Abstract
Introduction
Influenza epidemics and pandemics cause significant morbidity and mortality. An effective response to a potential pandemic requires the infrastructure to rapidly detect, characterise, and potentially contain new and emerging influenza strains at both an individual and population level. The objective of this study is to use data gathered simultaneously from community and hospital sites to develop a model of how influenza enters and spreads in a population.
Methods and analysis
Starting in the 2018–2019 season, we have been enrolling individuals with acute respiratory illness from community sites throughout the Seattle metropolitan area, including clinics, childcare facilities, Seattle-Tacoma International Airport, workplaces, college campuses and homeless shelters. At these sites, we collect clinical data and mid-nasal swabs from individuals with at least two acute respiratory symptoms. Additionally, we collect residual nasal swabs and data from individuals who seek care for respiratory symptoms at four regional hospitals. Samples are tested using a multiplex molecular assay, and influenza whole genome sequencing is performed for samples with influenza detected. Geospatial mapping and computational modelling platforms are in development to characterise the regional spread of influenza and other respiratory pathogens.
Ethics and dissemination
The study was approved by the University of Washington’s Institutional Review Board (STUDY00006181). Results will be disseminated through talks at conferences, peer-reviewed publications and on the study website (www.seattleflu.org).
Keywords: influenza, protocol, respiratory infection, virology, surveillance
Strengths and limitations of this study.
Large-scale multiple-arm study of respiratory illness characterisation with collection of samples from individuals in the community as well as in ambulatory care and hospital settings.
Multiplex molecular testing for multiple viral and bacterial pathogens and whole genome sequencing of influenza for detailed molecular epidemiological characterisation and transmission mapping.
Convenience sampling of participants from the community.
Undersampling of populations such as non-English speakers, older adults, and racial and ethnic minorities.
Study inclusion criteria do not allow for examining the role of asymptomatic respiratory viral transmission.
Introduction
In the USA, annual influenza epidemics cause 9–45 million illnesses, 140 000–810 000 hospitalisations and 12 000–67 000 deaths. The 1918 pandemic, the worst for which reliable records exist, resulted in an estimated 675 000 deaths in the USA and 50 million worldwide.1–3 More recently, the 2009 H1N1 pandemic caused 12 500 deaths and 60.8 million infections in the USA, with an estimated global burden of 284 500 deaths.4 5
Active surveillance for influenza is essential to monitor the impact of seasonal influenza and to detect and characterise emerging influenza viruses. In the USA, surveillance systems rely primarily on medically attended illnesses. This underestimates true influenza-related disease burden by 50% or more.6 Individuals may not seek care, diagnostic tests are often underused in the community and hospitalisations may be attributed to chronic conditions exacerbated by influenza.7 8
Community-wide studies provide a mechanism to identify influenza among individuals who may not present for care and who may provide the first signal of an impending pandemic.9 Previous community-wide studies of influenza, including family-based prospective studies conducted in the twentieth century, have provided important data on transmission dynamics, including the role of households and schools in driving seasonal epidemics. However, a common limitation of community-based studies is they do not routinely integrate their data with inpatient hospital and ambulatory care surveillance to understand the transmission and burden of influenza at a population level within a specific geographical area.
Despite substantial progress in the estimation of the burden of influenza through large-scale surveillance studies, there is an ongoing need for improved near-real-time monitoring that is coupled to pandemic preparedness. The integration of individual-level, community-based sampling is crucial to augment samples collected in clinical settings, particularly in the USA where care-seeking behaviour is impacted by many factors, including health insurance status.
Pandemic control requires the rapid accrual of actionable information from a diversity of sources. For example, rapid genome sequencing of influenza strains throughout a city could feed the creation of actionable maps that identify new and emerging influenza strains and associated transmission dynamics. With such a system established, we can more effectively develop and test strategies to rapidly deploy vaccines, antivirals and non-pharmaceutical interventions. This combination of rapid detection and intervention deployment provide a new paradigm for outbreak containment, which is directly translatable to the current SARS-CoV-2 pandemic.
This manuscript describes the protocol for the Seattle Flu Study (SFS), a multiarmed, regional study of influenza at a city-wide scale that integrates community, ambulatory care and inpatient surveillance at a large magnitude. The objectives of this study are to gather data from community and hospital sites to advance our understanding of how influenza and other respiratory pathogens enter and spread in a population, create a city-wide platform for testing novel interventions that may limit or contain outbreaks, and establish a deployable infrastructure for future pandemics.
