Abstract
Background
The case-fatality ratios (CFR) of coronavirus disease 2019 (COVID-19) and severe acute respiratory syndrome (SARS) appeared to differ substantially. We aimed to compare the CFR and its predictors of COVID-19 and SARS patients using a territory-wide cohort in Hong Kong.
Methods
This was a territory-wide retrospective cohort study using data captured from all public hospitals in Hong Kong. Laboratory-confirmed COVID-19 and SARS patients were identified. The primary endpoint was a composite endpoint of intensive care unit admission, use of mechanical ventilation, and/or death.
Results
We identified 1013 COVID-19 patients (mean age, 38.4 years; 53.9% male) diagnosed from 23 January to 14 April 2020 and 1670 SARS patients (mean age, 44.4 years; 44.0% male) from March to June 2003. Fifty-five (5.4%) COVID-19 patients and 432 (25.9%) SARS patients had reached the primary endpoint in 30 days. By 30 June 2003, 286 SARS patients had died (CFR, 17.1%). By 7 June 2020, 4 COVID-19 patients had died (CFR, 0.4%). After adjusting for demographic and clinical parameters, COVID-19 was associated with a 71% lower risk of primary endpoint compared with SARS (adjusted hazard ratio, 0.29; 95% confidence interval, .21–.40; P < .0001). Age, diabetes mellitus, and laboratory parameters (high lactate dehydrogenase, high C-reactive protein, and low platelet count) were independent predictors of the primary endpoint in COVID-19 patients, whereas use of antiviral treatments was not associated with primary endpoint.
Conclusions
The CFR of COVID-19 was 0.4%. Age and diabetes were associated with worse outcomes, whereas antiviral treatments were not.
Keywords: SARS-CoV-2, COVID-19, nCoV, death, pneumonia
The case-fatality ratio of COVID-19 observed in Hong Kong was 0.4%, much lower than that of SARS (17.1%) in 2003. Advanced age and presence of diabetes are 2 risk factors of death and intensive care unit admission in COVID-19 patients.
In late 2002 to mid-2003, an outbreak of severe acute respiratory syndrome (SARS), a newly emerged infectious disease caused by a novel coronavirus SARS-coronavirus (CoV), occurred in southern China [1]. SARS posed an enormous global public health threat in 2003 because 8098 patients were infected worldwide, resulting in 774 deaths in 26 countries, with an overall case fatality ratio (CFR) of 9.6% [2].
Another novel zoonotic coronavirus hit the world in late 2019 [3]. This novel virus, with the official name of SARS-CoV-2, is responsible for causing coronavirus disease 2019 (COVID-19), which has affected more than 180 countries and 30 territories in all 7 continents of the world. COVID-19 has resulted in more than 8.86 million cases and 465 000 deaths worldwide as of 22 June 2020, with a CFR of 5.3% [4]. The CFR related to COVID-19 appeared to be lower than to that of SARS.
Because SARS-CoV-2 is closely related to SARS-CoV [5], the exact reasons for the difference in CFRs between COVID-19 and SARS remained obscure. Moreover, because the number or proportion of infections that remained undiagnosed was unknown and varied widely across countries because of differences in testing strategies, the true CFR for COVID-19 was uncertain. With the evolving pandemic, the prediction of CFR and the risk factors of death are important for distribution of healthcare resources. In this study, we aimed to compare the CFR and its predictors of COVID-19 and SARS patients using a territory-wide cohort in Hong Kong.
METHODS
Setting and Study Design
We performed a territory-wide retrospective cohort study using data from the Clinical Data Analysis and Reporting System (CDARS) under the management of the Hospital Authority, Hong Kong [6]. CDARS is an electronic healthcare database that covers the patients’ demographic, death, diagnoses, procedures, drug prescription and dispensing history, and laboratory results from all public hospitals and clinics in Hong Kong. It represents inpatient data of about 90% of the 7.47 million population in Hong Kong [7]. Furthermore, all confirmed SARS and COVID-19 patients were hospitalized in public, not private, hospitals in Hong Kong. Patients were deidentified in CDARS to ensure confidentiality. Different territory-wide studies of various infectious diseases were previously conducted using CDARS [8–10].
Subjects
Consecutive SARS patients from March to June 2003 and consecutive COVID-19 patients from 23 January 2020 to 14 April 2020 were identified by International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) diagnosis codes and/or virological results (Supplementary Table 1). In Hong Kong, we performed testing for SARS-CoV-2 polymerase chain reaction (PCR) for both symptomatic patients presenting to outpatient clinics and hospitals as well as asymptomatic close contacts of confirmed patients and inbound travellers. All patients diagnosed with COVID-19 were hospitalized. Patients were followed until death, admission to an intensive care unit (ICU), use of invasive mechanical ventilation, last day of hospitalization or last clinic visit, date of data retrieval (7 June 2020), and up to 30 days of follow-up, whichever came first. The study protocol was approved by the Joint Chinese University of Hong Kong—New Territories East Cluster Clinical Research Ethics Committee.
