Abstract
Dynamin GTPases (Dyn1 and Dyn2) are indispensable proteins of the core clathrin-mediated endocytosis (CME) machinery. Best known for their role in fission at the late stages of CME, many studies have suggested that dynamin also plays a regulatory role during the early stages of CME; however, detailed studies regarding isoform-specific early regulatory functions of the dynamins are lacking. With a recent understanding of the regulation of Dyn1 in nonneuronal cells and improved algorithms for highly sensitive and quantitative analysis of clathrin-coated pit (CCP) dynamics, we have evaluated the differential functions of dynamin isoforms in CME using domain swap chimeras. We report that Dyn1 and Dyn2 play nonredundant, early regulatory roles during CME in nonneuronal cells. The proline/arginine-rich domain of Dyn2 is important for its targeting to nascent and growing CCPs, whereas the membrane-binding and curvature-generating pleckstrin homology domain of Dyn1 plays an important role in stabilizing nascent CCPs. We confirm the enhanced ability of dephosphorylated Dyn1 to support CME, even at substoichiometric levels compared with Dyn2. Domain swap chimeras also revealed previously unknown functional differences in the GTPase and stalk domains. Our study significantly extends the current understanding of the regulatory roles played by dynamin isoforms during early stages of CME.
INTRODUCTION
Dynamin GTPases are well known for their function during clathrin-mediated endocytosis (CME; van der Bliek, Redelmeier, et al., 1993; Damke et al., 1994; Hinshaw and Schmid, 1995). Vertebrates express three isoforms of modern/classical dynamins (Ramachandran and Schmid, 2018), namely, dynamin-1, dynamin-2, and dynamin-3 (Dyn1, Dyn2, and Dyn3). Dyn2 is uniformly expressed in all mammalian tissues (Cao et al., 1998). Dyn1 is the major neuronal isoform and its expression levels in the brain exceed by several orders of magnitudes those in any other tissue (Cao et al., 1998; Ferguson et al., 2007; Schmid, 2017). Nonetheless, Dyn1 is also expressed in most mammalian tissues at a level comparable to Dyn2 expression, but its activity is negatively regulated through phosphorylation by a constitutively active kinase, GSK3β (Clayton et al., 2010; Reis, Chen, et al., 2015; Mettlen et al., 2018). Dyn3 is mostly expressed in the brain, testes, heart, and lungs (Cao et al., 1998; Ferguson et al., 2007).
All three dynamin isoforms are composed of five domains, an N-terminal GTPase (G) domain attached to an α-helical stalk composed of the middle domain and GTPase effector domain (GED), a phospholipid-interacting pleckstrin homology (PH) domain, and a C-terminal proline/arginine-rich domain (PRD). A bundle signaling element composed of the N- and C-terminal α-helices of the G domain and the C-terminal α-helix of GED connects the G domain to the stalk and undergoes a dramatic rearrangement during cooperative GTP hydrolysis (Ramachandran and Schmid, 2018). Dynamin isoforms share a high degree of sequence identity across all domains (Figure 1A), the most divergent being the C-terminal PRD (Liu, Neumann, et al., 2011).
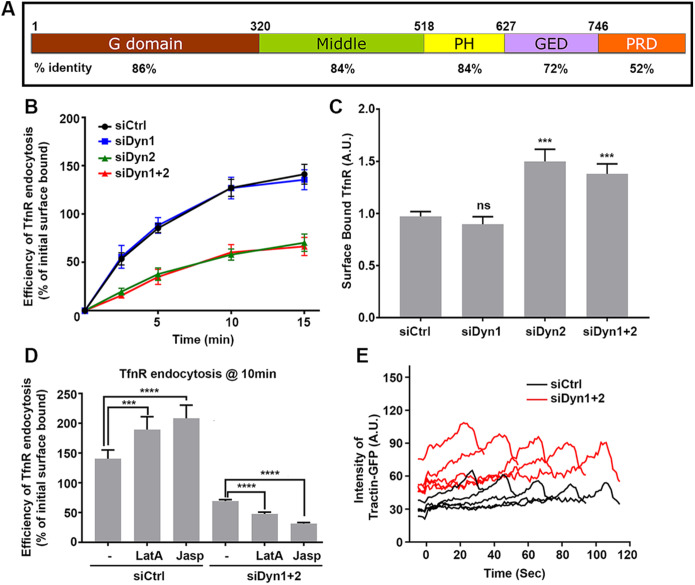
FIGURE 1:
Differential functions of dynamin isoforms in CME of nonneuronal cells. (A) A schematic representation of the domain structure and sequence identity of dynamin isoforms. (B) Effect of KD of dynamin isoforms on the efficiency of TfnR endocytosis in ARPE cells. The cells were treated with siRNAs as indicated and the efficiency of TfnR endocytosis was measured by in-cell ELISA assay and presented as the percentage of initial surface-bound TfnR. Data shown are average ± SD. (C) Quantification of surface accumulation of TfnR after KD of Dyn1, Dyn2, and Dyn1+2, as indicated (n = 3 independent biological repeats). (D) Effect on TfnR uptake efficiency in control and Dyn1+2 double KD cells with/without 30 min preincubation with chemicals affecting actin polymerization/depolymerization dynamics: latrunculin A (latA, 100 nM) and Jasp (1 µM). Statistical significance was calculated by t test. In this and subsequent figures, *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001; n = 3 independent biological repeats. (E) ARPE cells expressing mRuby-CLCa and Tractin-eGFP were plated on gelatin-coated coverslips and imaged by TIRFM. Shown are the intensity profiles of the recruitment of Tractin-eGFP (secondary channel) to the indicated lifetime cohorts of mRuby-CLCa containing CCPs (primary channel). The TIRFM data are representative of two independent biological repeats.
Although Dyn1 and Dyn2 share 79% sequence identity, based on knockout (KO) studies, the two dynamin isoforms cannot substitute for each other’s function (Ferguson et al., 2007; Liu et al., 2008; Reis, Chen, et al., 2015; Srinivasan, Burckhardt, et al., 2018). Dyn1 is important for rapid synaptic vesicle recycling in neurons (Ferguson et al., 2007), whereas Dyn2 is believed to perform housekeeping functions for CME in most other tissues. Dyn1 and Dyn3 appear to be at least, in part, functionally redundant, as Dyn3 null mice do not exhibit any pathological defects, whereas Dyn1 and Dyn3 double KO mice show more severe synaptic endocytosis defects than Dyn1 KO alone (Raimondi, Ferguson, et al., 2011). Recent studies have shown that the inability of Dyn1 to support CME in nonneuronal cells is, in part, a reflection of its inactivation through phosphorylation at the nonconserved serine residue (S774) in PRD1 by the constitutively active kinase, GSK3β (Reis, Chen, et al., 2015). GSK3β requires upstream priming by phosphorylation of Dyn1 at S778 which, at least in neurons, is mediated by CDK5 (Tan et al., 2003).
Most of the studies performed to understand the isoform-specific functions of dynamins during CME have relied on the measurement of CME using transferrin receptor (TfnR) uptake assays (Liu et al., 2008; Liu et al., 2011). This approach measures only the end result of CME and the important dynamics of early regulatory stages of clathrin-coated pit (CCP) assembly and maturation are missed. Given increasing evidence that dynamin isoforms are also involved in the regulation of the early stages of CME (Sever et al., 1999; Loerke, Mettlen, et al., 2009; Reis, Chen, et al., 2015; Mettlen et al., 2018), it is important to directly assess the differential effects of Dyn1 and Dyn2 knockdown (KD) and of Dyn1/2 chimeras on CCP dynamics using live cell imaging approaches.
In this study we first phenotypically characterize the effect of Dyn1 or Dyn2 KD in human retinal pigmented epithelial (ARPE) cells on TfnR uptake and by quantitative analysis of CCP dynamics using cmeAnalysis (Aguet et al., 2013) and a newly developed “disassembly asymmetry score classification” (DASC) analysis package to directly measure early stages of CME such as CCP initiation, stabilization, and maturation (Wang, Chen, et al., 2020). We then apply these methods to systematically investigate the domain requirements for their isoform-specific functions by examining the activities of homogenously expressed GTPase, middle, PH, GED, and PRD swap Dyn1/Dyn2 chimeras in ARPE cells after siRNA-mediated KD of endogenous Dyn1 and Dyn2. For PRD1-containing constructs, we examined the activities of both wild-type PRD1 and the nonphosphorylatable, constitutively active S774/778A PRD1 mutant. Together these studies provide structural insight into isoform-selective and nonredundant roles for dynamin during CME.
RESULTS
Dyn2, but not Dyn1, is required for bulk CME in ARPE cells
To study the isoform-specific functions of dynamins in detail, we selected ARPE cells as they express nearly equivalent levels of Dyn1 and Dyn2 isoforms (Dyn2:Dyn1 - 3:1; Supplemental Figure S1A) and predominantly exhibit dynamic, diffraction-limited CCPs with few static clathrin-labeled structures (CLS) when imaged on gelatin-coated coverslips by total internal reflection fluorescence microscopy (TIRFM). To evaluate the contribution of each dynamin isoform to CME, we knocked down endogenous Dyn1 or Dyn2 using 3′UTR siRNA individually, as well as together (see Supplemental Figure S1B for KD efficiency), and measured the uptake efficiency of TfnR, a well-known CME cargo. As previously reported, (Reis, Chen, et al., 2015), whereas siRNA-mediated Dyn2 KD strongly inhibited TfnR uptake efficiency (Figure 1B), Dyn1 KD did not show any effect, despite its abundance in ARPE cells (Supplemental Figure S1B). Moreover, double KD of Dyn1 and Dyn2 inhibited TfnR uptake to a similar extent as single Dyn2 KD. The single Dyn2 KD, as well as Dyn1 and Dyn2 double KD, resulted in a corresponding strong accumulation of surface TfnR (Figure 1C).
