Abstract
A new class of compounds formed by the linkage of −C(O)–NH– with pyridine and thiazole moieties was designed, synthesized, and characterized by various spectral approaches. The newly characterized compounds were evaluated for their antimicrobial as well as anti-inflammatory properties. The in vitro anti-inflammatory activity of these compounds was evaluated by denaturation of the bovine serum albumin method and showed inhibition in the range of IC50 values—46.29–100.60 μg/mL. Among all the tested compounds, compound 5l has the highest IC50 value and compound 5g has the least IC50 value. On the other hand, antimicrobial results revealed that compound 5j showed the lowest MIC values and compound 5a has the highest MIC values. Furthermore, molecular docking of the active compounds demonstrated a better docking score and interacted well with the target protein. Physicochemical parameters of the titled compounds were found suitable in the reference range only. The in silico molecular docking study revealed their COX-inhibitory action. Compound 5j emerged as a significant bioactive molecule among the synthesized analogues.
Introduction
Inflammation is provoked by injuring any tissue in the body or by infection through pathogens, such as bacteria and viruses, allergens, irritants, toxic compounds, and so forth, either exogenously or endogenously.1 This may also include injury, surgery, autoimmune disorder, adult respiratory distress syndrome, long-term exposure to industrial chemicals, and reoxygenation injuries. The inflammation process is catalyzed by easily accessible molecules in the body such as histamine, prostaglandins, leukotrienes, oxygen- and nitrogen-derived free radicals, serotonin, bradykinin, and interleukins. Extravagant inflammation may lead to stroke or heart diseases, lupus, cancer, tissue injury, physiological decompensation, organ damage, and death.2 The risks associated with the inflammation process pose a challenge to the medicinal chemists to search for more effective anti-inflammatory agents. A majority of the existing anti-inflammatory compounds, especially those with proven clinical efficiency, are acidic in nature, such as aspirin, indomethacin, flufenamic acid, ibuprofen, and so forth. Nonsteroidal anti-inflammatory drugs (NSAIDs) that act on the affected tissues3,4 by inhibiting the cyclooxygenase (COX) involved in the synthesis of prostaglandins are one of the major class of drugs used to treat inflammation.5−9
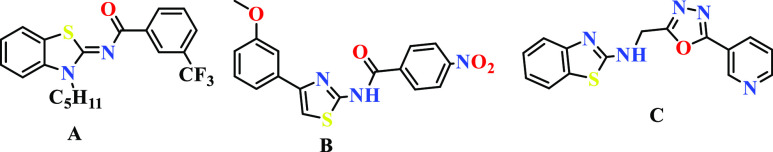
Various natural as well as synthetic compounds containing the thiazole core moiety have been explored for their antitumor,10,11 antimicrobial, anti-HIV,12 anticoagulant,13 anti-inflammatory,14 and antioxidant15 properties since many decades. The available literature also supports the bioactivities of thiazole scaffolds (Figure 1).
Figure 1.
Structure of thiazole-based molecules as anti-inflammatory agents.
Compound A demonstrated its selective cannabinoid CB2 agonists with anti-inflammatory activity.16 Compound B is a thiazole derivative exhibiting anti-inflammatory property,17 and compound C is a 1,3,4-oxadiazole-benzothiazole-pyridine conjugate which showed promising COX inhibition.18
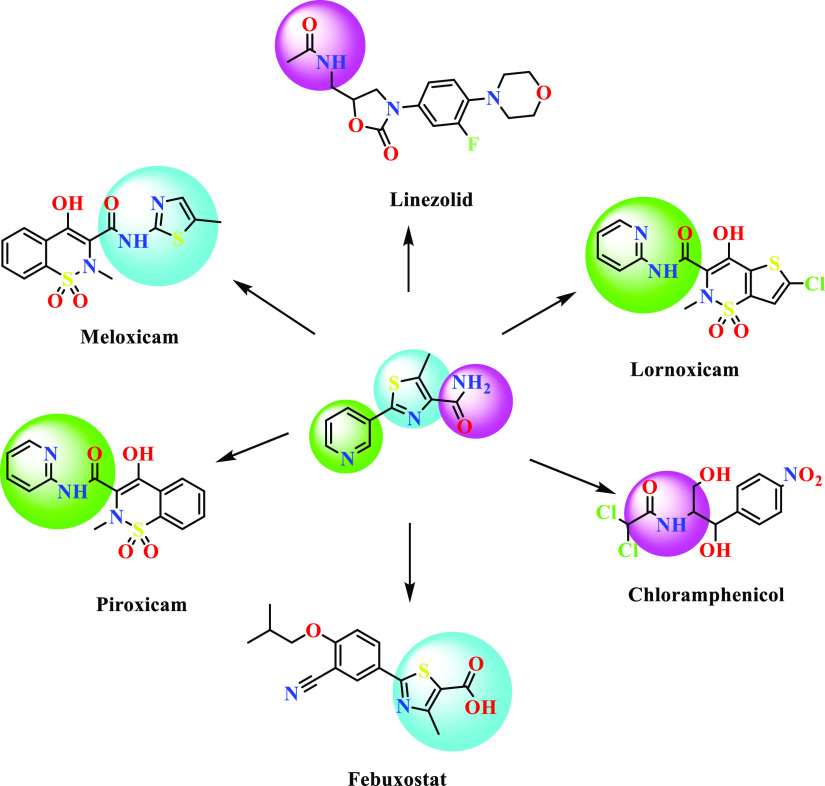
In recent decades, microbial infections are widespread and one of the life-threatening problems. To overcome these situations, researchers are trying hard to discover more efficient and effective antimicrobial candidates against both drug-sensitive as well as drug-resistant strains. Mahmoud et al.,19 Adole et al.,20 Wang et al.,21 and Eryılmaz et al.(22) reported various thiazole containing antimicrobial candidates. Some of the known reported bioactive compounds containing pyridine and thiazole having amide linkages are shown in Figure 2. Based on the available literature, the thiazole nucleus has been considered as one of the promising bioactive scaffolds in the wide range of biological systems.
Figure 2.
Structure of pyridine-, thiazole-, and amide-containing bioactive molecules.
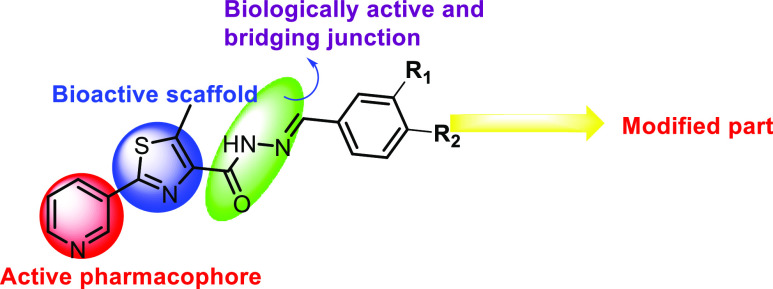
Various pyridine containing anti-inflammatory and antimicrobial agents are also reported in the literature. Researchers like Fan et al.,23 Bhila et al.,24 Desai et al.,25 Rani and Reddy,26 and Rashdan et al.(27) reported some antimicrobial agents. Laddha and Bhatnagar,28 Kumar et al.,29 and Thirumurugan et al.(30) reported some pyridine-based anti-inflammatory agents. In view of these observations,31 we also aimed to synthesize some hydrazides linked to pyridine-containing thiazole as potent bioactive scaffolds. The strategy used in this study is presented in Figure 3.
Figure 3.
Strategy used in designing the titled molecules.
Results and Discussion
Chemistry
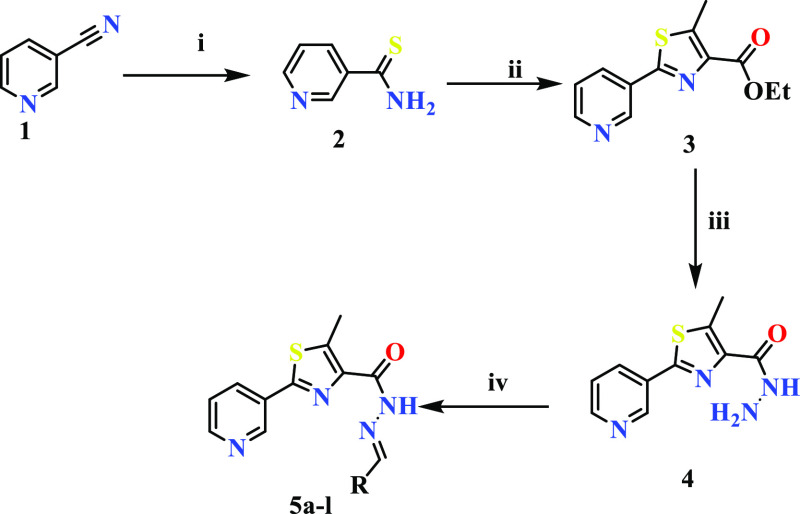
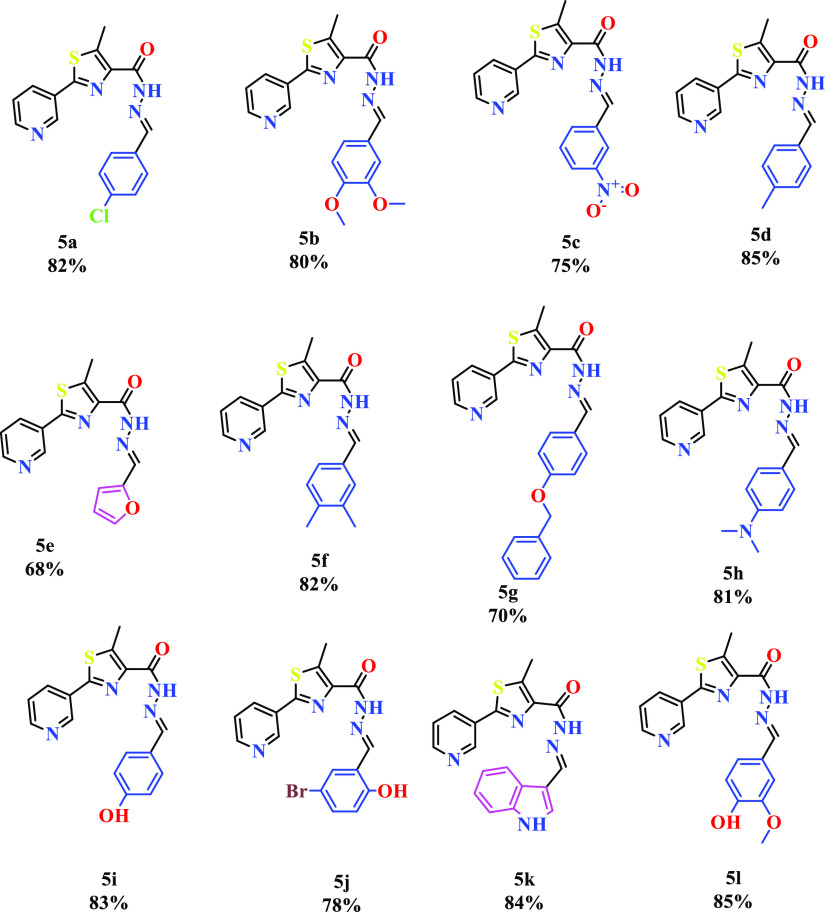
The detailed synthetic strategy for the target compounds is illustrated in Scheme 1. The structures and the yield of the synthesized compounds are represented in Figure 4. Initially, 3-cyanopyridine (1) was converted to pyridine-3-carbothiamide (2) by treating with P4S10. Compound 2 upon refluxing with ethyl-2-chloroacetoacetate resulted in ethyl-5-methyl-2-(pyridine-3-yl)thiazole-4-carboxylate (3). The resulting ester was treated with hydrazine hydrate to yield 5-methyl-2-(pyridine-3-yl)thiazole-4-carbohydrazide (4). Further, 5-methyl-2-(pyridine-3-yl)thiazole-4-carbohydrazide was condensed with an aromatic aldehyde to afford the target compounds (5a–l). The formation of 5a–l was confirmed by the absence of IR bands around 3330 cm–1 and scissoring at 1589 cm–1 due to the NH2 group and the presence of CH stretching at 2860 cm–1. A singlet at δ 8 ppm confirmed the formation of −CH=N.
