Abstract
Biological invasions have increased in the last few decades mostly due to anthropogenic causes such as globalization of trade. Because invaders sometimes cause large economic losses and ecological disturbances, estimating their origin and potential geographical ranges is useful. Drosophila subobscura is native to the Old World but was introduced in the New World in the late 1970s and spread widely. We incorporate information on adaptive genetic markers into ecological niche modelling and then estimate the most probable geographical source of colonizers; evaluate whether the genetic bottleneck experienced by founders affects their potential distribution; and finally test whether this species has spread to all its potential suitable habitats worldwide. We find the environmental space occupied by this species in its native and introduced distributions are notably the same, although the introduced niche has shifted slightly towards higher temperature and lower precipitation. The genetic bottleneck of founding individuals was a key factor limiting the spread of this introduced species. We also find that regions in the Mediterranean and north-central Portugal show the highest probability of being the origin of the colonizers. Using genetically informed environmental niche modelling can enhance our understanding of the initial colonization and spread of invasive species, and also elucidate potential areas of future expansions worldwide.
Keywords: chromosome arrangements, ecological niche models, origin of colonizers
1. Introduction
Biological invasions have increased in the last few decades mostly due to anthropogenic causes such as globalization of trade [1]. Invasions can originate from a single bottlenecked introduction, recurrent introductions from the same population or admixture from genetically differentiated source areas; these alternatives can differentially alter the ecological and evolutionary outcome of the colonization [2].
Ecological niche modelling (ENM) has been extensively used to predict the potential geographical ranges of several invasive species from different taxa [3–6]. For some species, the invaded ranges closely match predictions based on their native ecological niches; but for others, invaded and native ranges are somewhat ecologically distinct [7–9]. Niches are considered ‘conserved’ when invaders retain their native ecological associations (e.g. have similar averages or limits of environmental parameters such as temperature, precipitation): this might occur if the initial colonizers were genetically diverse and did not encounter dispersal barriers. Niches are considered ‘shifted’ when the introduced niche differs from the native one [10,11]. Shifts can reflect unfilled niche space, which is that section of the environmental space unoccupied by the introduced population. Niche unfilling can occur because dispersal is limited, suitable environments are inaccessible or the initial bottleneck reduced adaptive genetic variation necessary for broad colonization. On the other hand, an expanded niche space occurs when the invading populations spread beyond the habitat defined by the native niche, perhaps because invaders have adapted to novel environmental conditions [12] or have experienced ecological release from competitors, predators or disease vectors [13].
The geographical source of the colonizers—and thus the colonizer's adaptive genetic composition—can have a profound impact on the invasion of new regions. Species with wide native distributions encounter diverse climatic conditions, and rely on behaviour, phenotypic plasticity or local genetic adaptation to persist in diverse environments [14]. However, most genetic markers can be considered neutral, as evidenced by their low population differentiation [15], and only a few loci show geographical variation and strong genetic differentiation, suggesting local adaptation [16,17]. Consequently, only the adaptive genetic composition of source populations of a widespread species should affect the environmental tolerances and hence potential distribution of invaders. Including information of adaptive loci in ENM might well improve their ability to predict the distribution potential of invasive species.
Chromosomal arrangements (e.g. inversions) have been long studied for their abundant polymorphism in Drosophila species, and with the advances of comparative genomics, they have been detected to be widespread across taxa and a major force in ecological and evolutionary processes [18,19]. Combinations of alleles in those chromosomal arrangements are thought to be adaptive and shown to respond to different environmental cues [20]. For instance, chromosomal arrangements contribute to adaptive divergence in the sunflower Helianthus petiolaris [21], to alternative freshwater ecotypes of rainbow trout (Oncorhynchus mykiss) [22] and to desiccation resistance in the mosquito Anopheles gambiae [23], and are responsible for parallel evolution to freshwater adaptation of marine sticklebacks (Gasterosteus aculeatus) [24]. In some Drosophila species, chromosomal arrangements show compelling evidence of adaptation to different environmental conditions: species invading new continents independently evolved latitudinal clines in chromosomal arrangements that are similar to those in the native region [25,26]. Moreover, some chromosomal arrangements that are more frequent in low latitudes appear adapted to warm temperatures, as their frequencies have increased worldwide in step with global warming [27,28] or increase annually during warm seasons, even shortly after heat waves [29]. By contrast, other chromosomal arrangements are most common in high-latitude populations and during cool seasons, suggesting adaptation to cold conditions [30]. Thus, adaptive chromosomal arrangements in the founders of introduced species can affect their subsequent geographical spread in the new areas.
This effect can be evaluated in the Palaearctic species Drosophila subobscura, which colonized both American continents in the late 1970s [25]. This was a single (unique) invasion from the native region, and the number of initial founders was very small, inducing a major genetic bottleneck [31]. These flies first invaded South America (Chile) and from there spread quickly to the west coast of North America and later to Argentina and Uruguay [25,31,32]. Subsequent but minor bottlenecks were identified in the secondary introductions to North America and Argentina from Chile [33,34].
Despite the strong genetic bottleneck during the initial founding and despite the lack of evidence of additional introductions [31], D. subobscura spread rapidly over a wide area (24° of latitude in South America and 17° in North America) in less than 3 years [35]. This rapid colonization was probably achieved by passive dispersal (human aided), and its breadth was facilitated because the colonizers still had chromosomal arrangements considered cold- and warm-adapted [30]. Moreover, in only a few years, these flies rapidly and independently evolved latitudinal clines in those chromosomal arrangements in both North and South America; and these clines were in the same direction as in the native range, thereby demonstrating their adaptive role [25]. Specifically, chromosomal arrangements considered warm-adapted developed negative frequency clines with latitude independently in both South and North America, whereas putatively cold-adapted arrangements evolved positive frequency clines with latitude [25,30]. Moreover, the chromosome index—based on the proportion of chromosomal arrangements in old and new samples from the same localities worldwide—is highly correlated with climate in all areas, whether native (Europe) or introduced (South America and North America), corroborating the adaptive value of this chromosomal polymorphism and its relation to temperature [27]. Nevertheless, latitudinal clines in chromosomal arrangements in the Americas never became as steep as the native European ones and failed to continue to converge through time: for example, the frequency of warm-adapted arrangements has remained high even at high latitudes on both continents [30,36]. This could be related to the specific origin of colonizers because comparisons of chromosomal arrangements and microsatellite loci between native and invasive populations suggest that the western Mediterranean region was the most probable source of colonizers [31,37]. Thus, environmental conditions in this area could influence the potential distribution of the species in introduced areas by constraining the adaptive genetic composition of founders.