Clinical Evaluation
The clinical evaluation of SARS patients in 2003 was described in detail in our previous publications [11, 12]. All COVID-19 patients were admitted to medical wards or ICUs with isolation facilities. Initial investigations included a complete blood count (with a differential count), clotting profile (prothrombin time, activated partial-thromboplastin time, international normalized ratio), and serum biochemical measurements (including electrolytes, renal and liver biochemistries, C-reactive protein, lactate dehydrogenase, glucose, and procalcitonin). These laboratory assessments and chest radiography scans were performed regularly as clinically indicated. Microbiological workup, including sputum and blood bacterial culture, nasopharyngeal aspirate for respiratory viruses and atypical pathogens, and urine for Streptococcus pneumoniae and Legionella antigen tests, were performed as appropriate. A real-time reverse transcription PCR (RT-PCR) assay was used to detect a conserved region in the E gene of SARS-CoV and SARS-CoV-2.
Clinical Management of COVID-19 Patients
Supportive therapy, including supplemental oxygen, intravenous fluid, vasopressor support, mechanical ventilation, and renal replacement therapy, were given as appropriate. Patients were started on lopinavir-ritonavir (200 mg/50 mg twice daily) monotherapy or in combination with ribavirin (400 mg twice daily) and/or interferon beta-1b, for up to 14 days, according to local interim guidelines, if antiviral therapy was considered appropriate. Antibacterial therapy was given if bacterial coinfections were suspected clinically or confirmed by microbiological tests. Systemic corticosteroids were not given routinely, except for selected patients (eg, those with refractory shock). Patients were discharged when they improved clinically and when 2 consecutive clinical specimens tested negative for SARS-CoV-2 via RT-PCR.
Data Collection
Data were retrieved from CDARS in June 2020. Baseline date was defined as the date of hospitalization for the diseases in the corresponding periods. Demographic data including date of birth and sex were captured. At baseline, hematological and virologic parameters and liver and renal biochemistries were collected. Thereafter, serial liver and renal biochemistries as well as SARS-CoV or SARS-CoV-2 RT-PCR tests were collected until the last follow-up date (Supplementary Table 1). We also retrieved data on other relevant diagnoses, procedures, concomitant drugs, laboratory parameters, and exposure to antivirals, antibiotics and antifungals, corticosteroid, interferon-beta, and intravenous immunoglobulin (IVIG) during the hospitalization (Supplementary Table 2).
Definitions
The primary endpoint was a composite endpoint of ICU admission, use of invasive mechanical ventilation, and/or death. Death and its date were ascertained using data from both CDARS and the Hong Kong Death Registry. All deaths that happened during the study period from March 2003 to June 2020 were retrieved and analyzed. Use of invasive mechanical ventilation was defined by ICD-9-CM procedure codes (96.7). Significant comorbidities were defined as follows: hypertension was identified by any use of antihypertensive drugs and/or ICD-9-CM diagnosis codes; diabetes mellitus was defined by exposure to any antidiabetic agents, and/or hemoglobin A1c ≥ 6.5%, and/or fasting plasma glucose ≥7 mmol/L in 2 measurements or ≥11.1 mmol/L in 1 measurement, and/or the ICD-9-CM diagnosis codes for diabetes mellitus (250.00–250.93) [13]. Other comorbidities were identified based on ICD-9-CM codes (Supplementary Table 3). Leukopenia was defined by total white blood cell count <3.5 × 109/L, moderate lymphopenia by absolute lymphocyte count <1000 per cubic millimeter, and thrombocytopenia by platelet count <150 000 per cubic millimeter.
Statistical Analysis
Data were analyzed using Statistical Product and Service Solutions, version 25.0 (IBM Corp., Armonk, NY, USA), SAS (9.4; SAS Institute Inc., Cary, NC, USA), and R software (3.6.3; R Foundation for Statistical Computing, Vienna, Austria). Continuous variables were expressed in mean ± standard deviation or median (interquartile range [IQR]), as appropriate, while categorical variables were presented as number (percentage). Qualitative and quantitative differences between subgroups were analyzed by χ 2 or Fisher’s exact tests for categorical parameters and Student t test or Mann-Whitney U test for continuous parameters, as appropriate.
