Abstract
Objective
To identify Parkinson’s disease (PD)-associated deregulated pathways and genes, to further elucidate the pathogenesis of PD.
Methods
Dataset GSE100054 was downloaded from the Gene Expression Omnibus, and differentially expressed genes (DEGs) in PD samples were identified. Functional enrichment analyses were conducted for the DEGs. The top 10 hub genes in the protein–protein interaction (PPI) network were screened out and used to construct a support vector machine (SVM) model. The expression of the top 10 genes was then validated in another dataset, GSE46129, and a clinical patient cohort.
Results
A total of 333 DEGs were identified. The DEGs were clustered into two gene sets that were significantly enriched in 12 pathways, of which 8 were significantly deregulated in PD, including cytokine–cytokine receptor interaction, gap junction, and actin cytoskeleton regulation. The signature of the top 10 hub genes in the PPI network was used to construct the SVM model, which had high performance for predicting PD. Of the 10 genes, GP1BA, GP6, ITGB5, and P2RY12 were independent risk factors of PD.
Conclusion
Genes such as GP1BA, GP6, P2RY12, and ITGB5 play critical roles in PD pathology through pathways including cytokine−cytokine receptor interaction, gap junctions, and actin cytoskeleton regulation.
Keywords: Parkinson’s disease, deregulated pathway, differentially expressed gene, clustering analysis, protein–protein interactions, support vector machine model, genetic risk factors
Introduction
Parkinson’s disease (PD) is a common neurodegenerative disorder that primarily involves motor impairments that worsen over time. This disease is estimated to occur in 4% of the population aged over 80 years and 1% of the population aged over 65 years.1 The main pathological changes in PD are the progressive degeneration of dopaminergic neurons in the substantia nigra and the accumulation of intraneuronal inclusions that contain α-synuclein.2 During the disease progression, PD patients usually experience nonmotor symptoms such as sleep disturbances, anosmia, constipation, anxiety, depression, and cognitive decline. The motor symptoms of PD can be initially treated with dopaminergic therapies, but the disease continues to progress despite the use of these therapies.3 Unfortunately, the pathogenesis of PD remains unclear. There is an urgent need for more drug targets or diagnostic markers that may slow or even halt the progression of symptoms.For many years, PD was considered a nongenetic disorder caused by synergistic environmental factors.4 However, more recent evidence has revealed the complex and extensive genetic basis of PD.5–7 Six risk genes, including the genes encoding α-synuclein, leucine-rich repeat kinase 2, the VPS35 retromer complex component parkin, PTEN-induced putative kinase 1, and DJ-1, have been definitively associated with an autosomal recessive or dominant mode of PD inheritance.8 Additionally, family-based studies have also successfully identified genes related to PD, including those that encode F-box protein 7, phospholipase A2 group 6, and ATPase type 13A2.9 Although many genes have been demonstrated to be involved in PD progression, the identification of these genes is far from enough to understand the pathology of PD. The identification of additional PD-associated genes will enable the further elucidation of its pathogenesis, and may aid in the development of disease-modifying therapeutic strategies.
In the current study, the circulating mRNA expression profiling dataset GSE100054 was used to identify the functional association gene set associated with PD. This was then combined with a supervised classification algorithm to build a potential diagnostic model. Finally, the significantly correlated gene markers were screened out as characteristic genes, which may serve as potential drug targets or diagnostic markers in PD.
Data and methods
The database part of this study neither enrolled human participants nor involved animal experiments; therefore, ethics approval was not required.
Data collection
The mRNA expression profiling dataset GSE100054 was downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/).10 The chip platform was GPL23126 (Clariom_D_Human) Affymetrix Human Clariom D Assay (transcript (gene) version). This dataset came from samples of peripheral blood mononuclear cells (PBMCs) from both normal controls and patients with PD.
Data preprocessing
The original CEL data files were downloaded and preprocessed using the Robust Multichip Average method in the oligo package (version 1.42.0, http://www.bioconductor.org/packages/release/bioc/html/oligo.html) in R.11 Based on the platform annotation file, the probes were mapped to gene symbols. The probes that did not correspond to any gene symbol were removed. For different probes mapping to the same gene symbol, the mean value was calculated.
Differentially expressed gene (DEG) selection
The limma package (version 3.34.9, http://bioconductor.org/packages/release/bioc/html/limma.html)12 in R was used for selecting DEGs. Differences in the mean expression levels of genes between the two sample comparison groups were compared using the t-test. Genes with P < 0.05 and log fold change (FC) > 0.585 were considered as DEGs.