In this study, we incorporate information on the genetic composition of colonizers into an ENM approach, and thereby investigate potential niche shifts in introduced areas as well as help elucidate the potential geographical origin of colonizers. First, we evaluate whether flies have spread to all potentially suitable habitats in the invaded area. Second, we test whether model projections from the native range into the introduced area differ for three alternative groups of putative colonizers based on their frequency of chromosomal arrangements. Specifically, we considered (i) all occurrences in the native range, (ii) data from native localities where mostly warm-adapted chromosomal arrangements are present, and (iii) native occurrences from areas with mostly cold-adapted arrangements. Finally, we perform back-projection of the introduced model into the native area and evaluate whether ENM provides insights on the most probable area of origin for the colonizers.
2. Material and methods
(a). Datasets and study areas
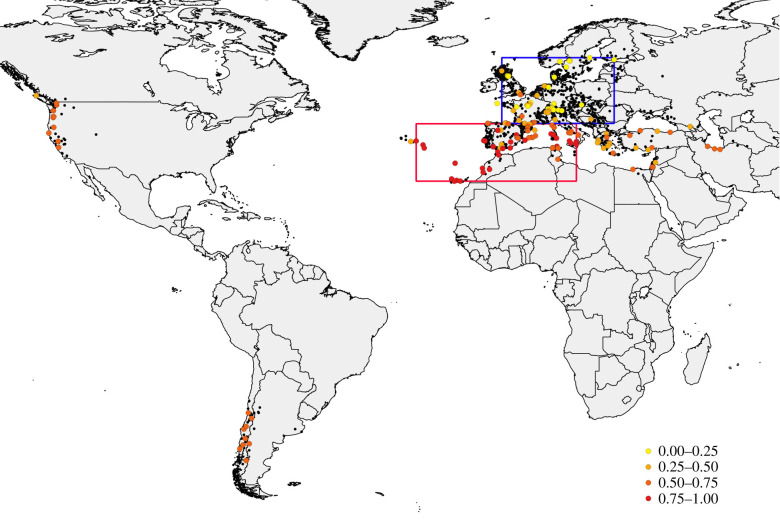
We extracted all worldwide records of presence data for D. subobscura from TAXODROS (www.taxodros.uzh.ch/). We validated data by reviewing the cited literature and removing a few detected errors where the species name was mentioned in the literature, indicating that it was not present but inadvertently considered as present in the database. We included 1467 presence data points in the species's native (1324 from Europe, 24 from Africa and 45 from Asia) and introduced areas (37 from South America and 37 from North America; electronic supplementary material, ‘Taxodros presence data’; figure 1). We compiled 207 published records on chromosomal arrangement polymorphisms (electronic supplementary material, ‘Chromosome inversion frequency’), 155 from locations in the native area and 52 in the introduced area [27,29,30,38,39]. We classified arrangements as cold-adapted if their frequencies in the native range increased with latitude, or warm-adapted if those frequencies decreased with latitude [30]. We estimated the mean frequency of warm-adapted arrangements present in America in native and introduced localities and plotted the results in figure 1 (electronic supplementary material, ‘Chromosome inversion frequency’).
Figure 1.
Map of the presence data points (black) of D. subobscura from TAXODROS for the Native area (Palaearctic region) and Introduced area (America). Colour points identify the mean frequency of warm-adapted chromosomal arrangements, present in America, in the Native and Introduced areas. Data points enclosed in the upper rectangle were included in the COLD chromosomal arrangement dataset and those enclosed in the lower rectangle in the WARM dataset. (Online version in colour.)
Localities in South and North America combined constituted the introduced range (INTRODUCED). We partitioned the presence sites in the native range into three datasets: one included all localities in the native area (NATIVE), a second included only locations having primarily cold-adapted arrangements (COLD) and a third included only locations having primarily warm-adapted arrangements (WARM). The chromosomal inversion polymorphism is very diverse in Drosophila subobscura with more than 80 identified arrangements in the native region, but only 23% of chromosomal arrangements are present in America, indicating a strong bottleneck [30]. Although in each chromosome, only one cold-adapted arrangement is present in the native and introduced areas, one to four warm-adapted arrangements per chromosome are present in both introduced areas; but many arrangements—especially from southern localities in the native region—are not found in the introduced region [25]. We made two datasets from the native area to focus on the areas with major frequencies of chromosomal arrangements present in America, while restricting to sites with mostly cold or mostly warm arrangements. To build the COLD dataset, we identified locations in the native area with a mean frequency of those cold-adapted arrangements present in America greater than or equal to 0.5 (electronic supplementary material, chromosomes data file). A total of 34 locations met this criterion and were used to partition a polygon demarcated between latitudes 43.4° N and 60.7° N and between longitudes −4.7° E and 25.0° E (figure 1). All presence records within this rectangle were used to build the COLD dataset (525 data points). To build the WARM dataset, we identified locations in the native area where the frequencies of the warm-adapted arrangements present in America were greater than or equal to 0.5 for each chromosome, with 32 locations meeting this criterion (electronic supplementary material, chromosomes data file). These locations partitioned a polygon for the WARM dataset demarcated between latitudes 28.1–43.2° N and longitudes −27.3° E–15.0° E (figure 1). All presence records in this rectangle were used to build the WARM dataset (274 data points). With these two partitions, we were assuming that if colonizers were from only a single area, then they would either have mostly cold- or mostly warm-adapted arrangements present in America, for all five chromosomes.
(b). Environmental data
We used two different study areas: (i) Eurasia, Africa and Oceania for the native distributional records and (ii) North and South America for the introduced records, including all the areas where the species could naturally disperse [40]. We used land climatic variables from WorldClim 2 [41]. We calculated pairwise correlations between 19 bioclimatic variables available. When two or more variables had Pearson correlation coefficients higher than 0.75, only one variable was retained to reduce co-linearity. This approach left six variables: TempDiurn = Bio2 (mean diurnal temperature range), TempWet = Bio8 (mean temperature of wettest quarter), TempDry = Bio9 (mean temperature of driest quarter), PrecipDry = Bio17 (precipitation of driest quarter), PrecipHot = Bio18 (precipitation of warmest quarter) and PrecipCold = Bio19 (precipitation of coldest quarter). The spatial resolution was 5 arc-minutes (approx. 10 km2).
