Abstract
Objective
MicroRNA-122 (miR-122) has been identified as a biomarker of liver diseases. However, the miR-122 detection accuracy in patients with hepatocellular carcinoma (HCC) associated with hepatitis C virus (HCV) is inconclusive.
Methods
We conducted a systematic literature search of Web of Science, Cochrane Library, PubMed, and Embase to identify studies related to the diagnostic value of miR-122 in HCV-related HCC. We analyzed the results and validated them using data from the Cancer Genome Atlas (TCGA).
Results
Six articles were included in this meta-analysis, comprising 354 cases and 420 controls. The pooled specificity, sensitivity, positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio, and area under the curve were 0.87, 0.83, 5.1, 0.16, 32, and 0.92, respectively. Additional sub-group analyses showed that results for plasma were more sensitive than those for serum. In addition, miR-122 was better at distinguishing between HCV-associated HCC and healthy people or those with HCV than between those with HCV-associated HCC and HCV-related cirrhosis. Small samples (≤100) had better diagnostic odds ratios than larger samples (>100). Analysis of data from TCGA confirmed that miRNA-122 had a high diagnostic value.
Conclusion
This meta-analysis demonstrates that miR-122 may be a useful diagnostic biomarker for HCV-associated HCC.
Keywords: microRNA-122, hepatocellular carcinoma, diagnosis, hepatitis C virus, meta-analysis, plasma
Background
Hepatocellular carcinoma (HCC) is associated with poor disease outcomes.1 Current statistics reveal that primary liver cancer affects 25.7 per 100,000 individuals in China, second only to lung cancer and gastric carcinoma,2 while HCC accounts for 75% of all cases of liver cancer.
HCC usually occurs in patients suffering from liver cirrhosis caused by hepatitis B virus or hepatitis C virus (HCV) infection.3 However, it may also develop as a result of other health and lifestyle factors e.g., alcoholism, non-alcoholic liver disease, eating aflatoxin-contaminated food, and genetics (e.g., hemochromatosis).4 HCC currently has a poor prognosis, with a 5-year survival rate of 40% to 80% if diagnosed early, but only 5% to 15% in patients with a late diagnosis.5 Current data indicates that about 170 million people worldwide suffer from HCV, leading to liver cirrhosis and a high prevalence of HCC.6 This suggests that screening for HCV should be conducted to ensure the early diagnosis of and timely intervention for HCC.7
Existing imaging techniques lack precision for detecting early HCC. Alpha-fetoprotein (AFP) is similarly unreliable for making an early diagnosis,8 because around 40% of HCC cells do not produce AFP, meaning that a patient with low levels of AFP may still have HCC.9 The lack of suitable and precise tumor markers for diagnosing HCC during the early stage has contributed to a low 5-year survival rate.10,11 It is therefore imperative to find novel, sensitive, precise, and less-invasive markers of the disease.
MicroRNAs (miRNAs) are non-coding RNAs comprising around 22 nucleotides, which have been shown to modulate post-transcriptional gene expression in nearly all cell types.12 Dysregulation of miRNA expression has been related to certain liver diseases, such as hepatitis, cirrhosis, and liver cancers. Some miRNAs have also been shown to have inverse regulatory effects on gene expression, and to act as either oncogenes or tumor inhibitors.13 MiR-122 is a major liver miRNA, accounting for about 70% of all miRNAs in the liver. Furthermore, studies have shown that miR-122 is dysregulated during HCV infection.6,13 MiR-122 has been shown to act as a tumor suppressor gene, and its expression is usually downregulated in HCC cells, thereby facilitating the progression of HCC.14
Recent studies6,14 suggested that miR-122 could have a potential diagnostic value in HCV-related HCC; however its diagnostic value in this context is still unclear, and studies have reported conflicting results, possibly attributed to variations among study designs and specimen types. Herein, we systematically reviewed the literature and performed a meta-analysis to confirm the diagnostic value of miR-122 in HCV-related HCC.
Materials and methods
Search strategy
We systematically searched Web of Science, the Cochrane Library, Embase, and PubMed, without language restrictions, to retrieve all articles that investigated the diagnostic value of miR-122 in HCV-related HCC (up to May 2020). The search was based on subject headings and random words, using the following search terms: (“miRNA-122” OR “microRNA-122” OR “miR-122” OR “has-miR-122”) AND (“HCV” OR “hepatitis C virus” OR “CHC”) AND (“Liver Cancer” OR “Hepatic carcinoma” OR “Hepatocellular carcinoma”) AND (“diagnose*” OR “ROC curve” OR “sensitivity” OR “specificity”).