Hierarchical clustering
To further explore the differences in genes between diseased and normal states, all samples were used to perform sample hierarchical clustering based on the expression profiles of DEGs. The hierarchical clustering of samples was conducted using the gplots package (version 2.14.1, https://cran.r-project.org/web/packages/gplots/) in R. The ConsensusClusterPlus algorithm13 was applied to cluster the genes based on the expression values of all DEGs in the two sample groups. The reasonable clustering number and corresponding gene sets were identified by the cumulative distribution function (CDF). These gene sets were considered to be characteristic genes.
Functional enrichment analysis
Using the common enrichment analysis tool DAVID (version 6.8, https://david.ncifcrf.gov/),14 the Gene Ontology (GO)15 biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways16 for gene sets in each cluster were analyzed. The significance level (P-value) of each function and pathway was calculated, and the criteria for significance screening were P < 0.05 and count>5.
Pathway deregulation analysis
The differentially expressed pathways between the PD and control groups were identified using the Pathifier17 algorithm, to analyze the effect of the expression level of a specific gene set on KEGG pathways. This method scores the deregulation degree of each sample in a specific pathway by calculating the specific gene expression values in normal and diseased samples. The resulting score is called the pathway deregulation score. This analysis converts gene-level information into pathway-level information, and can be used to identify the different pathways between normal and diseased samples. The pathogenic mechanisms of PD were illustrated according to this analysis.
Protein–protein interaction (PPI) network analysis
Protein network visualization plays an important role in analyzing protein interaction characteristics. The interactions among DEGs involved in the deregulated pathways were identified using the STRING database (version 10.5, https://string-db.org).18 The PPI score was set as 0.4 (medium confidence), and the interacted nodes needed to be DEGs. The PPI network was visualized using Cytoscape (version 3.6.1, http://www.cytoscape.org/).19 By analyzing the network topological properties, the interaction degree of each node was calculated, and the hub genes (the top 10) in the network were identified.
Support vector machine (SVM) model prediction
The expression values of characteristic genes in all samples were used as the characteristic values to predict the SVM model, using the e1071 package SVM classifier20 with the default parameters (kernel function: sigmoid kernel; cross: 10-fold cross validation). The analysis data were randomly sorted and 60% of the samples were taken as the training set, with the remaining 40% of samples as the validation set. The classification and prediction efficiency of the model was evaluated using receiver operating characteristic (ROC) curves.
Validation of the expression of hub genes
The expression profiles and FC of hub genes from the SVM model were validated using a second dataset (GSE49126 in the Gene Expression Omnibus database) and a patient cohort collected from our hospital. The GSE49126 dataset consisted of samples of PBMCs from normal controls and patients with PD. The expression values of DEGs in GSE49126 were identified using GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) with log2 transformation and Benjamini–Hochberg adjustment.
Patient collection
The validation patient samples were collected at the Department of Neurology, the First Affiliated Hospital of Soochow University, Suzhou, China, between January 2020 and May 2020. Blood samples were collected from 103 outpatients with PD after obtaining approval from the ethics committee of the hospital. Blood samples from 31 sex- and age-matched healthy individuals were used as controls. Written informed consent was obtained from all participants.
Real-time polymerase chain reaction (PCR) analysis
PBMCs were isolated from the blood samples, and total RNA was extracted using Trizol (Invitrogen, Shanghai, China). The RNA was reverse transcribed into cDNA and used for PCR analysis. The specific PCR primer pairs were purchased from Sangon (Shanghai, China). PCR amplification was performed on an Applied Biosystems 7500 Fast Real Time PCR System instrument (Foster City, CA, USA). Human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference. The relative expression level was calculated using the 2–ΔΔCt method.
Statistical analysis
Statistical analyses were performed using SPSS for Windows, version 22.0 (SPSS Inc., Chicago, IL, USA). Differences in variables between the PD and control groups were analyzed using the Mann–Whitney U test or χ2 test. Differences in gene expression levels between the two groups were analyzed using the Mann–Whitney U test. Logistic regression was performed to identify correlations between gene expression and PD diagnosis. Spearman’s correlation analysis was used to identify correlations between disease status and gene expression. Odds ratios (ORs) and 95% confidential intervals (CIs) were calculated. For all analyses, P < 0.05 was considered statistically significant.