Several studies have suggested the existence of both positive and negative feedback loops between dynamin and actin (Ferguson et al., 2009; Taylor et al., 2012). Interestingly, the residual uptake of TfnR observed in the absence of dynamins was sensitive to latrunulin A (LatA) and jasplakinolide (Jasp), inhibitors of actin dynamics, whereas these same compounds enhanced rates of CME in control cells (Figure 1D). Consistent with this differential dependence on actin, we observed an increased recruitment of Tractin-GFP to CCPs in Dyn1/2 double KD cells compared with control cells (Figure 1E). These findings are consistent with known roles for membrane tension in modulating endocytosis efficiency and actin dependence (Boulant et al., 2011; Gauthier et al., 2012). Thus, a decrease in cortical actin might reduce membrane tension in control cells resulting in increased rates of CME. In contrast, actin assembly might be engaged to drive invagination and fission in compensation for loss of the dynamin fission machinery, as occurs during dynamin-independent CME in yeast (Kaksonen et al., 2006).
Dynamin isoforms differentially regulate CCP dynamics
TfnR uptake measures only the end result of CME, that is, vesicle release into the cytoplasm. It does not give information about alterations in the early stages of CME, including CCP assembly, stabilization, and maturation efficiency. Thus, relying only on TfnR uptake to analyze CME function can mask any compensatory changes occurring in the early stages of CME (Aguet et al., 2013; Wang, Chen, et al., 2020). Using more sensitive and informative assays like TIRFM-based detection and tracking of CCP dynamics in live cells can offer detailed insight into possible isoform-specific roles for dynamin in these early regulatory stages of CME.
To visualize CCP dynamics, we used ARPE cells overexpressing mRuby-clathrin light chain A (mRuby-CLCa). Overexpression of mRuby-CLCa does not affect TfnR uptake efficiency and in fact, it down-regulates endogenous CLCa, thus allowing maximum labeling of triskelia for TIRFM analysis (Aguet et al., 2013). ARPE cells treated with Dyn1, Dyn2, or control siRNAs were seeded on gelatin-coated coverslips and imaged by TIRFM as described in Materials and Methods. Quantitative tracking and analysis of the live cell TIRFM movies by cmeAnalysis and DASC (Aguet et al., 2013; Wang, Chen, et al., 2020) revealed the isoform-specific regulation of CCP dynamics. Unexpectedly, cmeAnalysis revealed that KD of Dyn1, but not Dyn2, specifically increased initiation rates of subthreshold CLS (sCLS; Figure 2A). sCLSs are very dim clathrin assemblies that fail to grow past a threshold intensity and instead rapidly turn over as early abortive events. A similar increase in the early stages of CCP dynamics has been observed in response to the expression of AP2 mutants defective in endocytic accessory protein (EAP) recruitment (Aguet et al., 2013) or PI(4,5)P2-binding (Wang, Chen, et al., 2020) and interpreted as a “fail fast” compensatory mechanism that can restore CME, even when downstream events such as CCP stabilization and maturation are impaired (Aguet et al., 2013; Reis, Chen, et al., 2015; Wang, Chen, et al., 2020). Thus, although the rates of TfnR uptake are unaffected, the increase in sCLS initiation rates on Dyn1 KD likely reflects a compensatory mechanism in response to as yet undetected downstream defects in CCP maturation resulting from Dyn1 KD. Dyn2 KD, on the other hand, significantly reduced the rate of initiation of the bona fide CCPs (CLSs above the set intensity threshold) (Figure 2B), suggesting a role for Dyn2 in CCP stabilization and maturation. The lifetime frequency distribution generated by cmeAnalysis showed a characteristic Rayleigh-like distribution for siCtrl cells reflecting a multi-step regulated process (Aguet et al., 2013) that was unaltered in Dyn1KD cells. In contrast, in Dyn2 KD cells the curve was shifted leftward and a corresponding increase in the number of short-lived (<20s), presumably late, abortive pits (Figure 2C). Moreover, the lifetime distribution curve in Dyn2 KD cells exhibited quasi-exponential decay kinetics, which typically corresponds to a stochastic, dysregulated process (Aguet et al., 2013). These data support a role for Dyn2 in regulating early stages of CCP stabilization and maturation.
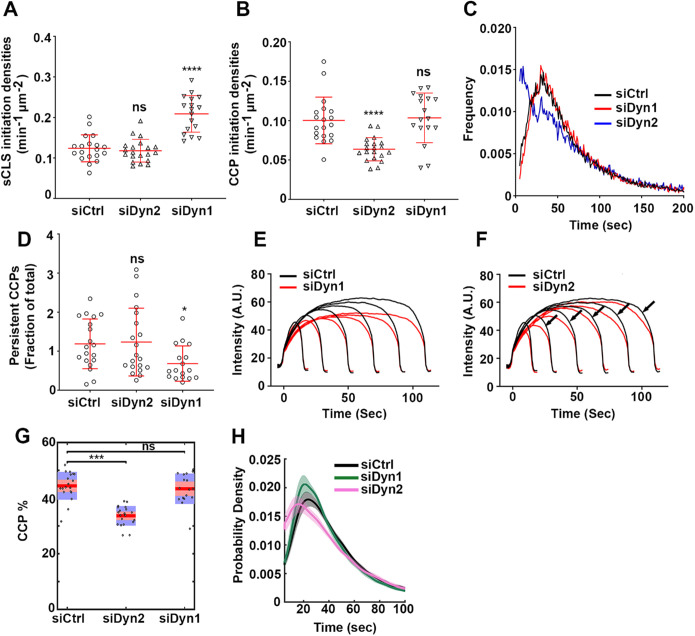
FIGURE 2:
Differential regulation of CCP dynamics by dynamin isoforms. ARPE mRuby-CLCa live cell TIRFM data were analyzed by cmeAnalysis (A–F) or an intensity threshold independent DASC analysis algorithm (G, H). (A–D) Effect of dynamin isoform KD on (A) the rate of initiation of the subthreshold CSs. (B) The rate of initiation of CCPs above threshold. (C) The lifetime distribution of CCPs analyzed by cmeAnalysis, and (D) the proportion of the persistent CCPs; t test was used to calculate the statistical significance of cmeAnalysis data. (E, F) Intensity profiles of lifetime cohorts of mRuby-CLCa in (E) siCtrl vs. siDyn1 cells and (F) siCtrl vs. siDyn2 cells (G) Change in the percentage of CCPs calculated from DASC analysis. Dots represent raw data points from individual movies, box plots show mean as a red line with 95%, and 1 SD as pink and blue blocks, respectively. Wilcoxon rank-sum test was used to calculate the statistical significance of changes in CCP%. (H) Lifetime distribution of CCPs is defined by DASC analysis (number of traces analyzed by cmeAnalysis for siCtrl: 59682; siDyn1:51084; siDyn2: 34284; number of traces analyzed by DASC for siCtrl: 142050; siDyn1: 162768; siDyn2:102856). Data shown are representative of three independent biological repeats.
As dynamin’s function during the fission step is well established, we hypothesized that loss of dynamin might result in an increased proportion of persistent structures because of compromised fission. To our surprise, we did not observe an increase in persistent CCPs after knocking down either Dyn1 or Dyn2 (Figure 2D). However, late effects in CCV formation could be detected by lifetime cohort analysis (Loerke, Mettlen, et al., 2011), which measures clathrin intensity changes during CCP maturation (Figure 2, E and F). Thus, we observed that CCPs in Dyn1 KD cells were dimmer, and presumably smaller, than in control cells (Figure 2E). Interestingly, when compared with siCtrl cells, the intensity cohorts from Dyn2 KD cells showed a slower, more gradual departure phase as evidenced by a gradual decline in clathrin fluorescence intensity prior to its abrupt disappearance corresponding to CCV detachment (arrows in Figure 2F). We interpret this change as reflecting a reduced rate of membrane scission during the inward budding of a CCV.
Although measurements from cmeAnalysis provide important information about CCP dynamics, the need to establish an intensity threshold makes them vulnerable to changes in CCP intensities. Since Dyn1 KD affected the intensities of CCPs, we also analyzed our data using our recently developed, threshold-independent DASC algorithm, which can accurately distinguish CCPs from abortive coats (ACs) despite their overlapping intensities and lifetimes (Wang, Chen, et al., 2020; see Supplemental Figure S2 for a brief desciption of DASC analysis). Using DASC, we calculated the percentage of CCPs (CCPs%) among all detected valid traces, which measures the growth and stabilization of nascent CCPs (Figure 2G). The CCP% was specifically reduced in Dyn2 KD cells but not in Dyn1 KD cells. We also plotted the lifetime distribution of DASC-defined CCPs (Figure 2H) and again observed a leftward shift and an increase in shorter-lived CCPs in Dyn2 KD cells, although the curve retained its Rayleigh-like shape. The cmeAnalysis lifetime curve differs from the DASC-based lifetime distribution curve as it includes a large fraction of very short-lived structures, which are identified by DASC as ACs and are not included when plotting the lifetime distribution curve of CCPs. Nonetheless, we tentatively identify the increase in DASC-defined CCPs with lifetimes of <20 s in Dyn2 KD cells (Figure 2H) as fully assembled, but late abortive CCPs.
Together, the above data on the effects of Dyn2 KD on CCP dynamics directly confirm earlier speculation that Dyn2 functions at the early stages of CME to regulate the CCP maturation (Sever et al., 1999, 2000; Loerke, Mettlen, et al., 2009; Mettlen et al., 2018), in addition to its role in membrane fission.
