Abstract
CD8 T cells are considered important contributors to the immune response against Mycobacterium tuberculosis, yet limited information is currently known regarding their specific immune signature and phenotype. In this study, we applied a cell population transcriptomics strategy to define immune signatures of human latent tuberculosis infection (LTBI) in memory CD8 T cells. We found a 41-gene signature that discriminates between memory CD8 T cells from healthy LTBI subjects and uninfected controls. The gene signature was dominated by genes associated with mucosal-associated invariant T cells (MAITs) and reflected the lower frequency of MAITs observed in individuals with LTBI. There was no evidence for a conventional CD8 T cell–specific signature between the two cohorts. We, therefore, investigated MAITs in more detail based on Vα7.2 and CD161 expression and staining with an MHC-related protein 1 (MR1) tetramer. This revealed two distinct populations of CD8+Vα7.2+CD161+ MAITs: MR1 tetramer+ and MR1 tetramer−, which both had distinct gene expression compared with memory CD8 T cells. Transcriptomic analysis of LTBI versus noninfected individuals did not reveal significant differences for MR1 tetramer+ MAITs. However, gene expression of MR1 tetramer− MAITs showed large interindividual diversity and a tuberculosis-specific signature. This was further strengthened by a more diverse TCR-α and -β repertoire of MR1 tetramer− cells as compared with MR1 tetramer+. Thus, circulating memory CD8 T cells in subjects with latent tuberculosis have a reduced number of conventional MR1 tetramer+ MAITs as well as a difference in phenotype in the rare population of MR1 tetramer− MAITs compared with uninfected controls.
INTRODUCTION
A considerable fraction (>20%) of the worldwide population is infected by Mycobacterium tuberculosis (1). M. tuberculosis infection can have a spectrum of outcomes in exposed individuals, ranging from life-long asymptomatic infection termed latent tuberculosis infection (LTBI), which is controlled by the host immune response, to various stages of active tuberculosis (TB) in which this control has broken down (2). To understand how most individuals can control the infection at the stage of LTBI, whereas others develop active disease, it is crucial to define the immune responses associated with LTBI.
The contribution of CD4+ T cells to the protection from active M. tuberculosis is well accepted (3, 4). Previous studies from our group have shown that transcriptomic analysis of isolated immune cell subsets increases the power to identify immune signatures for diseases (5–8), and we found that transcriptomic profiling of sorted memory CD4+ T cells can distinguish individuals with LTBI from uninfected (TB-negative [TB neg]) individuals (6). CD8+ T cells have also been shown to play a role in the cellular response against M. tuberculosis. However, the transcriptomic signature of LTBI infection in memory CD8+ T cells has not been defined.
Although most CD8 T cells respond to peptide Ags presented by classical MHC class Ia molecules, there is also a considerable fraction of T cells recognizing nonpeptidic Ags that are often restricted by nonclassical (MHC class Ib) molecules such as the cluster of differentiation 1 (CD1), HLA-E, or MHC-related protein 1 (MR1). Both classically and nonclassically restricted CD8 T cells are involved in the response against M. tuberculosis. Specifically, M. tuberculosis–reactive CD8+ T cells have been 1) identified in humans with LTBI (9), 2) shown to recognize several M. tuberculosis–derived peptide epitopes (10), 3) shown to recognize MHC class II cells heavily infected with M. tuberculosis (11), and 4) have the capability to induce apoptosis of infected cells (12) and control chronic infection (13).
One of the most frequent nonclassically restricted T cell populations in the blood are MR1-restricted T cells, which decrease in frequency in active TB (14, 15). For LTBI, there have been contradicting results regarding the frequency of MAITs in PBMCs (14, 16–18). MR1-restricted T cells represent a significant fraction of CD8+ and CD4−CD8− T cells in peripheral blood (19, 20). They express CD161, CD26, and a semi-invariant TCR α-chain, TRAV1–2 (Vα7.2), that is shared across genetically unrelated individuals (19–21). These Vα7.2+CD161+ T cells are called mucosal-associated invariant T cells (MAITs), and they are restricted by MR1 molecules, which can present bacterial metabolites such as vitamin B2 (riboflavin) metabolites, which are produced by most bacteria, including M. tuberculosis (22–26). Functionally diverse subsets of MAITs have been described, and the phenotypic heterogeneity of these cells is beginning to emerge (23, 27, 28).
Given the known role of CD8 T cells in M. tuberculosis infection, we hypothesized that similar to our previous studies in memory CD4 T cells (6), transcriptomic studies of sorted memory CD8 T cells would allow us to discover an immune signature of latent TB infection. Accordingly, we first defined the transcriptomic signature of memory CD8+ T cells and found several genes previously described to have high expression in MAITs. Given the known role of MAITs in M. tuberculosis infection, we also investigated the differential gene expression in MAIT subsets between individuals with LTBI and TB neg individuals. This comparison revealed that MR1 tetramer− MAITs have a TB-specific signature that is not found in MR1 tetramer+ MAITs. The results suggest that MAITs are more diverse than previously understood and advances the understanding of MAITs in the context of M. tuberculosis infection.
MATERIALS AND METHODS
Ethics statement
Blood samples were obtained from the University of California San Diego, Antiviral Research Center Clinic and the Universidad Peruana Cayetano Heredia. All samples were obtained for specific use in this study.
Ethical approval to carry out this work is maintained through the La Jolla Institute for Immunology and University of California San Diego Institutional Review Board and through Johns Hopkins School of Public Health Institutional Review Board (R.H.G. holds a dual appointment at Universidad Peruana Cayetano Heredia and Johns Hopkins University). All participants provided written informed consent prior to participation in the study.
Subjects and samples
We recruited 32 individuals with LTBI and 31 TB neg controls. LTBI was confirmed by a positive IFN-γ–release assay (QuantiFERON-TB Gold In-Tube; Cellestis or T.Spot-TB; Oxford Immunotec) and the absence of clinical and radiographic signs of active TB. TB neg control subjects were negative for IFN-γ–release assay. We also recruited three M. tuberculosis–infected individuals who were midtreatment (3–4 mo postdiagnosis) for active TB.
Venous blood was collected in heparin-containing blood bags or tubes. PBMCs were purified from whole blood or 100 ml of leukapheresis samples by density-gradient centrifugation (Ficoll-Hypaque; Amersham Biosciences) according to the manufacturer’s instructions. Cells were cryopreserved in liquid nitrogen suspended in FBS (Gemini Bio-Products) containing 10% (vol/vol) DMSO (Sigma-Aldrich).
Memory CD8 T cell sorting
Ten million PBMCs were stained with fixable viability dye eFluor 506 (eBioscience) and with anti-human CD3-Alexa Fluor 700 (UCHT1; BD Pharmingen), CD4-APCeFluor 780 (RPA-T4; eBioscience), CD8-BV650 (RPA-T8; BioLegend), CD45RA-eFluor 450 (HI100; eBioscience), and CCR7-PerCP Cy5.5 (UCHL1; BioLegend) as previously described (29). Briefly, cells were incubated in PBS containing 10% FBS for 10 min at 4°C and then stained with PBS containing the conjugated Abs for 30 min at 4°C. After two washes in PBS, cells were resuspended in PBS, and cell sorting was performed on a BD FACSAria III/Fusion Cell Sorter (Becton Dickinson). A total of 100,000 memory CD8 T cells (see Supplemental Fig. 1A for gating strategy) was sorted into TRIzol LS reagent (Invitrogen).
MAIT subset sorting
Ten million PBMCs were stained with 1:100 MR1 5-OP-RU or 6-FP (as a control) tetramer for 40 min at room temperature. The MR1 tetramer technology was developed jointly by Dr. J. McCluskey, Dr. J. Rossjohn, and Dr. D. Fairlie (30), and the material was produced by the National Institutes of Health Tetramer Core Facility, as permitted to be distributed by the University of Melbourne. After 40 min, incubation cells were also stained with fixable viability dye eFluor 506 (eBioscience) and with anti-human CD3-Alexa Fluor 700 (UCHT1; BD Pharmingen), CD4-allophycocyanin-eFluor 780 (RPA-T4; eBioscience), CD8-BV650 (RPA-T8; BioLegend), CD45RA-eFluor 450 (HI100; eBioscience), CCR7-PerCP Cy5.5 (UCHL1; BioLegend), Vα7.2-PE-Cy7 (3C10; BioLegend), CD161-allophycocyanin (HP-3G10; eBioscience), CD14-V500 (M5E2; BD Biosciences), and CD19-V500 (HIB19; BD Biosciences) for 30 min at room temperature. After two washes in PBS, cells were resuspended in MACS buffer and transferred into a 5-ml polypropylene FACS tube (BD Biosciences). Cell sorting was performed on a BD FACSAria III/Fusion Cell Sorter (Becton Dickinson; see Supplemental Fig. 1B, 1C for gating strategy). Cells were sorted into lysis buffer as described below.
The CD8+ MR1tet+ and CD8− MR1tet+ cell subsets were sorted into TRIzol LS reagent (Invitrogen) on a BD FACSAriaIII Cell Sorter. These cells were stained with anti-human CD3-BV421 (SK7; BioLegend), CD8-allophycocyanin-Cy7 (SK1; BioLegend), CCR7-PECy7 (G043H7; BioLegend), CD45RA-BV510 (HI100; BD Biosciences), CD4-FITC (SK3; BioLegend), CD19-FITC (HIB19; eBioscience), and CD14-FITC (HCD14; BioLegend), and propidium iodide to exclude dead cells.
RNA sequencing of memory CD8 T cells and CD8+/− MR1tet+ cells
RNA sequencing (RNA-seq) of memory CD8 T cells and CD8+/− MR1tet+ cells was performed as described previously (31). Briefly, total RNA was purified using an miRNeasy Micro Kit (QIAGEN) and quantified by quantitative PCR, as described previously (32). Purified total RNA (1–5 ng) was amplified following the Smart-Seq2 protocol (33). mRNA was captured using poly-dT oligonucleotides and directly reverse transcribed into full-length cDNA (amplified by PCR for 16 cycles). cDNA was purified using AMPure XP beads (Beckman Coulter). From this step, 1 ng of cDNA was used to prepare a standard Nextera XT sequencing library (Nextera XT DNA Sample Preparation Kit and Index Kit; Illumina). Whole-transcriptome amplification and sequencing library preparations were performed in a 96-well format to reduce assay-to-assay variability. Quality-control steps were included to determine total RNA quality and quantity, the optimal number of PCR preamplification cycles, and fragment size selection. Samples that failed quality control were eliminated from further downstream steps. Barcoded Illumina sequencing libraries (Nextera; Illumina) were generated using the automated platform (Biomek FXp). Libraries were sequenced on an HiSeq 2500 Illumina platform to obtain 50-bp single-end reads (TruSeq Rapid Kit; Illumina).
