Abstract
Background
The mitogen-activated protein kinases/extracelluar signal-regulated kinases pathway is involved in cell growth and proliferation, and mutations in BRAF have made it an oncogene of interest in pediatric cancer. Previous studies found that BRAF mutations as well as KIAA1549–BRAF fusions are common in intracranial low-grade gliomas (LGGs). Fewer studies have tested for the presence of these genetic changes in spinal LGGs. The aim of this study was to better understand the prevalence of BRAF and other genetic aberrations in spinal LGG.
Methods
We retrospectively analyzed 46 spinal gliomas from patients aged 1–25 years from Children’s Hospital Colorado (CHCO) and The Hospital for Sick Children (SickKids). CHCO utilized a 67-gene panel that assessed BRAF and additionally screened for other possible genetic abnormalities of interest. At SickKids, BRAFV600E was assessed by droplet digital polymerase chain reaction and immunohistochemistry. BRAF fusions were detected by fluorescence in situ hybridization, reverse transcription polymerase chain reaction, or NanoString platform. Data were correlated with clinical information.
Results
Of 31 samples with complete fusion analysis, 13 (42%) harbored KIAA1549–BRAF. All 13 (100%) patients with confirmed KIAA1549–BRAF survived the entirety of the study period (median [interquartile range] follow-up time: 47 months [27–85 months]) and 15 (83.3%) fusion-negative patients survived (follow-up time: 37.5 months [19.8–69.5 months]). Other mutations of interest were also identified in this patient cohort including BRAFV600E, PTPN11, H3F3A, TP53, FGFR1, and CDKN2A deletion.
Conclusion
KIAA1549–BRAF was seen in higher frequency than BRAFV600E or other genetic aberrations in pediatric spinal LGGs and experienced lower death rates compared to KIAA1549–BRAF negative patients, although this was not statistically significant.
Keywords: BRAF, low-grade glioma, spine
Key Points.
KIAA1549–BRAF fusion was the most common aberration identified in the low-grade glioma cohort.
Patients with KIAA1549–BRAF fusion had excellent long-term survival.
Spinal cord low-grade tumors may be good candidates for MEK inhibition.
Importance of the Study.
Intracranial low-grade glioma (LGG) in the pediatric population has been well researched for the genetic aberrations that define this population. They are known to have BRAF mutations as well as KIAA1549–BRAF fusions, which is helpful in predicting outcome and treatment responses in this group. Fewer studies have evaluated spinal cord LGG for similar genetic aberrations. This study retrospectively investigated the genetic landscape of a cohort of pediatric spinal LGGs for the presence of BRAF aberrations as well as other genetic mutations of interest. BRAF fusions were the most prevalent change identified. All patients with this fusion had excellent survival. This pediatric population suffers from severe complications as a result of treatment needed to maintain tumor control, and better understanding the genetic landscape of this tumor could potentially inform treatment and predict long-term outcomes in this population.
Low-grade gliomas (LGGs) of the spinal cord are rare in pediatric patients. Tumors arising in the central nervous system (CNS) make up 20% of all pediatric cancer, but intramedullary spinal cord tumors are only 2%–10% of CNS lesions.1 Patients diagnosed with spinal cord LGG have a high overall survival (OS) with progression-free survival (PFS) ranging from 34% to 88%.2 Despite low mortality, there is high morbidity associated with spinal LGG treatment. Specifically, patients can suffer from significant neurologic and orthopedic complications from surgery, chemotherapy, and radiation. Defining the mutational characteristics of spinal cord LGG could provide predictive information regarding clinical outcomes and potential therapy options.