Results
Data preprocessing and DEG analysis
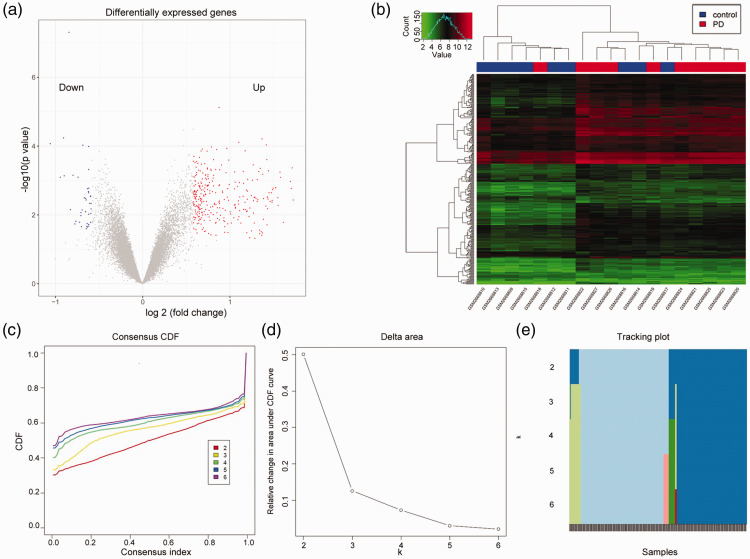
The GSE100054 dataset consisted of 19 samples (9 normal controls and 10 patients with PD). There were 24,233 genes in this dataset after processing. A total of 333 DEGs were identified, of which 293 genes were significantly upregulated and 40 were significantly downregulated (Figure 1a). These 333 DEGs were used for all further analyses. The hierarchical clustering analysis of the 19 samples based on DEG expression profiles is shown in Figure 1b. One PD sample was clustered into the normal group and three control samples were clustered into the PD group. After removing the four misclassified samples, the clustering of the other samples remained unchanged (data not shown).
Figure 1.
Statistical analysis and clustering of differentially expressed genes (DEGs) in Parkinson’s disease (PD). (a) Volcano plot of the DEGs. (b) Sample hierarchical clustering based on the expression of the DEGs. (c) Consensus cumulative distribution function (CDF) under different clustering numbers of DEGs. A larger CDF indicates a better gene clustering effect. When k = 2, the clustering result was the best. (d) The area difference value (delta area) between two CDF curves and the horizontal axis. k = 3 was the largest k with an appreciable increase in consensus. (e) Distribution of the clustering results shown by an item tracking plot. When k = 2, the number of mixed elements was the smallest.
Clustering analysis of DEGs
The clustering analysis of the DEGs based on the CDF revealed that the clustering result was the best when k = 2. As shown in Figure 1c and d, the increase rate of CDF significantly decreased and tended to reach consensus when the clustering number was >2. The largest k value with an appreciable increase in consensus was k = 3. Moreover, when k = 2, the number of mixed elements was the smallest (Figure 1e). Accordingly, the 333 DEGs were categorized into two clusters: cluster 1 genes (170 DEGs) and cluster 2 genes (163 DEGs).
Functions and pathway of gene sets
The cluster 1 genes were significantly enriched in one pathway, hsa04145:Phagosome, and in seven biological processes that related to innate immune and inflammatory responses. The cluster 2 genes were involved in 11 pathways, including hsa04510:Focal adhesion, hsa04060:Cytokine−cytokine receptor interaction, hsa04062:Chemokine signaling pathway, and hsa04512:ECM-receptor interaction (Figure 2a). They were also involved in 11 biological processes, including platelet degranulation, blood coagulation, cell adhesion, and inflammatory response (Figure 2b).
Figure 2.
Gene enrichment results of cluster 1 genes (a) and cluster 2 genes (b). The horizontal axis shows the number of genes. The gray solid line represents –lg(P-value).
Pathway deregulation analysis
The significant P-values of the aforementioned 12 pathways involving genes from the two clusters are shown in Table 1. Eight pathways were significantly different between the PD and control groups. These were hsa04060:Cytokine–cytokine receptor interaction, hsa04062:Chemokine signaling pathway, hsa04145:Phagosome, hsa04810:Regulation of actin cytoskeleton, hsa04144:Endocytosis, hsa04540:Gap junction, hsa04510:Focal adhesion, and hsa04611:Platelet activation. Thirty-nine genes involved in these pathways were obtained, and are listed in Table 2.
Table 1.