Ethical approval and informed consent requirements were not relevant, because this was a meta-analysis of previously published studies.
Inclusion/exclusion criteria
The inclusion criteria were: (1) studies that explored the diagnostic value of miR-122 in patients with HCV-related HCC; (2) cases and controls not limited by age or race, and confirmed by clinical gold standards; (3) data including sensitivity, specificity, and area under the curve (AUC) for miR-122 for independent diagnosis could be calculated from the literature or according to the data in the literature; and (4) studies not based on animal trials, abstracts, reviews, and editorial articles.
Data extraction and assessment of trial quality
The data were extracted from the included studies independently by two investigators (Xiao-yu Wei and Jing Ding) following a standard protocol and using a data-collection form based on the above inclusion criteria. Any disagreements were resolved by a third author (Wen-guang Tian) to reach a consensus. The data extracted from the selected articles included the name of the first author, country, publication year, basic information on the participants (e.g., age, ethnicity, numbers of cases and controls), miR-122 testing methods and sample types, and data. The risk of bias in the included studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) score system, which utilized the selection index of patients, index test, reference standard and flow, as well as timing, to analyze the bias risk and applicability. Seven items (four items on risk of bias and three items on applicability) were evaluated for all the included studies. Item classification was based on the risk of bias (“yes/no”) or (“unclear”) if there was inadequate data to evaluate bias risk. Two investigators independently verified the pilot QUADAS-2 items (Xiao-yu Wei and Yi-Chuan Yu), and inconsistencies were resolved by a third investigator (Jing Ding).
Statistical analysis
All analyses were conducted using Stata 14.0 software (Stata Corp, College Station, TX, USA). Values of P < 0.05 were considered to be statistically significant.
The pooled parameters sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and AUC, and their 95% confidence intervals (CIs) were calculated to assess the overall diagnostic accuracy based on a bivariate random-effects regression model. The heterogeneity of the findings among the studies was evaluated using a χ2-based Cochran’s Q test. Results with P < 0.10 and I2 > 50% were considered heterogeneous, and subgroup analysis was carried out to scrutinize the heterogeneity based on the features of the included studies. We also conducted a sensitivity analysis to evaluate the robustness of our analysis. Deeks’ funnel plot asymmetry test was used to determine the publication bias of the included articles.
Bioinformatics analysis
MiRNA data for HCV-associated HCC associated were downloaded from the Cancer Genome Atlas (TCGA) according to the following inclusion criteria: 1) samples of human HCC associated with HCV, including uncoded RNA chip data; 2) normal controls in at least two cases; and 3) chip data could be used for calculation and comparison. The exclusion criteria were: 1) no relevant data available for miRNA-122; 2) sample from a cancer cell line; and 3) data included samples other than from humans. The data were analyzed using R (R3.6.3) and receiver operating characteristic curves (ROCs) were used to test the diagnostic value of miRNA-122.
Results
Literature search
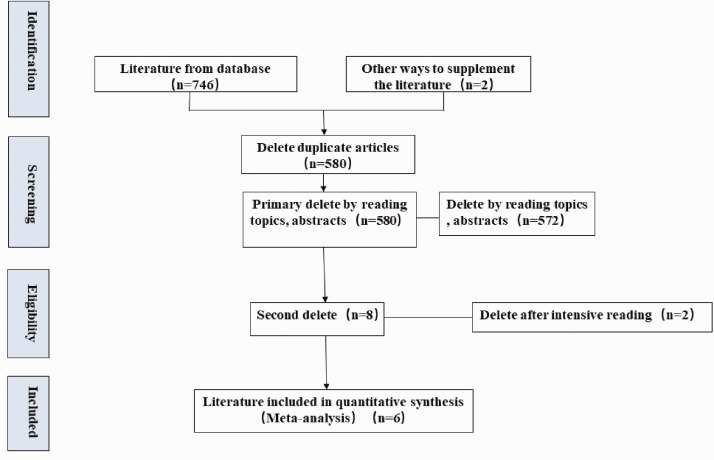
A total of 746 potentially relevant studies that utilized miR-122 for the diagnosis of patients with HCV-associated HCC were identified. An additional two eligible studies were included after scrutinizing the documents identified in our initial study. A total of 168 articles were removed as duplicates, an additional 572 studies were removed after reviewing the titles and abstracts, and two more studies were removed because they lacked the necessary data after checking the full text. Six studies,15–20 all in English, were finally selected for data extraction and analysis (Figure 1).