Aims and hypothesis
The primary aim of this study is to quantify community-wide prevalence transmission patterns through molecular epidemiology. Our approach combines cross-sectional surveillance with prospective cohort studies. In this study, we integrate data across all these sources to provide near-real-time analysis to guide disease control activities.
The primary hypothesis is that a large-scale cross-sectional design can detect early cases of influenza, and serve as a surveillance tool for monitoring and potentially reducing community-level transmission through viral genetic and spatial-demographic analyses, and be used to inform public health and clinical interventions.
Methods and analysis
Study design
The SFS is a multiarmed surveillance study. The arms include (1) community cross-sectional (2) clinical cross-sectional (3) prospective clinical cohort (4) prospective childcare cohort and (5) active clinical surveillance. This protocol describes the SFS as implemented in Year 1. Substudies and modifications in future years will be described in subsequent publications.
Study overview and setting
In this study, participants are eligible if they have two or more acute respiratory illness (ARI)-associated symptoms (table 1) or a medically collected respiratory specimen. The presence of a subjective or objective fever is not required. All participants have a respiratory specimen collected that is linked with demographic information, illness characteristics, behavioural and other clinical metadata. The primary outcome is influenza infection status as defined by molecular diagnostic testing. The study takes place during influenza season (October to May) annually, starting with 2018–2019.
Table 1.
Acute respiratory illness-associated symptoms used to trigger the collection of a mid-nasal swab in the community cross-sectional, prospective clinical cohort or childcare cohort arms of the Seattle Flu Study. Study participants must have at least two self-reported symptoms in the 7 days prior to swab collection
| Fever | Cough | Sore throat | Headache |
| Diarrhoea | Nausea or vomiting | Runny or stuffy nose | Rash |
| Fatigue (tiredness) | Muscle or body aches | Increased trouble with breathing | Ear pain or discharge |
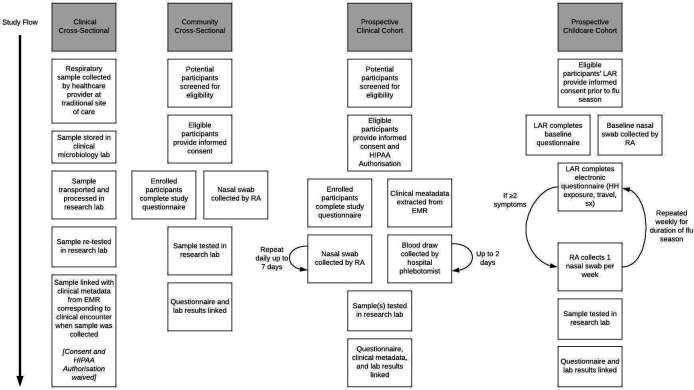
The study occurs in a variety of settings throughout the Seattle metropolitan area (figure 1). Participants in the community cross-sectional arm are recruited at stand-alone kiosks in public settings including clinical facilities, university campuses, airports, workplaces, homeless shelters and high-traffic tourist areas. Participants in the clinical cross-sectional and prospective clinical cohort arms are recruited from traditional sites of care such as hospitals and medical clinics. Participants in the prospective childcare cohort arm are recruited from childcare facilities. All collection sites are located in the greater Seattle metropolitan area, including King, Pierce, Snohomish, Skagit and Island Counties, Washington, USA.
Figure 1.
Outline of participant flow for different arms of the Seattle Flu Study. EMR, electronic medical record; HIPAA, Health Insurance Portability and Accountability Act; HH, household; LAR, legally authorised representative; RA, research assistant; sx, symptoms. Active clinical surveillance arm not included in figure. For methods, see Prevention USCfDCa. FluView and Prevention USCfDCa. FluView and Fong et al.13 14
Study arms
The community cross-sectional arm enrols individuals with two or more ARI-associated symptoms (table 1). One mid-nasal swab is collected at the time of enrolment and linked with questionnaire data. These individuals do not have follow-up. Individuals are eligible to re-enrol for new ARI episodes every 14 days. This arm aims to enrol community-dwelling individuals with varying degrees of care-seeking behaviour.
