Abstract
We evaluated severe acute respiratory syndrome coronavirus 2 RNA clearance in 22 children. The estimation of positivity at day 14 was 52% for nasopharyngeal swab and 31% for stool samples. These data underline the significance of nasopharyngeal and stoolsample for detecting infected children. Additional studies are needed for transmissibility.
Keywords: children, coronavirus, SARS-CoV-2, viral RNA shedding
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly spread worldwide since its onset in Wuhan, China, in December 2019. On 11 March 2020, the World Health Organization declared it a public health emergency of international concern.
Globally, the proportion of SARS-CoV-2 among children is small compared with other age groups. A recent review of 72 314 cases by the Chinese Center for Disease Control and Prevention showed that SARS-CoV-2 had affected only 1% of children aged ≤9 years and 1% aged 10 to 19 years [1].
In contrast with adults, children infected by SARS-CoV-2 seem to have a milder clinical course and good prognosis [2]. The most common signs and symptoms are cough, pharyngeal erythema, fever, upper respiratory symptoms, fatigue, and gastrointestinal symptoms including diarrhea and vomiting [3, 4].
At the pandemic onset, children were a possible source of spreading, even with a paucity of symptoms. Few data on viral RNA clearance and route of transmission of SARS-CoV-2 are available [5]. Still, debate is ongoing regarding oral–fecal transmission and the length of contagiousness. Considering the possible role of a “silent” source of infection [6], children need to be investigated as they relate to this issue in order to direct future actions when facing this pandemic.
Here, we present preliminary data on viral RNA clearance of SARS-CoV-2 in a series of 22 children admitted to the Bambino Gesù Pediatric Hospital (BGPH) coronavirus disease (COVID) Center.
METHODS
Twenty-two pediatric patients with infection confirmed by nasopharyngeal swab SARS-COV-2 nucleic acid test were included in the study. All patients were followed in an inpatient setting from 16 March 2020 to 8 April 2020 at a COVID center created for the SARS-CoV-2 pandemic in the first week of March 2020. Clinical records were reviewed to collect demographic information, contact history, previous history, clinical symptoms, and laboratory findings. Microbiological data consisted of reverse trascriptasi-polymerase chain reaction for SARS-CoV-2 RNA on nasopharyngeal conjunctival swabs and in stool and urine samples. These tests were repeated every 2–3 days until there were 2 consecutive negative results in the absence of new symptoms (see Supplementary Materials for SARS-CoV-2 nucleic acid detection). The follow-up date was moved to 12 April 2020; 13 patients were discharged on this date. The duration of symptoms and of RNA shedding was measured from the ilness onset to the date of symptom regression and to the date of first negative stwab or sample for viral shedding. The Kaplan-Meier method was used to estimate the duration of symptoms and viral RNA shedding for symptomatic patients; swab-positive patients were censored at the date of last swab. The study was approved by the local institutional review board.
