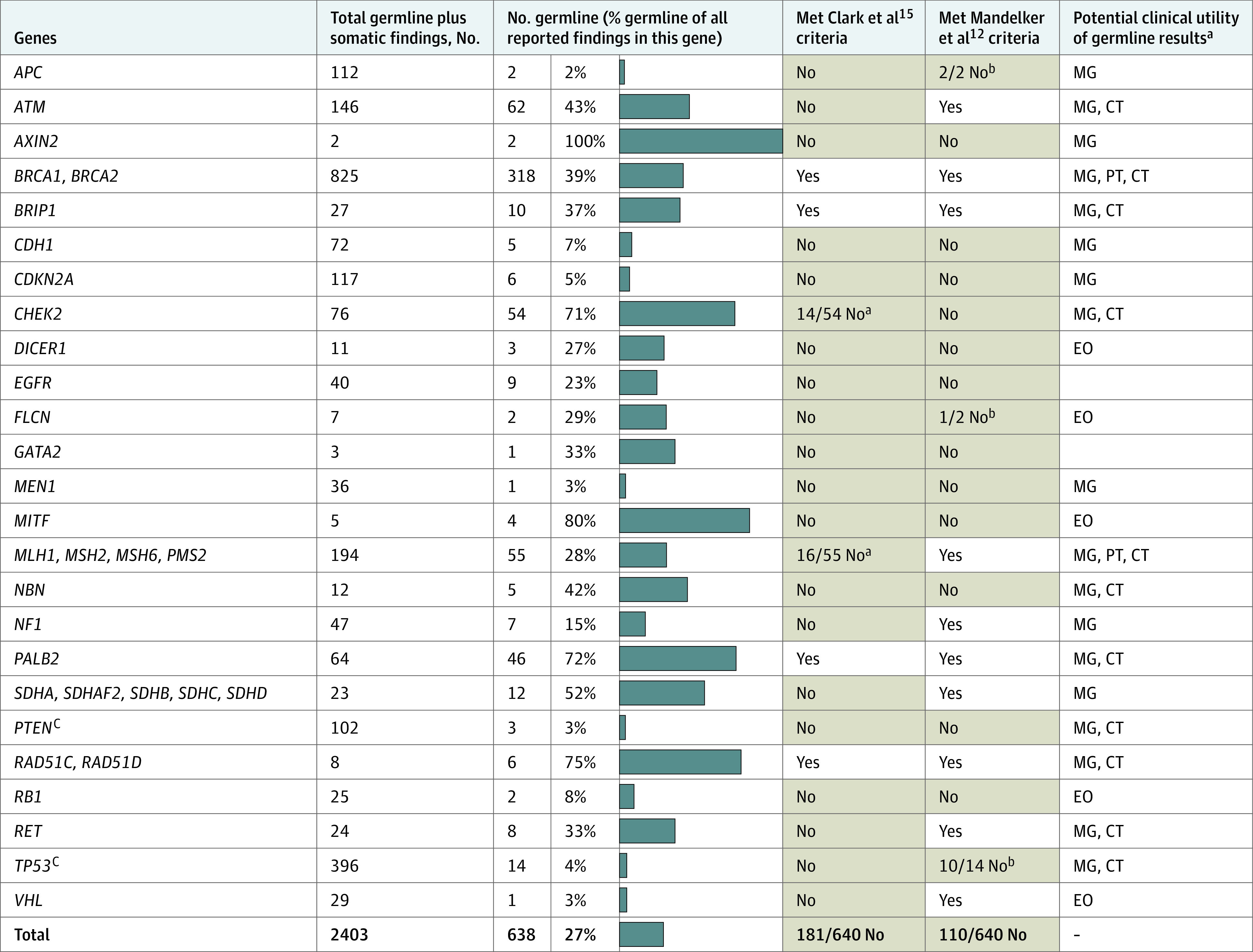
For each gene, the percentage of all reported findings that were of germline origin are shown by blue bars. Also indicated is whether the germline variants would (white background) or would not (light orange background) have met criteria for germline follow-up testing and whether the germline variants were potentially actionable. Only genes with at least 1 germline finding in this study are shown. Low-penetrance germline findings are excluded (eMethods in the Supplement). CT indicates gene-based clinical trial eligibility; EO, published expert opinion; MG, management guidelines; and PT, US Food and Drug Administration–approved precision-therapy labels. - indicates not applicable.