Scheme 1. Synthetic Pathway of the Thiazole-Based Hydrazides (5a–l).
Reagents: (i): P4S10, ethanol, reflux, 70 °C, 4 h; (ii): ethyl 2-chloroacetoacetate, ethanol, reflux, 70 °C, 8 h; (iii): hydrazine hydrate, ethanol, reflux, 70 °C, 4 h; (iv): Ar-CHO, ethanol, reflux, 70 °C, 12 h.
Figure 4.
Structure of the synthesized compounds.
Biological Activity
In Vitro Anti-inflammatory Activity (Denaturation of the Bovine Serum Albumin Method)
Inflammation is the biological response that takes place when the healthy tissues get damaged or injured by harmful pathogens or by the alteration of neighboring protein tissues. Because of the lack of groundwork, animal risk management, and ethical disputes, in vivo studies have potential complications in order to carry out the animal studies. The leading cause of inflammation is the alteration of the native state of the tissue proteins. In the in vivo studies, the denaturation of proteins takes place because of the synthesis of autoantigens, and the compounds which inhibit denaturation are potentially useful for the discovery of novel anti-inflammatory drugs. The inhibitory effect on denaturation of proteins can be accessed by using a heat-induced protein denaturation process using diclofenac sodium as the standard drug. For the protein denaturation process to take place, the main important property to be maintained in several NSAIDs evaluation is the physiological pH (6.2–6.5) range of the reaction medium. All the synthesized thiazole-based hydrazide derivatives were tested for their in vitro anti-inflammatory activity by inhibition of the protein (bovine albumin) denaturation method using diclofenac sodium as a standard drug. Percentage inhibition of the synthesized compounds was determined using varying concentrations that is, 20–100 μg/mL. The IC50 values were determined, and the results and graphical representations are tabulated in Table 1 and Figure 5, respectively. From the results, it is clear that some of the synthesized compounds showed better activity (compounds 5j, 5k, and 5l). IC50 values of the synthesized compounds were in the range of 46.29–100.60 μg/mL because of the structural variations of the substrate. By varying the substitutions, the potency of the compounds changed. Compound 5l has the highest inhibition among the synthesized compounds which have 4-hydroxy-3-methoxyphenyl, and it has more inhibition than 5b (which has 3,4-dimethoxy substitution) and 5i (which has a hydroxyl group at 4th position). The lowest inhibition was observed for 5g, which has the 4-benzyloxyphenyl group. The compounds containing heterocyclic rings, 5k, and 5e showed better inhibition when compared with the compounds having aromatic substituents such as 5a, 5c, and 5g. Compounds 5b, 5d, 5f, 5h, and 5i (3,4-dimethoxy, 4-methyl, 3,4-dimethyl, 4-N,N-dimethyl, and 4-hydroxy) showed moderate inhibition. It can be concluded finally that the presence of the hydroxyl group on the phenyl ring and/or the presence of heterocyclic moieties exhibit good inhibition.
Table 1. Percentage Inhibition of Egg Albumin Protein Denaturation by the Synthesized Compoundsa.
| inhibition of protein denaturation (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| con (μg/mL) | std* | 5a* | 5b* | 5c* | 5d* | 5e* | 5f* | 5g* | 5h* | 5i* | 5j* | 5k* | 5l* |
| 20 | 42.98 ± 1.28 | 11.66 ± 0.95 | 22.66 ± 1.05 | 12.76 ± 0.99 | 24.46 ± 1.19 | 28.63 ± 0.68 | 17.65 ± 1.34 | 9.94 ± 0.53 | 19.18 ± 0.71 | 25.49 ± 0.85 | 30.66 ± 1.04 | 31.42 ± 0.95 | 32.63 ± 1.07 |
| 40 | 57.08 ± 1.6 | 20.51 ± 0.82 | 30.51 ± 2.22 | 22.51 ± 2.33 | 35.06 ± 0.56 | 39.23 ± 1.37 | 27.27 ± 1.52 | 16.82 ± 1.74 | 27.92 ± 1.59 | 31.18 ± 1.28 | 40.01 ± 1.33 | 41.00 ± 1.24 | 43.20 ± 1.87 |
| 60 | 75.54 ± 1.26 | 28.25 ± 1.47 | 42.86 ± 2.43 | 28.91 ± 1.44 | 48.94 ± 1.53 | 51.18 ± 1.26 | 35.26 ± 1.42 | 26.00 ± 1.00 | 37.33 ± 1.43 | 49.24 ± 1.60 | 53.98 ± 1.78 | 56.72 ± 2.00 | 59.51 ± 2.46 |
| 80 | 83.56 ± 0.96 | 39.55 ± 0.92 | 52.54 ± 0.90 | 39.65 ± 1.08 | 61.43 ± 1.24 | 65.57 ± 1.33 | 49.58 ± 0.98 | 37.61 ± 1.99 | 49.89 ± 1.45 | 58.12 ± 0.56 | 68.39 ± 1.00 | 71.47 ± 0.90 | 78.24 ± 1.37 |
| 100 | 94.60 ± 0.90 | 52.56 ± 0.80 | 69.65 ± 1.21 | 58.28 ± 1.29 | 73.82 ± 1.45 | 77.37 ± 0.94 | 62.06 ± 1.57 | 49.70 ± 0.73 | 65.78 ± 1.64 | 71.07 ± 0.34 | 80.34 ± 0.95 | 83.46 ± 2.21 | 87.97 ± 1.37 |
| IC50 μg/mL | 35.03 | 95.11 | 76.12 | 85.79 | 61.29 | 58.62 | 80.68 | 100.60 | 80.18 | 62.74 | 55.58 | 52.89 | 46.29 |
Values are expressed as mean ± SD, n = 3, and * Correlation is significant at the 0.01 level.
Figure 5.
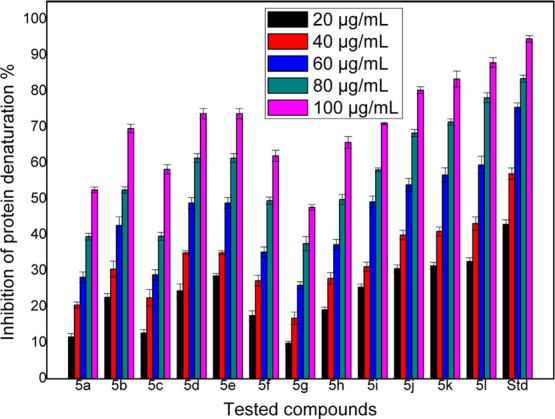
Graphical representation of in vitro protein denaturation results of the compounds.
The structure activity relationship study of the active compound 5l was deduced based on the IC50 value; it clearly reveals that the presence of hydroxyl and methoxy groups is responsible for the enhanced inhibition. The presence of the hydroxyl substituent on the 4th place and the methoxy substituent on the 3rd position enhanced the anti-inflammatory activity drastically.
In Vitro Antimicrobial Activity
Minimal inhibitory concentration of the synthesized compounds showed the highest to least values because of the structural modifications of the substrate. Minimal inhibitory concentration values along with mean ± SD and graphical representations of antibacterial and antifungal activities are tabulated in Tables 2 and 3 and in Figures S11 and S12, respectively.
Table 2. MIC Values (μg/mL) of Antibacterial Evaluation of the Titled Compoundsa.
| compound | S. aureus | B. subtilis | E. coli | P. aeruginosa |
|---|---|---|---|---|
| 5a | 127.66 ± 1.52 | 127.33 ± 1.15 | 128.33 ± 0.57 | 127.33 ± 1.15 |
| 5b | 127.00 ± 1.73 | 65.00 ± 1.73 | 128.33 ± 0.57 | 127.00 ± 1.73 |
| 5c | 64.00 ± 1.00 | 32.66 ± 1.15 | 64.33 ± 0.57 | 64.00 ± 1.00 |
| 5d | 63.00 ± 1.00 | 31.66 ± 0.57 | 65.00 ± 1.00 | 64.33 ± 1.52 |
| 5e | 15.66 ± 0.57 | 8.33 ± 0.57 | 17.33 ± 1.52 | 16.00 ± 1.00 |
| 5f | 33.00 ± 1.73 | 15.33 ± 1.15 | 31.66 ± 1.52 | 31.33 ± 1.15 |
| 5g | 63.66 ± 0.57 | 33.00 ± 1.73 | 64.66 ± 1.15 | 64.00 ± 1.00 |
| 5h | 31.66 ± 0.57 | 17.00 ± 1.73 | 32.66 ± 1.15 | 32.00 ± 2.00 |
| 5i | 15.33 ± 1.15 | 7.00 ± 1.00 | 17.00 ± 1.00 | 16.33 ± 0.57 |
| 5j | 1.66 ± 0.57 | 1.32 ± 0.57 | 2.66 ± 1.52 | 2.33 ± 0.57 |
| 5k | 3.66 ± 1.52 | 2.00 ± 1.00 | 4.66 ± 1.15 | 4.00 ± 1.00 |
| 5l | 4.00 ± 01.73 | 4.33 ± 0.57 | 7.00 ± 1.00 | 7.00 ± 1.00 |
| tetracycline | 0.68 ± 0.54 | 0.66 ± 0.28 | 1.66 ± 1.15 | 1.66 ± 1.15 |
| streptomycin | 1.00 ± 00 | 1.00 ± 00 | 1.66 ± 1.15 | 1.00 ± 0.86 |
Values are expressed as mean ± SD, n = 3, and * correlation is significant at the 0.01 level.