The clinical cross-sectional arm uses residual clinically collected respiratory specimens of all types from participating hospitals and clinics, regardless of clinical test result. Specimens are aliquoted and undergo the same laboratory processing as above. Demographic and clinical metadata are extracted from the electronic medical record (EMR) and linked to the corresponding respiratory specimen. There is no direct contact with these individuals. This arm aims to include a care-seeking population, which is the traditional backbone for respiratory viral surveillance.
The prospective clinical cohort enrols hospitalised individuals with a lab-confirmed respiratory virus as tested by the hospital’s clinical lab. Participants are enrolled by the same method as the community cross-sectional arm. They are surveyed daily for up to 7 days after diagnosis and may have additional respiratory and/or blood samples collected at these intervals. This arm aims to enrol individuals with more severe respiratory viral infections, and the longitudinal sampling allows for profiling of the humoral immune response to infection.
The prospective childcare cohort enrols children attending participating day cares. Children are enrolled prior to the local influenza season, and a baseline mid-nasal swab is collected at this time. They are then surveyed weekly for the duration of the local influenza season for development of ARI symptoms. If symptom criteria are met, an additional mid-nasal swab and corresponding data are collected. This arm aims to characterise respiratory virus epidemiology and transmission within the childcare setting, and help determine the contribution of respiratory illness in children in closed populations.
The active clinical surveillance arm uses specimens collected at participating medical clinics as well as through the Washington site of the US Flu Vaccine Effectiveness (US Influenza VE) network, details of which have been previously published.10 11 In the US Influenza VE Network, patients seeking ambulatory care for ARI are prospectively identified and recruited into the study, with collection of clinical data and a nasal swab. Clinical data and residual nasal swabs from individuals enrolled in the Influenza VE Study are further analysed for the SFS. This arm aims to enrol care-seeking individuals with mild or moderate respiratory illnesses and collect detailed illness characteristics that may not otherwise be extracted from EMR systems.
The environmental sampling arm involves specimen collection from high-touch surfaces and bioaerosol sampling at community enrolment sites and childcare sites to characterise the extent of environmental pathogen detection and the concordance between detection on environmental samples and participants’ respiratory samples at those sites.
Study population inclusion/exclusion criteria
Participants are eligible for inclusion if they meet all the following criteria for their study arm and do not meet any exclusion criteria. Prospectively recruited participants are excluded from the study if they are unable to provide consent themselves or through a legally authorised representative (LAR) or if they are incarcerated, wards of the state, or have any condition that, at the investigators’ discretion, may preclude or limit participation with study procedures.
Participants in the community cross-sectional study arm are eligible at any age, and can enrol at a stand-alone study kiosk if they have two or more new or worsening ARI-associated symptoms. In addition to overall study criteria, individuals are excluded if they have previously enrolled into the study within 14 days. For individuals under age 18 years, consent is obtained from an LAR.
There is no participant interaction in the clinical cross-sectional study arm. Data are used if a respiratory specimen is collected at clinician discretion from a participating site within the five-county study surveillance region during the study period.
Prospective clinical cohort participants are eligible if they are at least 18 years old, inpatient at a participating hospital, English-speaking or Spanish-speaking, have a laboratory-confirmed respiratory viral infection, and consent and provide Health Insurance Portability and Accountability Act (HIPAA) authorisation for themselves or through an LAR.
Prospective childcare cohort participants are eligible if they are children attending a participating childcare facility and have an LAR to provide consent.
Patients are eligible for US Influenza VE Network enrolment if they are aged 6 months or older as of 1 September 2018, have a cough of <8 days’ duration and have not used an antiviral medication in the past 7 days.10 11
Consent and recruitment
Recruitment methods vary by study arm. The community cross-sectional, prospective clinical cohort and prospective childcare cohort arms recruit participants through actively approaching potentially eligible individuals, digital media and flyers. For these study arms, eligible and interested participants are consented via an electronic consent form. For consent/assent details by participant age, see figure 2. In accordance with institutional review board (IRB) approval, consent and HIPAA authorisations for participants in the clinical cross-sectional study are fully waived.
Figure 2.