Dynamin1/2 chimeras reveal important domain-specific differences between isoforms
The above data show that despite their high degree of sequence identity, Dyn1 and Dyn2 have nonredundant functions in CME and cannot compensate for each other. To understand which domains of dynamin confer such differential functions, we studied Dyn1 and Dyn2 domain swap chimeras. Individual domain swap chimeras were generated as HA-tagged, GFP fusion proteins by seamless cloning in a retroviral vector (see Materials and Methods). A list of chimeras used in this study is provided in Supplemental Table S1 and the exact locations of their crossover points are indicated in Supplemental Figure S3. The chimeras were stably expressed in ARPE mRuby-CLCa cells and sorted by FACS for comparable levels of Dyn-eGFP expression, which was confirmed by Western blotting using a pan-dynamin antibody (Supplemental Figure S4, A and B). Despite our best efforts to sort low expressing cells, the expression levels of the dynamin chimera were approximately 10-fold higher than endogenous dynamin expression levels.
The activities of the Dyn1/2 chimeras, compared with comparably expressed Dyn1-eGFP and Dyn2-eGFP, were studied in the absence of endogenous dynamins following KD of endogenous Dyn1 and Dyn2 using 3′UTR-specific siRNA. We first measured the ability of these domain swap chimeras to rescue TfnR uptake efficiency (Figure 3A; see Supplemental Figure S5 for kinetics). The data are expressed relative to the residual TfnR uptake efficiency in Dyn1/2 double KD cells, which was set to 1. Exogenously expressed Dyn2-eGFP was able to rescue TfnR uptake efficiency to 80% of that of control cells. Control experiments established that the incomplete rescue by Dyn2 resulted from a partial dominant-negative effect potentially due to titration of Dyn2 effectors as a result of overexpression. Chimeras of Dyn2 bearing the GTP1, MID1, or GED1 domains of Dyn1 were less effective in restoring TfnR uptake, whereas the PH1-containing chimera was equally effective (Figure 3A, blue bars). Replacing PRD2 with PRD1, on the other hand, reduced the efficiency of TfnR uptake to an even lower level than that observed in the Dyn1/2 double KD cells, suggesting a dominant-negative effect. Interestingly, replacing PH2 with PH1 (i.e., Dyn2PH1PRD1) brought the uptake efficiency back to the level of Dyn1/2 double KD cells, suggesting that PH2 might have a role in this dominant-negative effect.
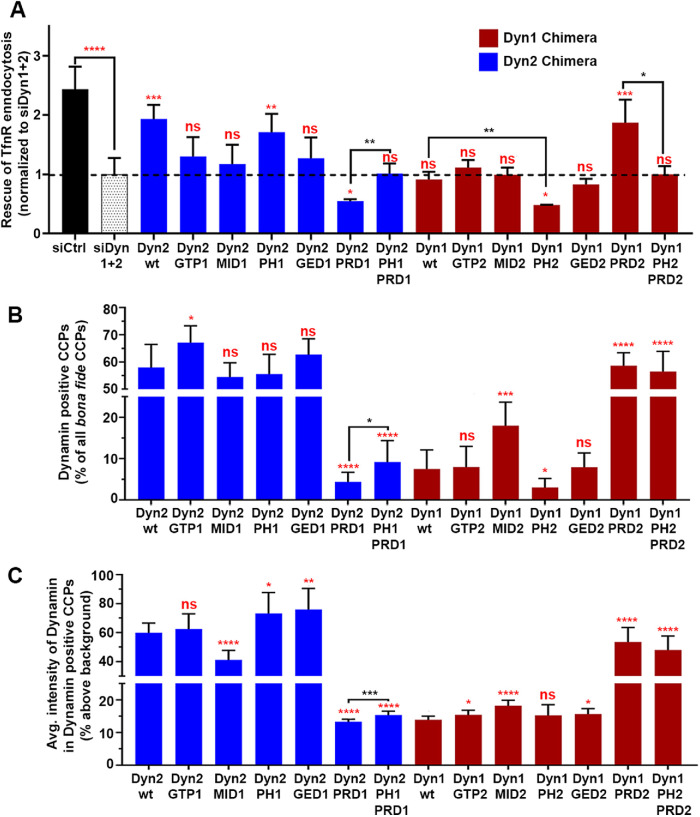
FIGURE 3:
Domain swap chimeras reveal important functional differences between dynamin isoforms. (A) The ability of dynamin chimera to rescue TfnR uptake in the absence of endogenous Dyn1 and Dyn2 compared with residual TfnR uptake in Dyn1+2 double KD cells (indicated by the dashed line). Red asterisks indicate statistical significance of CME rescue by dynamin chimera when compared with residual CME in Dyn1+2 double KD cells; black asterisks denote statistical significance of CME rescue by dynamin chimera when compared with each other. Data shown are average ± SD (n = 3 independent biological repeats). (B) Quantification of the percentage of Dyn-positive CCPs from dual-color TIRFM images of mRuby-CLCa cells expressing eGFP fusions of dynamin domain swap chimera in the absence of endogenous Dyn1 and Dyn2. (C) Quantification of average Dyn-eGFP intensity in Dyn-positive CCPs from dual-channel TIRF images in the absence of endogenous Dyn1 and Dyn2. The number of CCPs analyzed in B and C is 18,000 CCPs from 40–50 cells/condition in single imaging experiment. For B and C, red asterisks indicate statistical significance when Dyn1 and Dyn2 chimera are compared with their respective wild-type controls. Black asterisks are used when two chimeras are compared with each other. Error bars represent SD; t test was used to analyze statistical significance.
Consistent with the lack of functional redundancy, even when overexpressed, Dyn1-eGFP was unable to rescue TfnR uptake (Figure 3A, maroon bars). Chimeras containing GTP2, MID2, or GED2 were still unable to rescue TfnR uptake. The PH2-containing chimera resulted in further reduction of residual TfnR uptake efficiency, again suggesting a dominant-negative effect played by PH2. Strikingly, the Dyn1 chimera bearing PRD2 (Dyn1PRD2) was able to rescue TfnR uptake to a similar level as wtDyn2. In this context, replacing PH1 with PH2 (i.e., Dyn1PH2PRD2) again reduced TfnR uptake efficiency showing a detrimental effect of the presence of PH2. Together these data confirm previous findings in triple null mouse embryo fibroblasts (Liu, Neumann, et al., 2011) that PRD2 and PH1 play positive roles during CME, whereas PH2 plays a negative role.
Previous studies in different cell lines have shown the differential ability of dynamin isoforms to localize at CCPs (Liu et al., 2008; Srinivasan, Burckhardt, et al., 2018) and this property has been ascribed to the PRD of Dyn2 in the fibroblasts (Liu, Neumann, et al., 2011). Hence, we next checked whether the ability of dynamin chimeras to rescue TfnR uptake correlated with their ability to localize at CCPs. To this end, ARPE mRuby-CLCa cells expressing the dynamin chimera-GFP fusions were imaged by TIRFM and the GFP intensity was measured at mRuby–CLCa-labeled CCPs identified using the highly sensitive cmeAnalysis detection software (see Materials and Methods for details). At least 18,000 bona fide CCPs were analyzed from 10 images/condition (from 40–50 cells). We calculated the percentage of dynamin-positive CCPs (Figure 3B) as well as the intensity of dynamin in Dyn-positive CCPs (Figure 3C). As CCPs are dynamic structures, the former is dependent on the duration of dynamin association throughout the lifetime of a CCP, while the latter reflects the levels of recruited dynamin at individual CCPs captured at one time point during their lifetimes. Together these two measurements capture complementary aspects of the dynamic association of dynamin at CCPs.
As was previously observed in fibroblasts (Liu, Neumann, et al., 2011), PRD2 was the major determinant of efficient localization of dynamin chimeras to CCPs both in terms of %Dyn-positive CCPs (Figure 3B) and average Dyn intensity at Dyn-positive CCPs (Figure 3C). Chimeras expressing PRD2 uniformly show a clear punctate localization, whereas chimeras expressing PRD1 show diffused membrane localization with very few puncta (Supplemental Figure S6). Quantification of eGFP intensities revealed that in dynamin chimeras bearing PRD2, the dynamin-GFP signal colocalized with the mRuby-CLCa signal at 55–70% CCPs, (i.e., Dyn-positive CCPs), whereas in cells expressing chimeras bearing PRD1, only 4–20% of CCPs are Dyn-positive (Figure 3B). PRD2 also increased dynamin intensities at Dyn-positive CCPs by at least fourfold (Figure 3C).
Interestingly, the addition of PH1 to Dyn2PRD1, that is, Dyn2PH1PRD1 significantly increased the percentage of Dyn-positive CCPs, as well as the intensity of dynamin at Dyn-positive CCPs. Dyn2PH1 also showed increased average Dyn intensity in Dyn-positive CCPs compared with Dyn2wt. Conversely, replacing PH1 with PH2 further decreased the percentage of Dyn-positive CCPs in Dyn1PH2 compared with Dyn1wt. While not a major targeting determinant, these data suggest that the Dyn1 PH domain might stabilize dynamin’s association with CCPs. Surprisingly, Dyn1MID2 showed a significant increase in the percentage of Dyn-positive CCPs as well as the average Dyn intensity at Dyn-positive CCPs compared with Dyn1wt; whereas Dyn2MID1 showed a significant decrease in Dyn intensity at Dyn-positive CCPs but no significant change in the percentage of Dyn-positive CCPs. Both findings suggest a positive role for MID2 in regulating dynamin–CCP interactions. As the middle domain functions in dynamin assembly (Gao et al., 2010) and also as a docking site for intramolecular regulation of dynamin assembly through PH domain interactions (Kenniston and Lemmon, 2010; Reubold, Faelber, et al., 2015; Srinivasan et al., 2016), alterations in either or both functions could affect the lifetime and/or extent of dynamin interactions at CCPs. No other domains consistently affected dynamin–CCP interactions.