Table 3. MIC Values (μg/mL) of Antifungal Evaluation of the Titled Compoundsa.
| compound | A. flavus | T. atroviridae | P. citranum | C. albicans |
|---|---|---|---|---|
| 5a | 126.33 ± 2.08 | 126.00 ± 1.73 | 124.33 ± 2.30 | 128.33 ± 0.57 |
| 5b | 128.33 ± 2.08 | 129.00 ± 2.00 | 124.00 ± 1.00 | 126.00 ± 2.00 |
| 5c | 129.00 ± 1.73 | 127.33 ± 1.15 | 126.66 ± 2.30 | 62.33 ± 1.52 |
| 5d | 64.00 ± 1.00 | 65.66 ± 1.52 | 67.00 ± 2.64 | 32.00 ± 1.00 |
| 5e | 16.66 ± 1.15 | 15.66 ± 2.51 | 16.33 ± 1.52 | 8.33 ± 1.52 |
| 5f | 32.66 ± 3.05 | 33.33 ± 1.52 | 32.00 ± 1.00 | 14.33 ± 1.52 |
| 5g | 63.33 ± 1.15 | 62.66 ± 1.52 | 65.00 ± 1.73 | 130.33 ± 2.08 |
| 5h | 64.6 ± 3.05 | 65.00 ± 1.73 | 65.33 ± 1.52 | 33.66 ± 2.08 |
| 5i | 14.66 ± 2.30 | 15.00 ± 1.73 | 15.33 ± 1.15 | 7.66 ± 1.52 |
| 5j | 2.66 ± 1.15 | 2.33 ± 0.57 | 1.66 ± 0.57 | 1.66 ± 1.15 |
| 5k | 3.00 ± 1.00 | 5.33 ± 1.52 | 4.66 ± 1.15 | 3.33 ± 1.52 |
| 5l | 3.00 ± 1.00 | 5.33 ± 1.52 | 3.66 ± 1.52 | 4.00 ± 2.00 |
| fluconazole | 2.00 ± 0.00 | 2.00 ± 0.00 | 2.00 ± 0.00 | 1.00 ± 0.00 |
| nystatin | 3.33 ± 1.52 | 1.66 ± 1.15 | 2.00 ± 1.73 | 3.66 ± 2.08 |
Values are expressed as mean ± SD, n = 3, and * correlation is significant at the 0.01 level.
The variation of the position of the substituents and the introduction of the heterocyclic ring were explored to study the structure–activity relationship (SAR) of the synthesized compounds. Results obtained from the antimicrobial studies indicated that the compounds 5j, 5k, 5l, 5e, and 5i showed significant minimal inhibitory concentration (MIC) values, of which the compound 5j showed MIC values as that of the standard drugs against the tested bacterial and fungal strains. The enhanced antimicrobial activity is due to either the presence of a hydroxyl group on the aromatic ring and or the presence of the heterocyclic moiety in the synthesized derivatives when compared with other compounds having the aromatic moiety. The least activity was shown by compounds 5a, 5b, and 5c with 4-chloro, 3,4-dimethoxyphenyl, and 3-nitrophenyl moieties, respectively. The moderate antimicrobial property was observed in the case of compounds 5f, 5d, 5g, and 5h, which contain 3,4-dimethyl, 4-methyl, and 4-benzyloxyphenyl groups.
The structural activity relationship study deduced that the alterations in the aryl ring play the significant role for enhancing the activity. Compound 5j with the combinations of bromo substitution at the 5th position and hydroxyl substitution at the 2nd position produces effective and enhanced antimicrobial properties.
In silico prediction of physicochemical and ADME parameters: in the field of medicinal chemistry, Lipinski’s rule of five and Veber’s rule are the guidelines for medicinal chemists searching drug-like chemical compounds with effective oral potential. It is understood that fine-tuning the physicochemical properties (PCPs) of a lead molecule could alter the ADME property. Hence, close inspection was placed on the PCPs, by calculating the guiding parameters such as molecular weight (MW), number of hydrogen bond acceptors (HBAs), number of hydrogen bond donors (HBDs) calculated log P (clog P), number of rotatable bonds (NRBs), and polar surface area (PSA). Compounds with optimal PCPs (MW < 500, HBA < 10, HBD < 5, log P < 5, NRB < 10, and PSA < 140 Å) have higher probability for oral absorption. The predicted parameters of the titled compounds (Table 4) satisfied the Lipinski’s rule of five. Caco-2 permeability of all the predicted compounds lies within the range except compound 5c, which showed a less permeability of 91.567 nm/s. 50% of the compounds revealed the predicted results of 100% oral human absorption, and the rest of the compounds exhibited the values within the range of 77–96%. Overall, the predicted parameters of the compounds were satisfied with the physicochemical parameters, which are exhibited by most of the clinically approved drugs. Based on the prediction studies, these compounds may not face any problems in the mere future.
Table 4. In silico Predicted Physicochemical Parameters of the Titled Compounds.
| code | MWa | SASAb | donorHBc | accptHBd | log P o/we | PCacof | log Sg | #rotorh | % human oral absorptioni |
|---|---|---|---|---|---|---|---|---|---|
| 5a | 356.829 | 649.025 | 1 | 5.5 | 3.973 | 1059.54 | –5.876 | 4 | 100 |
| 5b | 382.436 | 705.878 | 1 | 7 | 3.667 | 839.026 | –5.698 | 6 | 100 |
| 5c | 367.381 | 671.77 | 1 | 6.5 | 2.701 | 91.567 | –5.446 | 5 | 77.874 |
| 5d | 336.411 | 663.689 | 1 | 5.5 | 3.875 | 951.014 | –5.852 | 4 | 100 |
| 5e | 312.345 | 571.87 | 1 | 6 | 2.687 | 976.977 | –4.332 | 4 | 96.192 |
| 5f | 350.437 | 689 | 1 | 5.5 | 4.093 | 982.991 | –7.296 | 4 | 100 |
| 5g | 428.508 | 788.776 | 1 | 6.25 | 5.283 | 1154.27 | –4.073 | 7 | 100 |
| 5h | 365.452 | 700.173 | 1 | 6.5 | 3.836 | 958.45 | –6.293 | 5 | 100 |
| 5i | 338.383 | 600.548 | 2 | 6.25 | 2.442 | 247.973 | –5.946 | 5 | 84.098 |
| 5j | 417.279 | 619.16 | 2 | 6.25 | 3.392 | 927.915 | –6.187 | 5 | 100 |
| 5k | 361.42 | 686.978 | 2 | 5.5 | 3.752 | 529.314 | –5.307 | 4 | 100 |
| 5l | 368.409 | 682.421 | 2 | 7 | 2.926 | 313.646 | –4.982 | 6 | 88.762 |
Molecular weight, in Da (range for 95% of drugs: 130–725 Da).
Total solvent-accessible volume in cubic angstroms using a probe with a 1.4 Å radius. (500–2000).
No. of hydrogen bonds donated by the molecule (range for 95% of drugs: 0–6).
No. of hydrogen bonds accepted by the molecule (range for 95% of drugs: 2–20).
Predicted octanol/water partition coefficient log P (acceptable range: −2.0 to 6.5).
Predicted aqueous solubility; S in mol/L (acceptable range: −6.5 to 0.5).
Apparent Caco-2 permeability (nm/s) (<25 poor, >500 great).
Number of nontrivial (not CX3) and nonhindered (not alkene, amide, small ring) rotatable bonds. (0–15).
% human oral absorption (>80 high and <25 poor).
Molecular Docking Studies
To find out the putative binding mode of the significantly active compound against the targeted protein COX-2, docking studies were performed. Before the docking studies of the test compound, root mean square deviation (RMSD) of the X-ray pose and redocked pose (Figure 6) of the co-crystallized ligand in the target protein was checked and found to be 0.20 Å, suggesting that the docking protocol could be relied on for the docking studies.
Figure 6.
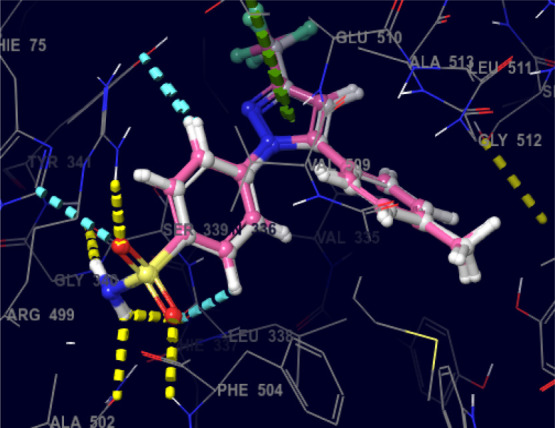
Superimposed view of the X-ray native pose of the ligand (Celecoxib) with its docking pose in the active site of the target (PDB-3LN1) [color interpretation: white—X-ray native pose of the ligand, pink—docked pose of the ligand].
In-depth scrutiny of the 3D and 2D poses of the co-crystal ligand (Celecoxib) was performed (Figures 7 and 8), and celecoxib unveiled four hydrogen bond interactions within the target with a highest docking score of −12.4 kcal/mol and energy of −62.50 kcal/mol (5). The amino acid residues such as GLN-178, SER-339, ARG-499, and PHE-504 were actively involved in the hydrogen bond formation with celecoxib. Apart from the hydrogen bond interaction, celecoxib also revealed two aromatic bond interactions with the amino acid residues HIS-75 and TYR-341; ARG-106 was involved in π–π-cationic interaction with the pyrazole moiety of celecoxib.