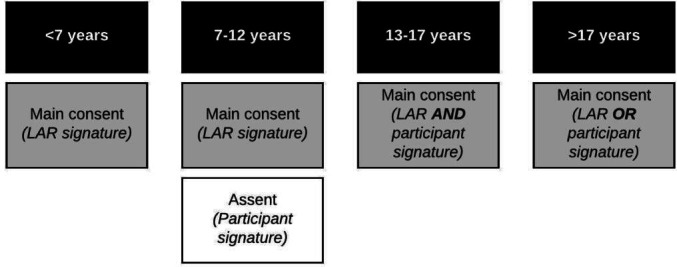
Documentation of written, informed consent by age of participant in the community cross-sectional, and prospective clinical and childcare cohorts of the Seattle Flu Study. If the participant is unable to provide informed consent due to cognitive impairment or because they have not attained the legal age for consent, a legally authorised representative (LAR) may sign the consent form on their behalf. Participants enrolled by Seattle Children’s Hospital staff or participants enrolled into the prospective clinical cohort sign a Health Insurance Portability and Accountability Act (HIPAA) authorisation in addition to the main consent.
Sample size
The goal of the study is to obtain samples and data from cases of influenza in the Seattle metropolitan and surrounding areas. In 2017–2018, 6953 positive influenza tests from Washington State were reported to the Centres for Disease Control and Prevention through participating clinical laboratories, which only represent a subset of the total cases diagnosed.12 13
Outcomes
Primary outcomes
Describe the clinical and sociodemographic characteristics and risk factors for individuals with ARI attributable to influenza.
Analyse the geospatial and temporal spread of influenza cases.
Evaluate the genetic diversity of circulating influenza strains in the Seattle metropolitan area, by variables including age, vaccine status, site of collection and home census tract.
Secondary outcomes
Determine the prevalence of ARI specifically attributable to influenza, as well as to respiratory syncytial virus (RSV), adenovirus, coronavirus, rhinovirus and other respiratory pathogens.
Describe the clinical, geospatial and sociodemographic characteristics of participants with ARI respiratory pathogens other than influenza.
Map geospatial and temporal trends of ARI cases attributable to other respiratory pathogens.
Assess genetic diversity of circulating strains of respiratory pathogens in the Seattle metropolitan area, by age, vaccine status, site of collection and home census tract.
Examine the impact of bacterial co-detection on viral ARI severity and outcomes.
Retrospectively track molecular epidemiology and transmission dynamics of respiratory pathogens within sites of collection.
Measure the impact of viral co-infection on viral ARI severity and outcomes.
Characterise the prevalence and concordance of environmental sampling by sample type and sampling location.
Compare viral kinetics of respiratory pathogens among individuals with longitudinal sample collection.
Ascertain prevalence and predictors of care-seeking and influenza vaccine receipt within participants.
Calculate viral load and clarify its relationship with clinical disease characteristics in both single and multiple virus infections.
Determine the strength and direction of the correlation of viral load with molecular markers of nasal swab sampling efficiency.
Model the probability of influenza based on individual-level factors, including symptoms.
Data collection methods
For participants enrolled in the clinical cross-sectional or prospective clinical arms, EMR data are obtained through the clinical data warehouse, which consolidates patient-level medical data from multiple sources. Survey data from eligible and enrolled participants in the community cross-sectional and prospective arms are collected through the FluTrack app (Audere, Seattle, Washington, USA), a mobile-enabled app created for the SFS and administered on a tablet. For participants unable or unfamiliar with the use of a tablet, questionnaires on the FluTrack app are administered by study staff verbally in English or Spanish. If the participant is under 7 years old, their LAR completes the questionnaire on their behalf. For participants aged 7–12 years, the LAR may decide whether to complete the survey on the participant’s behalf or in collaboration. Participants aged 13 years or older self-complete the questionnaire. Data are collected on US Influenza VE Network enrollees via interview and extraction from EMR and administrative healthcare databases.
Participant retention and compensation
Prospective clinical cohort participants receive a gift card for each longitudinal study specimen that is collected (nasal swabs and blood). Participants in the community cross-sectional arm receive a gift card for completing the study. If enrolled in the prospective childcare cohort, participants receive a gift card for each episode with collection of a mid-nasal swab. There is no remuneration for participants in the clinical cross-sectional arm since there is no participant interaction.