aThe Clark et al15 criteria include only certain variants in CHEK2 and exclude PMS2.
bThe Mandelker et al12 criteria for this gene specify patient age and/or tumor types.
cPTEN and TP53 frequently acquire somatic variations, which appear to be underreported in records available to this study (eResults in the Supplement).
Figure 2. Prevalence of Pathogenic Germline Variants (PGVs), by Cancer Type and Gene.
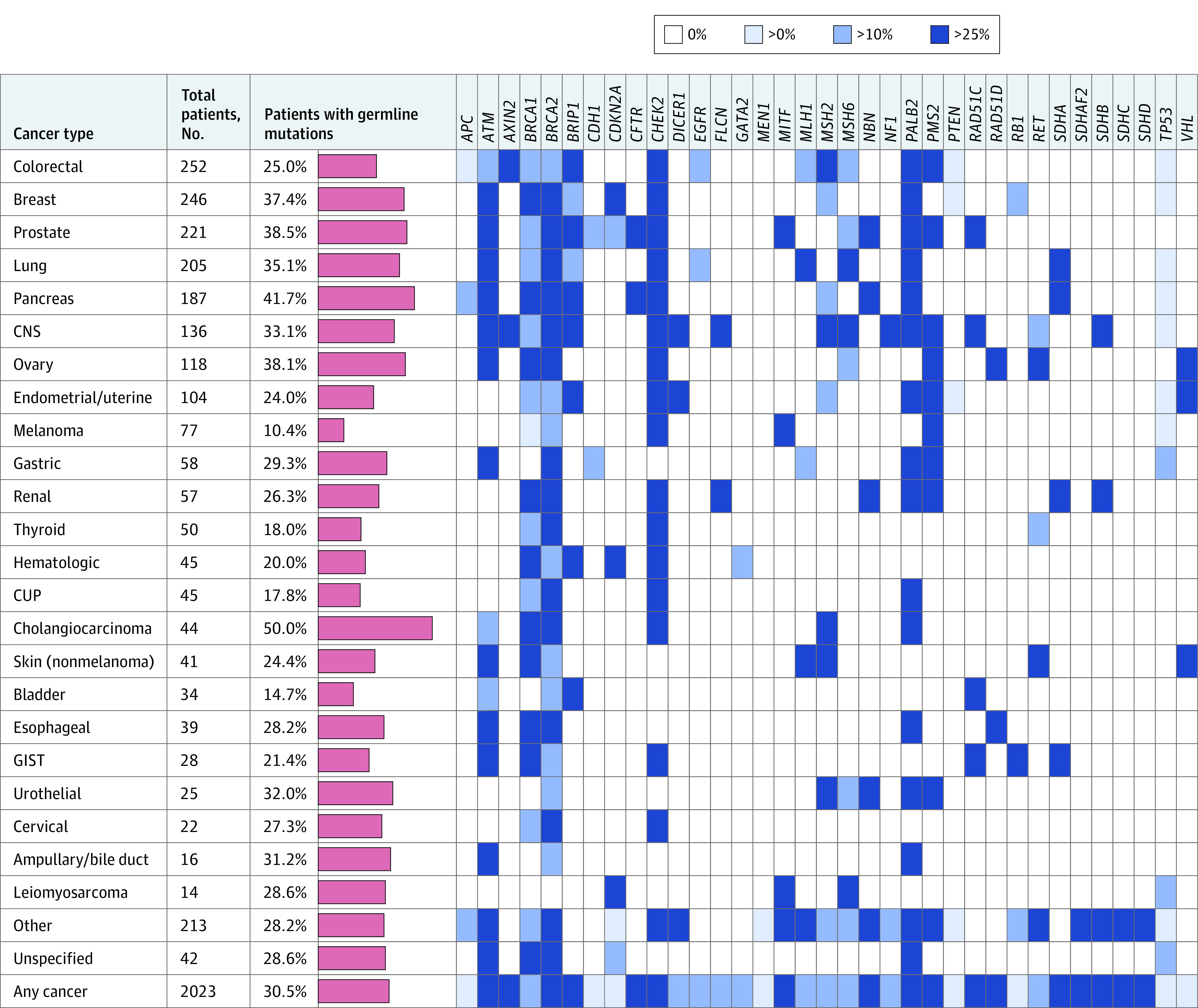
Number of patients with each cancer type and percentage of patients found to harbor 1 or more pathogenic germline variants (red bars, scale 0%-60%). Patients with more than 1 primary cancer were counted more than once in this figure. In addition, a heatmap of germline findings by gene and cancer type is shown. Blue cells indicate cancer/gene combinations in which PGVs were observed, with the shade indicating the fraction of findings determined to be of germline origin. CNS indicates central nervous system; CUP, cancer of unknown primary; and GIST, gastrointestinal stromal tumor.
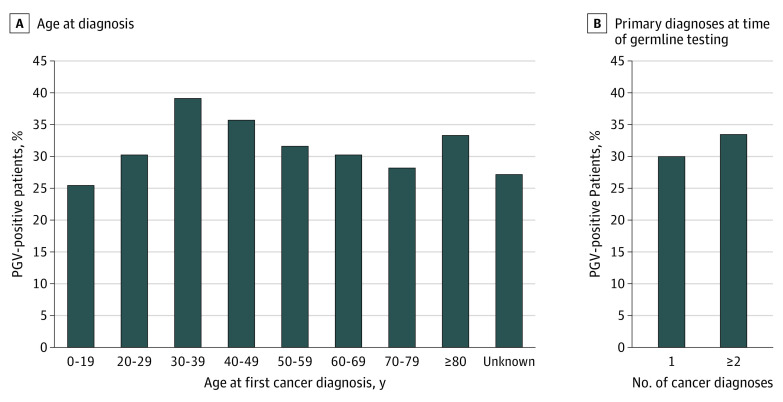
Figure 3. Prevalence of Pathogenic Germline Variants (PGVs), by Age and Number of Cancers.
Percentage of patients harboring PGVs by age at first cancer diagnosis (A) and number of primary cancer diagnoses at time of germline testing (B). The differences in PGV-positive rate between groups were not statistically significant or were of borderline significance (eg, age 0-19 vs 30-39 years, the largest difference between groups: pairwise P = .03).