Figure 7.
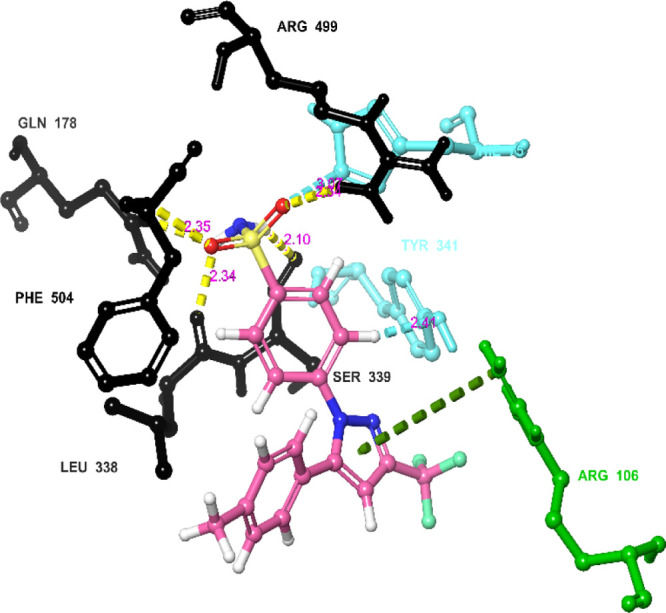
Networking of the co-crystal ligand exhibited various interactions in the active site of the protein (3LN1) [color interpretation: black—hydrogen bond, blue—aromatic bond, green—π–π cation interactions].
Figure 8.
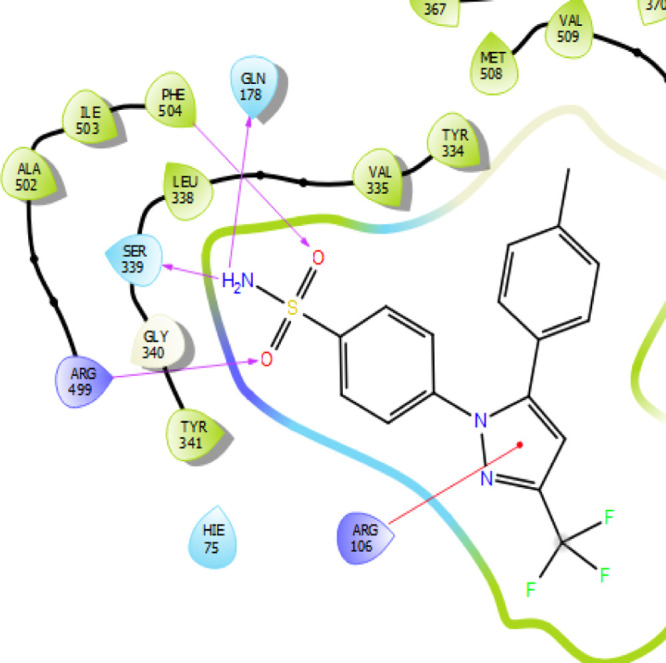
2D representation of the co-crystal ligand [color interpretation: magenta—hydrogen bond, red—cation interactions.
Table 5. Docking Analysis of Celecoxib and Significantly Active Compounds 5j, 5k, and 5l.
| compound code | Glide score (kcal/mol) |
|---|---|
| 5j | –9.80 |
| 5k | –7.80 |
| 5l | –7.10 |
| code (PDB-3LN1) | H-bond | aromatic bond | π–π interactions | Glide score (kcal/mol) | Glide energy (kcal/mol) |
|---|---|---|---|---|---|
| co-crystal ligand (Celecoxib) | GLN-178 | HIS-75 | –12.40 | –62.50 | |
| SER-339 | TYR-341 | ||||
| ARG-499 | |||||
| PHE-504 | |||||
| 5j | SER-516 | –9.80 | –46.80 |
Similarly, significantly active compound 5j (Figures 9 and 10) also exhibited only one aromatic bond interaction with the amino acid residue SER-516 through the pyridine moiety. The compound exhibited a docking score of −9.80 kcal/mol and docking energy of −46.80 kcal/mol, which are comparatively less than those of the standard drug celecoxib.
Figure 9.
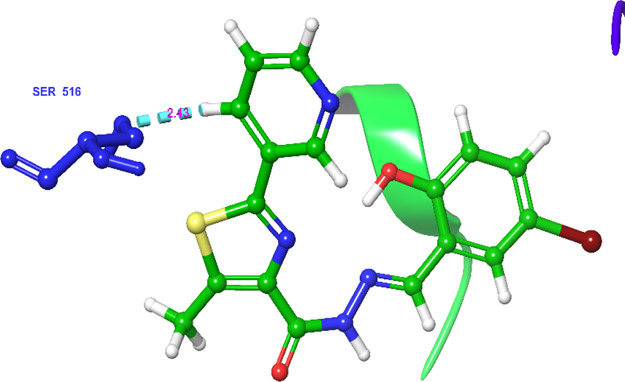
Networking of the significantly active compound 5j exhibited various interactions in the active site of the protein (3LN1) [color interpretation: blue—aromatic bond].
Figure 10.
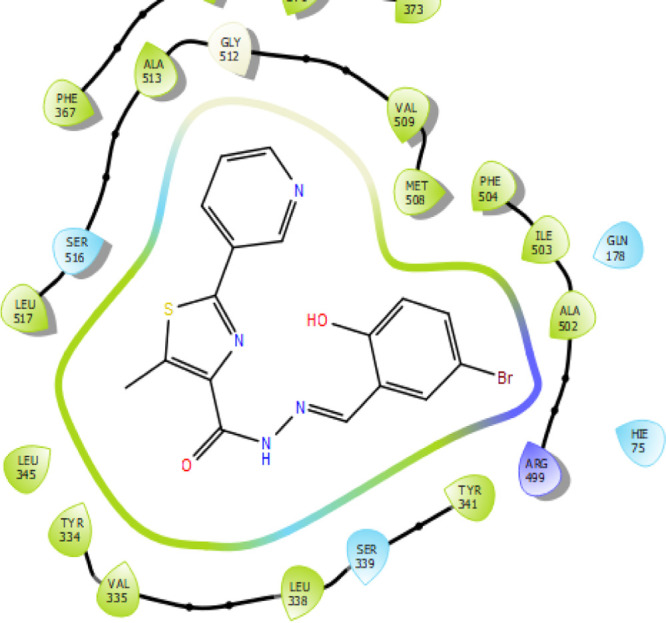
2D representation of the significantly active compound 5j.
Conclusions
In the present study, we developed thiazole-based hydrazides as anti-inflammatory and antimicrobial agents. The in vitro anti-inflammatory study of the target compounds revealed that compounds 5j, 5k, and 5l are significant inhibitors among the series. The in vitro antimicrobial assay showed that compounds 5j, 5k, and 5l are effective against all the tested strains. The phenolic group, along with methoxy or bromo substituents, showed better activity when compared with other substituents. Compounds 5j and 5l displayed better activity in the series. Therefore, compounds 5j and 5l can be used for further studies. Pharmacokinetic studies also underlined that the active compound has better absorption, distribution, metabolism, and elimination profiles. More interestingly, compounds 5j, 5k, and 5l showed significant biological activity with safe pharmacokinetic properties. The molecular docking study also supports the effective interactions with target molecules. Based on the present study, we can conclude that this is the potential route for the development of new effective antimicrobial as well as anti-inflammatory agents.
Experimental Section
Materials and Methods
All the chemicals and solvents used for the present study were purchased from commercial suppliers of Sigma-Aldrich, India, and Spectrochem Pvt. Ltd., India, and used without purification. Reactions were carried out using standard techniques. Melting points of the newly synthesized compounds were determined in an open capillary tube and were uncorrected. To monitor the progress of the reaction and purity of the product, thin layer chromatography (Merck silica gel 60 F254-coated aluminum plates) was used. The mixture of ethyl acetate and hexane solvents was used as the mobile phase and visualized under UV light at 254 nm. Shimadzu-FTIR was used to record FTIR spectra (γmax in cm–1). The Bruker AVANCE II 400 spectrometer (with 5 mm PABBO BB-1H tubes) was used to record NMR spectra, using DMSO-d6 as a solvent and TMS as an internal standard (the chemical shift in δ ppm and coupling constants (J) were expressed in parts per million (ppm) and hertz, respectively). VARIOEL-III (Elementar analysensysteme GmBH) was used for the elemental analysis.
Procedure for the Synthesis of Pyridine-3-carbothiamide (2)
A solution of P4S10 (2 equiv) and 3-cyanopyridine 1 (1 equiv) in ethanol was taken in a round-bottomed flask and stirred for 30 min at ambient temperature. The reaction mixture was then refluxed for 4 h. After the completion of the reaction [monitored by TLC (EtOAc/hexane 3:7)], the reaction mixture was cooled to room temperature, diluted with cold water, and extracted with chloroform (3 × 200 mL). The combined organic extract was dried over CaCl2 and concentrated to dryness. Further, the crude residue was recrystallized to afford a pure compound.32
Procedure for the Synthesis of Ethyl-5-methyl-2-(pyridine-3-yl)thiazole-4-carboxylate (3)
A mixture of ethyl-2-chloro acetoacetate (1.2 equiv) and compound 2 (1 equiv) dissolved in ethanol (50 mL) was refluxed for 8 h. After completion of the reaction [monitored by TLC (EtOAc/hexane 3:7)], the reaction mixture was cooled to ambient temperature. The reaction mixture was quenched with ice cold water. The solids precipitated out were collected by filtration, washed with ice cold water, and dried under reduced pressure to afford intermediate 3.33
Procedure for the Synthesis of 5-Methyl-2-(pyridine-3-yl)thiazole-4-carbohydrazide (4)
To the stirred solution of intermediate compound 3 (1.5 equiv) in ethanol (50 mL), hydrazine hydrate (2.5 equiv) was added at ambient temperature and heated to reflux for 4 h. The reaction progress was monitored by TLC (EtOAc/hexane 3:7). After completion of the reaction, the reaction mass was poured onto crushed ice and stirred for half an hour. The solid was filtered and dried under vacuum to get intermediate compound 4.34
General Procedure for the Synthesis of N′-Arylmethylidene-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazides (5)
A mixture of intermediate compound 4 (1 equiv) and aromatic aldehyde (1.2 equiv) in ethanol (50 mL) was stirred under reflux for about 12 h. The reaction mixture was cooled to 0–10 °C, filtered, and dried for 6 h. It was further recrystallized by the DMF–DMSO mixture to afford the pure titled target compounds.35 Purity of the synthesized compounds was in the range of 95–99%.