Data security and storage
All information from the study subjects is kept confidential. Participants are assigned an identification (ID) number (study ID) on enrolment that is used in place of names and other protected health information (PHI). Data are collected electronically in a Code of Federeal Regulations (CFR) Title 21 Part 11 compliant, password-protected and auditable database. The list linking participant PHI to the unique study identification number is stored separately from the database. Access to identifiable information is limited to the study staff. Electronic files are secured via log on password protection for study accounts. Any data sets containing PHI are stored in a HIPAA-compliant manner. No PHI is included with data sent to the broader study team or any other data-sharing repositories. Study data are transferred via encrypted software and the original files are kept in the database, hosted on a secure server at the Fred Hutchinson Cancer Research Institute.
Study IDs remain on biospecimens (nasal swabs, environmental samples, blood samples) during laboratory processing. All subjects who consent to participate are asked to approve the storage of their biospecimens and subsequent sample aliquots (nasal swabs and blood samples, where applicable). Persons who consent to participate in the study but do not want their biospecimens stored may still participate in the study. These biospecimens are tested as per protocol, but the remaining aliquots are destroyed.
Identifiers are kept on all data files until the study is closed out. Primary data collection sources will be maintained for at least 5 years following the publication of the primary study results. Once this time elapses and the electronic data files are fully cleaned, any paper forms will be destroyed.
Data quality
Data are checked for missing or unusual values and consistency in the centralised data capture system. Computerised checks are conducted weekly to identify missing, inconsistent or out-of-range data. Any suspect data are raised as data queries and are investigated by the study coordinators.
Protection against risks
All identifying data are stored using standard security techniques. Hard copies of data collection materials with PHI will be locked in an office with limited access by minimal members of the study team. When possible, redacted (deidentified) versions of the data will be used for coding and data analysis. PHI will be stored in the database on a password-protected network that is HIPAA-compliant, and only accessible to specific individuals. Transfer or storage on portable devices (eg, laptops, flash drives) will be encrypted, and only accessible to individuals who need access to the data.
Specimen collection methods
Respiratory specimens
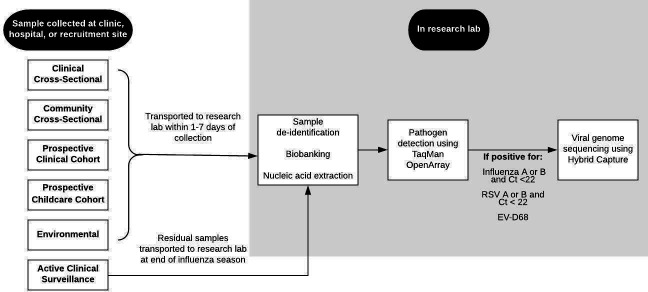
Respiratory specimens are collected in the prospective study arms using a sterile Copan flocked respiratory swab inserted and rotated mid-nasally. These specimens are then transported to the research laboratory in universal viral transport medium (UTM; Becton, Dickinson and Company, Franklin, New Jersey, USA). Clinically collected respiratory specimens are obtained from each participating hospital or clinic laboratory, maintained at 4℃, and transported on wet ice to the research laboratory within 3–7 days of specimen collection (figure 3).
Figure 3.
Laboratory pipeline for samples included in the Seattle Flu Study from time of collection through sequencing. CT, cycle threshold; EV-D68, enterovirus D68.
Environmental samples
Both bioaerosol and surface samples are collected longitudinally at select community cross-sectional and prospective childcare cohort sites. Prespecified high-touch surfaces are sampled by wiping a designated object or 10 cm2 area with a synthetic polyester swab and transported to the lab in UTM. Bioaerosol samples are collected over a 90 min sampling period using an SKC-West QuickTake 30 Air Pump with membrane filter paper. These samples are collected at prespecified locations with high foot traffic and low air ventilation. Environmental samples are transported in the same way as the mid-nasal swabs described above.
Laboratory methods
Respiratory specimens are aliquoted in triplicate and bar coded using a unique identifier that can be linked back to the participant and collection site. Samples are frozen at −80°C until thawed for extraction (figure 3). Total nucleic acids are extracted from 200 µl of UTM using Magna Pure 96 small total nucleic acids extraction kit (Roche). Extracted nucleic acids are screened for the presence of respiratory pathogens by TaqMan real-time PCR (RT-PCR) on the OpenArray platform (Thermo) (table 2). Specimens that test positive for influenza by RT-PCR are then sequenced using a modified oligo capture protocol. Total RNA is converted to cDNA and sequencing libraries are constructed using the Illumina TruSeq RNA Library Prep for Enrichment kit, as outlined by the manufacturer (Illumina, San Diego, California, USA). Library samples are ranked by TaqMan relative threshold (CRT) value, and 24 samples with similar CRT values are pooled prior to oligo capture. CRT values are further used to determine the viral load of each sample and inform the fraction of each capture pool that a given sample represents. Each pool of flu-positive samples is hybridised overnight to a custom pool of capture oligonucleotides following the manufacturer’s recommended protocol (Twist Bioscience, San Francisco, California, USA). Final libraries are sequenced on the Illumina Miseq, NextSeq or NovaSeq platform with paired end 150 bp reads.