(c). Environmental space overlap
To determine whether D. subobscura is using the same climatic niche in both native and introduced areas, we compared both ranges via equivalency and similarity tests [7,42], as implemented in Ecospat 3.0 R package [43]. Ecospat conducts a principal component analysis (PCA) on environmental values from both areas. Then, the density of occurrences for each study area over the two first PCA components is estimated with a kernel density function. Niche overlap is calculated using the D metric [44], which varies from 0 (no overlap) to 1 (complete overlap). Niche similarity between ranges can be tested by determining whether the observed D value is different from simulated D values based on 100 null models, which use records of one range as presence-only data and random records of the other range as background. The results are represented in a histogram: when the observed D metric falls above the 95% of the simulated values, both ranges occupy environments more similar than expected by chance. Ecospat splits the niche-overlap comparison into three components [10]: (i) stability is the proportion shared by both niches, (ii) unfilling is the proportion of the native niche that lies outside the introduced niche, and (iii) expansion is the proportion of the introduced niche that lies outside the native niche.
(d). Geographical space overlap
To analyse whether genetic composition and the bottleneck experienced might restrict the potential geographical range of colonizers, we calculated ecological niche models with all presence localities from the native area as the null model and then for localities having mostly cold- versus mostly warm-adapted chromosomal arrangements. We chose Maximum Entropy algorithm or Maxent [45], which requires presence and background records, to generate several ecological niche models [46]. Maxent compares the environmental data of presence points with the background, corresponding to the whole study area. By default, Maxent uses 10 000 background points selected randomly [47]. To allow model comparisons, we ran Maxent in logistic output format with default parameters and used 80% of presence records as training data and 20% as testing data. Duplicates and points falling in the ocean were eliminated. We calculated the arithmetic mean and standard deviation from a set of 10 iterations per dataset. In addition, we calculated a set of 100 null models [48]. Models were performed with Maxent 3.4.1 software (http://biodiversityinformatics.amnh.org/open_source/maxent/).
Four ecological niche models were calculated using the same modelling procedures: (i) model of all native records (NATIVE) and its projection to introduced areas (Americas), (ii) Model of native records with mostly cold arrangements (COLD) and its projection to introduced areas, (iii) Model of native records with mostly warm arrangements (WARM) and its projection to introduced areas, and (iv) Model of introduced records (INTRODUCED) and its back-projection to the native area. Each model was evaluated with receiver operated characteristics (ROC) plots, taking the area under the curve (AUC) as a measure of model fit [49]. Despite its dependence on the relationship between the extent of the study area and the species range [50], AUC remains an effective validation metric [51]. We identified the importance of each environmental variable in explaining species distributions.
To evaluate discrepancies among different groups of founders, we transformed each ecological niche model and projection into a habitat suitability map [46] with two categories (species presence versus absence): this involved applying the maximum training sensitivity plus specificity logistic threshold as provided by Maxent [45]. The habitat suitability map of the introduced area was compared with the projections of the three groups of putative colonizers (NATIVE, COLD and WARM). The same three comparisons were calculated between the models and the projections according to the geographical space: stability, unfilling and expansion. Finally, we estimated the most probable area of origin of colonizers by back projecting the introduced model to the native area and then by evaluating the resulting habitat suitability map.
3. Results
(a). Have the flies spread to fill all potentially suitable habitats in the invaded area?
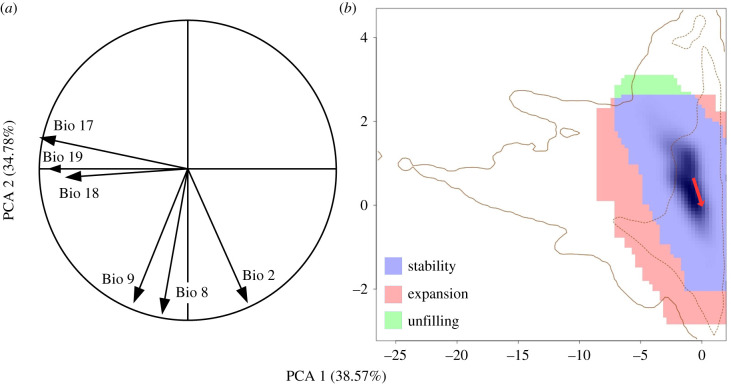
The first two principal components of the six bioclimatic variables for the combined study of the native and introduced areas explained about 73% of the total variation (figure 2). The first component (approx. 39%) grouped the three precipitation related variables, whereas the second component (approx. 35%) grouped the three temperature-related variables.
Figure 2.
Climatic niches of D. subobscura. Graphs of the first two principal components calculated from the six bioclimatic variables in the native and introduced areas. (a) Correlation analysis of the six bioclimatic variables for the two studied areas combined represented with the two first components. (b) Comparison of native and introduced climatic niches by the two main axes of the PCA representing the stability, unfilling and expansion in the introduced relative to the native area. The solid contour line represents 100% of the land climate of the whole world (excluding Antarctica) and the dotted inner line 50%. The arrow indicates centroid movement of the introduced relative to the native area. (Online version in colour.)
The introduced climatic niche had very high overlap with the native niche (high stability, 99.2%, figure 2), and the two climatic niches did not differ significantly, as niche similarity (D) was higher than expected by chance (p = 0.0198; electronic supplementary material, figure S1). Even so, the introduced niche has expanded slightly towards areas with higher temperature and lower precipitation than those in the native niche (0.8% expansion), but does not include some areas with low temperatures that are included in the niche of native flies (0.1% unfilling). Similarly, the centroid of the introduced niche was slightly shifted towards higher temperature and lower precipitation than that of the native area (figure 2).
(b). Would the invaded region be different if the founders had mostly cold- or warm-adapted arrangements?
The projections of the three native models (NATIVE, WARM and COLD) differed in the invaded region (figure 3). All Maxent models had both training and testing AUC values higher than 0.9 (electronic supplementary material, table S1). The variables with the highest contributions were always related to precipitation (electronic supplementary material, table S1). The most important variable was precipitation in the coldest quarter (PrecipCold) for both NATIVE and WARM models, precipitation of warmest quarter (PrecipHot) for the INTRODUCED model, but precipitation of driest quarter (PrecipCry) for the COLD model. Temperature-related variables were the second most important in all models (TempDry for NATIVE and COLD models and TempWet for the WARM model), except for the INTRODUCED one that was PrecipCold (electronic supplementary material, table S1).