RNA-seq of MAIT subsets
RNA-seq of MAIT subsets was performed as described previously (5). Briefly, for each condition, 200 cells were collected at 4°C in 8 μl of lysis buffer (0.2% Triton X-100, 2 U/μl rRNase inhibitor [Clontech Laboratories/Takara Bio], 5 mM dNTP mix [Life Technologies]) in a 96-well PCR plate (Bio-Rad Laboratories). Immediately after sorting, plates were spun for 1 min at 3000 rpm and stored at −80°C until RNA extraction. Four microliters of each sample were amplified following the Smart-seq2 protocol as described above.
RNA-seq analysis
RNA-seq analysis was performed as previously described (31). The single-end reads that passed Illumina filters were filtered for reads aligning to tRNA, rRNA, adapter sequences, and spike-in controls. The reads were then aligned to University of California, Santa Cruz hg19 reference genome using TopHat (v 1.4.1) (34). DUST scores were calculated with PRINSEQ Lite (v 0.20.3) (35), and low-complexity reads (DUST >4) were removed from the BAM files. The alignment results were parsed via SAMtools (36) to generate SAM files. Read counts for each genomic feature were obtained with the HTSeq-Count Program (v 0.6.0) (37) using the “union” option. After removing absent features (zero counts in all samples), the raw counts were imported to R/Bioconductor package DESeq2 (38) to identify differentially expressed genes among samples. Genes were considered differentially expressed for adjusted p values <0.05 and in some instances absolute log2 fold change >1 or <−1. Principal component analysis (PCA) was performed using Python SciPy package. Heat maps were created using Qlucore Omics Explorer 3.2 (Qlucore, Lund, Sweden). Volcano plots, PCA plots, and TCR analysis plots were constructed using Python Matplotlib package. The sequencing data presented in this study were submitted to the Gene Expression Omnibus under accession numbers GSE132790, GSE132931, and GSE132932 (https://www.ncbi.nlm.nih.gov/geo) and to ImmPort under study number SDY820 (http://www.immport.org).
ELISPOT assay
PBMCs were stimulated at 2 × 105 cells per well in triplicates with peptide pools (1 μg/ml per peptide), PHA (10 μg/ml; as a positive control for the assay), or medium containing 0.25% DMSO (percent DMSO in the pool stimulations as a control) in 96-well plates (Immobilion-P; Millipore) coated with 5 μg/ml anti–IFN-γ (1-D1K; Mabtech). After 20 h of incubation at 37°C, wells were washed with PBS/0.05% Tween 20 and incubated with biotinylated anti-IFN-γ (7-B6–1; Mabtech) for 2 h. Spots were developed using VECTASTAIN ELITE ABC Kits (Peroxidase) (Vector Laboratories) and 3-amino-9-ethylcarbazole (Sigma-Aldrich). Spots were counted by computer-assisted image analysis (AID iSpot; AID Autoimmun Diagnostika). Responses were considered positive if the net spot-forming cells mean of triplicate values of the response against relevant pools versus the DMSO control).
Peptide pools
Peptides were synthesized as crude material on a small (1 mg) scale by A and A (San Diego, CA). Multiepitope peptide pools (“megapools”) were prepared as previously described (39). Individual peptides were resuspended in DMSO, and equal amounts of each peptide were pooled to construct the peptide pool. After lyophilization, the peptide pool was resuspended in DMSO and stored at −20°C. A peptide pool containing 300 M. tuberculosis–derived 15- to −20-mer peptides (MTB300) primarily HLA class II restricted (39) was used.
To measure CD8 T cell reactivity, we constructed a peptide pool containing 113 M. tuberculosis–derived 8- to 11-mer peptides that had a confirmed HLA class I restriction as defined in the Immune Epitope Database Analysis and Resource (www.iedb.org; on September 2015). Out of 140 peptides, 47 were restricted by HLA-A*02:01 only. To get a more balanced set, we therefore removed any HLA-A*02:01–restricted peptides with a response frequency (RF) score below 0.28 (40). The RF score is calculated as (R−√R)/N, where N is the total number of subjects tested and R is the number of positive responses. The square root is a correction factor approximating one SD for the number of responding subjects.
Flow cytometry
PBMCs were stained with 1:100 MR1 5-OP-RU or 6-FP (as a control) tetramer for 40 min at room temperature. After 40 min, incubation cells were also stained with fixable viability dye eFluor 506 (eBioscience) and with combinations of anti-human CD3-Alexa Fluor 700 (UCHT1; BD Pharmingen), CD4-allophycocyanin-eFluor 780 (RPA-T4; eBioscience), CD8-BV650 (RPA-T8; BioLegend), Vα7.2-PE-Cy7 (3C10; BioLegend), CD161-allophycocyanin (HP-3G10; eBioscience), TIGIT-PerCP-eFluor710 (MBSA43; eBioscience), CXCR6-PE Dazzle594 (K041E5; BioLegend), CCR1-PerCP Cy5.5 (5F10B29; BioLegend), CD243-BV421 (UIC2; BD Biosciences), CXCR4-PE Dazzle 594 (12G5; BioLegend), CD127-FITC (eBioRDR5; eBioscience), CD14-V500 (M5E2; BD Biosciences), CD14-BV421 (HCD14; BioLegend), and CD19-V500 (HIB19; BD Biosciences) for 30 min at room temperature. After two washes in PBS, cells were resuspended in PBS and cells were acquired on a LSRII Cytometer (Becton Dickinson).
TCR analysis
MiXCR v2.1.5 (41) was used to extract TCR α- and β-chain VJ combinations and β-chain CDR3 repertoires from RNA-seq data of sorted T cell subsets according to guidelines for analysis of bulk RNA-seq data. TRBV genes were summarized by family without specifying the exact gene because of high similarity of genes in a single family.
MAIT-match (http://www.cbs.dtu.dk/services/MAIT_Match/) was used to determine the α-chain similarity with published MAIT CDR3α sequences (42).
RESULTS
The transcriptomic profile of memory CD8 T cells in LTBI versus TB neg individuals reveals a reduction in MAIT frequency
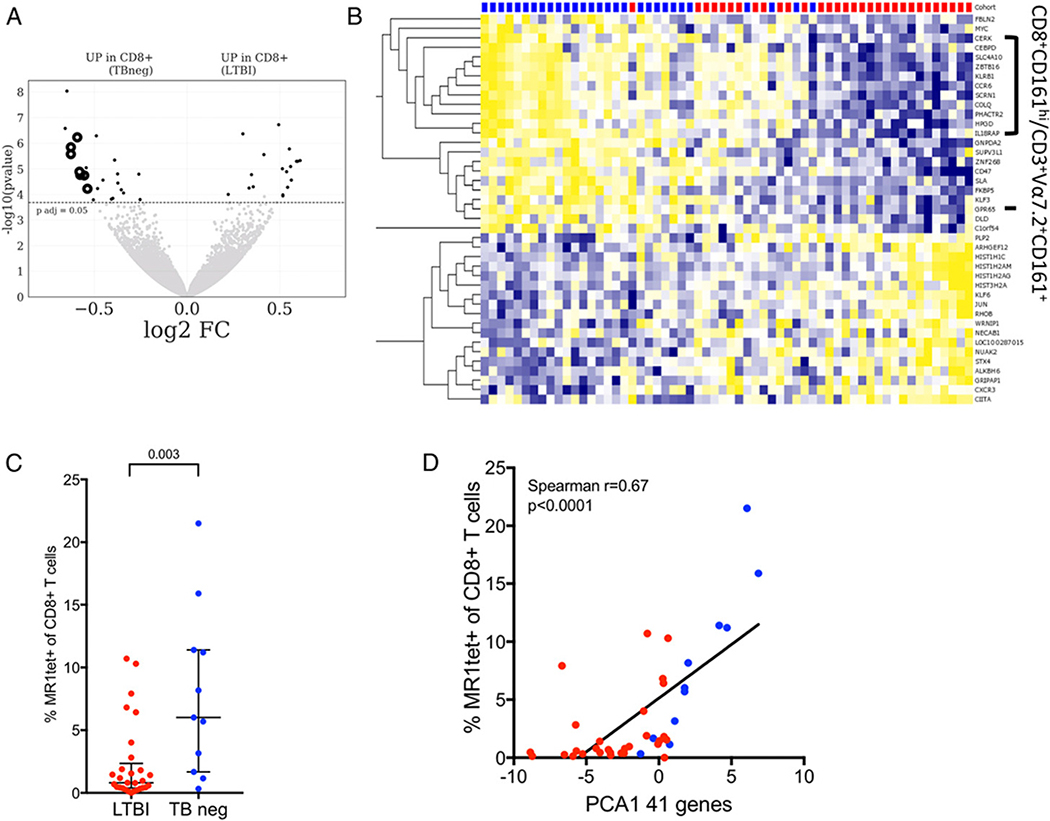
To define the immune signature of LTBI in memory CD8 T cells, we compared the genome-wide RNA expression of memory CD8 T cells (excluding CD45RA+CCR7+ cells; Supplemental Fig. 1A for gating strategy) isolated from 29 TB neg controls and 31 healthy M. tuberculosis–infected individuals (LTBI). We found a total of 41 differentially expressed genes (adjusted p value <0.05; Fig. 1A, Supplemental Table I) between the two cohorts. Of the 41 genes, 23 were upregulated and 18 were downregulated in the LTBI cohort compared with TB neg controls (Fig. 1B). Eleven of the downregulated genes in LTBI (CEBPD, SLC4A10, ZBTB16, KLRB1, CCR6, SCRN1, COLQ, PHACTR2, HPGD, GPR65, and IL18RAP) were previously described as part of a transcriptomic signature of CD8+CD161hi T cells and Vα7.2+CD161+ T cells (43–45), which are all populations enriched for MAITs. For example, both KLRB1 (CD161) and CCR6 have previously been described as surface markers of MAITs (15, 19, 20), and ZBTB16 (PLZF) has been identified as a transcription factor expressed by MAITs and other innate-like T cells (46).