Most PGVs we observed were potentially clinically actionable per current management guidelines, published expert opinion, approved precision therapy labels, and/or clinical trial eligibility criteria (Figure 1). Among those with germline findings, 69 patients (11.2%; 95% CI, 8.9%-14.0%) had PGVs identified only after presenting with a second, possibly preventable, primary cancer (eTable 1 in the Supplement). Thirty-two of these patients (46.4%) carried PGVs associated with specific screening or risk-reduction recommendations for their subsequent cancers.
Fifty PGVs (8.1%; 95% CI, 6.2%-10.5%) were not reported by tumor sequencing as either somatic or germline findings of clinical significance. These differences resulted from technical limitations of tumor sequencing, variant interpretation differences between the tumor and germline tests, or differences in the genes tested in the tumor and the germline (Table 2; eTable 2 in the Supplement). These PGVs were uncovered because clinicians ordered germline multigene panel tests rather than single gene tests for most patients in our study (Table 1). For the remaining PGVs (n = 567), the variant was reported by the tumor test as clinically significant but usually without any indication that it was (or even might be) a germline rather than somatic alteration. Measures that are often used to evaluate whether tumor findings are possibly of germline origin, specifically variant allele fractions (VAFs) and tumor cellularity (ie, neoplastic cell fraction within specimens), were usually not reported (eResults in the Supplement).