N′-(4-Chlorobenzylidene)-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5a)
Yellow solid; Yield: 82%; mp > 300 °C; FT-IR (KBr cm–1): 3270 (N–H), 3041 (Ar–H), 2964 (C–H), 1665 (NH–CO), 1610 (C=N), 653 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.48 (s, 3H, thz-CH3), 7.38–7.33 (d, 2H, Ar–H, J = 7.8 Hz), 7.60–7.57 (m, 1H, Pyr-H), 7.85 (d, 2H, Ar–H, J = 7.8 Hz), 8.14 (s, 1H, =CH), 8.39 (d, 1H, Pyr-H, J = 7.2 Hz), 8.73 (d, 1H, Pyr-H, J = 4 Hz), 9.21 (s, 1H, Pyr-H), 11.95 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 15.2, 115.4, 115.6, 127.5, 130.3, 130.4, 130.7, 131.1, 134.5, 134.9, 149.3, 151.4, 155.7, 157.2, 160.2, 162.3, 168.5; LCMS (m/z): 357.05 [M + H]+, 359.32 [(M + 2) + H]+; Elemental analysis of C17H13N4ClOS Calcd: C, 57.22; H, 3.67; N, 15.70. Found: C, 57.20; H, 3.65; N, 15.72.
N′-(3,4-Dimethoxybenzylidene)-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5b)
Pale yellow solid; Yield: 80%; mp 230–232 °C; FT-IR (KBr cm–1): 3447 (N–H), 3014 (Ar–H), 2839 (C–H), 1648 (−NHCO), 1597 (C=N), 651 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.79 (s, 3H, thz-CH3), 3.81 (s, 3H,–OCH3), 3.85 (s, 3H, −OCH3), 7.04 (d, 1H, Ar–H, J = 8.4 Hz), 7.21–7.23 (m, 1H, Ar–H), 7.45 (s, 1H, Ar–H), 7.56–7.59 (m, 1H, Pyr-H), 8.03 (s, 1H, =CH), 8.32 (d, 1H, Pyr-H, J = 8 Hz), 8.71 (d, 1H, Pyr-H, J = 4 Hz), 9.14 (s, 1H, Pyr-H), 11.80 (s, 1H, −NH); 13C NMR (DMSO-d6, 100 MHz): δ 15.2, 56.8, 110.1, 114.1, 122.1, 127.5, 129.4, 131.1, 134.5, 134.9, 148.6, 149.3, 149.8, 150.5, 151.4, 155.7, 157.2, 168.5, 181.7; LCMS (m/z): 383.10 [M + H]+; Elemental analysis of C19H18N4O3S Calcd: C, 59.67; H, 4.74; N, 14.65. Found: C, 59.65; H, 4.75; N, 14.63.
5-Methyl-N′-(3-nitrobenzylidene)-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5c)
Yellow solid; Yield: 75%; mp 266–268 °C; FT-IR (KBr cm–1): 3440 (N–H), 3041 (Ar–H), 2868 (C–H), 1651 (−NHCO), 1609 (C=N), 650 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.69 (s, 3H, thz-CH3), 7.56–7.59 (m, 1H, Pyr-H), 7.78 (d, 1H, Ar–H, J = 7.2 Hz), 8.04 (d, 1H, Ar–H, J = 7.6 Hz), 8.23 (d, 1H, Ar–H, J = 7.6 Hz), 8.35 (s, 1H, =CH), 8.4 (s, 1H, Ar–H), 8.42 (d, 1H, Pyr-H, J = 8 Hz), 8.75 (d, 1H, Pyr-H, J = 4 Hz), 9.24 (s, 1H, Pyr-H), 11.75 (s, 1H, −NH); 13C NMR (DMSO-d6, 100 MHz): δ 53.6, 107.1, 110.6, 120.1, 123.2, 125.1, 135.7, 144.7, 149.0, 150.5, 160.1, 160.8, 172.5; LCMS (m/z): 368.23 [M + H]+; Elemental analysis for C17H13N5O3S Calcd: C, 55.58; H, 3.57; N, 19.06. Found: C, 55.60; H, 3.60; N, 19.05.
5-Methyl-N′-(4-methylbenzylidene)-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5d)
Yellow solid; Yield: 85%; mp 216–218 °C; FT-IR (KBr cm–1): 3444 (N–H), 3049 (Ar–H), 2861 (C–H), 1655 (−NHCO), 1605 (C=N), 652 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.43 (s, 3H, CH3), 2.61 (s, 3H, thz-CH3), 7.32 (d, 2H, Ar–H, J = 7.8 Hz), 7.52–7.55 (m, 1H, Pyr-H), 7.76 (d, 2H, Ar–H, J = 7.8 Hz), 8.01 (s, 1H, =CH), 8.1 (d, 1H, Pyr-H, J = 7.8 Hz), 8.70 (d, 1H, Pyr-H, J = 4.6 Hz), 9.24 (s, 1H, Pyr-H), 11.72 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 28.1, 54.2, 108.3, 110.5, 123.1, 124.2, 126.5, 134.2, 146.8, 148.9, 152.6, 161.6, 174.2; LCMS (m/z): 337.42 [M + H]+; Elemental analysis for C18H16N4OS Calcd: C, 64.27; H, 4.79; N, 16.65. Found: C, 64.25; H, 4.80; N, 16.63.
N′-[(furan-2-ylmethylidene)-5-methyl]-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5e)
Yellow solid; Yield: 68%; mp 120–122 °C; FT-IR (KBr cm–1): 3434 (N–H), 3040 (Ar–H), 2860 (C–H), 1635 (−NHCO), 1609 (C=N), 658 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.62 (s, 3H, thz-CH3), 6.53 (t, 1H, furan-H, J = 3.6 Hz), 7.29 (d, 1H, furan-H, J = 3.6 Hz), 7.44–7.47 (m, 1H, Pyr-H), 7.65 (d, 1H, furan-H, of J = 1.8 Hz), 7.85 (s, 1H, =CH), 8.22 (d, 1H, Pyr-H, J = 7.4 Hz), 8.68 (d, 1H, Pyr-H, J = 4.7 Hz), 9.18 (s, 1H, Pyr-H), 11.12 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 55.5, 108.0, 111.8, 120.9, 124.3, 126.4, 134.3, 147.3, 149.1, 151.6, 160.1, 175.3; LCMS (m/z): 313.35 [M + H]+; Elemental analysis for C15H12N4O2S Calcd: C, 57.68; H, 3.87; N, 17.94. Found: C, 57.69; H, 3.88; N, 17.95.
N′-(3,4-Dimethylbenzylidene)-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5f)
Pale yellow solid; Yield: 82%; mp 210–212 °C; FT-IR (KBr cm–1): 3445 (N–H), 3024 (Ar–H), 2865 (C–H), 1621 (−NHCO), 1618 (C=N), 659 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.28 (s, 3H, −CH3), 2.31 (s, 3H, −CH3), 2.56 (s, 3H, thz-CH3), 7.01 (d, 1H, Ar–H, J = 8.4 Hz), 7.18–7.21 (m, 1H, Ar–H), 7.38 (s, 1H, Ar–H), 7.56–7.58 (m, 1H, Pyr-H), 8.01 (s, 1H, =CH), 8.30 (d, 1H, Pyr-H, J = 8 Hz), 8.68 (d, 1H, Pyr-H, J = 4 Hz), 9.13 (s, 1H, Pyr-H), 11.78 (s, 1H, −NH); 13C NMR (DMSO-d6, 100 MHz): δ 28.1, 54.5, 107.5, 110.8, 121.0, 123.4, 126.7, 134.5, 146.8, 148.1, 151.4, 161.5, 175.2; LCMS (m/z): 351.28 [M]+; Elemental analysis for C19H18N4OS Calcd: C, 65.12; H, 5.18; N, 15.99. Found: C, 65.10; H, 5.16; N, 15.97.
N′-(4-(Benzyloxy)benzylidene)-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5g)
Yellow solid; Yield: 70%; mp 208–210 °C; FT-IR (KBr cm–1): 3432 (N–H), 3073 (Ar–H), 2866 (C–H), 1653 (−NHCO), 1622 (C=N), 648 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.63 (s, 3H, thz-CH3), 5.16 (s, 2H, CH2), 7.18 (d, 2H, Ar–H, J = 7.8 Hz), 7.36 (m, 1H, Ar–H), 7.38 (d, 2H, Ar–H, J = 7.7 Hz), 7.47 (d, 2H, Ar–H, J = 7.7 Hz), 7.53–7.56 (m, 1H, Pyr-H), 7.87 (d, 2H, Ar–H, J = 7.8 Hz), 8.02 (s, 1H, =CH), 8.11 (d, 1H, Pyr-H, J = 7.6 Hz), 8.71 (d, 1H, Pyr-H, J = 4.6 Hz), 9.25 (s, 1H, Pyr-H), 11.73 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 55.6, 70.9, 108.3, 111.5, 114.8, 120.1, 124.4, 126.6, 127.1, 127.6, 128.9, 132.3, 133.7, 136.7, 146.8, 149.0, 151.5, 161.8, 164.8, 176.6; LCMS (m/z): 429.51 [M + H]+; Elemental analysis for C24H20N4O2S Calcd: C, 67.27; H, 4.70; N, 13.08. Found: C, 67.25; H, 4.72; N, 13.09.