RESULTS
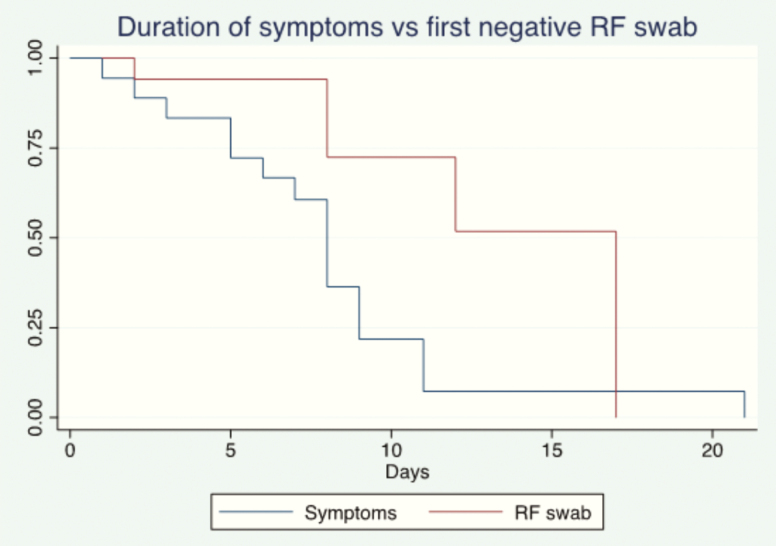
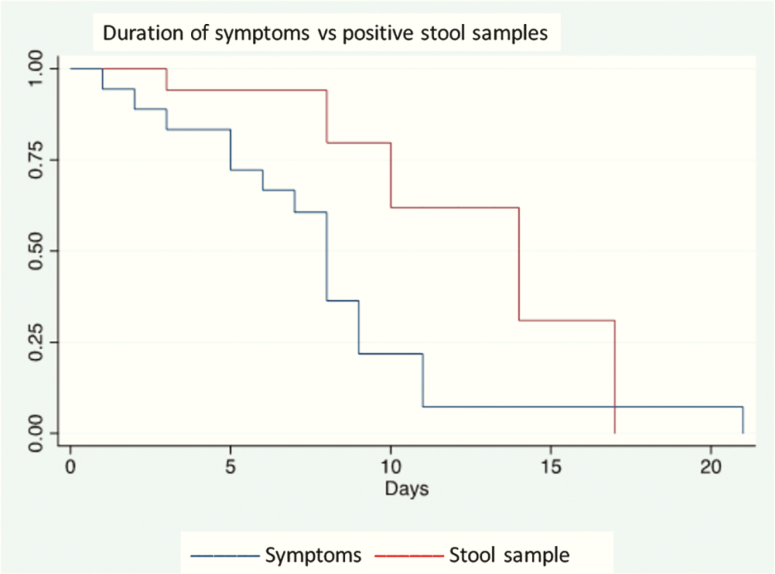
Twenty-two patients were followed in an inpatient setting at the BGPH COVID Center; the sex ratio (F/M) was 7/15 and median age was 84 months (range, 8 days to 210 months). The patients’ characteristics and symptoms are summarized in Table 1. Four patients were asymptomatic. None suffered immunodeficiency or received any immunosuppressive medication; 2 patients had an underlying condition (Angelman syndrome, suspected genetic syndrome, and autism). Of the 4 newborns, tests were performed after discovery of positive results for the mother (1 case) and 3 midwives; in all cases, the mother was positive. Two of the newborns were completely asymptomatic; the other 2 presented with low-grade fever and hyporexia, not initially related with SARS-CoV2 infection. At the last follow-up on 12 April 2020, symptoms had regressed in all 18 symptomatic patients; symptoms lasted for a median of 8 days (range, 1 to 21). A family cluster was identified in 19 patients. At diagnosis, stool samples were positive for SARS-CoV-2 in 15/22 (68%) patients, urine in 1/22 (4.5%), and conjunctival swabs in 2/22 (9.1%). At the last follow-up on 12 April 2020, 13 patients were discharged (median length of stay: 7 days, range, 3 to 15 days). Nasopharyngeal swabs remained positive in 7/13 patients, 54% (95% confidence interval [CI], 25–81), and the stool swabs remained positive in 6/9, 67% (95% CI, 30–93). Nasopharyngeal swabs were negative at a median of 8 days (range, 2 to 17 days) from the date of symptom onset, and stool samples were negative at a median of 14 days from the date of symptom onset (range, 10 to 15 days). Viral RNA shedding in stool samples and nasopharyngeal swabs at diagnosis and discharge is summarized in Table 1 where the differences in symptom occurrence according to positive stool swabs are summarized. Overall, the estimation of persistence of viral RNA shedding at day 14 from symptom onset was 52% (95% CI, 21–76) for nasopharyngeal swabs and 31% (95% CI 5–63) for stool samples (see Figures 1 and 2). Figures 1 and 2 show the estimation of symptom improvement and nasopharyngeal and stool RNA clearance over time in our population. At day 11, the estimated proportion of patients with symptoms was 6% (95% CI, 4%–22%); 94% of the population had no symptoms at day 11.