Table 2.
Pathogens for which all Seattle Flu Study respiratory specimens are tested using a TaqMan RT-PCR
| Viruses | Bacteria |
| Influenza A—H3N2 | Streptococcus pneumoniae |
| Influenza A—H1N1 | Mycoplasma pneumoniae |
| Influenza A—Pan | Chlamydia pneumoniae |
| Influenza B | |
| Influenza C | |
| Respiratory syncytial viruses A and B | |
| Parainfluenza viruses 1–4 | |
| Coronavirus 229E, NL63, OC43 and HKU1 | |
| Adenovirus | |
| Rhinovirus | |
| Human metapneumovirus | |
| Human parechovirus | |
| Enterovirus* | |
| Enterovirus D68 | |
| Human bocavirus |
*All enterovirus species A, B, C, D and G, including: all Coxsackie serotypes under species A, B, C; all Echovirus serotypes; all Poliovirus serotypes.1–3
Statistical methods
Respiratory pathogen prevalence is calculated as the number of cases detected out of the total number of episodes with testing. We do not make any statistical adjustments to account for specificity and sensitivity for the following reasons. Previous sensitivity analyses have verified that TaqMan and OpenArray are highly specific and so false positives are rare relative to true positives. For specificity, we have also compared our OpenArray results for samples with clinical test results and found them to be highly concordant with Cepheid and BioFire Film Array for detection of influenza (96%) and RSV (92%), with discordance most common for samples with concentrations near the detection thresholds for each assay. Thus, test prevalence without adjustment is comparable to standard reporting from clinical labs.
Risk ratios and associated 95% CIs are estimated for the cross-sectional study arms using Poisson or negative binomial regression. Similarly, risk ratios and associated 95% CIs are estimated for the prospective clinical and childcare cohorts. For questionnaire data, descriptive statistics are calculated and associations with respiratory pathogen prevalence analysed using parametrical and non-parametrical tests, as appropriate given the distributions. Geospatial incidence maps are estimated using generalised additive mixed models with spatial, temporal, age and non-structured random effects.14 Phylogenetic analyses are based on the Nextstrain pipeline.15
For longitudinal data such as viral kinetics, we will conduct a descriptive analysis of group means over time and use mixed models, general estimating equations and quantile regression to assess for change over time, adjusting for potential confounders, including age, comorbidities and vaccination status.
Missing data
For questionnaire data in the cross-sectional study, we do not anticipate substantial missing data because completion of the survey is a criterion for participation, though participants can state that they prefer not to answer specific questions. For longitudinal data, we are using complete case analysis. For analyses and subgroups where complete case analysis leads to loss of 10% or more of subjects, we are performing multiple imputation and sensitivity analyses.
Subgroup analyses
The main subgroups of interest are age, recruitment site (homeless shelter, childcare, clinical, other) and vaccination status.
Ethics and dissemination
Ethics approval
The SFS has been approved by the Human Subjects IRB at the University of Washington (STUDY00006181).
Monitoring
The SFS has a scientific advisory board to which it reports biannually. These reports include updates on enrolment, preliminary results, substudies and protocol modifications.
Assessment of harms and adverse events
Participants receive contact information for the study team and are encouraged to report any participation-related harm. Given the minimally invasive nature of the sample collection procedures, harms and adverse events are very unlikely. Should any significant adverse events arise related to the study procedures, they will be reported to the IRB at the University of Washington as well as any applicable participating site.
Access to data
Full, identifying study data will be accessed only by authorised individuals on the study team. Access to deidentified, aggregate data and analysis code will be publicly available on the study website (www.seattleflu.org).