PH1 contributes to CCP stabilization
The presence of PH1 improved the ability of dynamin chimeras to rescue TfnR uptake as well as their association with CCPs, whereas the presence of PH2 had the opposite effects. In vitro studies of purified dynamin isoforms have shown that Dyn1 and Dyn2 have considerable differences in their curvature generation and sensing abilities: Dyn1 is a potent curvature generator, whereas Dyn2 is a curvature sensor (Liu, Neumann, et al., 2011). To understand the differential CME functions of the PH domain chimeras, we focused on pairs of domain swap chimeras that differed only in their PH domain, namely, Dyn1PRD2 versus Dyn1PH2PRD2, Dyn2PRD1 versus Dyn2PH1PRD1, and wtDyn2 versus Dyn2PH1. Following KD of endogenous Dyn1 and Dyn2, these cells were imaged to acquire single-channel mRuby-CLCa movies using TIRF microscopy. The movies were analyzed by DASC to measure rates of initiation of all clathrin-coated structures (CSs) and the %CCPs. Replacement of PH1 with PH2 in Dyn1PRD2 strongly decreased the percentage of bona fide CCPs (CCP%), which is a measure of CCP stabilization (Figure 4B) without affecting the rate of initiation of all CLS (Figure 4A). Conversely, the replacement of PH2 with PH1 in Dyn2PRD1 significantly increased CCPs% (Figure 4D) without affecting the rate of initiation of all CSs (Figure 4C). We did not, however, detect an effect of swapping PH1 into wtDyn2, indicating that the PH domain functions may be less critical when other domain requirements are met (Figure 4, E and F).
FIGURE 4:
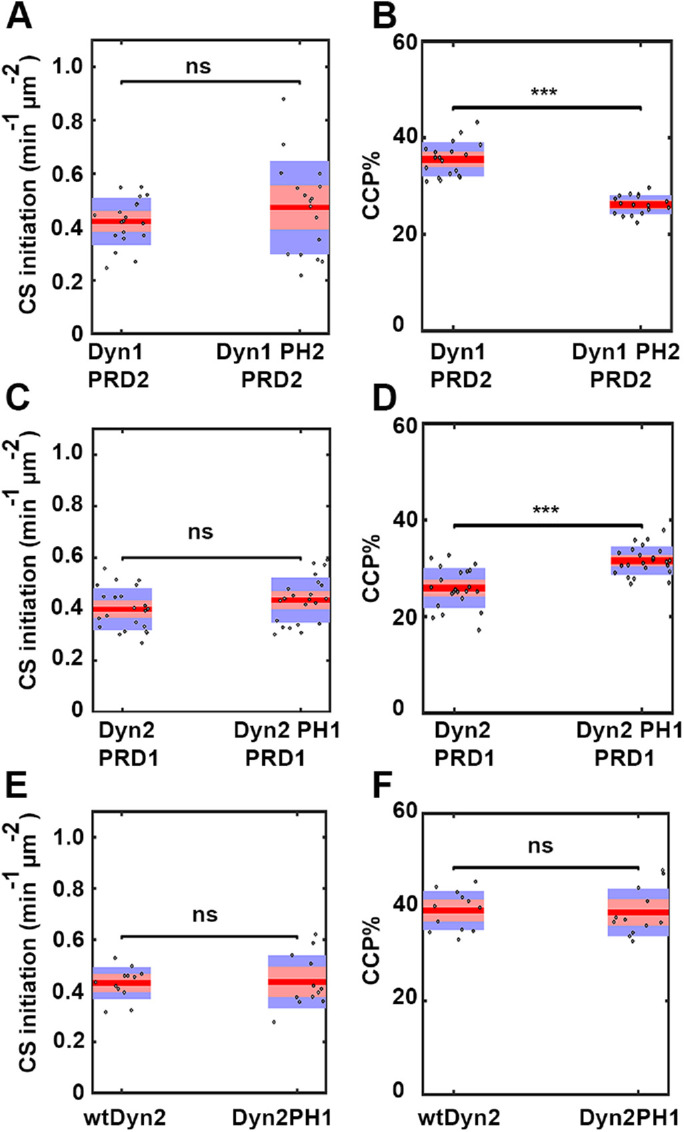
PH domains of dynamin isoforms play a differential role during CCP stabilization. DASC analysis of live cell TIRFM data from ARPE mRuby-CLCa cells stably overexpressing PH domain chimeras in the absence of endogenous Dyn1 and Dyn2. (A, C, E) Changes in the rate of initiation of all CSs. (B, D, F) Changes in the percentage of CCPs (CCP%). Dots represent raw data points from individual movies, box plots show mean as a red line with 95%, and 1 SD as pink and blue blocks, respectively. Wilcoxon rank-sum test was used to calculate statistical significance. The number of traces analyzed for above data (obtained from 1 experiment representive of 2–3 biological repeats) are: Dyn1PRD2-113430, Dyn1PH2PRD2-81527, Dyn2PRD1-114941, Dyn2PH1PRD1-162878, wtDyn2-69724, Dyn2PH1-65409.
Dephosphorylated and activated Dyn1 chimeras show enhanced ability to rescue CME
Previous studies in neurons (Liu et al., 1994a, b; Armbruster et al., 2013) and in nonneuronal cells (Reis, Chen, et al., 2015; Srinivasan, Burckhardt, et al., 2018) have shown that activation of Dyn1 by dephosphorylation of S774 and S778 is important for triggering and accelerating CME.
To determine whether dephosphorylated Dyn1 can rescue CME in ARPE cells in the absence of endogenous Dyn1 and Dyn2, we mutated Ser774 and Ser778 by site-directed mutagenesis to alanine (here onward referred to as AA chimera). ARPE mRuby-CLCa cells were transduced with retroviruses encoding the nonphosphorylatable Dyn1 chimeras. Cells were then FACS sorted for homogeneous eGFP signal and checked by Western blot to confirm equal expression levels of the wild-type and dephosphorylated chimeras (Supplemental Figure S7, A and B). We next measured their ability to rescue TfnR uptake after siRNA-mediated KD of endogenous Dyn1 and Dyn2. As expected, all of the dephosphorylated Dyn1 chimeras showed enhanced ability to rescue CME compared with their wild-type versions (Figure 5A). Moreover, the ability of Dyn1AA, Dyn2PRD1AA, and Dyn2PH1PRD1AA to rescue CME in the absence of endogenous dynamins was close to the level of rescue by wtDyn2. Also consistent with data in Figure 3, substitution of PH1 for PH2 increased the chimera’s ability to restore CME (compare Dyn2PRD1AA with Dyn2PH1PRDAA), while substitution of PH2 for PH1 (compare Dyn1AA with Dyn1PH2AA) had the opposite effect.
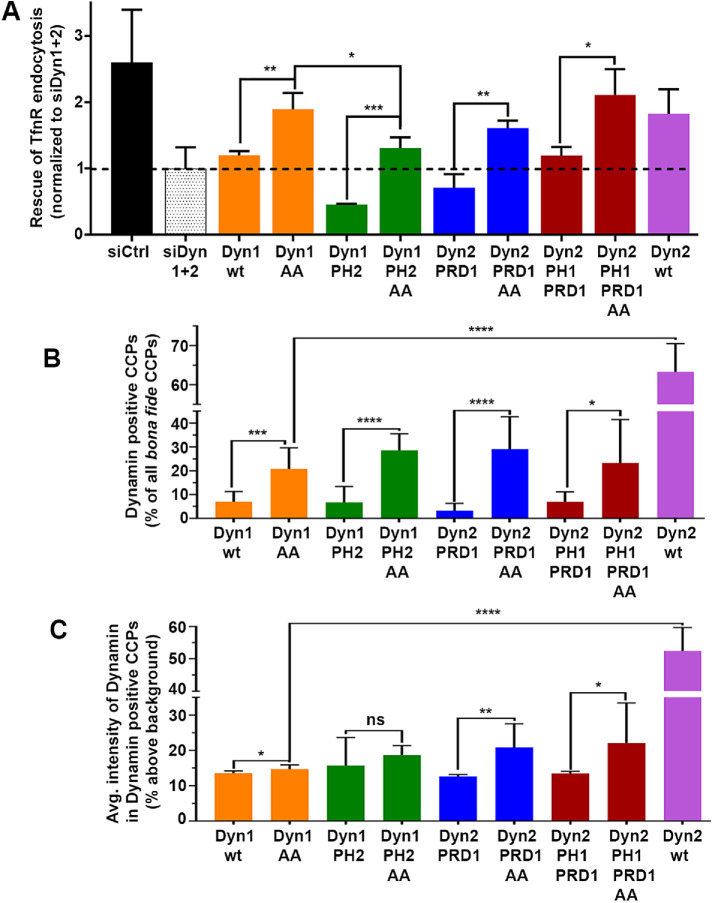
FIGURE 5:
Constitutively activating PRD1 enhances CME efficiency as well as dynamin recruitment at CCPs. (A) The ability of wild-type and constitutively active (S774/778A) dynamin chimera to rescue TfnR uptake in the absence of endogenous Dyn1 and Dyn2. Data shown are average ± SD (n = 3 independent biological repeats). (B) Quantification of the percentage of Dyn-positive CCPs from dual-color TIRF images of mRuby-CLCa cells expressing eGFP fusions of wild-type or constitutively active dynamin domain swap chimera in the absence of endogenous Dyn1 and Dyn2. (C) Quantification of average Dyn-eGFP intensity in Dyn-positive CCPs from dual-channel TIRF images in the absence of endogenous Dyn1 and Dyn2. The number of CCPs analyzed in B and C is 19,000 CCPs from 40–50 cells/condition in single imaging experiment. The error bars represent SD; t test was used to calculate the statistical significance.