Table 1.
Patient Characteristics
| Sex Ratio (F/M) | 7/15 | |
|---|---|---|
| Age (median months, range) | 84 (8 days to 210 months) | |
| Symptoms | None | 4/22 (18%) |
| Fever | 15/18 (83%) | |
| Respiratory symptoms | 10/18 (55%) | |
| Diarrhea and vomiting | 7/18 (39%) | |
| Seizure | 3/18 (17%) | |
| Symptoms in positive stool swaba | Respiratory symptoms | 7/15 (47%) |
| Diarrhea and vomiting | 3/15 (20%) | |
| Symptom length (median days, range) | 8 (2–21) | |
| Nasopharyngeal swab | Positive at admission | 22/22 (100%) |
| Positive at dischargeb | 7/13 (54%) | |
| Positive nasopharyngeal swab length (median days, range) | 8 (2–17) | |
| Stool swab | Positive at admission | 15/22 (68%) |
| Positive at dischargeb | 6/13 (46%) | |
| Persistent positive at dischargec | 6/9 (67%) | |
| Positive stool swab length (median days, range) | 14 (10–14) |
aOnly patients who were stool sample-positive were considered, and symptoms were calculated in this population.
bCalculated considering only patients discharged.
cPatients who were stool sample-positive at discharge out of patients with positive stool samples at admission to the hospital.
Figure 1.
Representation of viral RNA shedding from RF tract according to Kaplan-Meyer method. Day 0 represents the onset of symptoms. Asymptomatic patients were excluded from the Kaplan-Meier analysis. Abbreviation: RF, rhinopharyngeal.
Figure 2.
Representation of viral RNA shedding in stool samples according to Kaplan-Meyer method. Day 0 represents the onset of symptoms. Asymptomatic patients were excluded from the Kaplan Meier analysis.
DISCUSSION
Infants and young children are generally considered at high risk for viral respiratory tract infection from respiratory syncytial virus and influenza virus. The respiratory tract immaturity and immune system contribute to severe viral respiratory disease in this age group. The SARS-CoV-2 epidemiology and clinical course in childhood is still unclear to clinicians, epidemiologists, and scientists.
In our series, SARS-CoV-2 was confirmed to have lower incidence and milder symptoms [2–4] in children compared with adults, as was reported also in SARS-CoV and Middle East respiratory syndrome-CoV epidemics [7, 8].
The major concern relating to our data is that fecal RNA shedding was reported in 68% of patients independently from gastrointestinal symptoms and with relatively slow RNA clearance. Indeed, about 50% of patients were discharged with positive stool swabs. In children who initially tested positive for SARS-CoV-2 on feces, the median time to regression of fecal RNA shedding from symptoms onset was 14 days, with an estimated positive stool swab at day 14 from symptom onset in 31% of patients. For nasopharyngeal swabs, 52% of patients were still positive at day 14 from symptom onset. Considering the small patient population and incomplete observation, the Kaplan-Meier method was used for this estimation. Our data are consistent with the pediatric experience reported by Lo at al [9] who showed the common gastrointestinal involvement confirmed by fecal RNA shedding in the absence of specific symptoms. Moreover, the stool swabs remained positive even when nasopharyngeal swabs were negative; long-lasting RNA shedding was suggested by our estimation curve in one third of the population as observed by Xu et al [10]. Although both studies involved limited pediatric populations, viral RNA shedding from the digestive system raises the possibility of fecal–oral transmission. Fecal–oral transmission exists with other respiratory viruses, but evidence of replication-competent virus in fecal swabs is necessary to support extrapulmonary transmission. In adults, data related to fecal viral RNA shedding are more consistent [11, 12]. SARS-CoV-2 RNA and viral nucleocapsid were observed in a gut biopsy in intracellular staining, and vital viral load was confirmed in stool samples [13]. The clinical relevance of fecal viral RNA shedding both in terms of infectiousness and transmissibility needs to be confirmed. Indeed, as suggested by Yeo et al [14], future studies should investigate the oral–fecal transmission of SARS-CoV-2, including environmental analysis of sewage water to determine whether the virus remains viable in conditions favorable for transmission. Obviously, the oral–fecal transmission of SARS-CoV-2 is a main concern in pediatric populations.