The most common genetic changes in intracranial LGGs or glioneuronal tumors involve the mitogen-activated protein kinases/extracelluar signal-regulated kinases (MAPK/ERK) pathway,3–6 which drives a number of processes including cellular proliferation, differentiation, mortality, stress response, apoptosis, and survival.7 There are a number of mutations and fusions in this pathway that result in constitutive over-activation of the MAP/ERK pathway.8
Cerebellar pilocytic astrocytomas often exhibit MAPK/ERK hyperactivation as a consequence of a KIAA1549–BRAF fusion.3,4 LGGs occurring elsewhere in the brain have a higher percentage of tumors with a point mutation in BRAF at codon 600 (BRAFV600E and variants such as BRAFV600D). This mutation is also seen in many CNS tumors including desmoplastic infantile gangliogliomas, diffuse astrocytomas, gangliogliomas, pleomorphic xanthoastrocytomas, and epitheloid glioblastomas.9
At present, gross total surgical resection (GTR) is the first line of treatment for LGG, but adjuvant therapy could include chemotherapy (ie, carboplatin, vincristine, and vinblastine) and potentially radiation therapy. These are particularly considered if GTR is not feasible.10 BRAFV600E inhibitors like vemurafenib (NCT01748149) and dabrafenib (NCT01677741) and MEK inhibitors like trametinib (NCT03434262) are being explored to treat these tumors in phase 1 and 2 trials. Tumors with the KIAA1549–BRAF fusion are considered to be RAS-independent and are resistant to first-generation RAF inhibitors (vemurafenib and dabrafenib).11,12 This makes MEK inhibition a better option for these patients. In addition to the potential for the development of resistance to targeted therapies,5,11,13,14 progression following single- and multi-agent chemotherapy remains a concern in these patients with 5-year PFS estimated between 43% and 53.2%.15–17
To identify potential new therapy options in pediatric low-grade spinal cord tumors, we evaluated whether these tumors identified at 2 institutions (Children’s Hospital Colorado [CHCO] and SickKids) harbor targetable lesions such as the KIAA1549–BRAF fusion and BRAFV600E mutations. Sixty-seven secondary genes were also screened for mutations for the tumors from CHCO. We evaluated clinical data and performed genetic testing on 46 spinal LGG (WHO grade 1 or 2) with available tumor samples and mapped PFS and OS of these patients to see if an association between aberrations in BRAF, treatment, and OS could be identified.
Materials and Methods
Patients and Inclusion Criteria
Fifty-five patients were identified as having intramedullary spinal cord tumors between 1995 and 2016. Of these patients, 46 patients ranging in age from 1 to 25 years (median age at diagnosis: 9.5) had a confirmed WHO grade 1 or 2 spinal cord tumor at CHCO (Aurora, CO) and SickKids (Toronto, ON). Nine patients from CHCO were excluded from analysis after it was determined that they had high-grade spinal cord malignancies. Of note, samples included from SickKids were recently also analyzed in a wider study of LGG in multiple CNS locations.18
Patient charts were retrospectively analyzed for age, diagnosis, location of tumor, past genetic tests completed on a patient’s tumor, treatment, subsequent relapses, treatment at relapse, and date of last follow-up or death. Pathology reports and imaging for each patient were subsequently analyzed to verify an accurate diagnosis, location of tumor, and genetics run at the time of biopsy/resection at CHCO. SickKids considered initial diagnosis and subsequent treatments for each patient.
Institutional Review Board Approval of Patient Specimens
Primary patient samples were obtained and collected from CHCO and SickKids in accordance with local and federal human research protection guidelines and institutional review board (IRB) regulations. Ethical standards according to the Helsinki Declaration were followed and the work was approved by local IRB committees (CHCO approvals: COMIRB 95-500 and COMIRB 05-0149; SickKids approval: REB1000030563). Written informed consent was obtained for all specimens collected.
Statistical Analysis
Baseline characteristics were reported as count and proportion (%) or median (interquartile range [IQR]), separately by grade 1 and 2 tumors. Time to death was defined as the time from diagnosis to death from any cause or time from diagnosis to the last follow-up if a patient survived. Time to progression was defined as the time from diagnosis to tumor progression or death, with censoring also at the last follow-up date. OS and PFS were estimated at time points of interest using Kaplan–Meier curves. Cox proportional hazard models were fit for time to death and time to progression, and log-rank statistics were reported for differences between tumor grade and KIAA1549–BRAF positivity. All statistical analysis was performed in R version 3.6.1.
Mutational and Gene Fusion Analysis
Samples at CHCO were analyzed as follows. Total nucleic acid (TNA) was extracted in a CLIA-certified laboratory from formalin-fixed, paraffin-embedded (FFPE) processed material (n = 15) or from frozen material (n = 7) using the Agencourt FormaPure Kit (Beckman Coulter). For mutational analysis, TNA was then processed via the Archer VariantPlex Solid Tumor library preparation kit that is designed to amplify selected regions in 67 genes (ArcherDx). All manufacturer-recommended cutoffs for quality control were used and only samples that met appropriate quality levels were sequenced. Libraries were sequenced via the Illumina MiSeq or Illumina NextSeq (Illumina). Raw sequence data were processed for mutational calling by using the Archer Analysis software package (version 5.1.2.2; ArcherDx). For fusion analysis, 23 FFPE and 7 frozen samples had TNA processed via the Archer FusionPlex Solid Tumor library preparation kit that is designed to detect gene fusions and novel isoforms involving selected exons of 53 genes (ArcherDx). Libraries were sequenced via the Illumina MiSeq (Illumina). Raw sequence data were processed for fusion calling by using the Archer Analysis software package (version 4.1.1.7; ArcherDx). SickKids analyzed FFPE samples (n = 18) as previously described.18 In short, KIAA1549–BRAF fusion was tested by NanoString, reverse transcription polymerase chain reaction, and fluorescence in situ hybridization. BRAFV600E was tested by droplet digital polymerase chain reaction (ddPCR) and immunohistochemistry. Other genetic alterations were tested by Sanger and ddPCR.