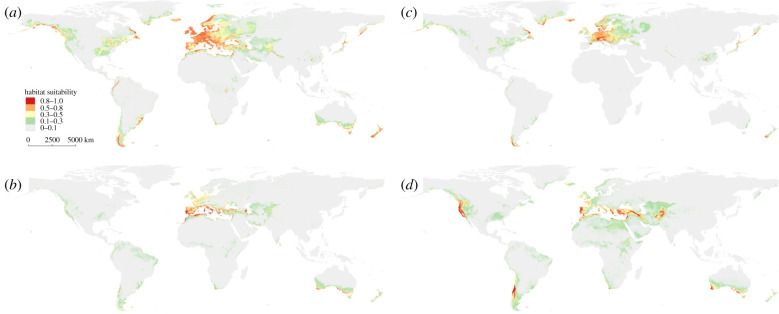
Figure 3.
Ecological niche models and projections. (a) NATIVE model and projection to the Americas, (b) WARM chromosomal arrangements model and projection to America, (c) COLD chromosomal arrangements model and projection to America and (d) INTRODUCED model (America) and projection back to the rest of the world. (Online version in colour.)
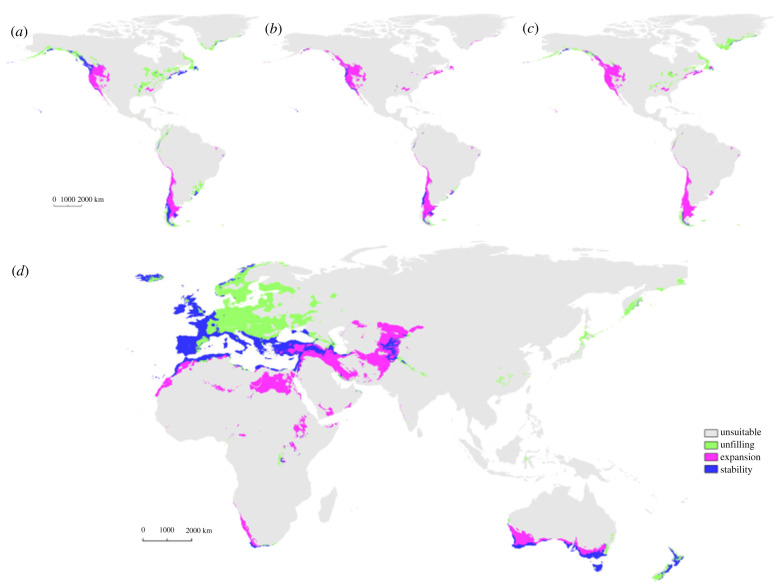
At the continental scale, most areas were considered unsuitable for both the model and the projection (figure 4; electronic supplementary material, table S2). The NATIVE projection yielded the largest stability when compared with the INTRODUCED model, whereas the WARM projection produced the smallest unfilling and the COLD projection the largest expansion. When compared with the NATIVE model, the INTRODUCED projection identified cold areas of central and northern Europe mostly as unfilled, and dryer areas in Africa, Middle East and Australia as expanded (figure 4).
Figure 4.
Geographical niche overlaps between the models and the projections. (a) INTRODUCED model overlapped with the projection of the NATIVE model in the Introduced area. (b) INTRODUCED model versus projection of the WARM model. (c) INTRODUCED model versus projection of the COLD model. (d) NATIVE model versus projection of the INTRODUCED model in the Native area. The stability represents the proportion of the niche that is shared by the model and the projection. The expansion is the proportion of the model non-overlapping the projection. The unfilling is the proportion of the projection non-overlapping the model. The unsuitable is the proportion of the niche predicted by the model and the projection as unsuitable. (Online version in colour.)
(c). Where was the most probable area of origin of colonizers?
The projection of the INTRODUCED model back to the NATIVE area identified the Mediterranean region (Libya, Lebanon, southern Turkey, southern Greece and around the Gibraltar strait) and north-central Portugal as regions with very high suitability (figure 3), suggesting that the colonizers may well have come from one of these regions. Stability was mostly detected around the Mediterranean region, unfilling was mostly identified in northern Europe, and expansion occurred in more desert environments. Thus, colonizers most likely carried a relatively high proportion of warm-adapted arrangements. Overall, stability, unfilling and expansion had similar proportions in these comparisons (figure 4; electronic supplementary material, table S2).
4. Discussion
Any prediction of the potential spread of invasive species makes a key assumption, namely that the niche of a species is largely conserved following the invasion. In fact, most invaders do seem to occupy climates similar to their source populations, but shifts have been observed in some taxa [7,8]. Niche shifts may result from novel biotic interactions and ecological release from competitors, predators and parasites in invaded areas [13,52], plasticity and behaviour [53,54], or from the ability of invasive species to thrive in human-altered environments in the introduced range [55]. Thus, human disturbances and habitat accessibility may be important to achieve invasion of all suitable space in the introduced area [8].
Here, we show that the genetic composition of founding individuals also affects the potential range of introduced species. In D. subobscura, the native and introduced environmental spaces are very similar, consistent with niche conservatism. Even so, introduced flies showed a small shift towards environments with relatively high temperature and low precipitation, but away from environments with relatively low temperature. This geographical pattern is consistent with independent genetic evidence, suggesting that colonizers likely came from the Mediterranean region rather than northern Europe and also that they suffered a large bottleneck during the colonization [31,37]. Despite this bottleneck, latitudinal clines in the frequency of many colonizing chromosomal arrangements quickly developed, although their slopes were less steep than in the native range [25]. The frequencies of cold-adapted arrangements in D. subobscura at high latitudes in the introduced area are lower than at equivalent latitudes in the native area [30]. These lower frequencies may be constrained by the small number and geographical origin (Mediterranean region) of colonizers [31,37], and by its low genetic diversity further maintained by gene flux reduction between arrangements [56]. In the geographical space, we found that unfilling was smallest for the projection of the WARM model, whereas stability was smallest for the projection of the COLD model. Thus, these two observations also suggest a Mediterranean source of the invaders. Moreover, the WARM model—but not the COLD model—correctly predicted areas of western US, such as Washington, Oregon and California, where the species has indeed spread. Therefore, the origin and genetic composition of the founders seem to have influenced the potential spread of this invasion.
The Maxent models also suggest that D. subobscura could potentially invade areas of the world beyond North and South America. Should these flies ever invade Greenland (extreme southern region) or eastern North America, their persistence will be more likely if the invading stock carries mostly cold-adapted arrangements. In this case, successful colonizers would more likely have to come from northern Europe than from western North America or southern Europe and Africa. Our models also predict that D. subobscura could invade Australia, especially if the colonizers carry mostly warm-adapted arrangements, as holds for flies from America, southern Europe or northern Africa. Thus, genetically informed ecological niche models [57–59] can improve predicting future introduced sites for invasive species and thus have the potential to guide prevention strategies.