FIGURE 1. Transcriptomic profile of memory CD8 T cells comparing LTBI subjects with TB neg controls reveal changes in MAIT frequency.
(A) Volcano plot obtained from the DEseq2 analysis showing log2 fold change versus −log10 p value. The 41 differentially expressed genes are represented in black (adjusted p value <0.05 indicated by dotted line). Known genes expressed by CD8+Vα7.2+CD161+ T cells are shown in black circles. Genes upregulated in TB neg controls (left) and genes upregulated in LTBI (right). (B) Heatmap displaying regularized logarithm–transformed raw counts of the 41 differentially expressed genes from (A), genes are ordered by hierarchical clustering, and subjects are ordered by principal component 1 (PC1). TB neg (blue), LTBI (red). (C) Frequency of MR1tet+ of CD8+ T cells as measured by flow cytometry LTBI (n = 29), TB neg (n = 11). Two-tailed Mann–Whitney U test. Median ± interquartile range is indicated. (D) Correlation between the PC1 of the combined expression of the 41 differentially expressed genes, identified in (A), and the frequency of MR1tet+ of CD8+ T cells in corresponding subjects. Correlation is indicated by Spearman r and associated two-tailed p value.
Due to this preponderance of MAIT genes in the transcriptomic signature of CD8 T cells in LTBI, we wanted to determine if there were differences in the frequency of MAITs in uninfected versus LTBI subjects. To achieve this, we determined the frequency of MR1 5-OP-RU tetramer+ (MR1tet; developed by Corbett et al. (30) and produced by the National Institutes of Health Tetramer Core Facility) CD8+ T cells for a subset of our subjects. Indeed, we found that the frequency of MR1tet+ CD8+ T cells were systematically lower in LTBIs as compared with TB neg controls (Fig. 1C). Furthermore, the combined expression of the 41 differentially expressed genes correlated positively with the frequency of MR1tet+ CD8+ T cells within each subject (Fig. 1D). Overall, this suggests that the differences in the gene expression profile of memory CD8+ T cells in LTBI are due to a reduced frequency of MAITs in this compartment.
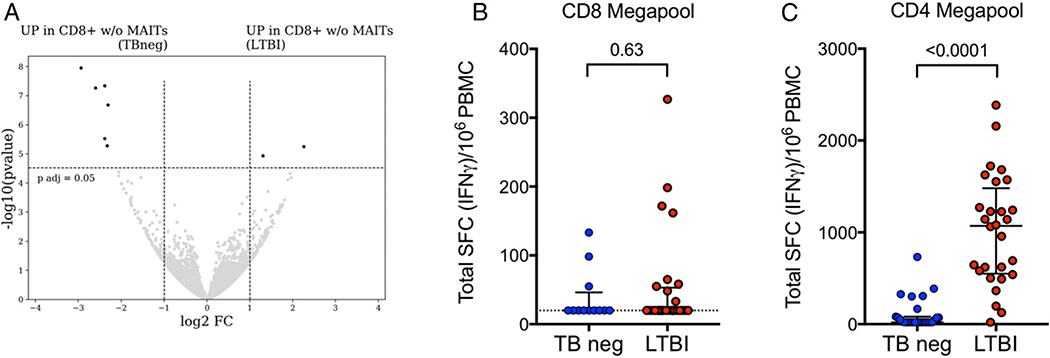
Our original goal was to identify markers of conventional CD8+ T cells that are upregulated in LTBI individuals to study M. tuberculosis epitope–specific CD8+ T cells as we have previously done for CD4+ T cells. Because of the dominant effect of differences in MAIT frequencies on CD8+ T cell gene expression, we hypothesized that any difference in classically restricted memory CD8+ T cells between LTBI and TB neg controls would become visible when excluding MAITs. Therefore, in a subset of subjects, we sorted memory CD8+ T cells excluding Vα7.2+CD161+ cells (gating strategy in Supplemental Fig. 1C) and compared the genome-wide expression profile of Vα7.2−CD161− memory CD8+ T cells isolated from six TB neg controls and 10 individuals with LTBI. This comparison showed only minor differences between the cohorts with RHOBTB3 and EFHA1 as the only two upregulated genes in LTBI (adjusted p value <0.05 and absolute log2 fold change >1; Fig. 2A, Supplemental Table I). Thus, our transcriptional analyses did not provide evidence for an expanded subset of conventionally restricted memory CD8+ T cells in LTBI subjects.
FIGURE 2. Minor differences between LTBI versus TB neg in M. tuberculosis–specific CD8+ T cell responses and transcriptomic analysis of memory CD8 T cells excluding Vα7.2+CD161+.
(A) Transcriptomic analysis of Vα7.2−CD161− memory CD8 T cells. Volcano plot obtained from the DEseq2 analysis showing log2 fold change versus −log10 p value. The eight differentially expressed genes are represented in black (adjusted p value <0.05 and absolute log2 fold change >1 are indicated by dotted lines). Genes upregulated in TB neg controls (left) and genes upregulated in LTBI (right). (B and C) Magnitude of epitope pool responses measured as total spot-forming cells (SCFs) per 106 PBMCs in an IFN-γ ELISPOT assay in individuals with LTBI and TB neg controls. Each dot represents one subject. Median ± interquartile range is indicated. Two-tailed Mann–Whitney U test. (B) CD8 megapool (LTBI, n = 28, red dots; TB neg, n = 12, blue dots), (C) CD4 megapool (LTBI, n = 28; TB neg, n = 27).
Peptide-specific CD8+ T cells have been shown to play an important role in the immune response against M. tuberculosis infection (47). To investigate if the absence of a transcriptional signature of such cells in LTBI was due to a low frequency, we compared M. tuberculosis–specific T cell epitope reactivity against a pool of peptides composed of known CD8- or CD4-restricted peptides in an ex vivo IFN-γ ELISPOT assay. We did not find evidence for an increase in CD8+ M. tuberculosis–specific T cell reactivity in LTBI compared with TB neg controls, either in RF (32 versus 33%, respectively) or magnitude of response (Fig. 2B). In contrast, we did see higher frequency and magnitude of responses against the CD4+ M. tuberculosis–specific T cell epitope pool (Fig. 2C), in agreement with previous results (39). Thus, our inability to identify a transcriptional signature of conventional CD8 T cells in LTBI can be explained by the absence of an expanded set of M. tuberculosis peptide-specific CD8+ T cells in circulating PBMCs of LTBI subjects.
CD8+ and CD8− MR1 tetramer+ T cells have a similar transcriptomic profile
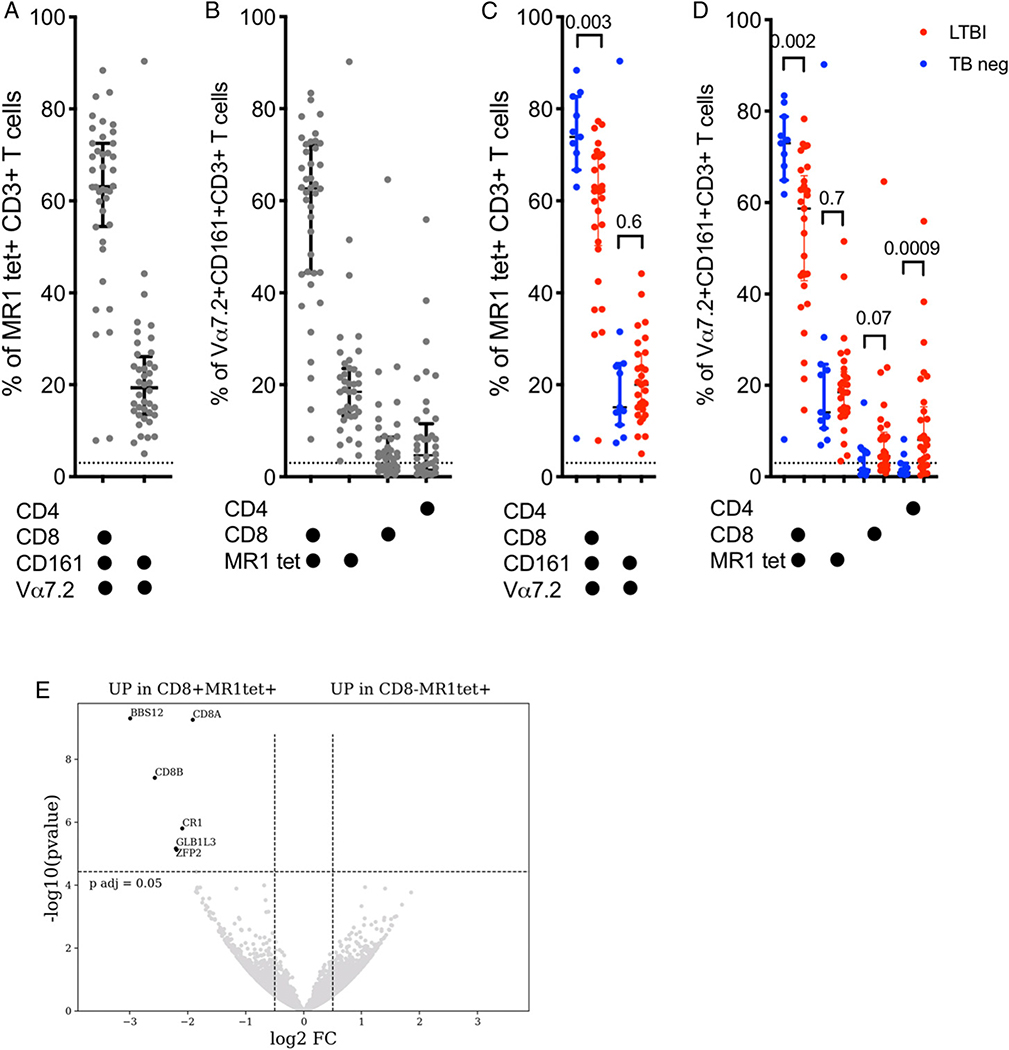
Given the difference in frequency of MAITs in the CD8 memory compartment of LTBI, we wanted to further define their transcriptional signature and determine if there are not only quantitative but also qualitative differences in MAITs associated with LTBI. There are different markers used to identify MAITs; therefore, we first performed flow cytometric analysis of T cells to determine how the expression of CD4, CD8, Vα7.2, CD161, and MR1tet correlate. Gating on MR1tet+ T cells revealed two dominant cell populations: Vα7.2+CD161+CD4−CD8+ and Vα7.2+CD161+CD4−CD8− (Fig. 3A, Supplemental Fig. 2A), as expected based on previously published results (48). Vice versa, gating on Vα7.2+CD161+ T cells revealed four populations: the expected MR1tet+ CD4−CD8+ cells and MR1tet+ CD4−CD8− but also MR1tet− CD8−CD4+ cells and interestingly MR1tet− CD4−CD8+ T cells (Fig. 3B, Supplemental Fig. 2B). MR1tet− CD8−CD4+ cells are potentially enriched for germline-encoded mycolyl-reactive cells (49) and were not studied further. As described in Fig. 1C, we found a significant higher proportion of Vα7.2+CD161+CD4−CD8+ and MR1tet+ CD4−CD8+ cells in the TB neg controls compared with LTBI (Fig. 3C, 3D). In contrast, there was no significant difference in the frequency of MR1tet− CD4−CD8+ T cell population between the two populations (Fig. 3D). In our subsequent studies, we investigated the two expected MR1tet+ cell populations, as well as MR1tet− CD4−CD8+ T cells because this population express the classical markers of MAITs, Vα7.2, and CD161, but does not bind the 5-OP-RU–loaded MR1 tetramer. Thus, suggesting that they may bind a different ligand.