Results
All patients diagnosed with an intramedullary spinal cord tumor between 1995 and 2016 at CHCO and patients with spinal LGG from SickKids were identified for potential inclusion and analysis (Table 1). Forty-six patients were identified with WHO grade 1 or 2 tumors. LGG patients were categorized into 6 different WHO grade 1 or 2 morphological diagnoses. The cohort was 59% males with a predominant diagnosis of pilocytic astrocytoma (57%). The majority of tumors were WHO grade 1 (78%). A significant number of patients had a subtotal resection (STR) or biopsy only (83%). Therapy provided included chemotherapy (50%) and radiation (17%). Additional demographics are given in Table 1.
Table 1.
Clinical Characteristics by Tumor Gradea
| Characteristic | All (n = 46) | Grade 1 (n = 36) | Grade 2 (n = 10) |
|---|---|---|---|
| Age at diagnosis, years | 9.5 (4.8–12) | 10 (6–12.2) | 5 (4.1–11.1) |
| Male | 27 (59%) | 20 (56%) | 7 (70%) |
| Diagnosis | |||
| Astrocytoma | 13 (28%) | 6 (17%) | 7 (70%) |
| Ganglioglioma | 4 (9%) | 4 (11%) | 0 (0%) |
| Gemistocitic astrocytoma | 1 (2%) | 0 (0%) | 1 (10%) |
| Glioneuronal tumor | 1 (2%) | 0 (0%) | 1 (10%) |
| Low-grade glioma with pilocytic/pilomyxoid features | 1 (2%) | 1 (3%) | 0 (0%) |
| Pilocytic astrocytoma | 26 (57%) | 25 (69%) | 1 (10%) |
| Region of spine | |||
| Cervical | 13 (28%) | 12 (33%) | 1 (10%) |
| Cervical/thoracic | 17 (37%) | 12 (33%) | 5 (50%) |
| Thoracic | 11 (24%) | 9 (25%) | 2 (20%) |
| Thoracic/lumbar | 5 (11%) | 3 (8%) | 2 (20%) |
| Treatment (surgery) | |||
| Gross total resection | 6 (13%) | 6 (17%) | 0 (0%) |
| Near total resection | 2 (4%) | 2 (6%) | 0 (0%) |
| Subtotal resection | 38 (83%) | 28 (78%) | 10 (100%) |
| Chemotherapy | 23 (50%) | 17 (47%) | 6 (60%) |
| Radiation | 8 (17%) | 7 (19%) | 1 (10%) |
| BRAF fusion (N = 31) | 13 (42%) | 12 (52%) | 1 (12%) |
| Clinical outcomes | |||
| Relapse | 17 (37%) | 13 (36%) | 4 (40%) |
| Months to relapse | 11 (3–24) | 14 (4–26) | 7 (3–14) |
| Death | 7 (15%) | 6 (17%) | 1 (10%) |
| Months to death | 26 (17–40) | 23 (15–41) | 34 (34–34) |
aNumbers reported are median (interquartile range) or count (proportion).
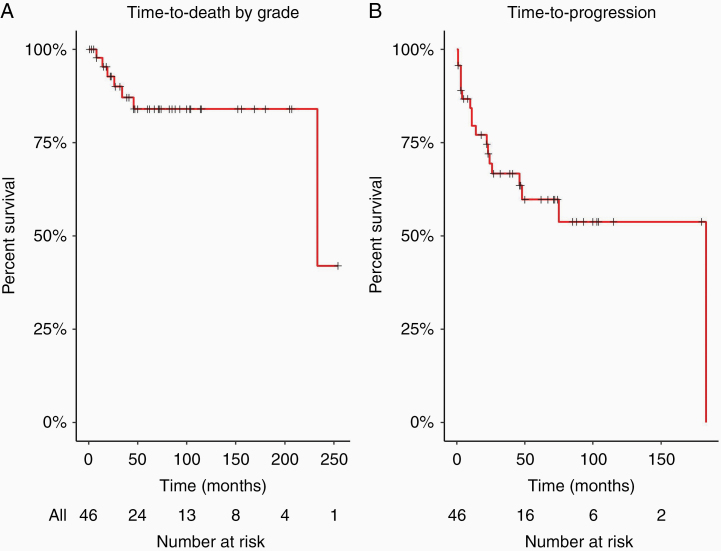
Among all participants, 85% survived the entirety of the study period (median follow-up time: 67 months, IQR [30–104]). Among the 7 (15%) who died, the median time to death was 26 months (IQR 17–40). The time-to-death and time-to-progression curves are shown in Figure 1. The estimated 5-year survival rate was 84% (95% confidence interval [CI]: 73%–97%), and the estimated 5-year PFS rate was 60% (95% CI: 46%–78%). Time to death and time to progression did not differ significantly by grade 1 versus grade 2 (P = .61 and P = .79, respectively).