N′-(4-Dimethylaminobenzylidene)-5-methyl-2-(pyridine-3-yl)thiazole-4-carbohydrazide (5h)
Yellow solid; Yield: 81%; mp 212–214 °C; FT-IR (KBr cm–1): 3435 (N–H), 3043 (Ar–H), 2849 (C–H), 1654 (−NHCO), 1610 (C=N), 653 (C–S–C); 1HNMR (DMSO-d6, 400 MHz): δ 2.79 (s, 3H, thz-CH3), 2.99 (s, 6H, N–Me), 6.81 (d, 1H, Ar–H, J = 8.8 Hz), 7.6 (d, 4H, Ar–H, J = 8.4 Hz), 8.0 (s, 1H, =CH), 8.39 (d, 1H, Pyr-H, J = 8.4 Hz), 8.73 (d, 1H, Pyr-H, J = 4.4 Hz), 9.19 (s, 1H, Pyr-H), 11.66 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 19.3, 52.1, 112.5, 120.4, 121.6, 124.9, 129.2, 134.3, 145.4, 147.5, 151.9, 161.6, 161.8, 167.5; LCMS (m/z): 366.46 [M + H]+; Elemental analysis for C19H19N5OS Calcd: C, 62.45; H, 5.24; N, 19.16. Found: C, 62.45; H, 5.25; N, 19.15.
N′-(4-Hydroxybenzylidene)-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5i)
Yellow solid; Yield: 83%; mp above 300 °C; FT-IR (KBr cm–1): 3434 (N–H), 2980 (Ar–H), 2832 (C–H), 1664 (−NHCO), 1615 (C=N), 641 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.65 (s, 3H, thz-CH3), 7.21 (d, 2H, Ar–H, J = 8.2 Hz), 7.76–7.78 (m, 1H, Pyr-H), 7.82 (d, 2H, Ar–H, J = 8.2 Hz), 8.10 (s, 1H, =CH), 8.42 (d, 1H, Pyr-H, J = 7.6 Hz), 8.72 (d, 1H, Pyr-H, J = 4.7 Hz), 9.15 (s, 1H, Pyr-H), 9.93 (s, 1H, O–H), 11.72 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 49.1, 112.3, 120.1, 121.3, 124.3, 129.1, 131.2, 133.1, 136.1, 146.7, 148.2, 151.3, 161.3, 171.6; LCMS (m/z): 339.39 [M + H]+; Elemental analysis for C17H14N4O2S Calcd: C, 60.34; H, 4.17; N, 16.56. Found: C, 60.32; H, 4.15; N, 16.57.
N′-(5-Bromo-2-hydroxybenzylidene)-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5j)
Yellow solid; Yield: 78%; mp 258–260 °C; FT-IR (KBr cm–1): 3432 (N–H), 2966 (Ar–H), 2866 (C–H), 1672 (−NHCO), 1612 (C=N), 651 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.69 (s, 3H, thz-CH3), 6.99 (d, 1H, Ar–H, J = 8.4 Hz), 7.48 (d, 1H, Ar–H, J = 8.4 Hz), 7.78–7.80 (m, 1H, Pyr-H), 7.82 (s, 1H, Ar–H), 8.02 (s, 1H, =CH), 8.19 (d, 1H, Pyr-H, J = 7.6 Hz), 8.68 (d, 1H, Pyr-H, J = 4.7 Hz), 9.20 (s, 1H, Pyr-H), 10.52 (s, 1H, O–H), 11.70 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 51.2, 112.8, 120.0, 120.6, 124.5, 129.5, 131.8, 133.3, 135.8, 146.6, 147.3, 152.3, 161.1, 170.6; LCMS (m/z): 418.28 [M + H]+, 420.21 [(M + 2) + H]+; Elemental analysis for C17H13N4BrO2S Calcd: C, 48.93; H, 3.14; N, 13.43. Found: C, 48.91; H, 3.15; N, 13.44.
N′-[(1H-indol-3-yl)methylidene]-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5k)
Yellow solid; Yield: 84%; mp above 300 °C; FT-IR (KBr cm–1): 3438 (N–H), 2981 (Ar–H), 2866 (C–H), 1651 (−NHCO), 1614 (C=N), 658 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.62 (s, 3H, thz-CH3), 7.04–7.14 (m, 2H, indolyl-H), 7.65 (m, 3H, indolyl-H), 7.79–7.81 (m, 1H, Pyr-H), 8.05 (s, 1H, =CH), 8.22 (d, 1H, Pyr-H, J = 7.4 Hz), 8.68 (d, 1H, Pyr-H, J = 4.7 Hz), 9.18 (s, 1H, Pyr-H), 10.23 (s, 1H, indole N–H), 11.12 (s, 1H, N–H); 13C NMR (DMSO-d6, 100 MHz): δ 54.5, 108.0, 111.8, 116.3, 118.4, 120.2, 123.9, 124.4, 126.2, 128.1, 134.1, 147.7, 148.8, 152.2, 161.2, 175.5; LCMS (m/z): 362.42 [M + H]+; Elemental analysis for C19H15N5OS Calcd: C, 63.14; H, 4.18; N, 19.38. Found: C, 61.90; H, 4.30; N, 20.03.
N′-(4-Hydroxy-3-methoxybenzylidene)-5-methyl-2-(pyridin-3-yl)thiazole-4-carbohydrazide (5l)
Yellow solid; Yield: 85%; mp 262–264 °C; FT-IR (KBr cm–1): 3441 (N–H), 2975 (Ar–H), 2865 (C–H), 1672 (−NHCO), 1619 (C=N), 655 (C–S–C); 1H NMR (DMSO-d6, 400 MHz): δ 2.76 (s, 3H, thz-CH3), 3.83 (s, 3H, −OCH3), 7.01 (m, 1H, Ar–H), 7.31–7.33 (m, 1H, Ar–H), 7.40 (s, 1H, Ar–H), 7.58–7.56 (m, 1H, Pyr-H), 8.08 (s, 1H, =CH), 8.34 (d, 1H, Pyr-H, J = 8 Hz), 8.69 (d, 1H, Pyr-H, J = 4 Hz), 9.15 (s, 1H, Pyr-H), 9.65 (s, 1H, −OH), 11.82 (s, 1H, −NH); 13C NMR (DMSO-d6, 100 MHz): δ 50.9, 114.4, 120.2, 121.2, 124.9, 126.0, 129.9, 131.1, 133.1, 135.2, 146.5, 147.2, 151.4, 161.9, 171.7; LCMS (m/z): [M + H]+; Elemental analysis for C18H16N4O3S Calcd: C, 58.68; H, 4.38; N, 15.21. Found: C, 58.69; H, 4.40; N, 15.22.
Biological Activity
In Vitro Anti-inflammatory Activity (Denaturation of the Bovine Serum Albumin Method)
The anti-inflammatory activity of the target compounds was determined by denaturation of the bovine serum albumin technique as per the Mizushima and Kobayashi36 and Sakat et al.(37) reported method. The test sample contains the test compound and 1% aqueous solution of the bovine albumin fraction, and the pH of the reaction mixture was adjusted to pH 7.4 using suitable stripping solutions. Further, test samples were incubated at 37 °C for 20 min, and then, they was heated to 51 °C for 20 min. Later, it was cooled to ambient temperature, and the obtained turbidity of the sample was measured at 660 nm using a UV–visible spectrophotometer. The experiment was performed in triplicates using diclofenac sodium as the standard drug.
The anti-inflammatory activity of the titled compounds was estimated based on the percentage of inhibition of albumin denaturation using the following equation
Antimicrobial Activity by the Macrodilution Broth Method
The antimicrobial susceptibility of the target compounds was evaluated by the NCCLS macrodilution broth method. The MIC of the target compounds was evaluated according to the Clinical and Laboratory Standards Institute (CLSI)38 protocol. As the definition of the Clinical and Laboratory Standards Institute document39 states, “the MIC is the lowest concentration of the antimicrobial agent that completely inhibits growth of the organism in the tubes or microdilution wells as detected by the unaided eye”. Hence, the standard procedure was followed. Antimicrobial evaluation was carried out for the synthesized novel thiazole-based hydrazides against fungal strains such as Aspergillus flavus (A. flavus) (MTCC 1316), Trichoderma atroviridae (T. atroviridae) (MTCC 28036), Penicillium citranum (P. citranum) (MTCC 9849), and Candida albicans (C. albicans) (MTCC 461) and bacterial strains such as Staphylococcus aureus (S. aureus) (MTCC 9660), Bacillus subtilis (B. subtilis) (MTCC 441), Escherichia coli (E. coli) (MTCC 443), and Pseudomonas aeruginosa (P. aeruginosa) (MTCC 424) at varying concentrations (0.125–128 μg/mL). Nystatin and tetracycline were used as a standard drug for antifungal and antibacterial evaluation, respectively. Sterile test tubes containing the target compounds were placed in sterile Mueller Hinton broth (MHB) medium for bacteria and sterile Saboraud dextrose broth (SDB) for the fungi for which the test microorganisms were added. The MHB and SDB tubes with and without target compounds were used as the control. The MIC was detected as the lowest concentration of the target compound containing the tube showing no visible growth of the test microorganism. The experiment was performed in triplicates, and the data were analyzed by SPSS 20.0 software.
Statistical Analysis
Statistical analysis was carried out using SPSS software, version 20.0. The experiments were carried out in triplicates, and the data were expressed as mean ± standard deviation by one-way ANOVA. Turkey’s multiple comparison test was used to determine significant differences between the standard and synthesized compounds. Correlation analysis was carried out using Pearson’s correlation analysis using p = 0.01.
In Silico Prediction of Physicochemical and ADME Parameters
Physicochemical parameters of the designed compounds were predicted in silico using the QikProp module of Schrödinger. Various parameters predicted were molecular weight (M.Wt.), total solvent-accessible volume (TSAV), number of hydrogen bond donors (HBDs), number of hydrogen bond acceptors (HBAs), van der Waals polar surface area (PSA) of nitrogen and oxygen atoms, octanol/water partition coefficient (log P), aqueous solubility (log S), predicted Caco-2 cell permeability in nm/s (PCaco), apparent Madin Darby canine kidney (MDCK) permeability, and percentage of human oral absorption.39−41 The details of the predicted values are presented in 5.
Molecular Docking Studies
Molecular docking of the significantly active compound was performed using the Glide module of Schrödinger software42 installed on the Intel Xenon W3565 processor and Ubuntu enterprise version 14.04 as an operating system. The selected target protein structure was retrieved from the RCSB protein data bank.43 The titled ligand was drawn using Chemdraw 18.0 PerkinElmer software.
Ligand Preparation
The ligands used as an input for the docking study was sketched by ChemDraw software, and the structure was cleaned up for bond alignment. Then, ligands were incorporated into the workstation, and the energy was minimized using the OPLS3e (Optimized Potentials for Liquid Simulations) force field in LigPrep43 (version 2019-1, Schrödinger). This minimization helps to assign bond orders, addition of hydrogens to the ligands, and conversion of 2D to the 3D structure for the docking studies. The generated output file (best conformations of the ligands) was further used for docking studies.