(a). Shift in environmental niche
Native species with wide distributions encounter diverse environmental regimes. Such species may show population structuring reflecting migration at neutral loci and selection at loci under local adaptation [15], with environmental fluctuations maintaining local polymorphism by balancing selection [29]. Low genetic differentiation within chromosomal arrangements but high differentiation between arrangements has been observed in D. subobscura across a wide environmental gradient in Europe both at neutral and candidate loci for thermal adaptation [60,61]. This suggests that gene flow is high, but that recombination is limited by inversions [56], which maintain specific allelic combinations that seemingly have positive effects on fitness [20]. Fitness of specific chromosomal arrangements is clearly related to environmental conditions, shown by the parallel intercontinental frequency clines seen in different insects [25,26].
Because of high overlaps in the environmental space of D. subobscura from native and invasive regions, the invasive flies appear to have already spread to most of their adjacent potential suitable habitat in the invaded region. Nonetheless, overlaps in the geographical space of the introduced model with the projections from the three native models showed that expansion was higher than stability, and with a shift towards hotter climate. The observed niche shift in the introduced area could be facilitated if founders came from a relatively warm part of the native range and thus arrived with predominately warm-adapted arrangements. Thus, pre-adaptation to abiotic conditions would be largely driving the spatial distribution of invading populations. Consequently, environmental variables could be used to predict the risk of—and potential sites of—invasion of alien species worldwide.
Ecological niche models can be improved by incorporating genetic information to forecast range shifts, as has been demonstrated in analyses of different plant species [57,59,62]. Moreover, in a few invasive species, genetically informed ecological models have been used to explore whether niche conservation or rapid adaptation occurs during invasion [58,63,64]. Niche expansions could occur if the founding stock was a mixture of genetically differentiated populations or if recurrent introductions from diverse sources occur [2]. Modelling the risk of invasion worldwide would be difficult if founders were a mixture of genetically distinct stocks, as is common in introduced populations [65,66]. For example, this could be the case for D. suzukii, which was recently introduced worldwide and genetic analyses indicate independent invasions from diverse locations, and admixture in some of the invaded regions [67]. Global distribution modelling has shown a shift in the ecological niche of D. suzukii [11] that could be mediated by the mixed origin of colonizers. Under complex invasion scenarios, information on adaptive genetic markers in the native and introduced areas, specially linked to chromosomal arrangements due to their recombination reduction in heterokaryotypes [56,68], can also improve the ability to predict species distributions. In these complex invasions, forward projections that use knowledge about the genetic composition of particular regions in the native range to predict the invaded areas, as done in the present study, can further improve our ability to predict the future geographical distribution of invasive species. In particular, by incorporating information on candidate loci for local adaptation, we show that genetically informed environmental niche modelling can help predict habitat suitability in the invaded range and thus aid in our understanding and ability to monitor the distribution of invasive species.
Drosophila subobscura, as well as many other invasive species, is most commonly found in human-modified environments in the introduced area and thus anthropogenic factors might contribute to dispersal as well as niche shifts [8]. Human-modified structures can create microhabitats suitable—or sometimes not—for the presence of the species within harsh environments. Unfortunately, these microhabitats cannot be detected by the low spatial resolution of environmental variables generally used for modelling distributions worldwide, being detected, therefore, as apparent niche shifts. Moreover, survival in harsh environments might be enhanced by thermoregulatory behaviour compensation [53]. Thus, the expansion predicted towards more deserted regions in America could be the result of a combination of behaviour and microhabitat suitable conditions associated with human modified structures, avoiding competition with native species [69] and explaining the secondary invasions already found in these regions [33].
(b). Origin of colonizers
The source of the original colonizers is of long-standing interest, and prior studies have compared the genetic composition of the invaders with that of flies from various regions in the Old World [31,37]. In general, these studies point towards the western Mediterranean as likely sources. Environmental niche modelling cannot only predict where introduced species can spread, but also identify from where they came. In D. subobscura, our reverse projections pinpoint six Old World regions (Libya, Lebanon, southern Turkey, southern Greece, around the Gibraltar strait and northern Portugal) with very high probabilities as being source populations (figure 3d). Some regions (such as Libya) still lack genetic analyses, and some others can be discarded as origins of the colonizers based on their chromosomal inversion polymorphisms. For instance, localities in Israel (as a proxy for Lebanon, where chromosomal arrangements have not been assayed) could be discarded because flies there lack the UST arrangement, which is frequent in all American localities, and because they have high frequencies (greater than 80%) of arrangements of the J chromosome not found in America [38,70]. Similarly, localities in Turkey have high frequencies of U chromosome arrangements that are not detected in America [9]. Similarly, lethal genes and gene sequences associated with chromosomal inversions do not support Greece as a source for colonizers [71].
The remaining region pinpointed by our reverse projections is the western Mediterranean, and this potential source is supported by chromosomal evidence [37]. The region around the Gibraltar strait has all the chromosomal inversions identified in South and North America, and microsatellite analyses were consistent with that site as a possible source of colonizers, although the Catalonia region, not predicted by our projections, had a higher likelihood of being the source based on microsatellites [31]. Finally, the region around north-central Portugal also has all chromosomal arrangements present in America [39], but molecular markers there have never been compared to American ones. Thus, these two regions are good candidates for future genetic studies of the possible origin of invaders.
This back-projection approach can also be used in species with complex invasion scenarios, as in D. suzukii [67]. In such complex invasions, reverse projections of groups of genetically independent colonized locations could provide an ecologically based assessment of the potential origin of colonizers in each event.
In conclusion, we show that environmental niche modelling in combination with a genetic perspective on candidate loci for local adaptation is a powerful tool to predict potential areas of introduction and expansion of alien species worldwide. We demonstrate that potential distributions in the invaded areas depend in part on the specific genetic composition of the colonizers, which in turn depends on the geographical origin of the colonizers as well as any genetic bottleneck associated with the invasion. Therefore, sound global legislative and policy responses on invasive species can take advantage of robust predictions concerning the likely origin and expansion of invasive alien species [72], and this goal may be best achieved by combining environmental and genetic information.
Supplementary Material
Supplementary Material
Acknowledgements
We dedicate this paper to the memory of our dear friend and colleague George W. Gilchrist. He was appreciated as a scientist, mentor, administrator and organizer of good times.