Figure 1.
Overall survival (OS) and progression-free survival (PFS) of low-grade spinal cord tumors. (A) Time to death of grade 1/2 spinal cord tumors (5-year OS 84%), N = 46, censoring denoted by “+”; and (B) PFS of grade 1/2 spinal cord tumors (5-year PFS 60%), N = 46.
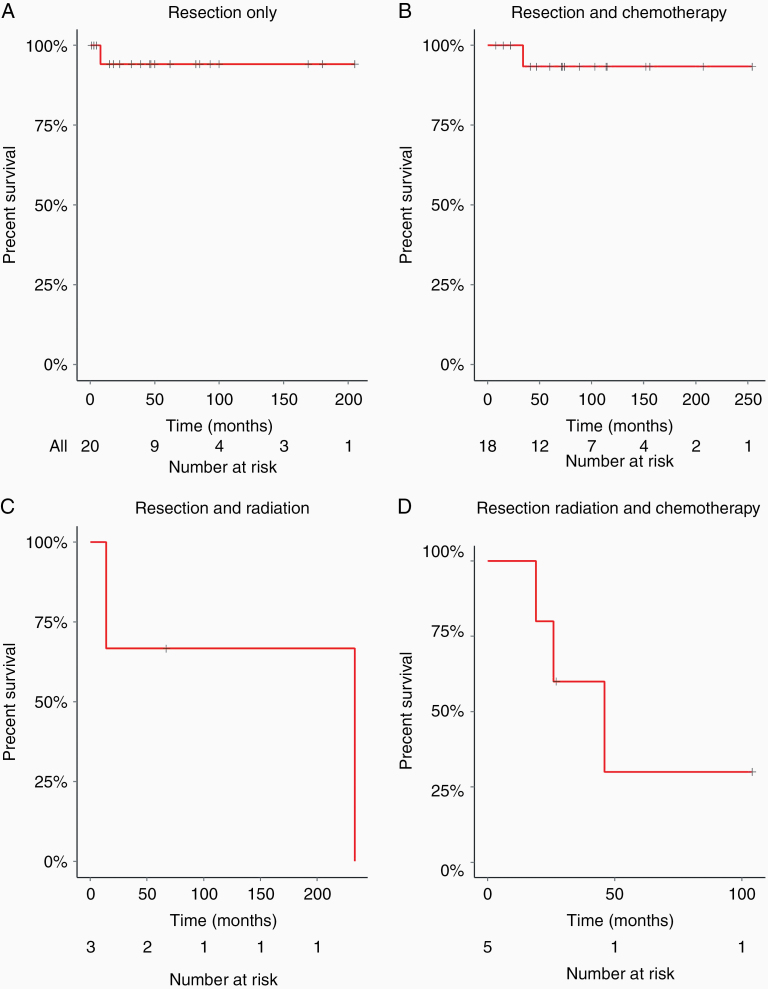
Patients treated with resection-only (Figure 2A) had the highest estimated survival rate of 94% (95% CI: 84%–100%) at 5 years. Surgical resection followed by a chemotherapy regimen resulted in an estimated 5-year survival rate of 93% (Figure 2B; 95% CI: 82%–100%). Those that underwent both surgical resection and radiation had an estimated 5-year survival rate of 67% (95% CI: 30%–100%), and those undergoing surgical resection, chemotherapy, and radiation had a 5-year survival rate of 30% (95% CI: 6%–100%).
Figure 2.
Overall survival (OS) in relation to a therapeutic treatment. (A) Time to death of grade 1/2 who had only undergone surgical resection (OS = 94%; n = 20). (B) Time to death of grade 1/2 who had resection and chemotherapy (OS = 93%; n = 18). (C) Time to death of grade 1/2 patients following resection and radiation (OS = 60%; n = 3). (D) Time to death of grade 1/2 patients following resection, chemotherapy, and radiation (OS = 30%; n = 5).
Sixteen patients at CHCO and 18 patients from SickKids had samples with results available for secondary mutation analysis. A 67-gene panel was used to test the frozen tumor specimens at CHCO (n = 7; Supplementary Figure 1). Failure of analysis in some of the samples (n = 12) was suspected to be due to samples being very old coupled with suboptimal tissue fixation resulting in TNA that did not pass the manufacturers quality control measures. Of those analyzed, there were 9 different genetic mutations and/or fusions identified in 21 patients including mutations in BRAF, PTPN11, H3F3A, TP53, FGFR1, and CDKN2A (Table 2).
Table 2.