Protein Preparation
The protein preparation wizard44 (version 2019–1, Schrödinger) was the main tool in Schrödinger to prepare and minimize the protein. The hydrogen atom was added to the protein, and charges were assigned. Het states were generated using Epik at pH 7.0 ± 2.0. The protein was preprocessed and refined, and the protein was modified by analyzing the workspace water molecules and others. The critical water molecules remained the same, and the rest of the molecules apart from heteroatoms was deleted. Finally, the protein was minimized using the OPLS3 force field. A grid was created by considering the co-crystal ligand, which was included in the active site of the protein of the selected target (PDB-3LN1).45 After the final step of docking with the co-crystal ligand in the XP mode, RMSD was checked to validate the protein, and the RMSD value was found to be 0.20 Å.
Receptor Grid Generation
A receptor grid was generated around the protein (PDB: 3LN1) by choosing the inhibitory ligand (X-ray pose of the ligand in the protein). The centroid of the ligand was selected to create a grid box around it, and the van der Waal radius of the receptor atoms was scaled to 1.00 Å with a partial atomic charge of 0.25.
Docking Validation
The most straightforward way of validating the accuracy of specified parameters for docking studies is to redock the co-crystallized ligand back into the binding site of the protein and calculate the RMSD value between the crystallographic orientation and the docking pose. The calculation of RMSD is a convenient method to study how much a structure has diverged from its initial geometry. Lower RMSD value between its docked pose and crystallographic orientation is an indication of the suitability of the docking protocol. Therefore, before the screening of all ligands, the co-crystal structure of 3LN1 was chosen, and their inhibitor was redocked back to their active site. The RMSD value of the crystallographic orientation and the best docked pose was found to be 0.20 Å.46,47
Docking Analysis
Docking and analysis of molecular docking were performed using the above prepared ligand and protein as the input. The results of the docking study were analyzed with the help of the XP visualizer (Version 2019-1, Schrödinger). All docking calculations were performed using the extra precision (XP) mode. A scaling factor of 0.8 and a partial atomic charge of less than 0.15 were applied to the atoms of the protein. The glide docking score was used to determine the best docked confirmation from the output. The interactions of these docked conformations were investigated further using the XP visualizer, and the results are depicted in the tables (Tables 1 and 5) and pictures (Figures 6–10).
Acknowledgments
Author Vinuta Kamat would like to thank DST (Award letter No.: DST/KSTePS/Ph.D.Fellowship/CHE-05:2018-19 dated 08.03.2019) for providing fellowship. Author Banoth Karan Kumar would like to thank Ministry of Tribal Affairs, Government of India Award no-201920-NFST-TEL-01497). Authors M.S. and F.F. gratefully acknowledge BITS-Pilani for providing the necessary facilities to do this work. This work was carried out under a grant from the Department of Biotechnology Indo-Spain, New Delhi. (Ref. No: BT/IN/Spain/39/SM/2017–2018).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c03386.
FTIR, NMR (1H and 13C), and LCMS spectra of the specific compounds and graphical representations of results of the antibacterial and antifungal evaluation of the compounds (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Lon H.-K.; Liu D.; Jusko W. J. Pharmacokinetic/pharmacodynamic modeling in inflammation. Crit. Rev. Biomed. Eng. 2012, 40, 295–312. 10.1615/critrevbiomedeng.v40.i4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood E. R.; Toliver-Kinsky T. Mechanisms of the inflammatory response. Best Pract. Res., Clin. Anaesthesiol. 2004, 18, 385–405. 10.1016/j.bpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Ferreira S. H.; Vane J. R. New aspects of the mode of action of nonsteroid anti-inflammatory drugs. Annu. Rev. Pharmacol. 1974, 14, 57–73. 10.1146/annurev.pa.14.040174.000421. [DOI] [Google Scholar]
- Vane J. R. Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nature, New Biol. 1971, 231, 232–235. 10.1038/newbio231232a0. [DOI] [PubMed] [Google Scholar]
- Jain H. K.; Mourya V. K.; Agrawal R. K. Inhibitory mode of 2-acetoxyphenyl alkyl sulfides against COX-1 and COX-2: QSAR analyses. Bioorg. Med. Chem. Lett. 2006, 16, 5280–5284. 10.1016/j.bmcl.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Anana R.; Rao P. N. P.; Chen Q.-H.; Knaus E. E. Synthesis and biological evaluation of linear phenylethynylbenzenesulfonamide regioisomers as cyclooxygenase-1/-2 (COX-1/-2) inhibitors. Bioorg. Med. Chem. 2006, 14, 5259–5265. 10.1016/j.bmc.2006.03.050. [DOI] [PubMed] [Google Scholar]
- Kovala-Demertzi D. Recent advances on non-steroidal anti-inflammatory drugs, NSAIDs: organotin complexes of NSAIDs. J. Organomet. Chem. 2006, 691, 1767–1774. 10.1016/j.jorganchem.2005.11.058. [DOI] [Google Scholar]
- Tacconelli S.; Capone M. L.; Sciulli M. G.; Ricciotti E.; Patrignani P. The biochemical selectivity of novel COX-2 inhibitors in whole blood assays of COX-isozyme activity. Curr. Med. Res. Opin. 2002, 18, 503–511. 10.1185/030079902125001335. [DOI] [PubMed] [Google Scholar]
- Tiwari A. D.; Panda S. S.; Girgis A. S.; Sahu S.; George R. F.; Srour A. M.; Starza B. L.; Asiri A. M.; Hall C. D.; Katritzky A. R. Microwave assisted synthesis and QSAR study of novel NSAID acetaminophen conjugates with amino acid linkers. Org. Biomol. Chem. 2014, 12, 7238–7249. 10.1039/c4ob01281j. [DOI] [PubMed] [Google Scholar]
- Morigi R.; Locatelli A.; Leoni A.; Rambaldi M. Recent patents on thiazole derivatives endowed with antitumor activity. Recent Pat. Anti-Cancer Drug Discovery 2015, 10, 280–297. 10.2174/1574892810666150708110432. [DOI] [PubMed] [Google Scholar]
- Ghorab M. M.; Al-Said M. S. Antitumor activity of novel pyridine, thiophene and thiazole derivatives. Arch. Pharmacal Res. 2012, 35, 965–973. 10.1007/s12272-012-0603-z. [DOI] [PubMed] [Google Scholar]
- Jadhavar S. C.; Bhansali S. G.; Patwari S. B.; Bhusare S. R.; Kasralikar H. M. Design, synthesis and docking studies of novel 1, 2, 3-triazolyl phenylthiazole analogs as potent anti-HIV-1 NNRT inhibitors. Med. Chem. 2017, 7, 268–275. 10.4172/2161-0444.1000467. [DOI] [Google Scholar]
- Amr A. E.-G. E.; Sabrry N. M.; Abdalla M. M.; Abdel-Wahab B. F. Synthesis, antiarrhythmic and anticoagulant activities of novel thiazolo derivatives from methyl 2-(thiazol-2-ylcarbamoyl) acetate. Eur. J. Med. Chem. 2009, 44, 725–735. 10.1016/j.ejmech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Maghraby M. T.-E.; Abou-Ghadir O. M. F.; Abdel-Moty S. G.; Ali A. Y.; Salem O. I. A. Novel class of benzimidazole-thiazole hybrids: The privileged scaffolds of potent anti-inflammatory activity with dual inhibition of cyclooxygenase and 15-lipoxygenase enzymes. Bioorg. Med. Chem. 2020, 28, 115403. 10.1016/j.bmc.2020.115403. [DOI] [PubMed] [Google Scholar]
- Ruberte A. C.; Ramos-Inza S.; Aydillo C.; Talavera I.; Encío I.; Plano D.; Sanmartín C. Novel N, N′-disubstituted acylselenoureas as potential antioxidant and cytotoxic agents. Antioxidants 2020, 9, 55. 10.3390/antiox9010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghonim A. E.; Ligresti A.; Rabbito A.; Mahmoud A. M.; Di Marzo V.; Osman N. A.; Abadi A. H. Structure-activity relationships of thiazole and benzothiazole derivatives as selective cannabinoid CB2 agonists with in vivo anti-inflammatory properties. Eur. J. Med. Chem. 2019, 180, 154–170. 10.1016/j.ejmech.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Hassan F. A. Synthesis, characterization, anti-inflammatory, and antioxidant activities of some new thiazole derivatives. Int. J. Sci. Tech. 2012, 2, 180–187. [Google Scholar]
- Iyer V. B.; Gurupadayya B.; Koganti V. S.; Inturi B.; Chandan R. S. Design, synthesis and biological evaluation of 1, 3, 4-oxadiazoles as promising anti-inflammatory agents. Med. Chem. Res. 2017, 26, 190–204. 10.1007/s00044-016-1740-6. [DOI] [Google Scholar]
- Mahmoud H. K.; Abbas A. A.; Gomha S. M. Synthesis, antimicrobial evaluation and molecular docking of new functionalized bis (1, 3, 4-thiadiazole) and bis (thiazole) derivatives. Polycyclic Aromat. Compd. 2020, 7, 1–13. 10.1080/10406638.2019.1709085. [DOI] [Google Scholar]
- Adole V. A.; More R. A.; Jagdale B. S.; Pawar T. B.; Chobe S. S. Efficient synthesis, antibacterial, antifungal, antioxidant and cytotoxicity study of 2-(2-hydrazineyl) thiazole derivatives. ChemistrySelect 2020, 5, 2778–2786. 10.1002/slct.201904609. [DOI] [Google Scholar]