We suggest close monitoring of SARS-CoV-2 patients after discharge with stool swabs added to nasopharyngeal swab testing in order to identify a child who has been cured and to clarify children’s role in the transmission chain. Obviously, an additional cost for the national health system has to be considered. The implication of potential oral–fecal transmission is more notable considering children’s specific characteristics, especially incontinent children. The viral spread via oral–fecal transmission is well known in school communities, primarily in younger children, with consequent implications on social and public health policies. Indeed, our data could suggest that a substantial proportion of viral RNA shedding likely occurred after symptoms resolved. More inclusive criteria for contact tracing should be considered for effective control of the outbreak and to capture potential transmission events the days before and following the onset of symptoms.
Based on results from Kelvin and Halperin [6], we support the idea that children are susceptible to SARS-CoV-2 infection but are less likely to be symptomatic or to present severe symptoms, raising the possibility that children could be facilitators of viral transmission. However, the importance of children in transmitting the virus remains uncertain. It is likely that pediatric infections partially contributed to “undocumented infections” [15] and explain the rapid geographic spread of SARS-CoV2 with its challenging containment.
Our data suggest the importance of fecal swabs along with nasopharyngeal swabs for monitoring viral RNA shedding in children and for clearly identifying patients who have been cured. Future studies are needed to clarify the potential infectiousness and the role of viral RNA shedding in pediatric populations. This will be important in limiting SARS-CoV-2 transmission and gaining an understanding of its relationship with clinical course.
Supplementary Material
Notes
Acknowledgments. We thank the medical and nursing staff members who cared for patients and parents at their personal risk during this pandemic.
Author contributions. All authors approved the final version of the paper.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2. Ludvigsson JF. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr 2020:10.1111/apa.15270. doi: 10.1111/apa.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hong H, Wang Y, Chung HT, Chen CJ. Clinical characteristics of novel coronavirus disease 2019 (COVID-19) in newborns, infants and children. Pediatr Neonatol 2020;61:131–2. doi: 10.1016/j.pedneo.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zimmermann P, Curtis N. Coronavirus infections in children including COVID-19. An overview of the epidemiology, clinical features, diagnosis, treatment and prevention options in children. Pediatr Infect Dis J 2020. doi: 10.1097/INF.0000000000002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu Y, Li X, Zhu B, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 2020. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kelvin AA, Halperin S. COVID-19 in children: the link in the transmission chain. Lancet Infect Dis 2020:S1473-3099(20)30236-X. doi: 10.1016/S1473-3099(20)30236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Denison MR. Severe acute respiratory syndrome coronavirus pathogenesis, disease and vaccines: an update. Pediatr Infect Dis J 2004; 23:S207–14. [DOI] [PubMed] [Google Scholar]
- 8. Al-Tawfiq JA, Kattan RF, Memish ZA. Middle East respiratory syndrome coronavirus disease is rare in children: an update from Saudi Arabia. World J Clin Pediatr 2016; 5:391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lo IL, Lio CF, Cheong HH, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci 2020; 16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Y, Li X, Zhu B, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med 2020. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID. medRxiv 2020. doi: 10.1101/2020.03.15.20036707. [DOI] [PubMed] [Google Scholar]
- 12. Cheung KS, Hung IF, Chan PP, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology 2020:S0016-5085(20)30448-0. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020:S0016-5085(20)30282-1. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal-oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol Hepatol 2020; 5:335–7. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science 2020:eabb3221. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.