Mutations Found in WHO Grade 1/2 Spinal Cord Tumors
| Histology, N = 34 | Pilocytic Astrocytoma, 20 (59%) | Gemistocitic Astrocytoma, 1 (3%) | Astrocytoma NOS, 7 (21%) | Ganglioglioma, 4 (12%) | Low-Grade Glioma Pilocytic/Pilomyxoid Features, 1 (3%) | Glioneuronal Tumor, 1 (3%) |
|---|---|---|---|---|---|---|
| Gene | Number (%) | |||||
| BRAF V600E | 0 (0%) | 0 (0%) | 1 (14%) | 1 (25%) | 0 (0%) | 0 (0%) |
| GOF | GOF | |||||
| PTPN11 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (25%) | 0 (0%) | 0 (0%) |
| Asp61Tyr | ||||||
| LOF | ||||||
| H3F3A | 0 (0%) | 0 (0%) | 0 (0%) | 1 (25%) | 0 (0%) | 0 (0%) |
| Lys28Met | ||||||
| LOF | ||||||
| Gln6Leu | ||||||
| US | ||||||
| TP53 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (25%) | 0 (0%) | 0 (0%) |
| Arg273Cys | ||||||
| LOF | ||||||
| FGFR1 | 1 (5%) | 0 (0%) | 1 (14%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Lys656Glu | Asp546Lys | |||||
| GOF | GOF | |||||
| CDKN2A | 1 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| LOF | ||||||
| BRAF– KIAA1549 Fusion | 12 (60%) | 0 (0%) | 1 (14%) | 0 (0%) | 0 (0%) | 0 (0%) |
| GOF | GOF |
US, unknown significant; GOF, gain of function; LOF, loss of function.
There were thirty-one total patients in which BRAF–KIAA1549 was successfully tested between CHCO (N = 13) and SickKids (N = 18). In total, 13 (42%) patients tested positive, 6 from CHCO and 7 from SickKids. Twelve of the patients diagnosed with the fusion had the diagnosis of pilocytic astrocytoma and one had an astrocytoma not otherwise specified (NOS; Table 2). In this patient cohort, there were 18 patients confirmed negative for the KIAA1549–BRAF fusion including the diagnoses: glioneuronal tumor, astrocytoma NOS, pilocytic astrocytoma, gemistocitic astrocytoma, ganglioglioma, and LGG with pilocytic/pilomyxoid features (Table 2). In agreement with previous publications, ganglioglioma is more frequently related to BRAFV600E mutations.19 Across all tumors, there were only 2 BRAFV600E mutations identified in patients diagnosed with astrocytoma NOS and ganglioglioma, respectively.
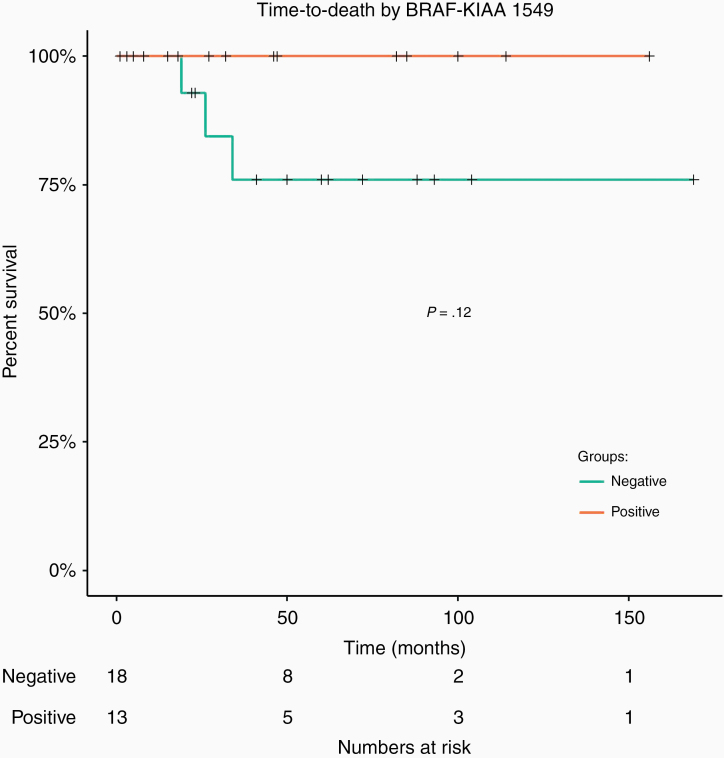
Time to death was not significantly different KIAA1549–BRAF fusion-positive and KIAA1549–BRAF fusion-negative tumors (P = .12; Figure 3). Interestingly, the estimated 5-year survival rate for patients who were confirmed to contain the KIAA1549–BRAF fusion was 100%, compared to 76% (95% CI: 56%–100%) who were confirmed to be negative for the KIAA1549–BRAF fusion, although this difference was not statistically significant.