- Wang L.-L.; Battini N.; Bheemanaboina R. R. Y.; Zhang S.-L.; Zhou C.-H. Design and synthesis of aminothiazolyl norfloxacin analogues as potential antimicrobial agents and their biological evaluation. Eur. J. Med. Chem. 2019, 167, 105–123. 10.1016/j.ejmech.2019.01.072. [DOI] [PubMed] [Google Scholar]
- Eryılmaz S.; Çelikoğlu E. T.; İdil Ö.; İnkaya E.; Kozak Z.; Mısır E.; Gül M. Derivatives of pyridine and thiazole hybrid: Synthesis, DFT, biological evaluation via antimicrobial and DNA cleavage activity. Bioorg. Chem. 2020, 95, 103476. 10.1016/j.bioorg.2019.103476. [DOI] [PubMed] [Google Scholar]
- Fan Z.; Shi J.; Luo N.; Ding M.; Bao X. Synthesis, crystal structure, and agricultural antimicrobial evaluation of novel quinazoline thioether derivatives incorporating the 1, 2, 4-triazolo [4, 3-a] pyridine moiety. J. Agric. Food Chem. 2019, 67, 11598–11606. 10.1021/acs.jafc.9b04733. [DOI] [PubMed] [Google Scholar]
- Bhila V. G.; Patel C. V.; Patel N. H.; Brahmbhatt D. I. One pot synthesis of some novel coumarins containing 5-(substituted-2-hydroxybenzoyl) pyridine as a new class of antimicrobial and antituberculosis agents. Med. Chem. Res. 2013, 22, 4338–4346. 10.1007/s00044-012-0437-8. [DOI] [Google Scholar]
- Desai N. C.; Bhatt N. B.; Joshi S. B.; Jadeja K. A.; Khedkar V. M. Synthesis, antimicrobial activity and 3D-QSAR study of hybrid oxazine clubbed pyridine scaffolds. ChemistrySelect 2019, 4, 7541–7550. 10.1002/slct.201901391. [DOI] [Google Scholar]
- Rani V. E.; Reddy P. R. Synthesis and antimicrobial activity of new pyridine containing substituted phenyl azetidine-2-one derivatives. Open J. Med. Chem. 2018, 08, 22–29. 10.4236/ojmc.2018.82003. [DOI] [Google Scholar]
- Rashdan H. R. M.; Abdel-Aziem A.; El-Naggar D. H.; Nabil S. Synthesis and biological evaluation of some new pyridines, isoxazoles and isoxazolopyridazines bearing 1, 2, 3-triazole moiety. Acta Pol. Pharm. 2019, 76, 469–482. 10.32383/appdr/103101. [DOI] [Google Scholar]
- Laddha S. S.; Bhatnagar S. P. A new therapeutic approach in Parkinson’s disease: Some novel quinazoline derivatives as dual selective phosphodiesterase 1 inhibitors and anti-inflammatory agents. Bioorg. Med. Chem. 2009, 17, 6796–6802. 10.1016/j.bmc.2009.08.041. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Chauhan A.; Drabu S. Synthesis of cyanopyridine and pyrimidine analogues as new anti-inflammatory and antimicrobial agents. Biomed. Pharmacother. 2011, 65, 375–380. 10.1016/j.biopha.2011.04.023. [DOI] [PubMed] [Google Scholar]
- Thirumurugan P.; Mahalaxmi S.; Perumal P. T. Synthesis and anti-inflammatory activity of 3-indolyl pyridine derivatives through one-pot multi component reaction. J. Chem. Sci. 2010, 122, 819–832. 10.1007/s12039-010-0070-3. [DOI] [Google Scholar]
- a Liu H.; Long S.; Rakesh K. P.; Zha G.-F. Structure-activity relationships (SAR) of triazine derivatives: Promising antimicrobial agents. Eur. J. Med. Chem. 2020, 185, 111804. 10.1016/j.ejmech.2019.111804. [DOI] [PubMed] [Google Scholar]; b Rakesh K. P.; Kumara H. K.; Ullas B. J.; Shivakumara J.; Channe Gowda D. Amino acids conjugated quinazolinone-Schiff’s bases as potential antimicrobial agents: Synthesis, SAR and molecular docking studies. Bioorg. Chem. 2019, 90, 103093. 10.1016/j.bioorg.2019.103093. [DOI] [PubMed] [Google Scholar]; c Xu M.; Wu P.; Shen F.; Ji J.; Rakesh K. P. Chalcone derivatives and their antibacterial activities: current development. Bioorg. Chem. 2019, 91, 103133. 10.1016/j.bioorg.2019.103133. [DOI] [PubMed] [Google Scholar]; d Rakesh K. P.; Gowda D. C. Schiff’s bases of quinazolinone derivatives: Synthesis and SAR studies of a novel series of potential anti-inflammatory and antioxidants. Bioorg. Med. Chem. Lett. 2015, 25, 1072–1077. 10.1016/j.bmcl.2015.01.010. [DOI] [PubMed] [Google Scholar]; e Rakesh K. P.; Ramesh S.; Kumar H. M.; Chandan S.; Gowda D. C. Quinazolinones linked amino acids derivatives as a new class of promising antimicrobial, antioxidant and anti-inflammatory agents. Eur. J. Chem. 2015, 6, 254–260. 10.5155/eurjchem.6.3.254-260.1233. [DOI] [Google Scholar]; f Rakesh K. P.; Gowda D. C.; Gowda D. C. Anti-inflammatory and antioxidant peptide-conjugates: Modulation of activity by charged and hydrophobic residues. Int. J. Pept. Res. Ther. 2019, 25, 227–234. 10.1007/s10989-017-9668-3. [DOI] [Google Scholar]; g Rakesh K. P.; Shivakumar S.; Gowda D. C. Effect of low charge and high hydrophobicity on antimicrobial activity of the quinazolinone-peptide conjugates. Russ. J. Bioorg. Chem. 2018, 44, 158–164. 10.1134/s1068162018020036. [DOI] [Google Scholar]; h Rakesh K. P.; Sridhara M. B.; Manukumar H. M.; Qin H.-L.; Qin H. L. Benzisoxazole: A privileged scaffold for medicinal chemistry. MedChemComm 2017, 8, 2023–2039. 10.1039/c7md00449d. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Manukumar H. M.; Chandrasekhar B.; Rakesh K. P.; Ananda A. P.; Nandhini M.; Lalitha P.; Sumathi S.; Qin H.-L.; Umesha S. Novel TC@ AgNPs mediated biocidal mechanism against biofilm associated methicillin-resistant Staphylococcus aureus (Bap-MRSA) 090, cytotoxicity and its molecular docking studies. MedChemComm 2017, 8, 2181–2194. 10.1039/c7md00486a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaboudin B.; Elhamifar D. Phosphorus pentasulfide: A mild and versatile reagent for the preparation of thioamides from nitriles. Synth 2006, 2006, 224–226. 10.1055/s-2005-918507. [DOI] [Google Scholar]
- Ali M. R.; Kumar S.; Afzal O.; Shalmali N.; Sharma M.; Bawa S. Development of 2-(substituted benzylamino)-4-methyl-1, 3-thiazole-5-carboxylic acid derivatives as xanthine oxidase inhibitors and free radical scavengers. Chem. Biol. Drug Des. 2016, 87, 508–516. 10.1111/cbdd.12686. [DOI] [PubMed] [Google Scholar]
- Han M. İ.; Bekçi H.; Uba A. I.; Yıldırım Y.; Karasulu E.; Cumaoğlu A.; Karasulu H. Y.; Yelekçi K.; Yılmaz Ö.; Küçükgüzel Ş. G. Synthesis, molecular modeling, in vivo study, and anticancer activity of 1, 2, 4-triazole containing hydrazide–hydrazones derived from (S)-naproxen. Arch. Pharm. 2019, 352, 1800365. 10.1002/ardp.201800365. [DOI] [PubMed] [Google Scholar]
- Sıcak Y.; Oruç-Emre E. E.; Öztürk M.; Taşkın-Tok T.; Karaküçük-Iyidoğan A. Novel fluorine-containing chiral hydrazide-hydrazones: Design, synthesis, structural elucidation, antioxidant and anticholinesterase activity and in silico studies. Chirality 2019, 31, 603–615. 10.1002/chir.23102. [DOI] [PubMed] [Google Scholar]
- Mizushima Y.; Kobayashi M. Interaction of anti-inflammatory drugs with serum proteins, especially with some biologically active proteins. J. Pharm. Pharmacol. 1968, 20, 169–173. 10.1111/j.2042-7158.1968.tb09718.x. [DOI] [PubMed] [Google Scholar]
- Sakat S.; Juvekar A. R.; Gambhire M. N. In vitro antioxidant and anti-inflammatory activity of methanol extract of Oxalis corniculata Linn. Int. J. Pharm. Pharm. Sci. 2010, 2, 146–155. [Google Scholar]
- National Committee for Clinical Laboratory Standards . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved standard, 7th ed.; NCCLS: Villanova PA, USA, 2006http://demo.nextlab.ir/getattachment/737fedd6-3926-4099-b6c8-19aa279bfd1f/CLSI-M7-A7.aspx.
- Clinical and Laboratory Standards Institute . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 9th ed.; CLSI; document M07-A9, 2012.
- van de Waterbeemd H.; Gifford E. ADMET in silico modelling: Towards prediction paradise. Nat. Rev. Drug Discovery 2003, 2, 192–204. 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- QikProp Descriptors and properties PISA, 2015; pp 2–4.
- Chander S.; Wang P.; Ashok P.; Yang L.-M.; Zheng Y.-T.; Sankaranarayanan M. Design, synthesis and anti-HIV-1 RT evaluation of 2-(benzyl(4-chlorophenyl)amino)-1-(piperazin-1-yl)ethanone derivatives. Bioorg. Med. Chem. Lett. 2017, 27, 61–65. 10.1016/j.bmcl.2016.11.030. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2019-1: Glide; Schrödinger, LLC: New York, NY.(5), 2019https://www.rcsb.org/.
- Schrödinger Release 2019-1; LigPrep, Schrödinger, LLC: New York, NY, 2019.
- Schrödinger Release 2019-1: Schrödinger Suite 2019-1 Protein preparation Wizard; Epik, Schrödinger, LLC, New York, NY, 2019.
- Wang J. L.; Limburg D.; Graneto M. J.; Springer J.; Hamper J. R. B.; Liao S.; Pawlitz J. L.; Kurumbail R. G.; Maziasz T.; Talley J. J.; Kiefer J. R.; Carter J. The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life. Bioorg. Med. Chem. Lett. 2010, 20, 7159–7163. 10.1016/j.bmcl.2010.07.054. [DOI] [PubMed] [Google Scholar]
- Ganesan M. S.; Raja K. K.; Narasimhan K.; Murugesan S.; Kumar B. K. Design, synthesis, α-amylase inhibition and in silico docking study of novel quinoline bearing proline derivatives. J. Mol. Struct. 2020, 1208, 127873. 10.1016/j.molstruc.2020.127873. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.