Figure 1.
Flowchart of selection of studies for inclusion in the meta-analysis.
Study characteristics and quality assessment
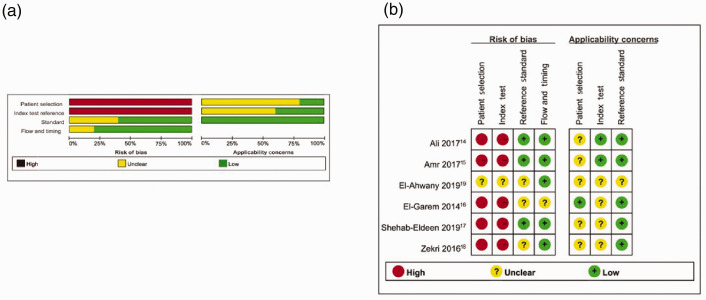
The six studies included 774 patients, comprising 354 cases and 420 controls. The controls included healthy individuals or patients with HCV or HCV-related cirrhosis. All articles were published between 2014 and 2019 and all were performed in Egypt. All the selected studies used real-time polymerase chain reaction. Five studies detected miR-122 in serum samples and one in plasma samples. The main features of the selected articles are outlined in Table 1. A summary of QUADAS-2-based evaluation of bias risk and applicability is provided in Figure 2.
Table 1.
Characteristics of studies included in the meta-analysis.
| Author | Year | Country | Detection method | Sample type | Control source | Age (years) |
Sample size |
Sensitivity (%) | Specificity (%) | AUC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case/control | Case | Control | |||||||||
| Ali et al.14 | 2017 | Egypt | RT-PCR | Serum | Healthy | NA | 34 | 25 | 94.3 | 92.9 | 0.959 |
| HCV | NA | 34 | 52 | 82.4 | 83.7 | 0.904 | |||||
| Amr et al.15 | 2017 | Egypt | RT-PCR | Serum | Healthy | 34–56/NA | 40 | 20 | 87.5 | 95 | 0.960 |
| HCV | 34–56/NA | 40 | 20 | 87.5 | 97.5 | 0.980 | |||||
| El-Ahwany et al.19 | 2019 | Egypt | RT-PCR | Serum | HCV | 48.1 ± 4.1/46.4 ± 7.3 | 38 | 42 | 85.71 | 83.33 | 0.897 |
| El-Garem et al.16 | 2014 | Egypt | RT-PCR | Serum | HCV-related cirrhosis | 60.27 ± 8.20/55.07 ± 7.35 | 30 | 30 | 82.9 | 65.7 | 0.646 |
| Shehab-Eldeen et al.17 | 2019 | Egypt | RT-PCR | Plasma | HCV-related cirrhosis | 56.95 ± 1.73/56.50 ± 2.1 | 20 | 20 | 90 | 70 | 0.980 |
| Healthy | 56.95 ± 1.73/52.35 ± 1.4 | 20 | 20 | 95 | 81 | 0.930 | |||||
| Zekri et al.18 | 2016 | Egypt | RT-PCR | Serum | Healthy | 56.7 ± 7.7/33.37 ± 11 | 192 | 95 | 90 | 80 | 0.617 |
| HCV-related cirrhosis | 56.7 ± 7.7/54.01 ± 8.3 | 192 | 96 | 80 | 82 | 0.617 | |||||
AUC, the area under curve; NA, data not available, RT-PCR, real-time polymerase chain reaction, HCV, hepatitis C virus.
Figure 2.
Quality of included studies and summary of bias risk. (a) Quality of included studies according to QUADAS-2 guidelines. (b) Summary of bias risk assessment results for QUADAS-2.