Dephosphorylation of S774,778 in Dyn1 PRD could alter its interaction with the SH3 domain-containing binding partners required for recruiting dynamin to CCPs. We therefore measured the association of these dephosphorylated dynamin chimeras with CCPs by dual-color TIRFM. At least 19,000 bona fide CCPs were analyzed from 10 images/condition (total 40–50 cells). The constitutively active versions of Dyn1 chimeras were indeed recruited more efficiently to CCPs as seen by enhanced punctate localization of the AA chimeras as compared with diffused membrane localization of the wild-type versions of the same chimeras (Supplemental Figure S7C). Quantification of intensities of Dyn chimeras at CCPs showed that the percentage of Dyn-positive CCPs as well as the average dynamin intensity at Dyn-positive CCPs were significantly greater for chimeras bearing the dephosphorylated PRD1 (Figure 5, B and C). However, the increased recruitment of AA chimeras at CCPs was still significantly lower (∼3-fold fewer Dyn-positive CCPs and ∼3-fold lower intensity of Dyn chimera per Dyn-positive CCP) than recruitment of Dyn2wt. Thus, dephosphorylated PRD1-containing chimeras were able to rescue TfnR uptake in dynamin-depleted ARPE cells, even though they were recruited less efficiently to CCPs as compared with Dyn2 chimera. This indicates that when activated by dephosphorylation, the “specific activity” of Dyn1 at CCPs may be greater than that of Dyn2.
The PRD determines differential recruitment kinetics of Dyn1 and Dyn2 to CCPs
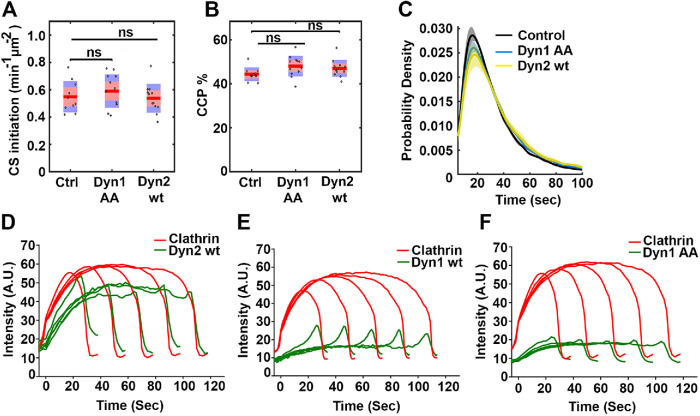
To further characterize the dephosphorylated Dyn chimeras, we next analyzed CCP dynamics using live cell TIRFM and DASC analysis. Consistent with the ability of Dyn1AA to restore CME in dynamin-deficient cells as efficiently as Dyn2, none of the parameters of CCP dynamics, such as CS initiations (Figure 6A), CCPs% (Figure 6B), or lifetime distribution (Figure 6C) showed significant differences. This was unexpected given that only ∼20% of CCPs exhibit detectable Dyn1AA-eGFP at steady-state (Figure 5B) These data suggest that even very low or very transient association of Dyn1AA is sufficient to drive efficient CME.
FIGURE 6:
Early recruitment of dynamin at CCPs correlates with successful CME. DASC analysis (A–C) or cmeAnalysis (D–F) results of live cell TIRFM data from ARPE mRuby-CLCa cells overexpressing the indicated eGFP-tagged dynamin isoforms in the absence of endogenous Dyn1 and Dyn2 compared with nonoverexpressing control cells (without KD of endogenous dynamins) (A) Changes in the rate of initiation of all clathrin-coated structures. (B) Change in the percentage of CCPs. Wilcoxon rank-sum test was used to calculate statistical significance. (C) The lifetime distribution of CCPs (number of traces analyzed for control: 90950; Dyn1AA: 103000; Dyn2wt: 91029). (D–F) Intensity cohorts for recruitment of mRuby2-CLCa (red, primary channel) and Dyn-eGFP (green, secondary channel) at CCPs in ARPE mRuby-CLCa cells overexpressing (D) Dyn2wt-eGFP, (E) Dyn1wt-eGFP of (F) Dyn1AA-eGFP (number of traces analyzed for Dyn2wt: 26771; Dyn1wt: 39022; Dyn1AA: 33075). Data are representative of three independent biological repeats.
The colocalization studies described above represent a single snapshot of dynamin’s recruitment to CCPs. Therefore, we next used two-color live cell TIRFM to measure the average kinetics of recruitment of Dyn2wt, Dyn1wt, and Dyn1AA chimera relative to the kinetics of mRuby-CLCa recruitment to CCPs for all lifetime cohorts (Figure 6, D–F, respectively). With the caveat that these dynamin constructs are overexpressed relative to endogenous, we nonetheless observed differential kinetics of recruitment. Dyn2wt appears early after CCP initiation and accumulates along with clathrin followed by a slight burst prior to CCV detachment. In contrast, Dyn1wt exhibits the characteristic burst of recruitment at late stages as described by others for cells overexpressing Dyn1 (Merrifield et al., 2002; Ehrlich et al., 2004; Soulet et al., 2005; Taylor et al., 2011). Interestingly the Dyn1AA chimera, although recruited to a lesser extent than Dyn2wt, is recruited early to CCPs along with clathrin. The observation that Dyn1AA and Dyn2wt can both rescue TfnR uptake and are present at CCPs from early stages again supports an essential early function(s) for dynamins to maintain CME in these cells.
DISCUSSION
Using a combination of traditional biochemical assays and state-of-the-art quantitative and analytical TIRFM, we show that Dyn1 and Dyn2 are not functionally redundant and their depletion selectively perturbs distinct, early stages of CME. By systematically comparing the activities of domain swap chimeras, we then identify domains that contribute to the isoform-specific functions of dynamins. In addition to confirming the previously reported functional differences in PH and PRD domains, we also report previously undetected functional differences in GTPase, middle, and GED domains of dynamin isoforms. While PRD2 plays a dominant role in efficiently targeting Dyn2 to CCPs, our study also highlights the importance of PH1, MID2, and dephosphorylated PRD1 in regulating the association of dynamin with CCPs. We also report that PH1 plays an important role in CME by stabilizing nascent CCPs. Finally, our studies support accumulating evidence that both dynamin isoforms are required for early stages of CME and that early effects on CCP stabilization and maturation, which precede the inhibition of late fission events, appear to account for the defect in CME in dynamin-deficient cells.
The TfnR uptake studies indicated that ARPE cells completely depend on Dyn2 for CME and that in the absence of dynamin, cells rely on actin for residual CME function. As expected, the increased dependence on actin dynamics for CME was observed on individual KD of Dyn2, but not on Dyn1 KD (data not shown). LatA sequesters actin monomers (Yarmola et al., 2000) as well as accelerates depolymerization of actin filaments (Fujiwara et al., 2018). Jasp is known to stabilize and induce polymerization of actin filaments (Bubb et al., 1994) and formation of actin aggregates in cells (Lee et al., 1998). Jasp also nucleates actin filaments de novo. Together, these effects deplete the monomeric pool of actin and consequently inhibit de novo actin filament assembly. Thus, it is not surprising that LatA and Jasp have similar effects on TfnR uptake (Bubb et al., 2000). Others have shown that actin is required for CME in mammalian cells only under conditions of high membrane tension (Boulant et al., 2011), or for the uptake of large cargoes (Cureton et al., 2010). Here we show that disruption of the dynamin fission machinery also results in a greater dependence on actin for CME.
Detailed TIRFM analysis of CCP dynamics using mRuby–CLCa-labeled ARPE cells confirmed previous speculation regarding the early function of Dyn2 during CME. Our data suggest that in the absence of Dyn2, CCP stabilization is greatly reduced and CCPs follow a quasi-exponential lifetime distribution supporting a role for Dyn2 as a fidelity monitor of CME (Loerke, Mettlen, et al., 2009; Mettlen et al., 2009; Aguet et al., 2013). Thus, in Dyn2 KD cells it is likely that most of the CCPs are unable to mature and reach the final fission step. Interestingly, those that complete the maturation appear to be more dependent on actin and undergo a slower departure phase prior to fission. Our data suggest that the strong inhibition of TfnR uptake in the absence of Dyn2 is a combination of early (reduced CCP stabilization) as well as widely accepted late-stage defects (delayed departure phase in Dyn2 KD cells). Consistent with an early role for Dyn2 and Dyn1AA in CME, both are recruited early to nascent CCPs, whereas the low level of recruitment of Dyn1wt occurs only at late stages. The mechanisms by which Dyn2 functions during early stages to stabilize nascent CCPs remain unknown but could relate to its ability to interact with multiple essential SH3 domain-containing EAPs via its PRD. Interestingly, we also detected a requirement for the G domain, which has been shown to interact with auxilin (Newmyer et al., 2003) and may regulate early stages of coat assembly (Chen et al., 2019). Further studies are needed to test these speculations.