Data accessibility
All presence data points and information on population chromosomal inversion frequency used to build the different datasets are available in the electronic supplementary material.
Authors' contributions
N.S., M.P., R.B.H. and G.G. designed the study. G.G. obtained all chromosome data. M.P. obtained all presence records. N.S. did all analyses. All authors contributed in writing and revising the manuscript and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by CEEC2017 contract (CEECIND/02213/2017) from FCT (Fundação para a Ciência e a Tecnologia, Portugal) to N.S., project CTM2017-88080 (AEI/FEDER, UE) to M.P. part of the research group 2017SGR-1120, NSF 1038016 and DEB-9981598 to R.B.H., NSF DEB-9981555 to G.G. The views expressed in this article do not necessarily reflect those of the National Science Foundation or the United States government.
References
- 1.Westphal MI, Browne M, MacKinnon K, Noble I. 2008. The link between international trade and the global distribution of invasive alien species. Biol. Invasions 10, 391–398. ( 10.1007/s10530-007-9138-5) [DOI] [Google Scholar]
- 2.Rius M, Darling JA. 2014. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 29, 233–242. ( 10.1016/j.tree.2014.02.003) [DOI] [PubMed] [Google Scholar]
- 3.Silva-Rocha I, Salvi D, Sillero N, Mateo JA, Carretero MA. 2015. Snakes on the Balearic Islands: an invasion tale with implications for native biodiversity conservation. PLoS ONE 10, e0121026 ( 10.1371/journal.pone.0121026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbet-Massin M, Rome Q, Villemant C, Courchamp F. 2018. Can species distribution models really predict the expansion of invasive species? PLoS ONE 13, e0193085 ( 10.1371/journal.pone.0193085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman D, Pescott OL, Roy HE, Tanner R. 2019. Improving species distribution models for invasive non-native species with biologically informed pseudo-absence selection. J. Biogeogr. 46, 1029–1040. ( 10.1111/jbi.13555) [DOI] [Google Scholar]
- 6.Briscoe RRD, Lake T, Tiffin P, Moeller DA. 2019. Species distribution models throughout the invasion history of Palmer amaranth predict regions at risk of future invasion and reveal challenges with modeling rapidly shifting geographic ranges. Sci. Rep. 9, 2426 ( 10.1038/s41598-018-38054-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petitpierre B, Kueffer C, Broennimann O, Randin C, Daehler C, Guisan A. 2012. Climatic niche shifts are rare among terrestrial plant invaders. Science 335, 1344–1348. ( 10.1126/science.1215933) [DOI] [PubMed] [Google Scholar]
- 8.Hill MP, Gallardo B, Terblanche JS. 2017. A global assessment of climatic niche shifts and human influence in insect invasions. Glob. Ecol. Biogeogr. 26, 679–689. ( 10.1111/geb.12578) [DOI] [Google Scholar]
- 9.Götz W. 1967. Untersuchungen über den chromosomalen Strukturpolymorphismus in kleinasiatischen und persischen Populationen von Drosophila subobscura Coll. Mol. Gen. Genet. 100, 1–38. ( 10.1007/BF00425773) [DOI] [PubMed] [Google Scholar]
- 10.Guisan A, Petitpierre B, Broennimann O, Daehler C, Kueffer C. 2014. Unifying niche shift studies: insights from biological invasions. Trends Ecol. Evol. 29, 260–269. ( 10.1016/j.tree.2014.02.009) [DOI] [PubMed] [Google Scholar]
- 11.Ørsted IV, Ørsted M. 2019. Species distribution models of the spotted wing Drosophila (Drosophila suzukii, Diptera: Drosophilidae) in its native and invasive range reveal an ecological niche shift. J. Appl. Ecol. 56, 423–435. ( 10.1111/1365-2664.13285) [DOI] [Google Scholar]
- 12.Leal M, Gunderson AR. 2012. Rapid change in the thermal tolerance of a tropical lizard. Am. Nat. 180, 815–822. ( 10.1086/668077) [DOI] [PubMed] [Google Scholar]
- 13.Tingley R, Vallinoto M, Sequeira F, Kearney MR. 2014. Realized niche shift during a global biological invasion. Proc. Natl Acad. Sci. USA 111, 10 233–10 238. ( 10.1073/pnas.1405766111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merilä J, Hendry AP. 2014. Climate change, adaptation, and phenotypic plasticity: the problem and the evidence. Evol. Appl. 7, 1–14. ( 10.1111/eva.12137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carreras C, Ordóñez V, Zane L, Kruschel C, Nasto I, Macpherson E, Pascual M. 2017. Population genomics of an endemic Mediterranean fish: differentiation by fine scale dispersal and adaptation. Sci. Rep. 7, 43417 ( 10.1038/srep43417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamichhaney S, et al. 2012. Population-scale sequencing reveals genetic differentiation due to local adaptation in Atlantic herring. Proc. Natl Acad. Sci. USA 109, 19 345–19 350. ( 10.1073/pnas.1216128109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreras C, García-Cisneros A, Wangensteen OS, Ordóñez V, Palacín C, Pascual M, Turon X. 2020. East is East and West is West: population genomics and hierarchical analyses reveal genetic structure and adaptation footprints in the keystone species Paracentrotus lividus (Echinoidea). Divers. Distrib. 26, 382–398. ( 10.1111/ddi.13016) [DOI] [Google Scholar]
- 18.Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8, e1000501 ( 10.1371/journal.pbio.1000501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellenreuther M, Bernatchez L. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 33, 427–440. ( 10.1016/j.tree.2018.04.002) [DOI] [PubMed] [Google Scholar]
- 20.Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434. ( 10.1534/genetics.105.047985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang K, Andrew RL, Owens GL, Ostevik KL, Rieseberg LH. 2020. Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype. Mol. Ecol. 29, 2535–2549. ( 10.1111/mec.15428) [DOI] [PubMed] [Google Scholar]
- 22.Arostegui MC, Quinn TP, Seeb LW, Seeb JE, McKinney GJ. 2019. Retention of a chromosomal inversion from an anadromous ancestor provides the genetic basis for alternative freshwater ecotypes in rainbow trout. Mol. Ecol. 28, 1412–1427. ( 10.1111/mec.15037) [DOI] [PubMed] [Google Scholar]