Table 2. Pathogenic Germline Variants Missed by Tumor Testing.
| No. | Likely reason for differencea |
|---|---|
| 13 | Gene not in the tumor testb |
| 14 | Technical limitation with tumor testc
|
| 11 | Variant interpretation differenced |
| 12 | Unknown reasone |
| 50 | Total |
Details are provided in eTable 2 in the Supplement.
In the case of hotspot assays (n = 2), the region of the gene containing the variation was not sequenced in the tumor.
Technical limitations detecting variants of these types are expected in tumor tests.
All of these variants met guidelines16 to be considered pathogenic in germline DNA. These variants, however, may not have met guidelines17 to be considered clinically significant if assumed incorrectly to be somatic. Most (n = 9) were missense variants that can require significant effort to research and interpret thoroughly.
Inadequate details were provided by the ordering clinician or tumor sequencing laboratory to determine a likely reason for the difference.
Discussion
Germline testing following tumor sequencing often produces findings that may impact patient care by influencing precision therapy selection, surgical decisions, additional cancer screening, reproductive choices, and genetic counseling in families. We observed PGVs across a wide range of genes, cancers, and patient ages. The high (>30%) germline-positive rate that we observed suggests that germline follow-up testing may be underused (ie, germline testing may have been preferentially applied to patients with the highest index of suspicion), although our study design did not allow us to explore this topic in detail (eMethods in the Supplement).
For comparison, Mandelker et al,12 on behalf of the European Society for Medical Oncology (ESMO), recommended criteria for germline analysis and follow-up testing intended to optimize the identification of actionable PGVs in tumor sequencing while avoiding “excessive diversion of effort” toward less productive germline follow-up testing. These criteria consider gene, tumor type, patient age, and VAF of the variants uncovered by tumor sequencing to determine which, if any, are recommended for follow-up. The ESMO criteria were designed to achieve a greater than 10% germline-positive rate per gene, although ESMO’s own analysis, using data from the Memorial Sloan Kettering (MSK) IMPACT study,9,12,18 demonstrated a higher overall positive rate of 63% among the 6.4% of variants meeting criteria.12 In our cohort, the ESMO criteria appeared to be effective, although conservative, as they would have recommended follow-up testing for only 82% of patients with PGV-positive findings (Figure 1)—slightly more than the 73% (653/890) of potentially actionable PGVs recommended for follow-up in the MSK-IMPACT cohort.12 The ESMO criteria thus miss 18% and 27% of PGVs in the 2 cohorts, respectively.
Our cohort analysis (Figure 1) did not use ESMO’s allele fraction criteria, however, as VAFs were not available for most of the tumor test results that we received (eResults in the Supplement). Rather, we considered all variants detected by tumor sequencing to be potentially of germline origin. Had we used VAFs, the fraction of PGVs with a follow-up testing recommendation in our cohort would have been less than 82%, as these additional criteria could only serve to exclude variants from follow-up, not add any. In the MSK-IMPACT data,12 VAFs were fairly informative (eg, 96.5% of PGVs met ESMO’s VAF minimums of 20%-30%), although other studies report limited correlation between VAF and somatic/germline status8,10,15 for both technical and biological reasons. This limitation is particularly true for insertion, deletion, and copy number variants, which together make up more than half of all PGVs in hereditary cancer genes.19,20 Thus, many current practice guidelines suggest caution in using tumor-only sequencing results, including VAF, to determine whether a variant is or is not potentially germline.1,2,13,17 The approach that we took in ignoring VAF (even when provided) may be clinically appropriate to maximize the yield of actionable germline findings, particularly for genes in which PGVs are relatively common, such as BRCA1 and MSH2 (Figure 1).