FIGURE 3. Phenotypic and transcriptomic profiling of MAITs reveals four distinct subsets and overlapping gene expression programs between CD8+ MR1tet+ and CD8− MR1tet+ cells.
(A) Percentage of the two dominant MR1 tetramer+ CD3+ T cell subsets expressing combinations of CD4, CD8, CD161, and/or Vα7.2 with a median expression above 3%. Each dot represents one subject (n = 40; 11 TB neg and 29 LTBI). Median ± interquartile range is indicated. Dotted line indicates arbitrary cut-off for median expression at 3%. (B) Percentage of the four dominant Vα7.2+CD161+CD3+ T cell subsets expressing or binding combinations of CD4, CD8, and MR1 tetramer with a median expression above 3%. Each dot represents one subject (n = 40; 11 TB neg and 29 LTBI). Median ± interquartile range is indicated. Dotted line indicates arbitrary cut-off for median expression at 3%. (C) Percentage of the two dominant MR1 tetramer+ CD3+ T cell subsets expressing combinations of CD4, CD8, CD161, and/or Vα7.2 with a median expression above 3% in 11 TB neg (blue) and 29 LTBI (red) individuals. Each dot represents one subject. Median ± interquartile range is indicated. Dotted line indicates arbitrary cut-off for median expression at 3%. Two-tailed Mann–Whitney U test. (D) Percentage of the four dominant Vα7.2+CD161+CD3+ T cell subsets expressing or binding combinations of CD4, CD8, and MR1 tetramer with a median expression above 3% in 11 TB neg (blue) and 29 LTBI (red) individuals. Each dot represents one subject. Median ± interquartile range is indicated. Dotted line indicates arbitrary cut-off for median expression at 3%. Two-tailed Mann–Whitney U test. (E) Volcano plot obtained from the DEseq2 analysis showing log2 fold change versus −log10 p value. The six differentially expressed genes are represented in black (adjusted p value <0.05 and absolute log2 fold change >1 are indicated by dotted lines). Genes upregulated in CD8+ MR1tet+ cells (left) and genes upregulated in CD8− MR1tet+ cells (right).
It is unclear if absence versus presence of CD8 expression in MR1tet+ cells defines two distinct cell populations or not. To investigate this, we sorted CD8+ MR1tet+ T cells and CD8− MR1tet+ T cells (Supplemental Fig. 1B) from six M. tuberculosis–exposed individuals (two LTBI, three individuals 3–4 mo postactive TB diagnosis and one bacillus Calmette-Guérin–vaccinated control), and compared their genome-wide RNA expression profile. Strikingly, we found only six differentially expressed genes, two of which were the α- and β-chain of the CD8 receptor itself (Fig. 3E). We therefore concluded that there were no major differences in the gene expression program of CD8+ versus CD8− MR1tet+ MAITs.
MR1tet+ MAITs have a distinct gene expression profile compared with memory CD8+ T cells and no evidence for a M. tuberculosis–specific signature
Based on the results above, we sorted three cell populations, Vα7.2+CD161+CD4−CD8+MR1tet+ (MR1tet+), tet− (MR1tet−) MAITs, as well as memory CD8+ T cells (Supplemental Fig. 1C), from 10 LTBI and seven TB neg subjects to define 1) differentially expressed genes between these cell populations and 2) whether TB infection changes the immune signature of MAITs.
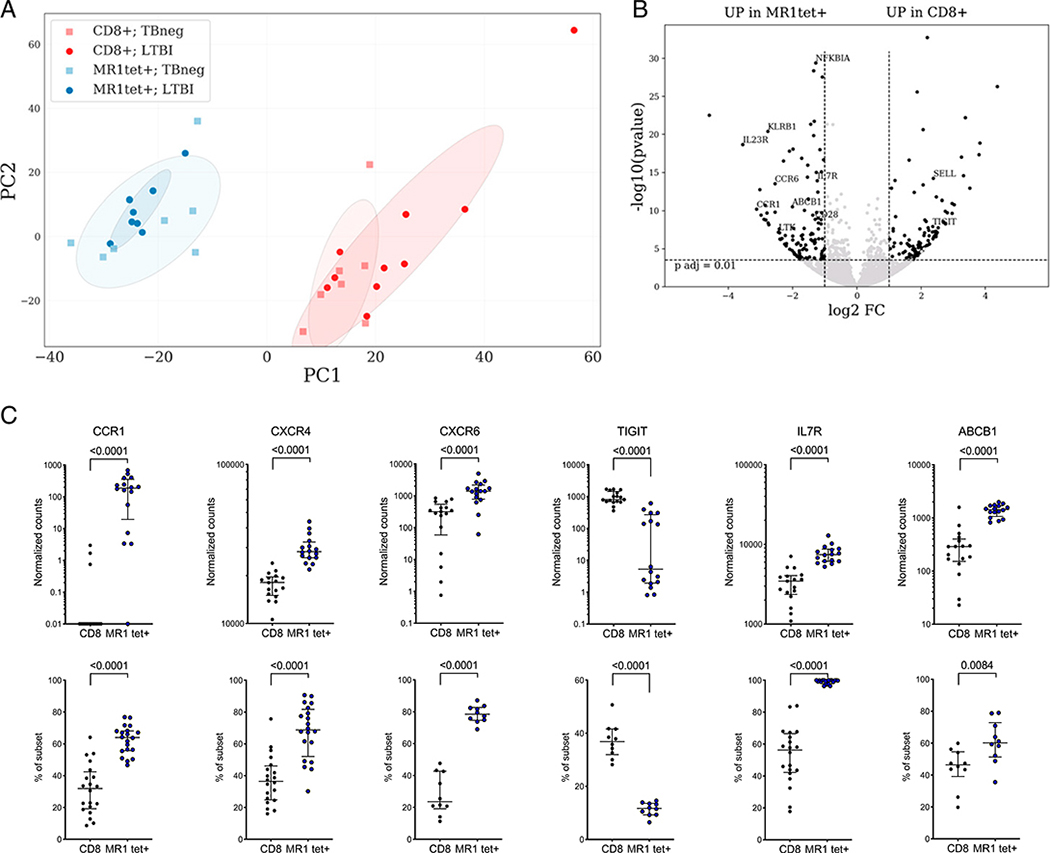
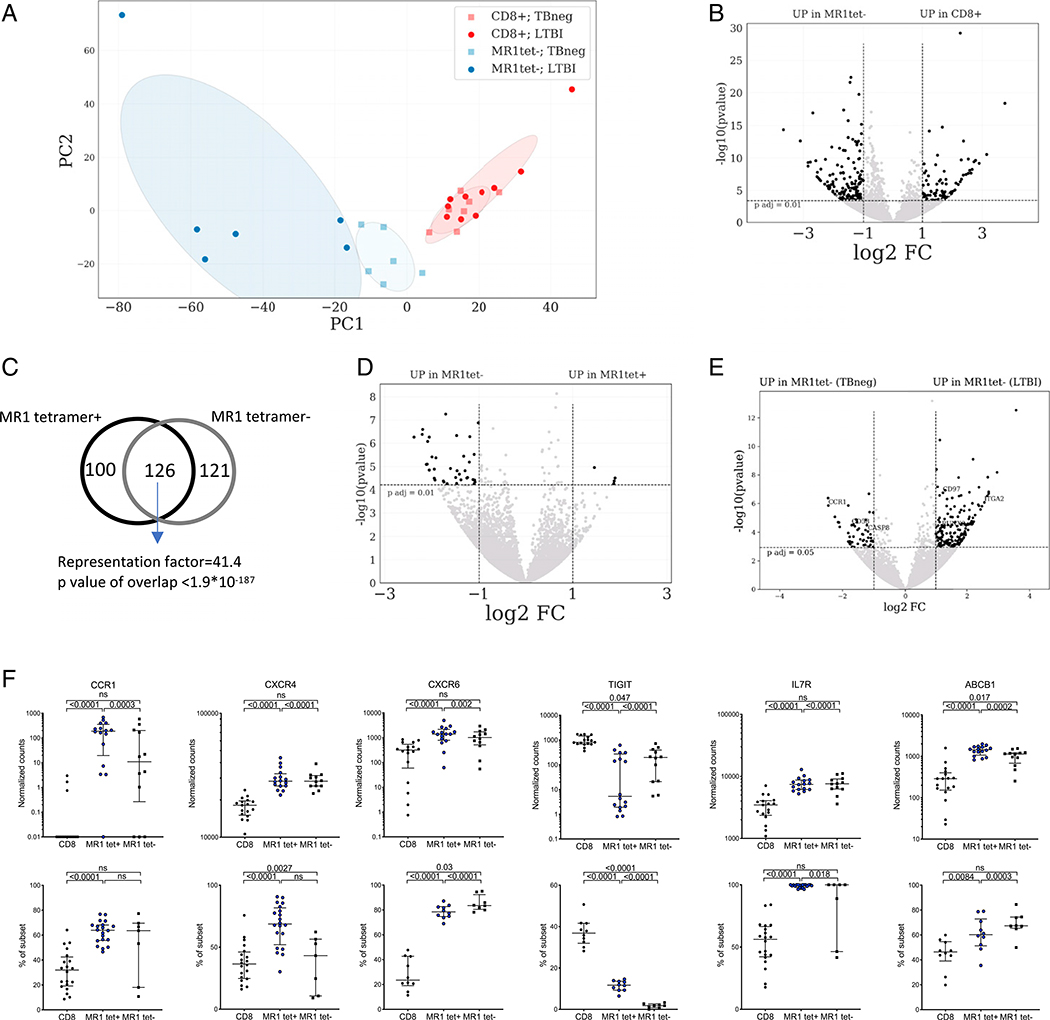
A PCA to visualize the global gene expression pattern of “classical” MR1tet+ MAITs and memory CD8+ T cells revealed systematic differences between these two populations (Fig. 4A). MR1tet+ cells formed a distinct cluster that was well separated from memory CD8 T cells as previously described (44, 45, 50). We found a total of 226 differentially expressed genes (adjusted p value <0.01 and absolute log2 fold change >1, Fig. 4B, Supplemental Table I) between MR1tet+ MAITs and memory CD8+ T cells. Several significant enrichments were found for gene ontology (GO) annotation analysis, the most relevant included genes involved in T cell differentiation (GO:0030217, false discovery rate [FDR]–adjusted p value 3.23 × 10−6), regulation of lymphocyte activation (GO:0051249, FDR-adjusted p value 8.00 × 10−10), and cytokine-mediated signaling pathways (GO:0019221, FDR-adjusted p value 0.00 × 100). Several transcription factors were upregulated in MR1tet+ MAITs, such as the canonical MAIT transcription factor ZBTB16 (PLZF) and MYC, ID2, BHLHE40, FOSL2, RORC, and IKZF2 (27, 44, 51, 52). These transcription factors are known to regulate the expression of IL-18RAP, IL-18R1, CXCR6, CCR6, LTK, IL-23R, and IFGR1. Some of these have been shown to be involved in tissue homing of T cells. For example, memory T cells residing in lung tissue have been shown to upregulate both CXCR6 and IL-23R and downregulate l-selectin and CX3CR1 (53), changes that are also observed in our MR1tet+ MAIT signature. Additionally, MR1tet+ cells have higher expression of MAIT markers such as KLRB1 (CD161), DPP4 (CD26), IL-7R, and ABCB1 (19, 20), as well as CCR1, CD40L, TNF, and TGFA. MR1tet+ MAITs also express higher levels of TCR-signaling–related genes, for example, CD28, LTK, and NFKBIA. We also found higher expression of the gene regulator CEBPD. TIGIT and SELL were among the genes downregulated in MR1tet+ MAITs compared with memory CD8 T cells.
FIGURE 4. Distinct gene expression profile of MR1tet+ MAITs compared with memory CD8+ T cells.
(A) PCA plot illustrating differences between memory CD8+ T cells and MR1tet+ MAITs and between LTBI and TB neg individuals. (B) Volcano plot obtained from the DEseq2 analysis showing log2 fold change versus −log10 p value. The differentially expressed genes are represented in black (adjusted p value <0.01, absolute log2 fold change >1 are indicated by dotted lines). (B) MR1tet+ cells compared with memory CD8+ T cells, (C) CCR1, CXCR4, CXCR6, TIGIT, IL7R, and ABCB1 expression at the mRNA (upper panels: gene expression values in counts normalized by sequencing depth calculated by the DEseq2 package) and protein (lower panels: protein expression as percent frequency of subset) levels in memory CD8+ T cells and MR1tet+ MAITs. Gene expression data were derived from memory CD8+ T cells from 17 individuals and MR1tet+ cells (n individuals = 16) using an Illumina sequencing platform. Protein expression data were derived from memory CD8+ T cells from 20 individuals and MR1tet+ cells (n individuals = 20) using flow cytometry. Median ± interquartile range is shown. Two-tailed Mann–Whitney U test.