- 23.Ayala D, Zhang S, Chateau M, Fouet C, Morlais I, Costantini C, Hahn MW, Besansky NJ. 2019. Association mapping desiccation resistance within chromosomal inversions in the African malaria vector Anopheles gambiae. Mol. Ecol. 28, 1333–1342. ( 10.1111/mec.14880) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61. ( 10.1038/nature10944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prevosti A, Ribo G, Serra L, Aguade M, Balaña J, Monclus M, Mestres F. 1988. Colonization of America by Drosophila subobscura: experiment in natural populations that supports the adaptive role of chromosomal-inversion polymorphism. Proc. Natl Acad. Sci. USA 85, 5597–5600. ( 10.1073/pnas.85.15.5597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mérot C, Berdan EL, Babin C, Normandeau E, Wellenreuther M, Bernatchez L. 2018. Intercontinental karyotype–environment parallelism supports a role for a chromosomal inversion in local adaptation in a seaweed fly. Proc. R. Soc. B 285, 20180519 ( 10.1098/rspb.2018.0519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balanyà J, Oller JM, Huey RB, Gilchrist GW, Raymond MO, Serra L. 2006. Global genetic change tracks global climate warming in Drosophila subobscura. Science 313, 1773–1775. ( 10.1126/science.1131002) [DOI] [PubMed] [Google Scholar]
- 28.Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. 2005. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691–693. ( 10.1126/science.1109523) [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez-Trelles F, Tarrío R, Santos M. 2013. Genome-wide evolutionary response to a heat wave in Drosophila. Biol. Lett. 9, 20130228 ( 10.1098/rsbl.2013.0228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balanyà J, Serra L, Gilchrist GW, Huey RB, Pascual M, Mestres F, Solé E. 2003. Evolutionary pace of chromosomal polymorphism in colonizing populations of Drosophila subobscura: an evolutionary time series. Evolution 57, 1837–1845. ( 10.1111/j.0014-3820.2003.tb00591.x) [DOI] [PubMed] [Google Scholar]
- 31.Pascual M, Chapuis MP, Mestres F, Balanyà J, Huey RB, Gilchrist GW, Serra L, Estoup A. 2007. Introduction history of Drosophila subobscura in the New World: a microsatellite-based survey using ABC methods. Mol. Ecol. 16, 3069–3083. ( 10.1111/j.1365-294X.2007.03336.x) [DOI] [PubMed] [Google Scholar]
- 32.Goñi B, Martínez ME. 1995. First record of Drosophila subobscura in Uruguay. Drosoph. Inf. Serv. 76, 164. [Google Scholar]
- 33.Noor MAF, Pascual M, Smith KR. 2000. Genetic variation in the spread of Drosophila subobscura from a nonequilibrium population. Evolution 54, 696–703. ( 10.1554/0014-3820(2000)054[0696:GVITSO]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 34.Fernández Iriarte PJ, Balanyà J, Pascual M, Mestres F, Hasson ER, Fontdevila A, Serra L. 2009. Tracking the origin of an invasive species: Drosophila subobscura in Argentina. J. Evol. Biol. 22, 650–658. ( 10.1111/j.1420-9101.2008.01659.x) [DOI] [PubMed] [Google Scholar]
- 35.Ayala FJ, Serra L, Prevosti A. 1989. A grand experiment in evolution: the Drosophila subobscura colonization of the Americas. Genome 31, 246–255. ( 10.1139/g89-042) [DOI] [Google Scholar]
- 36.Castañeda LE, Balanyà J, Rezende EL, Santos M. 2013. Vanishing chromosomal inversion clines in Drosophila subobscura from Chile: is behavioral thermoregulation to blame? Am. Nat. 182, 249–259. ( 10.1086/671057) [DOI] [PubMed] [Google Scholar]
- 37.Brncic D, Prevosti A, Budnik M, Monclus M, Ocana J. 1981. Colonization of Drosophila subobscura in Chile I. First population and cytogenetic studies. Genetica 56, 3–9. ( 10.1007/BF00126923) [DOI] [Google Scholar]
- 38.Menozzi P, Krimbas CB. 1992. The inversion polymorphism of D. subobscura revisited: synthetic maps of gene arrangement frequencies and their interpretation. J. Evol. Biol. 5, 625–641. ( 10.1046/j.1420-9101.1992.5040625.x) [DOI] [Google Scholar]
- 39.Santos J, et al. 2016. Tracking changes in chromosomal arrangements and their genetic content during adaptation. J. Evol. Biol. 29, 1151–1167. ( 10.1111/jeb.12856) [DOI] [PubMed] [Google Scholar]
- 40.Anderson RP, Raza A. 2010. The effect of the extent of the study region on GIS models of species geographic distributions and estimates of niche evolution: preliminary tests with montane rodents (genus Nephelomys) in Venezuela. J. Biogeogr. 37, 1378–1393. ( 10.1111/j.1365-2699.2010.02290.x) [DOI] [Google Scholar]
- 41.Fick SE, Hijmans RJ. 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315. ( 10.1002/joc.5086) [DOI] [Google Scholar]
- 42.Broennimann O, et al. 2012. Measuring ecological niche overlap from occurrence and spatial environmental data. Glob. Ecol. Biogeogr. 21, 481–497. ( 10.1111/j.1466-8238.2011.00698.x) [DOI] [Google Scholar]
- 43.Di Cola V, et al. 2017. ecospat: an R package to support spatial analyses and modeling of species niches and distributions. Ecography 40, 774–787. ( 10.1111/ecog.02671) [DOI] [Google Scholar]
- 44.Warren DL, Glor RE, Turelli M. 2008. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution 62, 2868–2883. ( 10.1111/j.1558-5646.2008.00482.x) [DOI] [PubMed] [Google Scholar]
- 45.Phillips SJ, Anderson RP, Schapire RE. 2006. Maximum entropy modeling of species geographic distributions. Ecol. Modell. 190, 231–259. ( 10.1016/j.ecolmodel.2005.03.026) [DOI] [Google Scholar]
- 46.Sillero N. 2011. What does ecological modelling model? A proposed classification of ecological niche models based on their underlying methods. Ecol. Modell. 222, 1343–1346. ( 10.1016/j.ecolmodel.2011.01.018) [DOI] [Google Scholar]
- 47.Phillips SJ, Dudík M, Elith J, Graham CH, Lehmann A, Leathwick J, Ferrier S. 2009. Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol. Appl. 19, 181–197. ( 10.1890/07-2153.1) [DOI] [PubMed] [Google Scholar]
- 48.Raes N, ter Steege H. 2007. A null-model for significance testing of presence-only species distribution models. Ecography (Cop.). 5, 727–736. ( 10.1111/j.2007.0906-7590.05041.x) [DOI] [Google Scholar]