Dissemination plans
Our study group will present results at national and international research conferences, through peer-reviewed publications, and on the study website (www.seattleflu.org). We will follow STrengthening the Reporting of OBservational studies in Epidemiology reporting guidelines, as applicable.16 Study materials, including site-specific questionnaires and FluTracker app screenshots, will be available on request.
Patient and public involvement
Most public involvement is as research participants. There are no current plans to involve participants in the study recruitment, conduct, analysis or dissemination, though aggregate data will be made publicly available.
Discussion
We present the study design and infrastructure for a large-scale assessment of the burden of ARI attributable to influenza and other respiratory pathogens. The overarching goal of this study is to develop and implement strategies for actionable pathogen surveillance in a major metropolitan area. Cumulatively, these strategies will facilitate the early identification of novel pathogens and allow for targeted deployment of public health resources and interventions at the community level during pandemics.
This study design has several strengths. Current respiratory virus-based transmission models are largely based on clinical data. The SFS integrates the sociodemographic, clinical and geospatial characterisations of individuals with respiratory illnesses from diverse sources, including community-based, ambulatory, hospital-based and environmental sampling. This large-scale, multiarm sampling strategy allows us to overcome biases introduced by influenza care-seeking trends in the USA and generates a more real-time mapping of community transmission dynamics. Further, the use of community-based ARI surveillance generates novel, individual-level respiratory pathogen data in individuals who may not seek care.
We use broad, multiplex molecular testing for viral and bacterial pathogens to characterise the molecular epidemiology of respiratory pathogens in the community. For samples with influenza detected, we perform whole genome sequencing to evaluate transmission patterns within a community and identify target populations for public health interventions. This strategy is translatable to the current SARS-CoV-2 pandemic. We used this platform to first identify community transmission of SARS-CoV-2 in the USA.15 17
This study has several limitations. We used convenience sampling for enrolment, and undersampled populations such as non-English speakers, older adults, and racial and ethnic minorities. In the second year of the SFS, we enhanced our sampling strategy so that it would be better representative of the diversity of the Seattle metropolitan population through targeted in-person and online marketing recruitment. Our community-based sampling required participants to have at least two ARI-associated symptoms, and therefore did not allow us to examine the role of asymptomatic respiratory viral transmission. To address this limitation, we broadened the community-based eligibility criteria to include individuals without symptoms.18 Finally, the in-person community-based recruitment restricted our geographical scope, thus resulting in data that may not be representative of suburban or rural transmission patterns. To account for this, we subsequently added an online-based, at-home enrolment strategy so that individuals could participate in the study from home through home-based delivery of self-collection kits.19
The first year of the SFS demonstrated the utility of an integrated surveillance system for pandemic preparedness. Our research team did not anticipate that a global pandemic would arise while the study was being conducted. However, with Seattle as an early hot spot of the SARS-CoV-2 pandemic, the SFS was well positioned to provide the infrastructure for early identification and mapping of transmission. During the pandemic, we rapidly developed an assay for SARS-CoV-2, eliminated in-person community-based sampling due to widespread ‘Stay-at-Home’ orders, and scaled up the online and at-home sampling strategies.18 20
In conclusion, the study design presented here may provide guidance for establishment of a respiratory pathogen surveillance system for current and future pandemics.
Supplementary Material
Footnotes
Twitter: @KiraNewmanMDPhD
Contributors: HYC, MB, JAE, MF, BL, DAN, MR, LMS, JS and TB designed and implemented the protocol. They also wrote, reviewed and edited the written protocol. EB, JL, KN and CW contributed to the design of the prospective clinical and cross-sectional community study arms and also wrote, reviewed and edited the written protocol. RKG, LEK, MJ assisted in the design of the overall protocol. AA and KL contributed to the design of the prospective childcare cohort and reviewed and edited the written protocol. TRS, JH, PDH, CDF and AK contributed to the development of the laboratory and data processing aspects of the protocol and reviewed and edited the written protocol, MZS and VRL contributed to the clinical data aspect of the protocol and reviewed and edited the written protocol.
Funding: The Seattle Flu Study is funded by Gates Ventures. The funder was not involved in the design of the study, does not have any ownership over the management and conduct of the study, the data, or the rights to publish. Award/Grant number is not applicable.