Next, we wanted to examine if there was not only a quantitative difference in MR1tet+ MAITs in LTBI versus uninfected controls, but also a qualitative difference in their gene expression profile. We thus compared the genome-wide expression profile of MR1tet+ cells isolated from seven TB neg controls and eight LTBI. Interestingly, we found no differentially expressed genes. This indicated that there are no substantial qualitative differences in the phenotype of MR1tet+ MAITs in TB neg versus LTBI individuals in memory CD8+ T cells.
Then, we validated the transcriptomic signature of MR1tet+ MAITs at the protein level. For this purpose, we selected six genes (CCR1, CXCR4, CXCR6, TIGIT, IL-7R, and ABCB1) with known protein expression on the cell surface and commercially available Abs for protein profiling. The protein expression patterns using flow cytometry largely matched what we observed at the gene expression level. Specifically, in comparison with memory CD8+ T cells, MR1tet+ cells had higher expression of CCR1, CXCR4, CXCR6, IL-7R, and ABCB1 and lower expression of TIGIT at the protein level (Fig. 4D). Taken together, MR1tet+ MAITs have a distinct gene expression profile and are clearly different from memory CD8+ T cells.
MR1tet− MAITs have a distinct gene expression profile compared with memory CD8+ T cells and evidence for an M. tuberculosis–specific signature
We next defined differentially expressed genes between MR1tet− cells and memory CD8+ T cells using the same approach as for the MR1tet+ MAITs above. Similarly, the PCA revealed differences between MR1tet− MAITs and memory CD8+ T cells (Fig. 5A). Using the same criteria as above, we found 247 differentially expressed genes between MR1tet− cells and memory CD8+ T cells (Fig. 5B, Supplemental Table I). This MR1tet− signature overlapped significantly by 126 genes with the signature of MR1tet+ MAITs (Fig. 5C). In contrast, genes uniquely upregulated in MR1tet− MAITs compared with memory CD8+ T cells, and not in MR1tet+ cells, included ABCA2 (a transporter in the same family as ABCB1), TNFSF14 (LIGHT), and the transcription factors RORA and RUNX3. RUNX3 has been shown to be required for tissue resident cells and is also associated with innate immunity (50, 51).
FIGURE 5. Gene expression profile and M. tuberculosis–specific signature of MR1tet− MAITs compared with memory CD8 T cells and MR1tet+ MAITs.
(A) PCA plot illustrating differences between memory CD8 T cells and MR1tet− MAITs and between LTBI and TB neg individuals. (B, D, and E) Volcano plots obtained from the DEseq2 analysis showing log2 fold change versus −log10 p value. The differentially expressed genes are represented in black [adjusted p value <0.01 (B and E) and p < 0.05 (D), absolute log2 fold change >1 are indicated by dotted lines]. (B) MR1tet− cells compared with memory CD8 T cells. (C) Venn-diagram showing overlap between the 226-gene signature identified in Fig. 4B and the signature in Fig. 5B, based on hypergeometric distribution test (considering the 18,315 transcripts detected within memory CD8 T cells as the total number of genes). (D) MR1tet− cells comparing individuals with LTBI versus TB neg. (E) Volcano plot comparing MR1tet− cells with MR1tet+ cells. (F) CCR1, CXCR4, CXCR6, TIGIT, IL-7R, and ABCB1 expression at the mRNA (upper panels: gene expression values in counts normalized by sequencing depth calculated by the DEseq2 package) and protein (lower panels: protein expression as percent frequency of subset) levels in memory CD8 T cells and MR1tet− MAITs. Gene expression data were derived from memory CD8 T cells from 17 individuals and MR1tet− cells (n individuals = 12) using an Illumina sequencing platform. Protein expression data were derived from memory CD8 T cells from 20 individuals and MR1tet− cells (n individuals = 7) using flow cytometry. Median ± interquartile range is shown. Two-tailed Mann–Whitney U test.
To study the differences between the two MAIT subsets in more detail, we performed a DEseq2 analysis of differentially expressed genes between MR1tet+ and MR1tet− cells. This revealed 45 differentially expressed genes with the majority being upregulated in MR1tet− cells (Fig. 5D, Supplemental Table I), including CD4 and A2M. A gene set enrichment analysis of these signature genes did not reveal significantly enriched GO categories (54), so we did not discover an obvious functional interpretation of the differences between these MAIT subtypes.
Next, we wanted to examine whether qualitative differences exist in the MR1tet− MAIT immune signature between TB neg controls and LTBI. We compared the genome-wide expression profile of MR1tet− cells isolated from six TB neg controls and six LTBI individuals. In this comparison, we found an M. tuberculosis–specific 217 MR1tet− MAIT gene signature (adjusted p value <0.05, absolute log2 fold change >1; Fig. 5E, Supplemental Table I), with 166 genes upregulated in LTBI and 52 genes upregulated in TB neg controls. No significantly enriched GO categories were identified in either gene set. Genes upregulated in LTBI included genes linked to tissue residency (RUNX3 and CD97) and genes with antimicrobial and homing properties [S100A9, CCL22, and ITGA2 (55)]. Genes that were downregulated in LTBI included CCR1, CD58, and CASP8.
The expression of the same genes, as for MR1tet+ cells above, were investigated at the protein level in MR1tet− cells (Fig. 5F; MR1tet+ included for comparison purposes). This analysis revealed that there are some variations in expression of these genes in MR1tet− cells as compared with MR1tet+ cells. For example, the MR1tet− cells had lower gene expression of CCR1 than MR1tet+ cells, but this was not as evident at the protein level. CXCR4 and CXCR6 were expressed at similar mRNA levels in both MR1tet− and tet+ cells; however, at the protein level, MR1tet− had lower CXCR4 and higher CXCR6 expression. Although the gene expression of TIGIT was higher in MR1tet− cells, its protein expression was significantly lower in both MR1tet− cell populations compared with MR1tet+ cells.