Significantly deregulated pathways between Parkinson’s disease and control samples.
| Pathway | P-value |
|---|---|
| hsa04060:Cytokine–cytokine receptor interaction | 0.025838 |
| hsa04062:Chemokine signaling pathway | 0.02773 |
| hsa04145:Phagosome | 0.032876 |
| hsa04810:Regulation of actin cytoskeleton | 0.03367 |
| hsa04144:Endocytosis | 0.03721 |
| hsa04540:Gap junction | 0.038855 |
| hsa04510:Focal adhesion | 0.042322 |
| hsa04611:Platelet activation | 0.049717 |
| hsa04640:Hematopoietic cell lineage | 0.053322 |
| hsa04022:cGMP–PKG signaling pathway | 0.054027 |
| hsa04512:ECM–receptor interaction | 0.058062 |
| hsa00230:Purine metabolism | 0.059821 |
Table 2.
The 39 differentially expressed genes in the eight significantly deregulated pathways between Parkinson’s disease patients and controls.
| Gene | logFC | Gene | logFC | Gene | logFC |
|---|---|---|---|---|---|
| MSR1 | 0.746 | PRKG1 | 0.688 | DNAJC6 | 0.856 |
| CD36 | 0.706 | MYLK | −0.195 | RAB11A | 0.690 |
| FCAR | 0.588 | GP9 | 0.631 | EHD3 | 0.589 |
| NCF2 | 0.622 | ITGA2B | 1.284 | TNFSF4 | 1.162 |
| TLR4 | 0.600 | PDGFA | 0.922 | PPBP | 0.885 |
| CD14 | 0.754 | ITGB5 | 0.827 | CXCL5 | 1.417 |
| P2RY12 | 1.720 | PARVB | 0.808 | IL1B | 1.232 |
| GP6 | 0.752 | MYL9 | 1.253 | PF4 | 1.185 |
| PTGS1 | 1.234 | VCL | 0.680 | MPL | 1.529 |
| GUCY1A3 | 1.020 | DNM3 | 1.454 | PF4V1 | 1.108 |
| GUCY1B3 | 1.096 | DAB2 | 1.059 | TUBA8 | 0.980 |
| GP1BA | 1.435 | PARD3 | 0.766 | TUBB1 | 1.531 |
| ITGB3 | 1.430 | PSD3 | 1.008 | GNG11 | 1.554 |
FC, fold change.
PPI network construction
The 39 genes involved in the eight significant pathways were extracted, and 85 interactions between 33 products were predicted. The PPI network included 33 nodes, and 85 edges were constructed accordingly (Figure 3a). The top 10 nodes with the highest degrees were considered as hub nodes, and were as follows: integrin subunit alpha 2b (ITGA2B, degree = 14), integrin subunit beta 3 (ITGB3, degree = 11), purinergic receptor P2Y12 (P2RY12, degree = 10), CD36 molecule (CD36, degree = 10), platelet factor 4 (PF4, degree = 10), glycoprotein IX platelet (GP9, degree = 10), glycoprotein Ib platelet subunit alpha (GP1BA, degree = 9), glycoprotein VI (GP6, also known as GPVI, degree = 7), integrin subunit beta 3 (ITGB5, degree = 7), and vinculin (VCL, degree = 7).
Figure 3.
The protein–protein interaction (PPI) network and analysis of the 10 hub genes. (a) PPI network of the genes enriched in the significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Pink nodes represent the 10 hub genes. A larger node size indicates a higher degree of interaction. (b) Receiver operating characteristic (ROC) curve analysis of the SVM model for predicting disease status in the GSE100054 dataset. (c) The log2(fold change) value of the 10 hub genes in the validation dataset (GSE49126) and study dataset (GSE100054). (d) Relative expression levels of the 10 genes in Parkinson’s disease (PD) and controls in the patient cohort.
Prediction and validation of the SVM model
The expression values of the top 10 genes in the 19 samples were regarded as the characteristic values, and the SVM model was constructed accordingly. Next, 60% of samples (n = 11) were taken as the training set, and the other 40% of samples (n = 8) were taken as the validation set and were used to validate the model. As shown in the ROC curves, the model fitting effect was the best in the training set, with an accuracy of 92%. The accuracy in the validation set was 75% (Figure 3b). Together, these results indicated that the SVM model was an independent predictive factor for PD.