Diagnostic accuracy of miR-122 for HCV-associated HCC
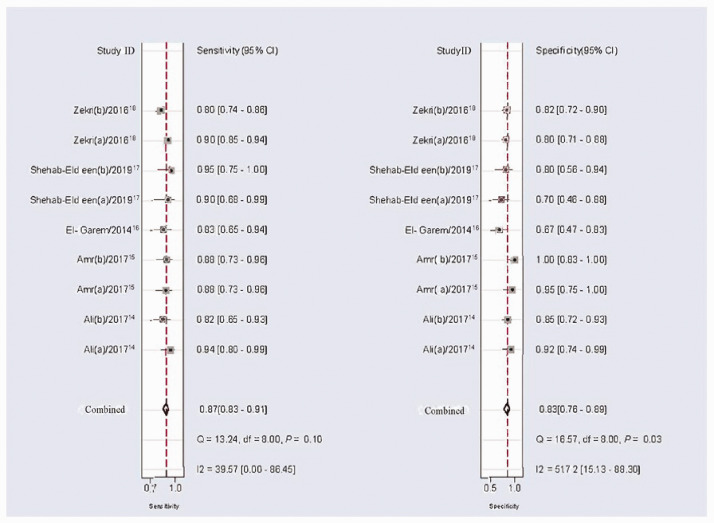
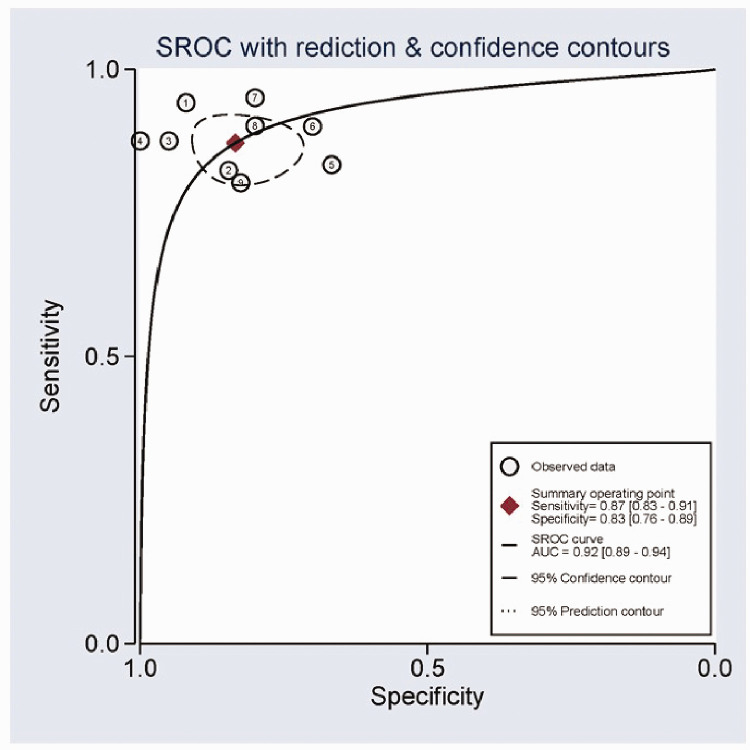
According to heterogeneity analysis, the I2 values for sensitivity and specificity were 31.19% (P = 0.16) and 45.40% (P = 0.06), respectively, suggesting a certain level of heterogeneity among the studies. The random-effects model was applied. The forest plots for specificity and sensitivity are shown in Figure 3. The pooled sensitivity was 0.87 (95%CI: 0.83–0.90), specificity was 0.83 (95%CI: 0.77–0.88), PLR was 5.1 (95%CI: 3.7–7.0), NLR was 0.16 (95%CI: 0.12–0.21), and DOR was 32 (95%CI: 19–54) (Table 2). The summary ROC (SROC) curve with an AUC of 0.92 (95%CI: 0.89–0.94) for the overall study is shown in Figure 4.
Figure 3.
Forest plots of summary sensitivities and specificity of circulating miR-122 for the diagnosis of hepatitis C virus-associated hepatocellular carcinoma.
df, degrees of freedom.
Table 2.