The National Comprehensive Cancer Network (NCCN) guidelines for follow-up germline testing are difficult to compare directly with the ESMO guideline, as the NCCN criteria are not as specific and vary by cancer type. However, we applied the NCCN-based algorithm proposed by Clark et al,15 which would have recommended follow-up testing for 78% of the PGV-positive findings in our study (Figure 1). Of note, in the Clark et al15 study, 31% of patients (37/118) were referred for germline testing using other clinical criteria external to this algorithm. Clearly, clinical judgment plays an important role in optimizing the sensitivity of germline follow-up testing, as the current guidelines capture many, but not all, patients who would potentially benefit.
Our observation that 8.1% of PGVs were missed by tumor sequencing was not unexpected. Diverse reasons for these omissions were observed (Table 2; eTable 2 in the Supplement), including (1) somatic variant interpretation guidelines17 differ from germline variant interpretation guidelines,16 and PGVs may not meet criteria to be considered clinically significant by tumor tests if these variants are incorrectly assumed to be somatic and interpreted as such; (2) high-quality germline tests can detect a broad spectrum of pathogenic variant types that are prevalent causes of inherited disease but present technical challenges,19,21,22,23 particularly when analyzing specimen types often encountered in oncology (eg, formalin-fixed, paraffin-embedded biopsy specimens or circulating tumor DNA [liquid biopsy]); and (3) tumor tests may not include all genes of potential germline relevance in any given patient.
These observations, together with our finding that 11.2% of PGVs were uncovered only after a second primary cancer was diagnosed in the patient, emphasize the importance of performing germline analysis independent of tumor sequencing in appropriate patients as early as possible. Clearly, tumor testing cannot substitute for germline testing in patients for whom a germline result is important to optimize care.
Genes that frequently acquire somatic alterations present a particular challenge1,12 because the yield of germline findings is relatively low, although the clinical impact can be substantial. TP53 (associated with Li-Fraumeni syndrome) and PTEN (Cowden syndrome) were specific examples in this study. The NCCN and ESMO criteria, which are intended to increase the germline-positive rate for these 2 genes, would have excluded most PGV-positive patients in our cohort from germline testing (Figure 1). The TP53 and PTEN variants identified by tumor sequencing were systematically underreported in the germline test requisitions that we received (eResults in the Supplement), presumably because clinicians sometimes assumed these variants to be somatic. Broader germline testing for these genes may be beneficial despite the low yield.
Limitations
This study had limitations. Like many cohort studies, a limitation of our study design is that we did not have a thorough understanding of the overall population from which our patients were drawn, ie, we did not have records for patients who were not referred for germline testing. This lack of data makes certain topics, such as the utilization patterns of germline follow-up testing or the positive rate in an unselected population, difficult to study in this cohort.
Conclusions
In this study, the high rate of actionable findings from germline testing following tumor sequencing suggests benefits for further integrating germline genetics into routine oncology testing and patient care. Current guidelines can help focus germline follow-up testing but at the expense of missing patients who might otherwise benefit. Additional considerations beyond those described herein apply to germline follow-up testing,15,24,25 and these can be particularly challenging in the community setting, where access to genetics professionals and time available for counseling may be limited. Additional clinical studies, including outcome and cost-effectiveness analysis, are warranted and may further justify addressing these challenges to continually improve precision oncology practices.