Validation of hub genes
The GSE49126 dataset consisted of 50 samples from 20 normal controls and 30 patients with PD. The expression profiles of the 10 hub genes are shown in Figure 3c and d. In this validation dataset, 9 of the 10 genes were upregulated in the PD group compared with the control group, while CD36 was downregulated (log2FC > 0; Figure 3c). The expression levels of the 10 genes were also validated in a patient cohort consisting of 103 patients with PD (67.10 ± 11.2 years old) and 31 healthy controls (65.51 ± 8.77 years old). There was no difference in age or the sex ratio between the two groups (Table 3). The PD patients had an average disease duration of 6.93 ± 2.23 years (range: 0.5–12 years) and an average Hoehn and Yahr stage of 2.61 ± 0.64 (range: 1–4; Table 3). Real-time PCR analysis revealed that 2 of the 10 hub genes (GP1BA and GP6) were upregulated in PD samples compared with controls in this validation patient cohort (n = 134; P < 0.01 in the Mann–Whitney U test; Figure 3d).
Table 3.
The demographic characteristics of the Parkinson’s disease patients and controls.
| Variables | Parkinson’s disease | Control | P-value |
|---|---|---|---|
| GSE100054 | (n = 10) | (n = 9) | |
| Age (years) | 67.00 ± 8.89 | 62.33 ± 7.50 | 0.211b |
| Sex ratio (male/female) | 5/4 | 5/5 | 1.000a |
| Validation cohort | n = 103 | n = 31 | |
| Age (years) | 67.10 ± 11.20 | 65.51 ± 8.77 | 0.463b |
| Sex ratio (male/female) | 54/49 | 16/15 | 1.000a |
| Duration (years) | 6.93 ± 2.23 | 0 | |
| Hoehn and Yahr stage | 2.61 ± 0.64 (range 1–4) | 0 |
aχ2 test; bMann–Whitney U test.
Screening of independent risk factors in PD patients
Logistic regression analysis revealed that the expression levels of GP1BA, GP6, ITGB5, and P2RY12 were independent predictors of PD in both the GSE100054 dataset and the validation cohort (Table 4). Spearman’s correlation analysis revealed that none of the 10 hub genes were associated with the Hoehn and Yahr stage of PD (data not shown).
Table 4.
Association between genes and Parkinson’s disease status in the GSE100054 dataset and validation cohort.
| Variable |
GSE100054 (n = 19) |
Validation cohort
(n = 134) |
||||||
|---|---|---|---|---|---|---|---|---|
| β | OR | 95% CI | P-value | β | OR | 95% CI | P-value | |
| CD36 | 2.191 | 12.075 | 1.159–125.786 | 0.037 | 0.175 | 2.574 | 0.286–2.470 | 0.752 |
| GP1BA | 1.325 | 3.761 | 1.106–12.793 | 0.034 | 2.568 | 6.526 | 2.885–13.455 | 0.019 |
| GP6 | 2.608 | 13.576 | 1.040–177.137 | 0.047 | 2.365 | 8.636 | 1.630–15.676 | <0.0001 |
| GP9 | 1.740 | 5.699 | 0.966–33.618 | 0.055 | 0.366 | 1.998 | 0.354–2.809 | 0.996 |
| ITGA2B | 0.815 | 2.260 | 0.982–5.199 | 0.055 | 0.533 | 1.704 | 0.581–5.002 | 0.332 |
| ITGB3 | 0.719 | 2.051 | 0.968–4.346 | 0.061 | 0.063 | 1.725 | 0.873–40.162 | 0.435 |
| ITGB5 | 1.926 | 6.862 | 1.173–40.162 | 0.033 | 1.260 | 3.526 | 1.249–9.957 | 0.017 |
| P2RY12 | 1.341 | 3.822 | 1.260–11.592 | 0.018 | 2.612 | 13.628 | 4.343–42.758 | <0.0001 |
| PF4 | 1.182 | 3.260 | 1.051–10.108 | 0.041 | 0.645 | 1.907 | 0.635–5.727 | 0.250 |
| VCL | 2.185 | 8.888 | 1.054–74.996 | 0.045 | 1.208 | 3.449 | 0.805–2.874 | 0.371 |
OR, odds ratio; CI, confidence interval.Significant difference (p < 0.05) is highlighted by bold.