Competing interests: HYC receives research support from Sanofi, Cepheid and Genentech/Roche and is a consultant for Merck. JAE receives research support to her institution from AstraZeneca, GlaxoSmithKline, Merck and Novavax and is a consultant for Sanofi Pasteur and Meissa Vaccines.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Glezen WP. Emerging infections: pandemic influenza. Epidemiol Rev 1996;18:64–76. 10.1093/oxfordjournals.epirev.a017917 [DOI] [PubMed] [Google Scholar]
- 2.Jester B, Uyeki T, Jernigan D. Readiness for responding to a severe pandemic 100 years after 1918. Am J Epidemiol 2018;187:2596–602. 10.1093/aje/kwy165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prevention CfDCa Disease burden of influenza 2020. Available: https://www.cdc.gov/flu/about/burden/index.html [Accessed 1 Oct 2020].
- 4.Dawood FS, Iuliano AD, Reed C, et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 2012;12:687–95. 10.1016/S1473-3099(12)70121-4 [DOI] [PubMed] [Google Scholar]
- 5.Shrestha SS, Swerdlow DL, Borse RH, et al. Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009-April 2010). Clin Infect Dis 2011;52 Suppl 1:S75–82. 10.1093/cid/ciq012 [DOI] [PubMed] [Google Scholar]
- 6.Reed C, Chaves SS, Daily Kirley P, et al. Estimating influenza disease burden from population-based surveillance data in the United States. PLoS One 2015;10:e0118369. 10.1371/journal.pone.0118369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GBD 2017 Influenza Collaborators Mortality, morbidity, and hospitalisations due to influenza lower respiratory tract infections, 2017: an analysis for the global burden of disease study 2017. Lancet Respir Med 2019;7:69–89. 10.1016/S2213-2600(18)30496-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng H, Liao Q, Suen LKP, et al. Healthcare seeking behavior of patients with influenza like illness: comparison of the summer and winter influenza epidemics. BMC Infect Dis 2016;16:499. 10.1186/s12879-016-1821-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fragaszy EB, Warren-Gash C, Wang L, et al. Cohort profile: the flu watch study. Int J Epidemiol 2017;46:e18. 10.1093/ije/dyv370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson ML, Chung JR, Jackson LA, et al. Influenza vaccine effectiveness in the United States during the 2015-2016 season. N Engl J Med 2017;377:534–43. 10.1056/NEJMoa1700153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohmit SE, Thompson MG, Petrie JG, et al. Influenza vaccine effectiveness in the 2011-2012 season: protection against each circulating virus and the effect of prior vaccination on estimates. Clin Infect Dis 2014;58:319–27. 10.1093/cid/cit736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jester B, Schwerzmann J, Mustaquim D, et al. Mapping of the US domestic influenza virologic surveillance landscape. Emerg Infect Dis 2018;24:1300–6. 10.3201/eid2407.180028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prevention USCfDCa FluView : National, regional, and state level outpatient illness and viral surveillance. Atlanta,GA, 2019. https://gis.cdc.gov/grasp/fluview/fluportaldashboard.html [Google Scholar]
- 14.Fong Y, Rue H, Wakefield J. Bayesian inference for generalized linear mixed models. Biostatistics 2010;11:397–412. 10.1093/biostatistics/kxp053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018;34:4121–3. 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Elm E, Altman DG, Egger M, et al. Strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ 2007;335:806–8. 10.1136/bmj.39335.541782.AD [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedford T, Greninger A, Roychoudhury P, et al. Cryptic transmission of SARS-CoV-2 in Washington state, 2020. Available: https://www.medrxiv.org/content/10.1101/2020.04.02.20051417v1 [DOI] [PMC free article] [PubMed]
- 18.Chu HY, Englund JA, Starita LM, et al. Early detection of Covid-19 through a citywide pandemic surveillance platform. N Engl J Med 2020;383:185–7. 10.1056/NEJMc2008646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim A, Brandstetter E, Graham C, et al. Seattle Flu Study - Swab and Send: Study Protocol for At-Home Surveillance Methods to Estimate the Burden of Respiratory Pathogens on a City-Wide Scale.medRxiv, 2020. Available: https://www.medrxiv.org/content/10.1101/2020.03.04.20031211v1
- 20.Burnstein R, Cowgill K. Scan technical report, 2020. Available: https://publichealthinsider.com/wp-content/uploads/2020/04/SCAN-Technical-Report-v1-17-APR-2020.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.