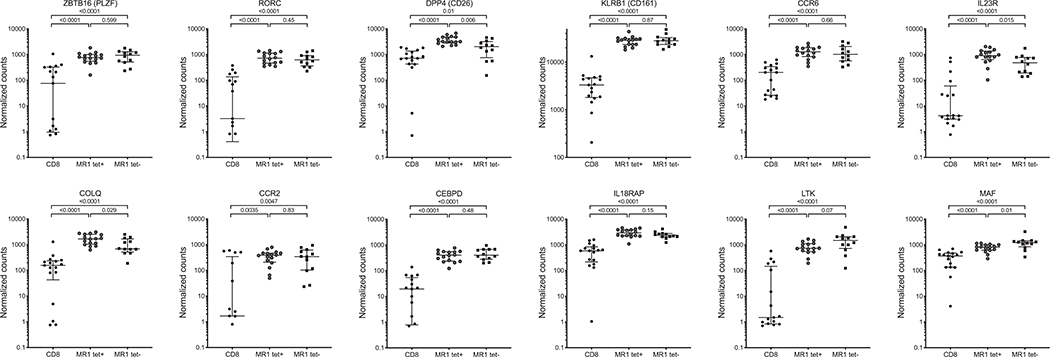
Finally, we wanted to investigate the expression of genes previously described as the transcriptomic signature of CD8+CD161hi T cells or described as genes expressed by MAITs (44, 45, 50) in our two MAIT subsets. Both MR1tet− and tet+ cells had similar expressions of ZBTB16 (PLZF transcription factor), RORC, KLRB1, CCR6, CCR2, CEBPD, IL-18RAP, and LTK (Fig. 6). MR1tet− cells had lower expression of DPP4, IL-23R, and COLQ and higher expression of MAF than MR1tet+ cells, albeit still significantly different as compared with memory CD8 T cells.
FIGURE 6. Expression of previously described MAIT signature genes in MR1tet+ and tet− cells.
Expression at the mRNA level (gene expression values in counts normalized by sequencing depth calculated by the DEseq2 package) in memory CD8 T cells, MR1tet+, and MR1tet− MAITs. Gene expression data were derived from memory CD8 T cells from 17 individuals, MR1tet+ (n individuals = 16), and MR1tet− cells (n individuals = 12) using an Illumina sequencing platform. Median ± interquartile range is shown. Two-tailed Mann–Whitney U test.
Taken together, LTBI results in a lower frequency of MR1tet+ MAITs as compared with TB neg controls, but these cells do not differ in their gene expression program. MR1tet− MAITs, in contrast, represent a more diverse cell population and exhibit a differential gene expression as a function of M. tuberculosis infection.
TCR repertoire of MAIT populations
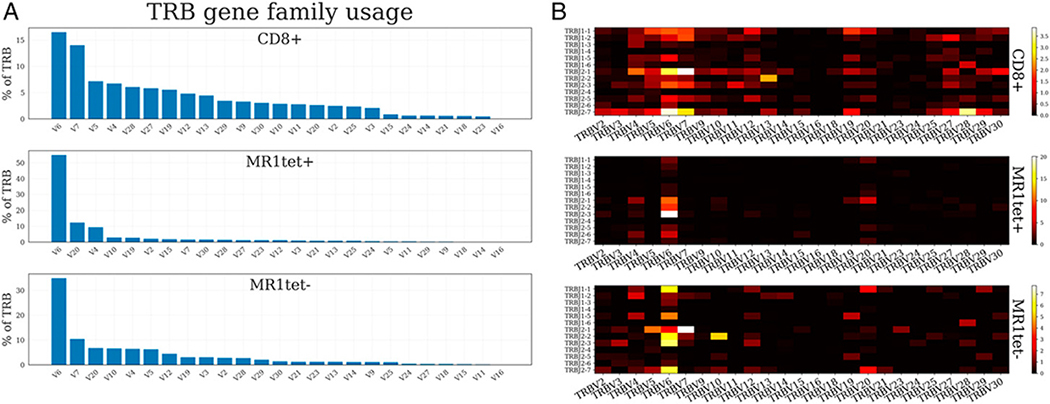
Previous studies have shown that MAITs have a restricted TCR repertoire compared with CD8 T cells (21, 56). Given that MR1tet− MAITs had a more diverse immune signature than MR1tet+ MAITs, we hypothesized that MR1tet− cells might also have a more diverse TCR repertoire as compared with MR1tet+ cells. To test this hypothesis, we investigated the TCR repertoire characteristics of both subpopulations of Vα7.2+CD161+ cells, MR1tet+ and MR1tet− cells, along with memory CD8+ T cells. We extracted TCR α- and β-chain sequences from bulk RNA-seq data using MiXCR (41). As expected, based on our sorting strategy (Supplemental Fig. 1C), both MR1tet+ and MR1tet− had Vα7.2 (TRAV1–2 in the IMGT nomenclature) as the most abundant α-chain V gene, 75 and 71%, respectively (Supplemental Fig. 3). TRAV1–2 was also the most abundant α-chain V gene in memory CD8+ T cells (9%), which reflects the impact of MAITs on the memory CD8+ T cells compartment (that contain an average of 4% MAITs).
We next analyzed the pattern of TCR β-chain VDJ-gene usage. As anticipated, for MR1tet+ cells the β-chain preference was generally concordant with previously described repertoires (56). Although no V or J segment was found to dominate, we found that 54.9% of β-chains contained TRBV6 family gene segments in MR1tet+ cells versus 16.5% in memory CD8+ cells. Additionally, TRBV20 was the second most abundant TRBV gene segment family (Fig. 7A). The most commonly used J gene segments were TRBJ2–3 (25.4%) and TRBJ2–1 (23.9%).
FIGURE 7. TCR β-chain gene segment usage.
(A) TRBV family usage in CD8+, MR1tet+, and MR1tet− cells summing over J combinations. (B) Heatmap illustrating frequency of a particular V-J combination in CD8+, MR1tet+, and MR1tet− cells. Averaging is done across the subjects. V gene segments are grouped by family by summing up family gene segments prior to plotting.
For MR1tet− cells, we observed a more diverse β-chain gene usage (Fig. 7A, 7B). The TRBV6 gene family segments constituted a relatively large proportion (34.8%) of all rearrangements. However, increased diversity in the MR1tet− cells was found by significantly less frequent expression of TRBV20xTRBJ2–1 (p value 2.85 × 10−3), TRBV6xTRBJ2–6 (p value 1.74 × 10−3), TRBV6xTRBJ2–1 (p value 8.29 × 10−5), TRBV6xTRBJ2–2 (p value 3.41 × 10−3), and TRBV6xTRBJ2–3 (p value 5.32 × 10−4) V-J combinations as compared with MR1tet+ cells. The increased diversity was also highlighted by our finding that no V-J combinations were enriched in the MR1tet− cells.
Finally, we investigated the extracted β-chains using the MAIT-match web application (42), in which a score reflects the likelihood that a particular α-chain CDR3 sequence corresponds to a previously described MAIT-associated sequence. We averaged MAIT-match score by taking a median of scores of α-chains detected within an individual, and then calculating the median of the considered individuals. As expected, the average score was very high, 0.97, for MR1tet+ cells. MR1tet− cells had an average score of 0.91, and memory CD8+ T cells had an average score of 0.85. Thus, the MR1tet− cells likely reflects MAITs with a more diverse TCR repertoire.
Based in the recent publication by Wong et al. (42), we used a threshold of 0.95 to distinguish MAIT-like α-chains. For each cell type and subject combination, we calculated the frequency of detected α-chains with a MAIT-match score higher than 0.95 and then averaged (by median) across the investigated subjects. The majority of detected α-chains in MR1tet+ cells (54.2%) were MAIT-like. In MR1tet−, 39.7% of α-chains were MAIT-like and, in contrast, only 6.0% of α-chains were detected in memory CD8+ T cells.
In summary, these results suggest that MR1tet− MAITs are different from MR1tet+ MAITs with a larger diversity in gene expression, more diverse TCR repertoire, and they also correlate with M. tuberculosis infection status.
DISCUSSION
This study describes the systematic transcriptomic analysis of sorted human memory CD8+ T cells and MAIT subsets, namely Vα7.2+CD161+CD4−CD8+MR1 tetramer+ and MR1 tetramer− cells, in the context of LTBI. We provide an in-depth characterization through integration of transcriptional and phenotypic profiling combined with TCR repertoire analysis.
We were able to show that a specific immune signature in memory CD8+ T cells is linked to a lower frequency of MAITs in LTBI. Our study demonstrates a decrease in MAIT frequencies in LTBI individuals compared with TB neg controls. However, this finding is in concordance with previous findings that have found a decrease in MAIT frequencies in individuals with active TB (14, 15). Wong et al. (17) found a trend toward decreased frequency of CD161hi CD8+ T cells in PBMCs from LTBI compared with controls in a TB-endemic African setting. Other studies have found no difference in MAIT frequencies in LTBI compared with TB neg, or even an increase (16, 18). These contradictory results may be explained by variations in M. tuberculosis exposure rates in endemic versus nonendemic TB areas, how well the latent infection is controlled, as well as a difference in how MAIT populations were defined. The frequency of MAITs in the blood is also decreased in patients with a large variety of infectious diseases other than TB such as in cystic fibrosis with Pseudomonas infection (57), during infection with Vibrio cholera (58), and Helicobacter pylori (59). In parallel, MAITs have been detected in infected tissues during microbial infections (14, 15, 60), thus leading to the hypothesis that MAITs have been recruited to infected tissues. In the context of M. tuberculosis infection, they might provide an early defense against M. tuberculosis infection particularly before the arrival of conventional effector T cells in the lungs. Although experiments in nonhuman primates have not yet been able to prove this hypothesis (61, 62), it was strengthened by recent findings that MAITs are increased in bronchoalveolar lavage from individuals with active TB as compared with uninfected controls (42). Performing transcriptional studies on MAITs in the lung would be highly relevant in the context of M. tuberculosis infection and may reveal differences between individuals with LTBI and TB neg individuals.