Figure 3.
Overall survival in KIAA1549–BRAF fusion patients. Patients had to be tested for the KIAA1549–BRAF fusion (n = 31) in order to be included in this figure. No difference in time to death by KIAA1549–BRAF status (P = .12).
Discussion
Previous studies have revealed that KIAA1549–BRAF fusions are a hallmark mutation in pediatric pilocytic, pilomyxoid, and diffuse astrocytomas of the brain.20 Other studies have demonstrated that BRAFV600E mutations are common in grade 2 pleomorphic xanthoastrocytomas, pleomorphic xanthoastrocytomas with anaplasia, grade 1 gangliogliomas, grade 3 anaplastic gangliogliomas, and pilocytic astrocytomas in the brain. More specifically, BRAF rearrangement has been found in 75%–80% of cerebellar pilocytic astrocytomas, and BRAFV600E mutations have been identified in non-cerebellar regions of the brain.21 Most recently, a large analysis of LGG (which includes the spinal LGG analyzed in this study) demonstrated how molecular studies can be used to stratify these tumors into risk categories.18 To date, there are few studies specifically addressing the presence of KIAA1549–BRAF fusions and BRAFV600E point mutations in low-grade spinal cord tumors. One study by Gessi et al.22 found that spinal gangliogliomas do not harbor the BRAFV600E mutation in high frequency. Another study by Wang et al.23 acknowledged that very little work has been done in regards to understanding the role of BRAF mutations in spinal cord tumors. Ryall et al.18 reported the presence of both BRAFV660E, FGFR1 single nucleotide variants, as well as KIAA1549–BRAF and FGFR1–TACC1 fusions in spinal cord LGG. Our data support these previous studies and add to the overall understanding of the genetic landscape of these tumors.
BRAF fusions are often correlated with a favorable prognosis in cerebellar LGG. One group, in particular, found that pediatric patients who underwent STR but harbored KIAA1549–BRAF fusion had a positive clinical outcome in comparison to patients who had STR and were negative for KIAA1549–BRAF fusion.20 Our results support these findings. Of the 13 patients with confirmed KIAA1549–BRAF fusion, none died during follow-up, regardless of treatment group. Of the 18 patients who confirmed negative for the KIAA1549–BRAF fusion, 3 of the patients died putting the 5-year estimated survival rate at 76% with 7 patients experiencing a relapse. This further supports that identification of genetic mutations and fusions in pediatric spinal LGG may be useful in prognostication18 and provide information for therapy choices as more targeted therapies are being studied.
There is increasing use of targeted therapy against BRAF alterations. A number of clinical trials are evaluating the efficacy of inhibitors of the RAS/RAF/MEK/ERK signaling pathways as well as small molecule inhibitors of BRAFV600E in these tumor types (NCT01748149).19 The specific BRAF alteration is correlated to response to treatment; KIAA1549–BRAF fusions are RAF-independent and BRAFV600E mutations are responsive to autophagy and small molecule inhibitors.11,12,14 Two of the tumors evaluated in this study were found to harbor BRAFV600E and would have the potential for BRAF-targeted inhibition. Identification of KIAA1549–BRAF fusion mutations could help to inform treatment decisions to include MEK inhibition. Recent data have demonstrated significant responses to MEK inhibition in this patient population.24
The primary aim of this study was to investigate the prevalence of BRAF mutations and fusions in low-grade spinal cord tumors. Additional mutations were found in FGFR1, CDKN2A, H3F3A, TP53, and PTPN11. This is consistent with previous reports of alternate mutations found in LGG, but in far less frequency.3–6,25 Research has found that some low-grade intracranial tumors harbor alterations in FGFRs which involve fusions with TACC genes and FGFR1 tyrosine kinase domain duplication (FGFR1-TKDD) resulting in upregulation of MAPK/ERK and the PI3K pathway.6,18,26 Similarly, our data found a FGFR1 mutation in a patient diagnosed with grade 2 astrocytoma NOS who experienced a relapse of this primary tumor 3 months from diagnosis but has survived to 60 months since diagnosis. Another patient with a pilocytic astrocytoma had a confirmed FGFR1 mutation. Genetic screening studies have found that FGFR1-TKDDs are found predominantly in diffuse gliomas,6,27 although it can also be seen in other tumors such as pilocytic astrocytomas26 and dysembryoplastic neuroepithelial tumors.28,29 Recently, the Consortium to Inform Molecular and Practical Approaches to CNS tumor Taxonomy—Not Official WHO (cIMPACT-NOW) reported that diffuse gliomas characterized by FGFR1 alterations occur primarily in children and that these should be classified as diffuse glioma, FGFR1-mutant tumors.30 A comprehensive evaluation of LGG of the CNS found FGFR1 mutations in 1.5% of tumors analyzed and that these often co-occurred with other genetic alterations.18 FGFR1-activating mutations and fusions have also been reported in pediatric spinal tumors.18,31 At present, there are FGFR inhibitors in a clinical trial including erdafitinib which is being studied in the current pediatric MATCH Trial (NCT03210714).