Discussion
A previous study by Miki et al.21 identified that the downregulation of six core regulators of autophagy and proteins upstream of autophagy may play crucial roles in PD, and are associated with increased α-synuclein levels in PBMCs. In the present study, using different analytical methods and criteria for selecting DEGs, we identified 333 circulating DEGs in the PBMCs of PD patients. These genes were clustered into two gene sets and were significantly enriched in 12 pathways, of which 8 were significantly different between the PD and control groups, including hsa04060:Cytokine–cytokine receptor interaction, hsa04062:Chemokine signaling pathway, hsa04810:Regulation of actin cytoskeleton, and hsa04540:Gap junction. These results are in line with a previous study by Liu et al.,22 who reported that DEGs in the GSE100054 dataset are mainly enriched in KEGG pathways, including gap junctions and platelet activation. Furthermore, 10 hub nodes in the PPI network, including GP1BA, ITGB3, GP6, and ITGB5, were used to construct the SVM model, which had high accuracy in predicting PD in the present study. Among the 10 hub genes, 4 (GP1BA, ITGB3, GP6, and P2RY12) were independent risk factors of PD. However, none of these genes were associated with Hoehn and Yahr scores. PD is a relatively common neurological disorder characterized by the degeneration of motor and cognitive functions, and is caused by a loss of neurons in the central nervous system. Inflammation is a key part of the immune response that protects the human body against injury.23 Inflammatory components, such as cytokines and chemokines, are related to neuroinflammation in the central nervous system. Notably, studies have demonstrated the important role of neuroinflammation in PD pathology.24 For example, Mutez et al.25 used PBMC transcriptome profiles to show that the deregulation of eukaryotic initiation factor 2 (EIF2) signaling, endocytosis, and the immune system are the main transcriptome features of sporadic or hereditary PD. Pro-inflammatory cytokines not only contribute to neuronal death, but also influence neurodegenerative pathways such as tau phosphorylation and amyloid precursor protein processing.24 In the present study, pathways including hsa04060:Cytokine–cytokine receptor interaction and hsa04062:Chemokine signaling pathway were significantly deregulated, and were enriched in PF4, which is crucial for platelet death and activation as well as for monocyte migration.26–28 Other upregulated genes, including P2RY12, GP9, GP6, and GP1BA, were also enriched in these pathways. These findings indicate the critical roles of these genes in PD pathology via inflammation-related pathways.
The three upregulated platelet glycoproteins (GP9, GP6, and GP1BA) are physiological collagen receptors that activate the collagen receptor α2β1 to bind collagen.29–31 GP6 is a major platelet receptor, and its clustering or aggregation plays crucial roles in promoting platelet activation, thrombus growth, and atherosclerosis by interacting with exposed collagen on injury vessel walls.31,32 Interestingly, PD is associated with increased deep vein thrombosis.33 The purinergic G-protein coupled receptor P2RY12 is a therapeutic target of the clinically approved anti-thrombotic drugs cangrelor and clopidogrel.34 Kloss et al.34 demonstrated that P2RY12 activation triggers the migration of macrophages to necrotic tumor areas and modulates chemotaxis and the chemokine secretion of macrophages. Our research revealed that the genes for all four of these proteins (P2RY12, GP6, GP9, and GP1BA) were upregulated in PD samples in the GSE49126 and GSE100054 datasets, although only GP1BA and GP6 were upregulated in our clinical cohort of PD patients. However, all four of these genes (P2RY12, GP6, GP9, and GP1BA) were independent risk factors of PD. These findings highlight the roles of macrophage migration and platelet activation in the pathology of PD. Furthermore, they suggest that P2RY12 might be a therapeutic target in PD.
As described earlier in this article, the loss of nigrostriatal dopaminergic neurons and the formation of α-synuclein-rich inclusions (Lewy bodies) are the neuropathological hallmarks of PD.35 The present study indicates that abnormal stabilization of the actin cytoskeleton plays a key role in mediating α-synuclein neurotoxicity, and that modifiers exert their effects by modulating the actin cytoskeleton.36 Genetic modification of the actin cytoskeleton may control both α-synuclein and tau neurotoxicity.37 Moreover, GP6 clustering depends on a dynamic actin cytoskeleton,31 and platelet activation is also actin-dependent.38,39 In addition, some studies have reported that β3 integrins play a dominant role in plasma fibronectin fibrillogenesis via β3 integrin–cytoskeleton interaction-mediated fibronectin unfolding and assembly.40 Interestingly, our results showed that the hsa04810:Regulation of actin cytoskeleton pathway, which contained enriched hub genes such as upregulated ITGB5, ITGB3, and ITGA2B, was significantly deregulated in the blood samples from PD patients compared with controls. The association of ITGB5 with the diagnosis of PD in our study highlights the role of the actin cytoskeleton as well as integrin–cytoskeleton interactions in the pathology of PD.