Results of subgroup analyses.
| Analysis | Sensitivity (95%CI) | Specificity (95%CI) | PLR (95%CI) | NLR (95%CI) | DOR (95%CI) |
|---|---|---|---|---|---|
| Control source | |||||
| Healthy | 0.91 (0.86–0.94) | 0.84 (0.72–0.92) | 5.7 (3.0–10.7) | 0.11 (0.07–0.17) | 51 (21–124) |
| HCV | 0.86 (0.78–0.92) | 0.87 (0.79–0.92) | 10.31 (1.09–97.52) | 0.17 (0.11–0.26) | 54.26 (14.59–83.44) |
| HCV-related cirrhosis | 0.81 (0.76–0.86) | 0.77 (0.69–0.84) | 3.74 (2.64–5.22) | 0.23 (0.17–0.31) | 16.83 (9.71–29.19) |
| Sample type | |||||
| Serum | 0.87 (0.82–0.90) | 0.85 (0.77–0.90) | 5.60 (3.70–8.60) | 0.16 (0.12–0.22) | 34 (19–67) |
| Plasma | 0.93 (0.8–0.98) | 0.75 (0.59–0.87) | 3.57 (2.08–6.13) | 0.11 (0.04–0.33) | 33.7 (8.41–135.08) |
| Sample size | |||||
| ≤100 | 0.88 (0.83–0.91) | 0.85 (0.76–0.91) | 5.90 (3.50–9.70) | 0.14 (0.10–0.20) | 41 (20–85) |
| >100 | 0.85 (0.81–0.89) | 0.81 (0.75–0.87) | 4.54 (3.33–6.18) | 0.81 (0.09–0.34) | 26.28 (13.96–49.48) |
| Overall | 0.87 (0.83–0.90) | 0.83 (0.77–0.88) | 5.1 (3.7–7.0) | 0.16 (0.12–0.21) | 32 (19–54) |
CI, confidence interval; DOR, diagnostic odds ratio; NLR, negative likelihood ratio; PLR, positive likelihood ratio.
Figure 4.
Summary receiver operating characteristic curves for miR-122 in the diagnosis of hepatitis C virus-associated hepatocellular carcinoma.
SROC, summary receiver operating characteristic curve; AUC, area under the curve.
Threshold effect and subgroup analyses
Spearman’s correlation coefficient was 1, reflecting a lack of a threshold effect in our study. Subgroup analyses were performed according to control source, sample type, and sample size. The pooled DOR, NLR, PLR, specificity, and sensitivity for each subgroup are shown in Table 2. The healthy control group and HCV group showed higher overall accuracies than the HCV-related cirrhosis group, suggesting that miR-122 had a better ability to distinguish between HCV-associated HCC and healthy people or those with HCV than between patients with HCV-associated HCC and HCV-related cirrhosis. Moreover, analysis of miR-122 expression profiles in plasma and serum revealed that serum levels were less sensitive than plasma-based tests. Analysis based on sample size demonstrated that small samples (≤100) had a better diagnostic odds ratio than larger samples (>100).
Publication bias and sensitivity analysis