Although we were able to distinguish individuals with LTBI from TB neg individuals based on their gene expression in memory CD8+ T cells, we found no evidence for a distinct immune signature in classically restricted CD8+ T cells. Recently, Huang et al. (63) provided evidence for extensive clonal expansion of CD8+ T cells in asymptomatic M. tuberculosis–infected individuals in response to M. tuberculosis lysate. These T cells exhibited a senescent phenotype when reactivated, thus suggesting continuous activation in vivo (63). M. tuberculosis–specific CD8+ T cells recognize several epitopes (10), but the cross-reactivity with environmental mycobacteria has not been studied in detail. We used a peptide pool of previously defined M. tuberculosis–specific MHC class I–restricted epitopes available in the Immune Epitope Database Analysis and Resource and found no evidence of increased RF or IFN-γ production in LTBI as compared with TB neg individuals. No difference in magnitude or frequency of responses of M. tuberculosis–specific CD8 T cells suggests that the existing reactivity can be attributed to cross-reactivity with Ags that the individual has experienced in the past. We have previously found evidence that M. tuberculosis–specific epitope reactivity in TB neg individuals correlates with conservation in nontuberculous mycobacteria (64). Pre-exposure to nontuberculous mycobacteria may have contributed to the lack of differential M. tuberculosis–specific responses between LTBI and TB neg seen in this study.
We discovered that CD8 expression in MR1 tetramer+ MAITs does not correlate with a broader gene expression program because only six genes were differentially expressed between CD8+ and CD8− MR1 tetramer+ MAITs. Kurioka et al. (28) also found, through phenotypic analysis and functional assays, that most features are shared between CD8+ and CD8− Vα7.2+CD161+ MAITs.
Our current study found that the transcriptional program of CD8+ MAITs, either MR1tet+ or tet−, is very distant from memory CD8+ T cells. This has been reported previously in studies that have investigated the transcriptional signature of subsets enriched for MAITs: Vα7.2+CD161+CD3+ T cells and CD161hiCD8+ T cells (43–45, 50). Park et al. (45) compared transcriptomics of Vα7.2+CD161+ T cells, Vα7.2+CD161− T cells, and Vα7.2− T cells in a small cohort of three individuals. Our study systematically characterized the immune signatures of MAIT Vα7.2+CD161+ subsets in comparison with memory CD8+ T cells. The expression of several genes was confirmed at the protein level by flow cytometry. Previously described genes that are upregulated in MAITs include SLC4A10, LTK, FLT4, and DUSP2 (43, 45) as well as downregulated genes LEF1, KLRC4, TBC1D4, ITK, and FYB as compared with memory CD8+ T cells.
We found that MAITs (both tetramer+/−) express CCR1, which is a protein primarily expressed on monocytes and NK cells [according to dice-database.org (65)]. Interestingly, activated MAITs are thought to facilitate recruitment of CCR1+ monocytes through their production of CCL3 and CCL4 (66). The expression of CCR1 in MAITs suggests a role for paracrine and autocrine stimulation.
Our study and others thus indicate that MAITs have distinctive phenotypic and transcriptional characteristics that set them apart from memory CD8+ T cells and conventional T cells in general. They do not fit neatly into traditional paradigms of adaptive or innate immunity. In support of this, a recent publication found distinct gene expression profiles comparing CD8+ T cells and MAIT and invariant NKT (as defined by their TCRs) cells following activation with M. tuberculosis lysate (63). However, no differences were found between activated MAITs and invariant NKT cells at the transcriptional level. The study by Fergusson et al. (44) suggested that there is a shared gene expression signature between CD161+ cell subsets, including MAITs, CD161+CD4+, and CD161+ γδ T cells. Furthermore, MAITs exhibit concomitant expression of transcription factors that have been linked to both adaptive and innate immune functions (51). Expression of these transcription factors are shared between subsets of innate T cells (67). Side-by-side comparisons or single-cell analysis of different subsets will likely reveal subset-specific differences.
MR1tet− cells do not recognize the 5-OP-RU-loaded tetramer, but they have a large number of genes overlapping with MR1tet+ cells, including genes previously associated with MAITs. This common gene expression program suggests that the MR1tet− cells in our study are MAITs that may be specific for a different ligand. This was also strengthened by the TCR repertoire analysis, which revealed that MR1tet+ MAITs have a distinct use of β-chain genes including TRBV6 and TRBV20 family gene segments. However, MR1tet− cells have an increased β-chain diversity. Recent studies have demonstrated the existence of TRAV1–2-negative MR1-restricted T cells, suggesting the presence of a larger group of MR1-restricted T cells (23, 26, 68). Recent studies have also provided evidence for a more diverse MAIT TCR repertoire for both TRAJ gene usage and TRBV gene usage (14, 23, 24, 48, 56). These findings indicate that MAITs, similar to other T cell subsets, are more diverse than previously understood. Furthermore, recent reports (23, 26, 68) suggest the presence of MR1 ligands that are not derived from riboflavin biosynthesis. For example, Streptococcus pyogenes was recognized in an MR1-dependent manner, despite the fact that this bacterium lacks the enzymatic pathway for riboflavin biosynthesis (68). Further studies will investigate whether expansion of MR1tet− cells retain their phenotype, confirm if they are restricted by MR1, as well as investigate their ligand for MR1tet− cells by cloning and screening against different MAIT ligands.
Taken together, our results identified the gene expression signature of Vα7.2+CD161+ MAIT subsets and, to our knowledge, discovered a population of MAITs with an M. tuberculosis–specific signature that do not bind the 5-OP-RU MR1 tetramer. This furthers our understanding of immune responses and the differentiation of MAITs involved in the context of M. tuberculosis infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Flow Cytometry Core Facility and the Bioinformatics Core Facility at the La Jolla Institute for Immunology for technical assistance.
This study was supported by National Institutes of Health National Institute of Allergy and Infectious Diseases Grants U19 AI118626, S10 RR027366, and S10 OD016262.
Abbreviations used in this article
- FDR
false discovery rate
- GO
gene ontology
- LTBI
latent tuberculosis infection
- MAIT
mucosal-associated invariant T cell
- MR1
MHC-related protein 1
- PCA
principal component analysis
- RF
response frequency
- RNA-seq
RNA sequencing
- TB
tuberculosis
- TB neg
TB-negative
Footnotes
DISCLOSURES
The authors have no financial conflicts of interest.
The sequencing data presented in this study have been submitted to the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE132790, GSE132931, and GSE132932 and to ImmPort (http://www.immport.org) under study number SDY820.
The online version of this article contains supplemental material.
REFERENCES
- 1.World Health Organization. 2018. Global Tuberculosis Report 2018. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Russell DG, Barry CE III., and Flynn JL. 2010. Tuberculosis: what we don’t know can, and does, hurt us. Science 328: 852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes PF, Bloch AB, Davidson PT, and Snider DE Jr. 1991. Tuberculosis in patients with human immunodeficiency virus infection. N. Engl. J. Med. 324: 1644–1650. [DOI] [PubMed] [Google Scholar]
- 4.Flynn JL, and Chan J. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19: 93–129. [DOI] [PubMed] [Google Scholar]
- 5.Tian Y, Babor M, Lane J, Seumois G, Liang S, Goonawardhana NDS, De Silva AD, Phillips EJ, Mallal SA, da Silva Antunes R, et al. 2019. Dengue-specific CD8+ T cell subsets display specialized transcriptomic and TCR profiles. J. Clin. Invest. 130: 1727–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burel JG, Lindestam Arlehamn CS, Khan N, Seumois G, Greenbaum JA, Taplitz R, Gilman RH, Saito M, Vijayanand P, Sette A, and Peters B. 2018. Transcriptomic analysis of CD4+ T cells reveals novel immune signatures of latent tuberculosis. J. Immunol. 200: 3283–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Silva Antunes R, Babor M, Carpenter C, Khalil N, Cortese M, Mentzer AJ, Seumois G, Petro CD, Purcell LA, Vijayanand P, et al. 2018. Th1/Th17 polarization persists following whole-cell pertussis vaccination despite repeated acellular boosters. J. Clin. Invest. 128: 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, and Peters B. 2014. Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features. J. Immunol. 193: 2931–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behar SM 2013. Antigen-specific CD8(+) T cells and protective immunity to tuberculosis. Adv. Exp. Med. Biol. 783: 141–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewinsohn DA, Swarbrick GM, Park B, Cansler ME, Null MD, Toren KG, Baseke J, Zalwango S, Mayanja-Kizza H, Malone LL, et al. 2017. Comprehensive definition of human immunodominant CD8 antigens in tuberculosis. NPJ Vaccines 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewinsohn DA, Heinzel AS, Gardner JM, Zhu L, Alderson MR, and Lewinsohn DM. 2003. Mycobacterium tuberculosis-specific CD8+ T cells preferentially recognize heavily infected cells. Am. J. Respir. Crit. Care Med. 168: 1346–1352. [DOI] [PubMed] [Google Scholar]
- 12.Behar SM, Divangahi M, and Remold HG. 2010. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 8: 668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewinsohn DA, Gold MC, and Lewinsohn DM. 2011. Views of immunology: effector T cells. Immunol. Rev. 240: 25–39. [DOI] [PubMed] [Google Scholar]
- 14.Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, Winata E, Swarbrick GM, Chua W-J, Yu YYL, et al. 2010. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8: e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, Lévy E, Dusseaux M, Meyssonnier V, Premel V, et al. 2010. Antimicrobial activity of mucosal-associated invariant T cells. [Published erratum appears in 2010 Nat. Immunol. 11: 969.] Nat. Immunol. 11: 701–708. [DOI] [PubMed] [Google Scholar]
- 16.Paquin-Proulx D, Costa PR, Terrassani Silveira CG, Marmorato MP, Cerqueira NB, Sutton MS, O’Connor SL, Carvalho KI, Nixon DF, and Kallas EG. 2018. Latent Mycobacterium tuberculosis infection is associated with a higher frequency of mucosal-associated invariant T and invariant natural killer T cells. Front. Immunol. 9: 1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong EB, Akilimali NA, Govender P, Sullivan ZA, Cosgrove C, Pillay M, Lewinsohn DM, Bishai WR, Walker BD, Ndung’u T, et al. 2013. Low levels of peripheral CD161++CD8+ mucosal associated invariant T (MAIT) cells are found in HIV and HIV/TB co-infection. [Published erratum appears in 2014 PLoS One 9: e95115.] PLoS One 8: e83474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suliman S, Gela A, Mendelsohn SC, Iwany SK, Tamara KL, Mabwe S, Bilek N, Darboe F, Fisher M, Corbett AJ, et al. South African Tuberculosis Vaccine Initiative (SATVI) Clinical Immunology Team. 2020. Peripheral blood mucosal-associated invariant T (MAIT) cells in tuberculosis patients and healthy Mycobacterium tuberculosis-exposed controls. J. Infect. Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, Milder M, Le Bourhis L, Soudais C, Treiner E, and Lantz O. 2011. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117: 1250–1259. [DOI] [PubMed] [Google Scholar]