Moreover, the hsa04540:Gap junction pathway was also significantly deregulated in the present study, with enriched protein kinase CGMP-dependent 1 (PRKG1). Gap junctions contribute to the formation of intercellular channels that connect the cytoplasmic compartments of neighboring cells.41 It has been reported that glial gap junctions play critical roles in maintaining homeostasis in the central nervous system under physiological conditions. These structures also contribute to the onset and progression of pathological conditions.42 Importantly, a recent study demonstrated that neurotoxic activated microglia secrete glutamate through gap junction hemichannels.43 Thus, glial and neuronal communication via gap junctions might amplify neuroinflammation and neurodegeneration.43 Understanding the pathological roles of gap junctions may therefore contribute to new therapeutic strategies against neurodegenerative diseases, including PD.
In conclusion, our results suggest that inflammation-associated pathways, including cytokine−cytokine receptor interaction and chemokine signaling pathways, as well as pathways regulating the actin cytoskeleton and gap junctions, play critical roles in PD pathology. The hub genes, including GP1BA, GP6, P2RY12, and ITGB5, that were enriched in these pathways may serve as potential diagnostic markers of PD. However, these findings need to be further validated in clinical studies.
Availability of data and material
All data generated or analyzed during this study are included in this published article. The mRNA expression profiling datasets GSE100054 and GSE46129 are available from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/).
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author contributions
Xue SR conceived and designed the research. Chen XC, Chen C, Li WJ, Shen J, Han Q, Zhen JY, and Lu JM acquired, analyzed and interpreted data. Shen J drafted the manuscript. Xue SR revised the manuscript for important intellectual content. All authors have read and approved the manuscript.
ORCID iD
Shou-Ru Xue https://orcid.org/0000-0001-9178-0433
References
- 1.Elbaz A, Carcaillon L, Kab S, et al. Epidemiology of Parkinson's disease. Rev Neurol (Paris) 2016; 172: 14–26. [DOI] [PubMed] [Google Scholar]
- 2.Jellinger KA. Neuropathology of sporadic Parkinson's disease: evaluation and changes of concepts. Mov Disord 2012; 27: 8–30. [DOI] [PubMed] [Google Scholar]
- 3.Shihabuddin LS, Brundin P, Greenamyre JT, et al. New frontiers in Parkinson's disease: from genetics to the clinic. J Neurosci 2018; 38: 9375–9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verstraeten A, Theuns J, Van Broeckhoven C. Progress in unraveling the genetic etiology of Parkinson disease in a genomic era. Trends Genet 2015; 31: 140–149. [DOI] [PubMed] [Google Scholar]
- 5.Do CB, Tung JY, Elizabeth D, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson's disease. PLoS Genet 2011; 7: e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pihlstrøm L, Axelsson G, Bjørnarå KA, et al. Supportive evidence for 11 loci from genome-wide association studies in Parkinson's disease. Neurobiol Aging 2013; 34: 1708.e7–e13. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez DG, Nalls MA, Ylikotila P, et al. Genome wide assessment of young onset Parkinson's disease from Finland. PLos One 2012; 7: e41859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalinderi K, Bostantjopoulou S, Fidani L. The genetic background of Parkinson's disease: current progress and future prospects. Acta Neurol Scand 2016; 134: 314–326. [DOI] [PubMed] [Google Scholar]
- 9.Spatola M, Wider C. Genetics of Parkinson's disease: the yield. Parkinsonism Relat Disord 2014; 20: S35–S38. [DOI] [PubMed] [Google Scholar]
- 10.Barrett T, Troup D, Wilhite S, et al. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res 2007; 35: D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry R, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie M, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010; 26: 1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang DW, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball C, Blake J, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drier Y, Sheffer M, Domany E. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci U S A 2013; 16: 6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2014: gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Wang Q. Screening of feature genes in distinguishing different types of breast cancer using support vector machine. Onco Targets Ther 2015; 8: 2311–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miki Y, Shimoyama S, Kon T, et al. Alteration of autophagy-related proteins in peripheral blood mononuclear cells of patients with Parkinson's disease. Neurobiol Aging 2018; 63: 33–43. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Chen J, Guan T, et al. miRNAs and target genes in the blood as biomarkers for the early diagnosis of Parkinson’s disease. BMC Syst Biol 2019; 13: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finch CE, Morgan TE. Systemic inflammation, infection, ApoE alleles, and Alzheimer disease: a position paper. Curr Alzheimer Res 2007; 4: 185–189. [DOI] [PubMed] [Google Scholar]