When activated by dephosphorylation, Dyn1 can rescue CME as efficiently as Dyn2wt, even when recruited to CCPs much less efficiently than Dyn2. Thus, the lack of effect of Dyn1 KD on CME in ARPE and other nonneuronal cells (Liu et al., 2008) is likely not due to its lower levels of expression (∼1/3 of Dyn2 expression in ARPE cells), but rather that Dyn1 is constitutively inactivated by phosphorylation of S774/778 in PRD1. The fact that Dyn1 is detected in virtually all tissues begs the question: under what conditions is Dyn1 activated by dephosphorylation of its PRD? Studies of Dyn1 KO mice, which are born but die postnatally, have shown that under moderate neuronal activity, synaptic connections are still able to form and function, but that high levels of Dyn1 are essential under conditions of high neuronal activity due to an increased load of endocytosis under these conditions (Ferguson et al., 2007; Lou et al., 2008). Similarly, although Dyn1 is not required for CME in ARPE cells under control conditions, when CCP maturation is impaired, for example, on expression of a truncated α-adaptin subunit defective in EAP recruitment (Aguet et al., 2013), Dyn1 is activated by dephosphorylation of its PRD and is able to restore CME (Reis, Chen, et al., 2015). These observations support the idea that Dyn2 performs a housekeeping role in CME and that Dyn1 can be activated by dephosphorylation of its PRD to sustain the essential CME functions under conditions of stress. Interestingly, up-regulation of CDK5, the kinase thought to phosphorylate S778, creating a recognition site for the inhibitory phosphorylation of S774 by constitutively active GSK3β (Tan et al., 2003; Clayton et al., 2010), is associated with many diseases, including diabetes, immune responses, and angiogenesis among others (Sharma and Sicinski, 2020). While CDK5 has many substrates, whether inactivation of Dyn1 has a role in any of these disease phenotypes remains to be determined. Moreover, whether CDK5 is responsible for Dyn1 phosphorylation at S778 in nonneuronal cells has yet to be demonstrated. On the other hand, recent studies have shown rewiring of CME by selective dephosphorylation and activation of Dyn1 in a cargo-specific manner, as well as under specific cellular conditions like cancer progression (Reis, Chen, et al., 2015; Chen et al., 2017; Reis, Chen, et al., 2017; Schmid, 2017; Srinivasan, Burckhardt, et al., 2018; Lakoduk et al., 2019). Future studies will be necessary to determine under what other conditions (e.g., in response to changes in membrane tension, or for the engulfment of larger cargo molecules or particles) Dyn1 can be activated by dephosphorylation of its PRD to maintain and/or accelerate CME.
Dynamin domain swap chimeras highlighted important positive and negative roles of PH1 and PH2 domains, respectively, on the recruitment of dynamin to CCPs, as well as their ability to rescue CME in the absence of endogenous dynamins. Dyn1 has a conserved Tyr at residue 600 in variable loop3 of PH1, whereas Dyn2 has a Leu at that same position. Strikingly, single amino acid substitutions—Y600L in Dyn1 or L600Y in Dyn2—were shown to completely reverse the differential fission and curvature generating activities of the two dynamin isoforms (Liu, Neumann, et al., 2011). The aromatic ring of tyrosine can insert into the lipid bilayer, while its hydroxyl moiety interacts with lipid headgroups. Thus, this tyrosine could contribute to tighter membrane association accounting for the enhanced ability of PH1-containing chimeras to be recruited to and stabilized on CCPs, as well as enhanced curvature generation, which has been shown to be required to stabilize nascent CCPs (Wang, Chen, et al., 2020). More direct experiments of the effects of wild-type and mutant PH1-containing chimeras on curvature generation during CCP maturation would be necessary to test this hypothesis (Liu, Neumann, et al., 2011).
We observed that the ability of dynamin chimeras to rescue TfnR uptake is positively correlated with their recruitment to CCPs, which is largely determined by the PRD (Liu, Neumann, et al., 2011). For Dyn1, recruitment to CCPs and its ability to rescue TfnR uptake are regulated by phosphorylation/dephosphorylation of S774/778 in its PRD. These data suggest that at least some of the motifs in PRD1 important for the recruitment to CCPs are located near S774, S778. Indeed, previous studies have shown that the phosphorylation status of Dyn1 at S774 and S778 determines its ability to interact with syndapin1 (Anggono et al., 2006; Cheung and Cousin, 2019). During synaptic vesicle endocytosis, endophilin1 and syndapin1 bind to an overlapping sequence in PRD1 that includes these phosphorylation sites (amino acids 778–796 and 772–796, respectively) (Anggono and Robinson, 2007). Which SH3 domain-containing protein(s) function in recruiting dynamin to CCPs remains to be unambiguously determined.
Recent studies in nonneuronal cells point to GSK3β as a kinase responsible for Dyn1 phosphorylation at S774 (Reis, Chen, et al., 2015). However, the ratio of phosphorylated:dephosphorylated Dyn1 at steady state in nonneuronal cells is not known. Whether dephosphorylated Dyn1 is enriched at a specific location and/or population of CCPs is also not known. From studies of synaptic vesicle endocytosis, it is clear that Dyn1 is constitutively maintained in a phosphorylated state, dephosphorylated briefly on nerve terminal depolarization-dependent calcium influx (Cousin and Robinson, 2001), and rephosphorylated immediately after stimulus loss (Tan et al., 2003). Other phosphorylation sites identified in the PRD of Dyn1 include S822, S851, and S857 (Graham et al., 2007), and we have preliminary evidence suggesting the phosphorylation of Dyn1 by Dyrk1A at S857, which regulates its interaction with amphiphysin and Grb2 (Huang et al., 2004), also regulates its recruitment to CCPs (Srinivasan and S.L.S., unpublished results). It will be interesting to identify binding partners of Dyn2 and phosphorylation status-dependent binding partners of Dyn1 that are essential for their differential regulation of CME.
Since its discovery almost 30 years ago (van der Bliek and Meyerowitz, 1991; Vallee, 1992), dynamin’s essential role in CME has been extensively studied, with a focus on membrane fission. Our findings suggest that there exist many unknown aspects of dynamin function that remain to be understood.
MATERIALS AND METHODS
Cell culture and generation of stable Dynamin chimera cell lines
The normal human retinal pigmental epithelial-19 (herein referred to as ARPE) cell line was acquired from ATCC and cultured in DMEM/F12 (1:1) (Life Technologies 11330-032) supplemented with 10% fetal bovine serum (FBS; Sigma F0926). ARPE19 cells were infected with pLVX-Puro-mRuby2-CLCa lentiviral expression vector (Srinivasan, Burckhardt, et al., 2018) followed by FACS sorting to select mRuby–CLC-expressing cells. Incorporation of mRuby-CLCa in complex with clathrin heavy chain was confirmed by immunoprecipitation of clathrin heavy chain and analyzing the percentage of mRuby-CLCa pulled down with clathrin heavy chain. ARPE mRuby-CLCa cells were then used to generate stable dynamin chimera cell lines. All the dynamin chimeras (Supplemental Table S2) were cloned in a retroviral expression vector pMIEG3 using seamless cloning. The seamless cloning is a method that uses PCR to amplify the insert. The insert and the vector fragment are then treated with exonuclease to create overhangs. This is followed by treatment with a ligase or in vivo repair in bacteria to allow the joining of insert and vector. Such cloning method is scarless and sequence independent compared with the restriction enzyme-based traditional cut–paste method. pMIEG3 is a modified form of pMIG vector (Williams et al., 2000) with enhanced GFP fluorescence. For the purpose of this study, the pMIEG3 backbone was modified to insert an N-terminal HA tag and the IRES was removed to make C-terminal GFP fusions of dynamin chimeras. To achieve this, dynamin chimera vectors were PCR amplified using the primers listed in Supplemental Table S2 so as to exclude the IRES region and generate a PCR product with homologous overhanging regions to allow for subsequent homologous recombination when transformed in competent bacterial cells. The removal of IRES was confirmed by sequencing. ARPE CLCa-mRuby cells were transduced with retroviral dynamin chimera constructs and the cells were FACS sorted to select GFP-positive cells, while maintaining CLCa-mRuby expression. The constitutively active Dyn1 chimera plasmids were generated by site-directed mutagenesis of wild-type chimeras to replace serine at positions 774 and 778 to alanine (here onward referred to as AA chimeras) using primers listed in Supplemental Table 2 and confirmed by sequencing. Expression levels of chimera were checked by Western blotting using pan-dynamin antibody 4003 (in-house Rabbit).
siRNA KD
We used lipofectamine RNAi Max (ThermoFisher Scientific 13778150) transfection reagent for transfecting all siRNAs. As a control siRNA for all our experiments, we used Allstars negative control siRNA (Qiagen 1027281). The siRNA sequences used in this study to KD endogenous dynamin isoforms were designed against 3′UTRs of Dyn1 and Dyn2 specifically so as not to affect the expression of the overexpressed dynamin chimera. To maximize KD efficiency, a siRNA mixture of two different targeting sequences was used to knock down each of the endogenous dynamin isoforms. Dyn1 3′ UTR siRNA sense used were (1) 5′-CAUCUGUGACUUAAUCUGUUGUAGT-3′ and (2) 5′-GGUGAGCUGAUACAUUCAGGUGUGA-3′. Dyn2 3′ UTR siRNA sense used were (1) 5′-CUGCACUCUGUAUUCUAUUAAUAAA-3′ and (2) 5′-GGUAUAUCAACUUCCCAUUAGCAGG-3′. All were obtained from IDT. The day before adding siRNA (day 0), 280,000 cells/well of a 6-well plate were seeded for each condition. Cells were treated with two rounds of siRNA treatment on day 1 and day 3. On the day of siRNA addition, the existing medium was replaced with fresh medium 30 min before siRNA addition; 100 µl of OptiMEM containing 1 μl each of Dyn1 and Dyn2 siRNA mix from 50 μM stock of siRNA were added to 100 μl OptiMEM containing 6.5 μl of transfection reagent. The mixture was gently vortexed and incubated at RT for 20–30 min. This mixture was then pipetted up and down gently before adding drop by drop to the cells. Cells were incubated for 5–6 h at 37°C before the medium was replaced with fresh medium. The KD efficiency was checked by Western blotting. Antibodies used were Rabbit monoclonal anti-Dyn1 (Abcam ab52852) and Goat polyclonal anti-Dyn2 (Santa Cruz SC6400).