- 20.Sharma PK, Wong EB, Napier RJ, Bishai WR, Ndung’u T, Kasprowicz VO, Lewinsohn DA, Lewinsohn DM, and Gold MC. 2015. High expression of CD26 accurately identifies human bacteria-reactive MR1-restricted MAIT cells. Immunology 145: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porcelli S, Yockey CE, Brenner MB, and Balk SP. 1993. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4–8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J. Exp. Med. 178: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491: 717–723. [DOI] [PubMed] [Google Scholar]
- 23.Gherardin NA, Keller AN, Woolley RE, Le Nours J, Ritchie DS, Neeson PJ, Birkinshaw RW, Eckle SBG, Waddington JN, Liu L, et al. 2016. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity 44: 32–45. [DOI] [PubMed] [Google Scholar]
- 24.Gold MC, McLaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbrick GM, Yu YY, Hansen TH, Lund O, Nielsen M, et al. 2014. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J. Exp. Med. 211: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harriff MJ, McMurtrey C, Froyd CA, Jin H, Cansler M, Null M, Worley A, Meermeier EW, Swarbrick G, Nilsen A, et al. 2018. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci. Immunol. 3: eaao2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M, Narang V, Zolezzi F, Poidinger M, Mori L, and De Libero G. 2017. Functionally diverse human T cells recognize nonmicrobial antigens presented by MR1. [Published erratum appears in 2017 Elife 6.] Elife 6: e24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dias J, Leeansyah E, and Sandberg JK. 2017. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc. Natl. Acad. Sci. USA 114: E5434–E5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurioka A, Jahun AS, Hannaway RF, Walker LJ, Fergusson JR, Sverremark-Ekström E, Corbett AJ, Ussher JE, Willberg CB, and Klenerman P. 2017. Shared and distinct phenotypes and functions of human CD161++ Vα7.2+ T cell subsets. Front. Immunol. 8: 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burel JG, Qian Y, Lindestam Arlehamn C, Weiskopf D, Zapardiel-Gonzalo J, Taplitz R, Gilman RH, Saito M, de Silva AD, Vijayanand P, et al. 2017. An integrated workflow to assess technical and biological variability of cell population frequencies in human peripheral blood by flow cytometry. J. Immunol. 198: 1748–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, Chen Z, Reantragoon R, Meehan B, Cao H, et al. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509: 361–365. [DOI] [PubMed] [Google Scholar]
- 31.Tian Y, Babor M, Lane J, Schulten V, Patil VS, Seumois G, Rosales SL, Fu Z, Picarda G, Burel J, et al. 2017. Unique phenotypes and clonal expansions of human CD4 effector memory T cells re-expressing CD45RA. Nat. Commun. 8: 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seumois G, Vijayanand P, Eisley CJ, Omran N, Kalinke L, North M, Ganesan AP, Simpson LJ, Hunkapiller N, Moltzahn F, et al. 2012. An integrated nano-scale approach to profile miRNAs in limited clinical samples. Am. J. Clin. Exp. Immunol. 1: 70–89. [PMC free article] [PubMed] [Google Scholar]
- 33.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, and Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9: 171–181. [DOI] [PubMed] [Google Scholar]
- 34.Trapnell C, Pachter L, and Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmieder R, and Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Pyl PT, and Huber W. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, and Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindestam Arlehamn CS, McKinney DM, Carpenter C, Paul S, Rozot V, Makgotlho E, Gregg Y, van Rooyen M, Ernst JD, Hatherill M, et al. 2016. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis infected South Africans. PLoS Pathog. 12: e1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y, Vaughan K, Greenbaum J, Peters B, Law M, and Sette A. 2012. A meta-analysis of the existing knowledge of immunoreactivity against hepatitis C virus (HCV). PLoS One 7: e38028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM. 2015. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12: 380–381. [DOI] [PubMed] [Google Scholar]
- 42.Wong EB, Gold MC, Meermeier EW, Xulu BZ, Khuzwayo S, Sullivan ZA, Mahyari E, Rogers Z, Kløverpris H, Sharma PK, et al. 2019. TRAV1–2+ CD8+ T-cells including oligoconal expansions of MAIT cells are enriched in the airways in human tuberculosis. Commun. Biol. 2: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billerbeck E, Kang YH, Walker L, Lockstone H, Grafmueller S, Fleming V, Flint J, Willberg CB, Bengsch B, Seigel B, et al. 2010. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc. Natl. Acad. Sci. USA 107: 3006–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, Marchi E, Björkander S, Kang YH, Swadling L, et al. 2014. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. 9: 1075–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park D, Kim HG, Kim M, Park T, Ha HH, Lee DH, Park KS, Park SJ, Lim HJ, and Lee CH. 2019. Differences in the molecular signatures of mucosal-associated invariant T cells and conventional T cells. Sci. Rep. 9: 7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, and Bendelac A. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29: 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin PL, and Flynn JL. 2015. CD8 T cells and Mycobacterium tuberculosis infection. Semin. Immunopathol. 37: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, Eckle SB, Uldrich AP, Birkinshaw RW, Patel O, et al. 2013. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210: 2305–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Door-enspleet ME, de Vries N, Godfrey DI, Altman JD, de Jager W, et al. 2013. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat. Immunol. 14: 706–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, Richer W, Goubet AG, Daviaud C, Menger L, et al. 2019. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med. 216: 133–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gutierrez-Arcelus M, Teslovich N, Mola AR, Polidoro RB, Nathan A, Kim H, Hannes S, Slowikowski K, Watts GFM, Korsunsky I, et al. 2019. Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nat. Commun. 10: 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leeansyah E, Svärd J, Dias J, Buggert M, Nyström J, Quigley MF, Moll M, Sönnerborg A, Nowak P, and Sandberg JK. 2015. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PLoS Pathog. 11: e1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, et al. 2017. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20: 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pomaznoy M, Ha B, and Peters B. 2018. GOnet: a tool for interactive gene ontology analysis. BMC Bioinformatics 19: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan X, and Rudensky AY. 2016. Hallmarks of tissue-resident lymphocytes. Cell 164: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, Lee B, Poidinger M, Zolezzi F, Quagliata L, et al. 2014. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβ repertoire. [Published erratum appears in 2014 Nat. Commun. 5: 4493.] Nat. Commun. 5: 3866. [DOI] [PubMed] [Google Scholar]
- 57.Smith DJ, Hill GR, Bell SC, and Reid DW. 2014. Reduced mucosal associated invariant T-cells are associated with increased disease severity and Pseudomonas aeruginosa infection in cystic fibrosis. PLoS One 9: e109891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leung DT, Bhuiyan TR, Nishat NS, Hoq MR, Aktar A, Rahman MA, Uddin T, Khan AI, Chowdhury F, Charles RC, et al. 2014. Circulating mucosal associated invariant T cells are activated in Vibrio cholerae O1 infection and associated with lipopolysaccharide antibody responses. PLoS Negl. Trop. Dis. 8: e3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Booth JS, Salerno-Goncalves R, Blanchard TG, Patil SA, Kader HA, Safta AM, Morningstar LM, Czinn SJ, Greenwald BD, and Sztein MB. 2015. Mucosal-associated invariant T cells in the human gastric mucosa and blood: role in Helicobacter pylori infection. Front. Immunol. 6: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liuzzi AR, Kift-Morgan A, Lopez-Anton M, Friberg IM, Zhang J, Brook AC, Roberts GW, Donovan KL, Colmont CS, Toleman MA, et al. 2016. Unconventional human T cells accumulate at the site of infection in response to microbial ligands and induce local tissue remodeling. J. Immunol. 197: 2195–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bucsan AN, Rout N, Foreman TW, Khader SA, Rengarajan J, and Kaushal D. 2019. Mucosal-activated invariant T cells do not exhibit significant lung recruitment and proliferation profiles in macaques in response to infection with Mycobacterium tuberculosis CDC1551. Tuberculosis (Edinb). 116S: S11–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kauffman KD, Sallin MA, Hoft SG, Sakai S, Moore R, Wilder-Kofie T, Moore IN, Sette A, Arlehamn CSL, and Barber DL. 2018. Limited pulmonary mucosal-associated invariant T cell accumulation and activation during Mycobacterium tuberculosis infection in rhesus macaques. Infect. Immun. 86: e00431–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang H, Sikora MJ, Islam S, Chowdhury RR, Chien YH, Scriba TJ, Davis MM, and Steinmetz LM. 2019. Select sequencing of clonally expanded CD8+ T cells reveals limits to clonal expansion. Proc. Natl. Acad. Sci. USA 116: 8995–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindestam Arlehamn CS, Paul S, Mele F, Huang C, Greenbaum JA, Vita R, Sidney J, Peters B, Sallusto F, and Sette A. 2015. Immunological consequences of intragenus conservation of Mycobacterium tuberculosis T-cell epitopes. Proc. Natl. Acad. Sci. USA 112: E147–E155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, Ha B, Altay G, Greenbaum JA, McVicker G, et al. 2018. Impact of genetic polymorphisms on human immune cell gene expression. Cell 175: 1701–1715.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakala IG, Kjer-Nielsen L, Eickhoff CS, Wang X, Blazevic A, Liu L, Fairlie DP, Rossjohn J, McCluskey J, Fremont DH, et al. 2015. Functional heterogeneity and antimycobacterial effects of mouse mucosal-associated invariant T cells specific for riboflavin metabolites. J. Immunol. 195: 587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, Packe GE, Davidson RN, Griffiths CJ, and Wilkinson RJ. 2007. Neutrophil-mediated innate immune resistance to mycobacteria. J. Clin. Invest. 117: 1988–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meermeier EW, Laugel BF, Sewell AK, Corbett AJ, Rossjohn J, McCluskey J, Harriff MJ, Franks T, Gold MC, and Lewinsohn DM. 2016. Human TRAV1–2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat. Commun. 7: 12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.