- 24.Alam Q, Alam MZ, Mushtaq G, et al. Inflammatory process in Alzheimer and Parkinson's diseases: central role of cytokines. Curr Pharm Des 2015; 22: 541–548. [DOI] [PubMed] [Google Scholar]
- 25.Mutez E, Nkiliza A, Belarbi K, et al. Involvement of the immune system, endocytosis and EIF2 signaling in both genetically determined and sporadic forms of Parkinson's disease. Neurobiol Dis 2014; 63: 165–170. [DOI] [PubMed] [Google Scholar]
- 26.Fox JM, Kausar F, Day A, et al. CXCL4/platelet factor 4 is an agonist of CCR1 and drives human monocyte migration. Sci Rep 2018; 8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord MS, Cheng B, Farrugia BL, et al. Platelet factor 4 binds to vascular proteoglycans and controls both growth factor activities and platelet activation. J Biol Chem 2017; 292: 4054–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones CG, Pechauer SM, Curtis BR, et al. A platelet factor 4-dependent platelet activation assay facilitates early detection of pathogenic heparin-induced thrombocytopenia antibodies. Chest 2017; 152: e77–e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz MA, Chen J, Whitelock JL, et al. The platelet glycoprotein Ib–von Willebrand factor interaction activates the collagen receptor α2β1 to bind collagen: activation-dependent conformational change of the α2-I domain. Blood 2005; 105: 1986–1991. [DOI] [PubMed] [Google Scholar]
- 30.Li R. The Glycoprotein Ib-IX-V complex In: Michelson AD, editor. Platelets. 4th ed Massachusetts: Academic Press, 2019, pp.193–211. [Google Scholar]
- 31.Poulter N, Pollitt AY, Owen D, et al. Clustering of glycoprotein VI (GPVI) dimers upon adhesion to collagen as a mechanism to regulate GPVI signaling in platelets. J Thromb Haemost 2017; 15: 549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao Y, Chen Y, Adili R, et al. Plant-based food cyanidin-3-glucoside modulates human platelet glycoprotein VI signaling and inhibits platelet activation and thrombus formation. J Nutr 2017; 147: 1917–1925. [DOI] [PubMed] [Google Scholar]
- 33.Tana C, Lauretani F, Ticinesi A, et al. Molecular and clinical issues about the risk of venous thromboembolism in older patients: a focus on Parkinson’s disease and parkinsonism. Int J Mol Sci 2018; 19: 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloss L, Dollt C, Schledzewski K, et al. ADP secreted by dying melanoma cells mediates chemotaxis and chemokine secretion of macrophages via the purinergic receptor P2Y12. Cell Death Dis 2019; 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci 2001; 2: 492–501. [DOI] [PubMed] [Google Scholar]
- 36.Ordonez DG, Lee MK, Feany MB. α-synuclein induces mitochondrial dysfunction through spectrin and the actin cytoskeleton. Neuron 2018; 97: 108–124e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tudor AF, Ilan ES, Vikram K, et al. Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat Cell Biol 2007; 9: 139–148. [DOI] [PubMed] [Google Scholar]
- 38.Dasgupta SK, Le A, Da Q, et al. Wdr1-dependent actin reorganization in platelet activation. PLoS One 2016; 11: e0162897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayr S, Hauser F, Peterbauer A, et al. Localization microscopy of actin cytoskeleton in human platelets. Int J Mol Sci 2018; 19: 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen H, Huynh K, Stoldt VR. Shear-dependent fibrillogenesis of fibronectin: impact of platelet integrins and actin cytoskeleton. Biochem Biophys Res Commun 2018; 497: 797–803. [DOI] [PubMed] [Google Scholar]
- 41.Yeager M, Harris AL. Gap junction channel structure in the early 21st century: facts and fantasies. Curr Opin Cell Biol 2007; 19: 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orellana JA, Sáez PJ, Shoji KF, et al. Modulation of brain hemichannels and gap junction channels by pro-inflammatory agents and their possible role in neurodegeneration. Antioxid Redox Signal 2009; 11: 369–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi H, Suzumura A. Gap junctions and hemichannels composed of connexins: potential therapeutic targets for neurodegenerative diseases. Front Cell Neurosci 2014; 8: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The mRNA expression profiling datasets GSE100054 and GSE46129 are available from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/).