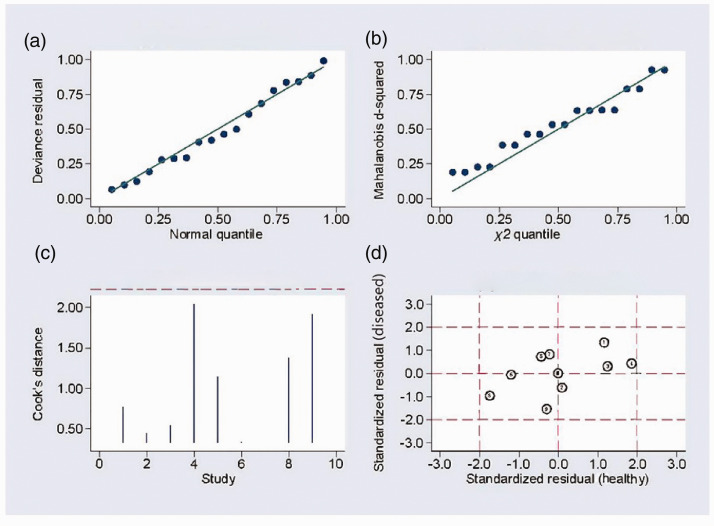
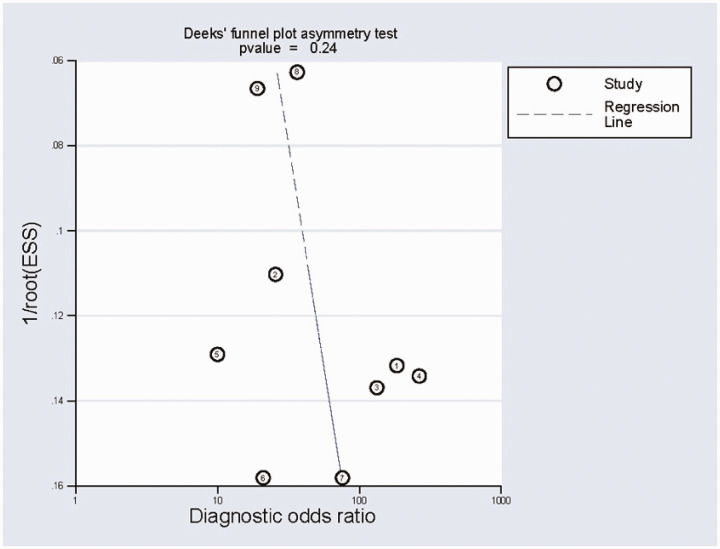
Sensitivity analysis (Figure 5a, b) using goodness-of-fit and bivariate normality analysis implied that the random-effects model was suitable. Influence analysis and outlier detection (Figure 5c, d) found no outlier studies. The results of this meta-analysis were therefore considered to be relatively stable. The results of Deeks’ test were not significant (bias=1.34) (Figure 6), indicating the absence of publication bias in this meta-analysis.
Figure 5.
Sensitivity analysis. (a) Goodness-of-fit, (b) bivariate normality analysis, (c) influence, and (d) outlier detection analysis.
Figure 6.
Deeks’ funnel plot asymmetry test for the assessment of potential publication bias.
Bioinformatics analysis
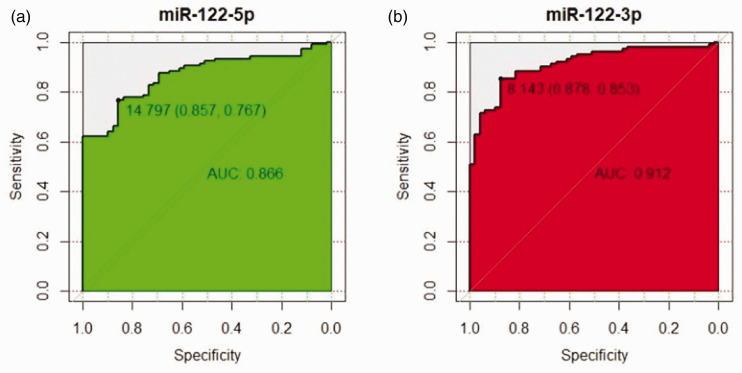
Based on TCGA data, the AUCs for miRNA-122-5p and miRNA-122-3p were 0.866 and 0.912, respectively, indicating that miRNA-122 had a high diagnostic value (Figure 7).
Figure 7.
Receiver operating characteristic curve for data from The Cancer Genome Atlas. (a) MiR-122-5p; (b) miR-122-3p.
AUC, area under the curve.
Discussion
HCV infection causes HCC.21 Although AFP is used as a biomarker for the early detection of HCC, it is limited by low sensitivity and specificity.22 There is thus an urgent need to explore and identify new non-invasive diagnostic biomarkers with high accuracy and feasibility. MiRNAs that are differentially expressed between normal and tumor tissues and which show stable expression in serum and plasma, could be useful, non-invasive biomarkers.23 The liver-specific miRNA, miR-122, plays a vital role in physiological and pathological mechanisms in the liver,24,25 and several studies have examined the use of miR-122 as a biomarker for HCV-associated HCC. Shehab-Eldeen et al.18 found that miR-122 expression was substantially decreased in patients with HCC relative to those with cirrhosis and healthy controls. Other studies11,16,26,27 have reported similar results. These studies thus suggested that miR-122 might be a diagnostic marker for HCV-associated HCC. However, variations among study designs and subjects meant that the results were inconsistent. We therefore conducted the present meta-analysis to explore the diagnostic and clinical values of miR-122 as a novel biomarker for HCV-associated HCC.