In a previous study, 1.9% of patients with low-grade spinal cord tumors demonstrated mutations in H3F3A.6 H3F3A K27M has proven to be a hallmark of many HG midline gliomas and occurs in about 20% of pediatric glioblastomas.32 It was detected in one of our patients diagnosed with a WHO grade 1 ganglioglioma. This patient experienced a relapse and was subsequently diagnosed with an anaplastic ganglioglioma WHO grade 3 and ultimately passed away 19 months from the original diagnosis. This would suggest a concern for a poor clinical outcome with this mutation regardless of primary pathologic diagnosis. At present, there are no treatments to specifically target this mutation but could have potential importance in the prognosis of these tumors.33
Limitations of this study include the small patient cohort. Another limitation was that as a retrospective study, some samples were old and that was coupled with suboptimal fixation methods resulting in poor quality DNA and RNA for mutational and fusion assays.34 A last potential limitation to this study is that diagnoses were made strictly based off of histology rather than based on other molecular profile data.
The genetic aberrations reported here add to the available information to better understand the molecular underpinnings of low-grade spinal cord tumors. Knowledge about the distinct genetic landscape of these tumors could potentially help to inform treatment choices and predict overall prognosis in patients with pediatric spinal tumors.
Supplementary Material
Funding
This work was supported by National Institute of Health/National Cancer Institute (NIH/NCI; K08CA193982 to J.M.M.L.) and National Institutes of Health/National Institute of Neurological Disorders and Stroke (R01NS107313 to J.M.M.L.), and The Morgan Adams Foundation (to S.T.G., L.H., S.Z., A.M., A.N., A.L.G., N.F., R.V., T.C.H., M.H.H., and J.M.M.L.). NIH/NCI R03 CA235200 (to T.C.H.), GN-000522—The Brain Tumour Charity (to T.C.H.), NIH/NCI R03 CA212800 (to T.C.H.), and University of Colorado Shared Resources Cancer Center Support Grant (P30CA046934; Molecular Pathology and Functional Genomics).
Conflict of interest statement. K.D.D. has received sponsored travel from ArcherDx. All other authors have no conflicts to report.
Authorship Statement. Conception/design of the work: S.T.G. and J.M.M.L. Data collection: S.T.G., L.N., K.D.D., D.L.A., L.H., S.Z., A.M., A.N., M.C., A.L.G., N.F., R.V., T.C.H., M.H.H., S.R., C.H., B.K.D., U.T., and J.M.M.L. Data analyses/interpretation: S.T.G., L.N., K.D.D., K.R.C., D.L.A., C.H., and J.M.M.L. Drafting of the article: S.T.G., K.R.C., and J.M.M.L. Critical revision of the article: S.T.G., K.R.C., and J.M.M.L. Final approval of the article: All authors.
References
- 1. Hassall TE, Mitchell AE, Ashley DM. Carboplatin chemotherapy for progressive intramedullary spinal cord low-grade gliomas in children: three case studies and a review of the literature. Neuro Oncol. 2001;3(4):251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scheinemann K, Bartels U, Huang A, et al. Survival and functional outcome of childhood spinal cord low-grade gliomas. Clinical article. J Neurosurg Pediatr. 2009;4(3):254–261. [DOI] [PubMed] [Google Scholar]
- 3. Forshew T, Tatevossian RG, Lawson AR, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218(2):172–181. [DOI] [PubMed] [Google Scholar]
- 4. Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sievert AJ, Jackson EM, Gai X, et al. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19(3):449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang J, Wu G, Miller CP, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project . Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. 2013;45(6):602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6(4):313–319. [DOI] [PubMed] [Google Scholar]
- 8. Tian Y, Rich BE, Vena N, et al. Detection of KIAA1549-BRAF fusion transcripts in formalin-fixed paraffin-embedded pediatric low-grade gliomas. J Mol Diagn. 2011;13(6):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dougherty MJ, Santi M, Brose MS, et al. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol. 2010;12(7):621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Packer RJ, Ater J, Allen J, et al. Carboplatin and vincristine chemotherapy for children with newly diagnosed progressive low-grade gliomas. J Neurosurg. 1997;86(5):747–754. [DOI] [PubMed] [Google Scholar]