TfnR endocytosis assay
In cell ELISA assays were used to determine the efficiency of TfnR uptake as described previously (Reis, Chen, et al., 2015) using anti-TfnR mAb HTR-D65 (Schmid and Smythe, 1991) as ligand. Cells were seeded at 70–80% confluency in 96-well Stripwell plates (Costar 9102) a day before the experiment. On the day of the experiment, cells were starved in phosphate-buffered saline (PBS)4+ (PBS supplemented with 1 mM MgCl2, 1 mM CaCl2, 5 mM glucose, and 0.2% bovine serum albumin) by incubating at 37°C for 30 min in the CO2 incubator. After starvation, PBS4+ was removed and cells were treated with ice-cold 5 µg/ml D65 diluted in PBS4+ and then transferred to a 37°C water bath for indicated time points (herein referred to as the uptake wells). Simultaneously, some of the cells were treated with prechilled 5 µg/ml D65 in PBS4+ and incubated at 4°C to measure the amount of TfnRs on the cell surface (herein referred to as the totals) and some as a negative control (blanks). For the uptake wells, after the incubation time was over, the ligand was the removed and immediately replaced with cold PBS4+ and the cells were transferred to 4°C to stop the uptake, after which remaining surface-bound D65 (for uptake and blank wells only) was removed by washing with a cold acetic acid solution (0.2 M acetic acid, 0.2 M NaCl, pH 2.5). All the cells (uptake, blank, and totals) were then washed with cold PBS4 and fixed with prechilled 4% PFA. After fixation, cells were permeabilized with 0.1% Triton X100 for 10 min followed by blocking with 5% powdered milk for 1 h at RT. To detect surface-bound and internalized D65, cells were then treated with 1:5000 dilution of the secondary antibody against D65 (HRP–conjugated goat anti-mouse from ThermoFisher G21040) overnight at 4°C. The signal was developed by adding the OPD solution (25 ml of Citrate buffer, pH 5, 10 mg OPD [Sigma P1526], 10 µl of 30% H2O2 [Alfa Aesar L14000]). After the color was developed, 5 M H2SO4 was added to stop the reaction. Absorbance was read at 490 nm using BioTek Synergy H1 Hybrid Reader. To avoid errors due to loss of cells in different wells, the BCA assay was performed in the same wells and BCA reading was used to normalize the OPD reading. TfnR endocytosis efficiency was plotted as the percentage of internalized TfnR over the initial total surface-bound TfnR.
TIRFM
The siRNA-treated cells were seeded on gelatin-coated 22 × 22-mm coverslips (Corning 2850-22) overnight. Before imaging, cells were checked for appropriate spreading. Fresh media was added 30 min before starting the imaging. Coverslips were mounted on glass slides and sealed using Valap. During TIRF imaging sessions, cells were maintained at 37°C in DMEM/F12 1:1 with 10% FBS. TIRF imaging was performed using motorized Nikon Eclipse Ti-E inverted microscope with 60× Nikon 1.49 NA TIRF DIC objective. The microscope was also equipped with an integrated second-generation Perfect Focus System (Nikon) and an additional 1.8× tube lens which resulted in total magnification of 108×. TIRF illumination was generated using the Diskovery platform (Andor) with motorized laser incident angle adjustment. Illumination was obtained using Andor laser launch with 50 mW 488 nm, 50 mW 560 nm solid state smart lasers. The microscope was also equipped with ASI controller for ultra-fast excitation and emission filter wheels as well as 2 × PCO-Edge 16 bit, 100 fps, 2560 × 2160 px sCMOS cameras for image acquisition, OKO lab custom built full body environmental chamber for temperature control and CO2 stage incubator operated by Bold Line controller. The penetration depth of 80 nm was used for all TIRF imaging experiments. Time-lapse movies for all conditions were acquired as 7.5-min movies with 1-s intervals between frames.
TIRFM analysis and quantification of CCP dynamics
For TIRFM, 30–40 cells from 10–12 movies were imaged/condition per experiment. cmeAnalysis detects all CLSs irrespective of their size/intensity due to its sensitive detection algorithm (Mettlen and Danuser, 2014). Since single triskelia can bear as many as three fluorescently tagged CLCa, along with detecting bona fide CCPs, cmeAnalysis also detects the spurious nucleation and assembly of only a few triskelia, which provide enough signal for successful detection by most modern imaging platforms. Consequently, these dim and transient sCLSs dominate the total detections. To be considered a bona fide CCP, our software package requires that 1) structures are detected for at least five consecutive frames (i.e., 5 s); 2) within these 5 s, structures are detected within at least three frames (our software package is equipped with a powerful gap-closing algorithm); and 3) grow above the user-defined intensity threshold (i.e., above a certain size). Bona fide CCPs that meet these criteria and mature beyond the threshold intensity represent 30–40% of all detections (this number may vary based on the cell line being used). To reduce the effects of day-to-day variability in signal intensities, controls and conditions were imaged on the same day and the same intensity threshold was applied to all datasets for cmeAnalysis. Briefly, using the control dataset, an intensity threshold was applied which segregates all detected valid traces into sCLSs and CCPs. This same threshold is then applied to all other conditions imaged on the same day and then CCP dynamics parameters are measured. The cmeAnalysis software package (Aguet et al., 2013) as well as a recently published, threshold-independent DASC analysis package (described in Supplemental Figure S2) (Wang, Chen, et al., 2020) were used for the analysis of TIRF data and calculation of CCP dynamics parameters.
Quantification of dynamin colocalization with CCPs
Recruitment of dynamin chimeras to CCPs was quantified using a combination of our cmeAnalysis software package and custom-written Fiji and Excel macros. In brief, for each condition, 10 dual-color (mRuby-CLCa/Dyn-eGFP) images were first analyzed by cmeAnalysis, using mRuby-CLCa as a fiduciary to detect all CSs. The resulting binary detection masks were imported into Fiji, converted into individual regions of interest (ROIs) and transferred back onto the raw image data. In each channel, for each individual detection, the mean maximum intensity was measured within a 3 × 3 pixel box drawn around the brightest pixel of that ROI and the mean background intensity was measured within a 1-pixelwide band drawn around the corresponding ROI. Per dual-channel image, this procedure yielded several thousand values which were automatically imported into Excel. Within Excel, data were automatically analyzed and summarized; CCPs were considered “bona fide” if their mean maximum intensity in the clathrin channel was 20% above the local clathrin background, and finally, if these bona fide CCPs displayed a dynamin signal that was 10% above their local dynamin background, they were classified as Dyn-positive. We also measured the average intensity of Dyn-eGFP at each, so designated, Dyn-positive CCP.
Mass-Spec analysis for relative dynamin isoform levels
ARPE-mRuby-CLCa cells were allowed to grow to 70–80% confluency in a 100-mm dish. After washing twice with PBS, cells were detached by incubating at 37°C in 2 ml of 50 mM EDTA in PBS until they could be readily resuspended by pipetting. Cells were then spun down and washed once with cold PBS, spun down again, and finally resuspended in cold lysis buffer (25 mM HEPES, 150 mM KCl, 0.5% Triton X-100, 1 mM MgCl2, 1 mM EGTA, 1 tablet of protease inhibitor cocktail/50 ml lysis buffer, pH 7.4) in low-binding Eppendorfs. Cell lysis was facilitated by rotating the cells resuspended in lysis buffer at 4°C for 30 min with intermittent vortexing. The lysates were then heat-denatured and loaded on SDS–PAGE gel before sending to the proteomics core facility for Mass-spec sample preparation and processing.
Quantification and statistical analysis of data
All biochemical and TIRF data were acquired from multiple biological repeats.The t test was used to calculate statistical significance using Graphpad Prism 8 software (Graphpad Software, La Jolla, CA). DASC analysis used the Wilcoxon rank-sum test to calculate statistical significance. Error bars represent standard deviations. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; ns, not significant.
Supplementary Material
Acknowledgments
We acknowledge Zhiming Chen for providing dual labeled ARPE mRuby-CLCa/ Tractin-GFP cell line and help with sample preparation for studying relative dynamin isoform expression by Mass-spec. We thank the UT Southwestern proteomics core facility for Mass-spec sample preparation and analysis. We thank all members of the Schmid lab for critical scientific discussions. We acknowledge Aparna Mohanakrishnan for generating GFP dynamin fusions and site directed mutagenesis to generate AA chimeras and Heather Grossman for help with FACS sorting of dynamin-expressing cells. This work was supported by National Institutes of Health R01 grants GM42455 and GM73165 to SLS.
Abbreviations used:
- AC
abortive coat
- CCP
clathrin-coated pit
- CLC
clathrin light chain
- CLS
clathrin-labeled structure
- CME
clathrin-mediated endocytosis
- CS
clathrin-coated structure
- DASC
disassembly asymmetry score classification
- Dyn
dynamin
- EAP
endocytic accessory protein
- FBS
fetal bovine serum
- G
GTPase
- GED
GTPase effector domain
- Jasp
jasplakinolide
- KD
knockdown
- KO
knockout
- LatA
latrunulin A
- PBS
phosphate-buffered saline
- PH
Pleckstrin homology
- PRD
proline/arginine-rich domain
- ROI
regions of interest
- sCLS
subthreshold cathrin-labeled structure
- TfnR
transferrin receptor
- TIRFM
total internal reflection fluorescence microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-06-0363) on June 24, 2020.
REFERENCES
Boldface names denote co–first authors.
- Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. (2013). Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev Cell , 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V, Robinson PJ. (2007). Syndapin I and endophilin I bind overlapping proline-rich regions of dynamin I: role in synaptic vesicle endocytosis. J Neurochem , 931–943. [DOI] [PubMed] [Google Scholar]
- Anggono V, Smillie KJ, Graham ME, Valova VA, Cousin MA, Robinson PJ. (2006). Syndapin I is the phosphorylation-regulated dynamin I partner in synaptic vesicle endocytosis. Nat Neurosci , 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster M, Messa M, Ferguson SM, De Camilli P, Ryan TA. (2013). Dynamin phosphorylation controls optimization of endocytosis for brief action potential bursts. Elife , e00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. (2011). Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol , 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Senderowicz AM, Sausville EA, Duncan KL, Korn ED. (1994). Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J Biol Chem , 14869–14871. [PubMed] [Google Scholar]
- Bubb MR, Spector I, Beyer BB, Fosen KM. (2000). Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J Biol Chem , 5163–5170. [DOI] [PubMed] [Google Scholar]
- Cao H, Garcia F, McNiven MA. (1998). Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell , 2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PH, Bendris N, Hsiao YJ, Reis CR, Mettlen M, Chen HY, Yu SL, Schmid SL. (2017). Crosstalk between CLCb/Dyn1-mediated adaptive clathrin-mediated endocytosis and epidermal growth factor receptor signaling increases metastasis. Dev Cell , 278–288.e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yong J, Martinez-Sanchez A, Yang Y, Wu Y, De Camilli P, Fernandez-Busnadiego R, Wu M. (2019). Dynamic instability of clathrin assembly provides proofreading control for endocytosis. J Cell Biol , 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G, Cousin MA. (2019). Synaptic vesicle generation from activity-dependent bulk endosomes requires a dephosphorylation-dependent dynamin-syndapin interaction. J Neurochem , 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton EL, Sue N, Smillie KJ, O’Leary T, Bache N, Cheung G, Cole AR, Wyllie DJ, Sutherland C, Robinson PJ, Cousin MA. (2010). Dynamin I phosphorylation by GSK3 controls activity-dependent bulk endocytosis of synaptic vesicles. Nat Neurosci , 845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin MA, Robinson PJ. (2001). The dephosphins: dephosphorylation by calcineurin triggers synaptic vesicle endocytosis. Trends Neurosci , 659–665. [DOI] [PubMed] [Google Scholar]
- Cureton DK, Massol RH, Whelan SP, Kirchhausen T. (2010). The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog , e1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL. (1994). Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol , 915–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. (2004). Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell , 591–605. [DOI] [PubMed] [Google Scholar]
- Ferguson SM, Brasnjo G, Hayashi M, Wolfel M, Collesi C, Giovedi S, Raimondi A, Gong LW, Ariel P, Paradise S, et al. (2007). A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science , 570–574. [DOI] [PubMed] [Google Scholar]
- Ferguson SM, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, et al. (2009). Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell , 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I, Zweifel ME, Courtemanche N, Pollard TD. (2018). Latrunculin A accelerates actin filament depolymerization in addition to sequestering actin monomers. Curr Biol , 3183–3192, e3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. (2010). Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature , 502–506. [DOI] [PubMed] [Google Scholar]
- Gauthier NC, Masters TA, Sheetz MP. (2012). Mechanical feedback between membrane tension and dynamics. Trends Cell Biol , 527–535. [DOI] [PubMed] [Google Scholar]
- Graham ME, Anggono V, Bache N, Larsen MR, Craft GE, Robinson PJ. (2007). The in vivo phosphorylation sites of rat brain dynamin I. J Biol Chem , 14695–14707. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. (1995). Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature , 190–192. [DOI] [PubMed] [Google Scholar]
- Huang Y, Chen-Hwang MC, Dolios G, Murakami N, Padovan JC, Wang R, Hwang YW. (2004). Mnb/Dyrk1A phosphorylation regulates the interaction of dynamin 1 with SH3 domain-containing proteins. Biochemistry , 10173–10185. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. (2006). Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol , 404–414. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Lemmon MA. (2010). Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J , 3054–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakoduk AM, Roudot P, Mettlen M, Grossman HM, Schmid SL, Chen PH. (2019). Mutant p53 amplifies a dynamin-1/APPL1 endosome feedback loop that regulates recycling and migration. J Cell Biol , 1928–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Shelden EA, Knecht DA. (1998). Formation of F-actin aggregates in cells treated with actin stabilizing drugs. Cell Motil Cytoskeleton , 122–133. [DOI] [PubMed] [Google Scholar]
- Liu JP, Powell KA, Sudhof TC, Robinson PJ. (1994a). Dynamin I is a Ca(2+)-sensitive phospholipid-binding protein with very high affinity for protein kinase C. J Biol Chem , 21043–21050. [PubMed] [Google Scholar]
- Liu JP, Sim AT, Robinson PJ. (1994b). Calcineurin inhibition of dynamin I GTPase activity coupled to nerve terminal depolarization. Science , 970–973. [DOI] [PubMed] [Google Scholar]
- Liu YW, Neumann S, Ramachandran R, Ferguson SM, Pucadyil TJ, Schmid SL. (2011). Differential curvature sensing and generating activities of dynamin isoforms provide opportunities for tissue-specific regulation. Proc Natl Acad Sci USA , E234–E242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. (2008). Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells. Mol Biol Cell , 5347–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke D, Mettlen M, Schmid SL, Danuser G. (2011). Measuring the hierarchy of molecular events during clathrin-mediated endocytosis. Traffic , 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL. (2009). Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol , e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Paradise S, Ferguson SM, De Camilli P. (2008). Selective saturation of slow endocytosis at a giant glutamatergic central synapse lacking dynamin 1. Proc Natl Acad Sci USA , 17555–17560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W. (2002). Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol , 691–698. [DOI] [PubMed] [Google Scholar]
- Mettlen M, Chen PH, Srinivasan S, Danuser G, Schmid SL. (2018). Regulation of clathrin-mediated endocytosis. Annu Rev Biochem , 871–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Danuser G. (2014). Imaging and modeling the dynamics of clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol , a017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. (2009). Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochem Soc Trans , 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmyer SL, Christensen A, Sever S. (2003). Auxilin-dynamin interactions link the uncoating ATPase chaperone machinery with vesicle formation. Dev Cell , 929–940. [DOI] [PubMed] [Google Scholar]
- Raimondi A, Ferguson SM, Lou X, Armbruster M, Paradise S, Giovedi S, Messa M, Kono N, Takasaki J, Cappello V, et al. (2011). Overlapping role of dynamin isoforms in synaptic vesicle endocytosis. Neuron , 1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL. (2018). The dynamin superfamily. Curr Biol , R411–R416. [DOI] [PubMed] [Google Scholar]
- Reis CR, Chen PH, Bendris N, Schmid SL. (2017). TRAIL-death receptor endocytosis and apoptosis are selectively regulated by dynamin-1 activation. Proc Natl Acad Sci USA , 504–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis CR, Chen PH, Srinivasan S, Aguet F, Mettlen M, Schmid SL. (2015). Crosstalk between Akt/GSK3beta signaling and dynamin-1 regulates clathrin-mediated endocytosis. EMBO J , 2132–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Faelber K, Plattner N, Posor Y, Ketel K, Curth U, Schlegel J, Anand R, Manstein DJ, Noe F, et al. (2015). Crystal structure of the dynamin tetramer. Nature , 404–408. [DOI] [PubMed] [Google Scholar]
- Schmid SL. (2017). Reciprocal regulation of signaling and endocytosis: Implications for the evolving cancer cell. J Cell Biol , 2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Smythe E. (1991). Stage-specific assays for coated pit formation and coated vesicle budding in vitro. J Cell Biol , 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S, Damke H, Schmid SL. (2000). Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J Cell Biol , 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever S, Muhlberg AB, Schmid SL. (1999). Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature , 481–486. [DOI] [PubMed] [Google Scholar]
- Sharma S, Sicinski P. (2020). A kinase of many talents: non-neuronal functions of CDK5 in development and disease. Open Biol , 190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulet F, Yarar D, Leonard M, Schmid SL. (2005). SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell , 2058–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Burckhardt CJ, Bhave M, Chen Z, Chen PH, Wang X, Danuser G, Schmid SL. (2018). A noncanonical role for dynamin-1 in regulating early stages of clathrin-mediated endocytosis in non-neuronal cells. PLoS Biol , e2005377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Dharmarajan V, Reed DK, Griffin PR, Schmid SL. (2016). Identification and function of conformational dynamics in the multidomain GTPase dynamin. EMBO J , 443–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TC, Valova VA, Malladi CS, Graham ME, Berven LA, Jupp OJ, Hansra G, McClure SJ, Sarcevic B, Boadle RA, et al. (2003). Cdk5 is essential for synaptic vesicle endocytosis. Nat Cell Biol , 701–710. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Lampe M, Merrifield CJ. (2012). A feedback loop between dynamin and actin recruitment during clathrin-mediated endocytosis. PLoS Biol , e1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. (2011). A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol , e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee RB. (1992). Dynamin: motor protein or regulatory GTPase. J Muscle Res Cell Motil , 493–496. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM, Meyerowitz EM. (1991). Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature , 411–414. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. (1993). Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol , 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen Z, Mettlen M, Noh J, Schmid SL, Danuser G. (2020). DASC, a sensitive classifier for measuring discrete early stages in clathrin-mediated endocytosis. Elife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, et al. (2000). Dominant negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood , 1646–1654. [PubMed] [Google Scholar]
- Yarmola EG, Somasundaram T, Boring TA, Spector I, Bubb MR. (2000). Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J Biol Chem , 28120–28127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.