This meta-analysis included six studies,15–20 covering 354 cases and 420 controls. The pooled sensitivity ( 0.87), specificity (0.83), and DOR (32) indicated a relatively high level of overall accuracy. The pooled PLR was 5.1 and the pooled NLR was 0.16, while the AUC of the SORC curve was 0.92, indicating good diagnostic accuracy for miR-122 as a biomarker for HCV-associated HCC. Sensitivity analysis indicated that the results of this meta-analysis were relatively stable. Overall, these findings suggest that miR-122 was a promising novel and valuable biomarker for the diagnosis of HCV-associated HCC, with relatively high specificity and sensitivity.
We explored the sources of heterogeneity among the studies by subgroup analyses based on control source, sample type, and sample size. Subgroup analysis according to control source suggested that miR-122 was better able to distinguish between patients with HCV-associated HCC and healthy people or those with HCV than between patients with HCV-associated HCC and those with HCV-related cirrhosis. Regarding sample type, miRNA expression profiles are affected by the blood-coagulation process.28 The current analysis indicated that plasma was a better matrix than serum for diagnostic profiling of miR-122 in HCV-related HCC; however, only one study used plasma, and this conclusion needs further verification. In terms of sample size, a small sample (≤100) produced a better diagnostic odds ratio than a larger sample (>100). We did not conduct subgroup analyses for other factors, including ethnicity and age, because of limited information.
We verified the results of our meta-analysis using relevant data from TCGA. The AUCs for miRNA-122-5p and miRNA-122-3p were 0.866 and 0.912, respectively, indicating that miRNA-122 had a high diagnostic value.
The relatively simple procedures mean that the detection of serum biomarkers is of great clinical value in the early screening and diagnosis of HCC. Although AFP is a common serum marker for clinical use, its sensitivity and accuracy are not ideal, and related research18 demonstrated an AUC of 0.68, sensitivity of 70%, and specificity of 70%. Regarding other potential biomarkers, the sensitivity and specificity of Golgi glycoprotein 73 for the diagnosis of HCC were 82% and 80%, respectively,29 and the sensitivity and specificity of des-γ-carboxy prothrombin were 51.7% and 86.7%, respectively.30 The findings of the current meta-analysis showed that miRNA-122 had a better diagnostic performance than other current biomarkers for diagnosing HCC.
This study had several limitations. First, only studies in English were included and findings reported from non-English speaking regions were therefore excluded. Second, all the included studies were from Egypt, which reduced the inclusiveness of the results. Third, some studies had small sample sizes, and finally, most studies involved the use of healthy subjects as controls.
In conclusion, the present meta-analysis verified the use of miR-122 as a unique and valuable marker for the diagnosis of HCV-related HCC, with high specificity and sensitivity. However, extensive functional assessments and prospective population-based studies involving larger sample sizes and diverse ethnic groups are needed to validate these results.
Data availability statement
All data gathered and included in the present study are available and can be provided by the corresponding author upon reasonable request.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Program of Technology of Science and Technology Committee of Yuzhong District in Chongqing (Ycst2015nc5029) and Program of Yongchuan Hospital of Chongqing Medical University (YJJC201704).