- 49.Liu C, Berry PM, Dawson TP, Pearson RG. 2005. Selecting thresholds of occurrence in the prediction of species distributions. Ecography (Cop.). 28, 385–393. ( 10.1111/j.0906-7590.2005.03957.x) [DOI] [Google Scholar]
- 50.Lobo JM, Jiménez-valverde A, Real R. 2008. AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17, 145–151. ( 10.1111/j.1466-8238.2007.00358.x) [DOI] [Google Scholar]
- 51.VanDerWal J, Shoo LP, Graham C, Williams SE. 2009. Selecting pseudo-absence data for presence-only distribution modeling: how far should you stray from what you know? Ecol. Modell. 220, 589–594. ( 10.1016/j.ecolmodel.2008.11.010) [DOI] [Google Scholar]
- 52.Mitchell CE, et al. 2006. Biotic interactions and plant invasions. Ecol. Lett. 9, 726–740. ( 10.1111/j.1461-0248.2006.00908.x) [DOI] [PubMed] [Google Scholar]
- 53.Huey RB, Pascual M. 2009. Partial thermoregulatory compensation by a rapidly evolving invasive species along a latitudinal cline. Ecology 90, 1715–1720. ( 10.1890/09-0097.1) [DOI] [PubMed] [Google Scholar]
- 54.Foucaud J, Moreno C, Pascual M, Rezende EL, Castañeda LE, Gibert P, Mery F. 2016. Introduced Drosophila subobscura populations perform better than native populations during an oviposition choice task due to increased fecundity but similar learning ability. Ecol. Evol. 6, 1725–1736. ( 10.1002/ece3.2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.González-Moreno P, Diez JM, Richardson DM, Vilà M. 2015. Beyond climate: disturbance niche shifts in invasive species. Glob. Ecol. Biogeogr. 24, 360–370. ( 10.1111/geb.12271) [DOI] [Google Scholar]
- 56.Pegueroles C, Ordóñez V, Mestres F, Pascual M. 2010. Recombination and selection in the maintenance of the adaptive value of inversions. J. Evol. Biol. 23, 2709–2717. ( 10.1111/j.1420-9101.2010.02136.x) [DOI] [PubMed] [Google Scholar]
- 57.Ikeda DH, Max TL, Allan GJ, Lau MK, Shuster SM, Whitham TG. 2017. Genetically informed ecological niche models improve climate change predictions. Glob. Chang. Biol. 23, 164–176. ( 10.1111/gcb.13470) [DOI] [PubMed] [Google Scholar]
- 58.Sotka EE, et al. 2018. Combining niche shift and population genetic analyses predicts rapid phenotypic evolution during invasion. Evol. Appl. 11, 781–793. ( 10.1111/eva.12592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marcer A, Méndez-Vigo B, Alonso-Blanco C, Picó FX. 2016. Tackling intraspecific genetic structure in distribution models better reflects species geographical range. Ecol. Evol. 6, 2084–2097. ( 10.1002/ece3.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simões P, Calabria G, Picão-Osório J, Balanyà J, Pascual M. 2012. The genetic content of chromosomal inversions across a wide latitudinal gradient. PLoS ONE 7, e51625 ( 10.1371/journal.pone.0051625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simões P, Pascual M. 2018. Patterns of geographic variation of thermal adapted candidate genes in Drosophila subobscura sex chromosome arrangements. BMC Evol. Biol. 18, 60 ( 10.1186/s12862-018-1178-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Theodoridis S, Patsiou TS, Randin C, Conti E. 2018. Forecasting range shifts of a cold-adapted species under climate change: are genomic and ecological diversity within species crucial for future resilience? Ecography 41, 1357–1369. ( 10.1111/ecog.03346) [DOI] [Google Scholar]
- 63.Herrando-Moraira S, Nualart N, Herrando-Moraira A, Chung MY, Chung MG, López-Pujol J. 2019. Climatic niche characteristics of native and invasive Lilium lancifolium. Sci. Rep. 9, 14334 ( 10.1038/s41598-019-50762-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rey O, et al. 2012. Where do adaptive shifts occur during invasion? A multidisciplinary approach to unravelling cold adaptation in a tropical ant species invading the Mediterranean area. Ecol. Lett. 15, 1266–1275. ( 10.1111/j.1461-0248.2012.01849.x) [DOI] [PubMed] [Google Scholar]
- 65.Kolbe JJ, Larson A, Losos JB, de Queiroz K. 2008. Admixture determines genetic diversity and population differentiation in the biological invasion of a lizard species. Biol. Lett. 4, 434–437. ( 10.1098/rsbl.2008.0205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rius M, Turon X, Ordóñez V, Pascual M. 2012. Tracking invasion histories in the sea: facing complex scenarios using multilocus data. PLoS ONE 7, e35815 ( 10.1371/journal.pone.0035815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fraimout A, et al. 2017. Deciphering the routes of invasion of Drosophila suzukii by means of ABC random forest. Mol. Biol. Evol. 34, 980–996. ( 10.1093/molbev/msx050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pegueroles C, Ferrés-Coy A, Martí-Solano M, Aquadro CF, Pascual M, Mestres F. 2016. Inversions and adaptation to the plant toxin ouabain shape DNA sequence variation within and between chromosomal inversions of Drosophila subobscura. Sci. Rep. 6, 23754 ( 10.1038/srep23754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pascual M, Serra L, Ayala FJ. 1998. Interspecific laboratory competition of the recently sympatric species Drosophila subobscura and Drosophila pseudoobscura. Evolution 52, 269–274. ( 10.2307/2410944) [DOI] [PubMed] [Google Scholar]
- 70.Goldschmidt E. 1956. Chromosomal polymorphism in a population of Drosophila subobscura from Israel. J. Genet. 54, 474–496. ( 10.1007/BF02982962) [DOI] [Google Scholar]
- 71.Araúz PA, Peris-Bondia F, Latorre A, Serra L, Mestres F. 2011. Molecular evidence to suggest the origin of a colonization: Drosophila subobscura in America. Genetica 139, 1477–1486. ( 10.1007/s10709-012-9647-8) [DOI] [PubMed] [Google Scholar]
- 72.Turbelin AJ, Malamud BD, Francis RA. 2017. Mapping the global state of invasive alien species: patterns of invasion and policy responses. Glob. Ecol. Biogeogr. 26, 78–92. ( 10.1111/geb.12517) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All presence data points and information on population chromosomal inversion frequency used to build the different datasets are available in the electronic supplementary material.