eMethods. Detailed Methods
eResults. Detailed Results
eTable 1. Patients With Multiple Sequential Primary Cancers (N = 69)
eTable 2. Germline Findings Not Reported by Tumor Tests (N = 50)
References
- 1.National Comprehensive Cancer Network . Genetic/familial high-risk assessment: breast, ovarian and pancreatic (version 1.2020). Published December 4, 2019. Accessed March 27, 2020. https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf
- 2.National Comprehensive Cancer Network . Genetic/familial high-risk assessment: colorectal (version 3.2019). Published December 13, 2019. Accessed March 27, 2020. https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf
- 3.Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017;168(4):584-599. doi: 10.1016/j.cell.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703-713. doi: 10.1038/nm.4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spencer DH, Ley TJ. Sequencing of tumor DNA to guide cancer risk assessment and therapy. JAMA. 2018;319(14):1497-1498. doi: 10.1001/jama.2018.2281 [DOI] [PubMed] [Google Scholar]
- 6.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523-533. doi: 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 7.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379(8):753-763. doi: 10.1056/NEJMoa1802905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meric-Bernstam F, Brusco L, Daniels M, et al. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol. 2016;27(5):795-800. doi: 10.1093/annonc/mdw018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandelker D, Zhang L, Kemel Y, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318(9):825-835. doi: 10.1001/jama.2017.11137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery ND, Selitsky SR, Patel NM, Hayes DN, Parker JS, Weck KE. Identification of germline variants in tumor genomic sequencing analysis. J Mol Diagn. 2018;20(1):123-125. doi: 10.1016/j.jmoldx.2017.09.008 [DOI] [PubMed] [Google Scholar]
- 11.National Comprehensive Cancer Network . Prostate cancer (version 1.2020). Published March 16, 2020. Accessed April 9, 2020. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- 12.Mandelker D, Donoghue M, Talukdar S, et al. Germline-focused analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. 2019;30(8):1221-1231. doi: 10.1093/annonc/mdz136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li MM, Chao E, Esplin ED, et al. ; ACMG Professional Practice and Guidelines Committee . Points to consider for reporting of germline variation in patients undergoing tumor testing: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020;22(7):1142-1148. doi: 10.1038/s41436-020-0783-8 [DOI] [PubMed] [Google Scholar]
- 14.National Comprehensive Cancer Network . Non-small cell lung cancer (version 3.2020). Published February 11, 2020. Accessed April 1, 2020. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- 15.Clark DF, Maxwell KN, Powers J, et al. Identification and confirmation of potentially actionable germline mutations in tumor-only genomic sequencing. JCO Precis Oncol. 2019;3:10.1200/PO.19.00076. doi: 10.1200/PO.19.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4-23. doi: 10.1016/j.jmoldx.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251-264. doi: 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lincoln SE, Kobayashi Y, Anderson MJ, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17(5):533-544. doi: 10.1016/j.jmoldx.2015.04.009 [DOI] [PubMed] [Google Scholar]
- 20.Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34(13):1460-1468. doi: 10.1200/JCO.2015.65.0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandelker D, Schmidt RJ, Ankala A, et al. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet Med. 2016;18(12):1282-1289. doi: 10.1038/gim.2016.58 [DOI] [PubMed] [Google Scholar]
- 22.Eichler EE. Genetic variation, comparative genomics, and the diagnosis of disease. N Engl J Med. 2019;381(1):64-74. doi: 10.1056/NEJMra1809315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lincoln SE, Hambuch T, Zook JM, et al. One in seven pathogenic variants can be challenging to detect by NGS: an analysis of 450,000 patients with implications for clinical sensitivity and genetic test implementation. medRXiv 20159434. Preprint. July 25, 2020. doi: 10.1101/2020.07.22.20159434 [DOI] [PMC free article] [PubMed]
- 24.Robson ME, Bradbury AR, Arun B, et al. American Society of Clinical Oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2015;33(31):3660-3667. doi: 10.1200/JCO.2015.63.0996 [DOI] [PubMed] [Google Scholar]
- 25.DeLeonardis K, Hogan L, Cannistra SA, Rangachari D, Tung N. When should tumor genomic profiling prompt consideration of germline testing? J Oncol Pract. 2019;15(9):465-473. doi: 10.1200/JOP.19.00201 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Detailed Methods
eResults. Detailed Results
eTable 1. Patients With Multiple Sequential Primary Cancers (N = 69)
eTable 2. Germline Findings Not Reported by Tumor Tests (N = 50)