- 11. Mulcahy Levy JM, Foreman NK, Thorburn A. Using BRAF(V600E) as a marker of autophagy dependence in pediatric brain tumors. Autophagy. 2014;10(11):2077–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A. 2013;110(15):5957–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yao Z, Torres NM, Tao A, et al. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell. 2015;28(3):370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mulcahy Levy JM, Zahedi S, Griesinger AM, et al. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. eLife. 2017;6:e19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lassaletta A, Scheinemann K, Zelcer SM, et al. Phase II weekly vinblastine for chemotherapy-naïve children with progressive low-grade glioma: a Canadian pediatric brain tumor consortium study. J Clin Oncol. 2016;34(29):3537–3543. [DOI] [PubMed] [Google Scholar]
- 16. Dodgshun AJ, Maixner WJ, Heath JA, Sullivan MJ, Hansford JR. Single agent carboplatin for pediatric low-grade glioma: a retrospective analysis shows equivalent efficacy to multiagent chemotherapy. Int J Cancer. 2016;138(2):481–488. [DOI] [PubMed] [Google Scholar]
- 17. Ater JL, Zhou T, Holmes E, et al. Randomized study of two chemotherapy regimens for treatment of low-grade glioma in young children: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(21):2641–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ryall S, Zapotocky M, Fukuoka K, et al. Integrated molecular and clinical analysis of 1,000 pediatric low-grade gliomas. Cancer Cell. 2020;37(4):569–583 e565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121(3): 397–405. [DOI] [PubMed] [Google Scholar]
- 20. Hawkins C, Walker E, Mohamed N, et al. BRAF-KIAA1549 fusion predicts better clinical outcome in pediatric low-grade astrocytoma. Clin Cancer Res. 2011;17(14):4790–4798. [DOI] [PubMed] [Google Scholar]
- 21. Horbinski C, Nikiforova MN, Hagenkord JM, Hamilton RL, Pollack IF. Interplay among BRAF, p16, p53, and MIB1 in pediatric low-grade gliomas. Neuro Oncol. 2012;14(6):777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gessi M, Dörner E, Dreschmann V, et al. Intramedullary gangliogliomas: histopathologic and molecular features of 25 cases. Hum Pathol. 2016;49:107–113. [DOI] [PubMed] [Google Scholar]
- 23. Wang JL, Hong CS, Otero J, Puduvalli VK, Elder JB. Genetic characterization of a multifocal ganglioglioma originating within the spinal cord. World Neurosurg. 2016;96:608.e1–608.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Banerjee A, Jakacki RI, Onar-Thomas A, et al. A phase I trial of the MEK inhibitor selumetinib (AZD6244) in pediatric patients with recurrent or refractory low-grade glioma: a Pediatric Brain Tumor Consortium (PBTC) study. Neuro Oncol. 2017;19(8):1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfister S, Hartmann C, Korshunov A. Histology and molecular pathology of pediatric brain tumors. J Child Neurol. 2009;24(11):1375–1386. [DOI] [PubMed] [Google Scholar]
- 26. Jones DT, Hutter B, Jäger N, et al. ; International Cancer Genome Consortium PedBrain Tumor Project . Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet. 2013;45(8):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qaddoumi I, Orisme W, Wen J, et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016;131(6):833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fina F, Barets D, Colin C, et al. Droplet digital PCR is a powerful technique to demonstrate frequent FGFR1 duplication in dysembryoplastic neuroepithelial tumors. Oncotarget. 2017;8(2):2104–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rivera B, Gayden T, Carrot-Zhang J, et al. Germline and somatic FGFR1 abnormalities in dysembryoplastic neuroepithelial tumors. Acta Neuropathol. 2016;131(6):847–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ellison DW, Hawkins C, Jones DTW, et al. cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAFV600E mutation. Acta Neuropathol. 2019;137(4):683–687. [DOI] [PubMed] [Google Scholar]
- 31. Bruzek AK, Zureick AH, McKeever PE, et al. Molecular characterization reveals NF1 deletions and FGFR1-activating mutations in a pediatric spinal oligodendroglioma. Pediatr Blood Cancer. 2017;64(6). doi: 10.1002/pbc.26346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gielen GH, Gessi M, Hammes J, Kramm CM, Waha A, Pietsch T. H3F3A K27M mutation in pediatric CNS tumors: a marker for diffuse high-grade astrocytomas. Am J Clin Pathol. 2013;139(3):345–349. [DOI] [PubMed] [Google Scholar]
- 33. Grob ST, Levy JMM. Improving diagnostic and therapeutic outcomes in pediatric brain tumors. Mol Diagn Ther. 2018;22(1):25–39. [DOI] [PubMed] [Google Scholar]
- 34. Davies KD, Le AT, Sheren J, et al. Comparison of molecular testing modalities for detection of ROS1 rearrangements in a cohort of positive patient samples. J Thorac Oncol. 2018;13(10):1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.