ORCID iD
Xiao-yu Wei https://orcid.org/0000-0001-5780-5756
References
- 1.Liu XF, Long HJ, Miao XY, et al. Fisetin inhibits liver cancer growth in a mouse model: relation to dopamine receptor. Oncol Rep 2017; 38: 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Zhang J, Wang C, et al. Safety of implanting sustained-release 5-fluorouracil into hepatic cross-section and omentum majus after primary liver cancer resection. Int J Immunopathol Pharmacol 2016; 29: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai K, Takai K, Hanai T, et al. Homeostatic Model Assessment of insulin resistance for predicting the recurrence of hepatocellular carcinoma after curative treatment. Int J Mol Sci 2019; 20: 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizuguchi Y, Takizawa T, Yoshida H, et al. Dysregulated miRNA in progression of hepatocellular carcinoma: a systematic review. Hepatol Res 2016; 46: 391–406. [DOI] [PubMed] [Google Scholar]
- 5.Duan F, Wang YY, Xu DG, et al. Feasibility of terahertz imaging for discrimination of human hepatocellular carcinoma. World J Gastrointest Oncol 2019; 11: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ono C, Fukuhara T, Motooka D, et al. Characterization of miR-122-independent propagation of HCV. PLoS Pathog 2017; 13: e1006374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanda T, Yasui S, Nakamura M, et al. Real-world experiences with the combination treatment of ledipasvir plus sofosbuvir for 12 weeks in HCV genotype 1-infected Japanese patients: achievement of a sustained virological response in previous users of peginterferon plus ribavirin with HCV NS3/4Ai. Int J Mol Sci 2017; 18: 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun L, Su Y, Liu X, et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer 2018; 9: 2631–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao S, Xu X, Wang Y, et al. Diagnostic utility of plasma lncRNA small nucleolar RNA host gene 1 in patients with hepatocellular carcinoma. Mol Med Rep 2018; 18: 3305–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang JT, Liu SM, Ma H, et al. Systematic review and meta-analysis: circulating miRNAs for diagnosis of hepatocellular carcinoma. J Cell Physiol 2016; 231: 328–335. [DOI] [PubMed] [Google Scholar]
- 11.Qi J, Wang J, Katayama H, et al. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma 2013; 60: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi Y, Hwang DW, Kim MY, et al. Transgenic mouse expressing optical microRNA reporter for monitoring microRNA-124 action during development. Front Mol Neurosci 2016; 9: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim GW, Lee SH, Cho H, et al. Hepatitis C virus core protein promotes miR-122 destabilization by inhibiting GLD-2. PLoS Pathog 2016; 12: e1005714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulouarn C, Factor VM, Andersen JB, et al. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009; 28: 3526–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali HEA, Abdel Hameed R, Effat H, et al. Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2017; 41: e51–e62. [DOI] [PubMed] [Google Scholar]
- 16.Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, et al. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis 2017; 4: 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Garem H, Ammer A, Shehab H, et al. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol 2014; 6: 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shehab-Eldeen S, Nada A, Abou-Elela D, et al. Diagnostic performance of microRNA-122 and microRNA-224 in hepatitis C virus-induced hepatocellular carcinoma (HCC). Asian Pac J Cancer Prev 2019; 20: 2515–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zekri AN, Youssef AS, El-Desouky ED, et al. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol 2016; 37: 12273–12286. [DOI] [PubMed] [Google Scholar]
- 20.El-Ahwany EGE, Mourad L, Zoheiry MMK, et al. MicroRNA-122a as a non-invasive biomarker for HCV genotype 4-related hepatocellular carcinoma in Egyptian patients. Arch Med Sci 2019; 15: 1454–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwana K, Ichida T, Kamimura T, et al. Risk factors and the effect of interferon therapy in the development of hepatocellular carcinoma: a multivariate analysis in 343 patients. J Gastroenterol Hepatol 1997; 12: 149–155. [DOI] [PubMed] [Google Scholar]
- 22.Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol 2013; 1: 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gougelet A, Colnot S. [microRNA: new diagnostic and therapeutic tools in liver disease?]. Med Sci (Paris) 2013; 29: 861–867. [DOI] [PubMed] [Google Scholar]
- 24.Tsai WC, Hsu SD, Hsu CS, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 2012; 122: 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang J, Guo JT, Jiang D, et al. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol 2008; 82: 8215–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Wu C, Che X, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog 2011; 50: 136–142. [DOI] [PubMed] [Google Scholar]
- 27.Weis A, Marquart L, Calvopina DA, et al. Serum microRNAs as biomarkers in hepatitis C: preliminary evidence of a microRNA panel for the diagnosis of hepatocellular carcinoma. Int J Mol Sci 2019; 20: 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Yuan Y, Cho JH, et al. Comparing the microRNA spectrum between serum and plasma. PLoS One 2012; 7: e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y, Chen W, Zhao Y, et al. Quantitative analysis of elevated serum Golgi protein-73 expression in patients with liver diseases. Ann Clin Biochem 2009; 46: 38–43. [DOI] [PubMed] [Google Scholar]
- 30.Cui R, He J, Zhang F, et al. Diagnostic value of protein induced by vitamin K absence (PIVKAII) and hepatoma-specific band of serum gamma-glutamyl transferase (GGTII) as hepatocellular carcinoma markers complementary to alpha-fetoprotein. Br J Cancer 2003; 88: 1878–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data gathered and included in the present study are available and can be provided by the corresponding author upon reasonable request.