Missing data were assumed missing at random and replaced with substituted values by multiple imputation by chained equations to create 20 complete data sets after the first 10 burn-in iterations [14]. The variables (percentage of missing data) included in the imputation model were age, sex, hemoglobin (1.6%), white blood cell (1.7%), platelet (1.7%), alanine aminotransferase (1.8%), alkaline phosphatase (1.8%), albumin (1.8%), total bilirubin (1.8%), international normalized ratio (25.3%), creatinine (1.8%), urea (1.8%), sodium (2.9%), potassium (2.9%), C-reactive protein (17.0%), lactate dehydrogenase (LDH) (4.1%), and comorbidities at baseline (Supplementary Table 4). The primary endpoint measure and the corresponding Nelson-Aalen estimator of the cumulative hazard at the time of primary endpoint or censoring were also included in the imputation model [15]. Imputed values were constrained within plausible ranges.
Cumulative probabilities of the primary endpoint were estimated by Kaplan-Meier method with 95% confidence interval (CI); log-rank test was used to compare the cumulative probability in COVID-19 and SARS patients. On univariate and multivariable analysis, hazard ratios and adjusted hazard ratios (aHRs) with 95% CI were estimated with the Cox proportional hazard model. We included the following covariates: COVID-19 versus SARS, age, sex, presence of comorbidities, laboratory parameters including baseline complete blood count, liver and renal functions, C-reactive protein, and LDH. The overall coefficient estimates and standard errors were computed by combining the estimates obtained on each individual multiple imputation data set using Rubin’s rules [16]. On multivariable analysis, backward elimination was performed by repeated use of Rubin’s rules to select important covariates [17]. Schoenfeld’s global test was used to test the proportional hazards assumption, which did not detect any significant violations. All statistical tests were 2-sided. Statistical significance was taken as P < .05.
RESULTS
Demographic Characteristics
We identified 1013 COVID-19 patients (all COVID-19 patients reported to the Department of Health from 23 January to 14 April 2020) and 1670 SARS patients (95.2% of all reported SARS patients) from March to June 2003. Among COVID-19 patients, 705 (69.6%) were imported cases and 222 patients (21.9%) were secondary cases of imported and local cases, respectively [18]. The number of COVID-19 patients with coexisting conditions were: diabetes mellitus in 80 (7.9%), cardiovascular disease in 145 (14.3%), chronic liver disease in 37 (3.7%), respiratory disease in 11 (1.1%), and kidney disease in 8 (0.8%). One of them was a healthcare worker. At baseline, compared with patients with SARS, patients with COVID-19 were younger, more likely to be male, and had a lower prevalence of various comorbidities, including cardiovascular diseases and diabetes (Table 1 and Supplementary Table 5).
Table 1.
Baseline Clinical Characteristics Before Multiple Imputation of Patients With SARS-CoV-2 Infection/COVID-19 or SARS-CoV Infection/SARS
| Baseline clinical characteristics | All N = 2683 | COVID-19 N = 1013 | SARS N = 1670 | P Value |
|---|---|---|---|---|
| Age, y | 42.2 ± 19.5 | 38.4 ± 17.7 | 44.4 ± 20.1 | <.0001 |
| Male, n (%) | 1280 (47.7) | 546 (53.9) | 734 (44.0) | <.0001 |
| Comorbidities, n (%) | ||||
| Cardiovascular diseases | 539 (20.1) | 145 (14.3) | 394 (23.6) | <.0001 |
| Hypertension | 493 (18.4) | 138 (13.6) | 355 (21.3) | <.0001 |
| Ischemic heart disease | 61 (2.3) | 6 (0.6) | 55 (3.3) | <.0001 |
| Cardiac dysrhythmias | 93 (3.5) | 10 (1.0) | 83 (5.0) | <.0001 |
| Heart failure | 58 (2.2) | 2 (0.2) | 56 (3.4) | <.0001 |
| Digestive diseases | 272 (10.1) | 42 (4.1) | 230 (13.8) | <.0001 |
| Peptic ulcer | 47 (1.8) | 1 (0.1) | 46 (2.8) | <.0001 |
| Chronic liver disease | 179 (6.7) | 37 (3.7) | 142 (8.5) | <.0001 |
| Liver failure, cirrhosis, or cirrhotic complications | 8 (0.3) | 0 (0) | 8 (0.5) | .028 |
| Biliary disease | 24 (0.9) | 3 (0.3) | 21 (1.3) | .010 |
| Gastrointestinal hemorrhage | 49 (1.8) | 1 (0.1) | 48 (2.9) | <.0001 |
| Diabetes mellitus | 357 (13.3) | 80 (7.9) | 277 (16.6) | <.0001 |
| Malignant tumor | 75 (2.8) | 13 (1.3) | 62 (3.7) | <.0001 |
| Nervous system diseases | 130 (4.8) | 13 (1.3) | 117 (7.0) | <.0001 |
| Cerebrovascular events | 87 (3.2) | 8 (0.8) | 79 (4.7) | <.0001 |
| Other nervous system diseasesa | 83 (3.1) | 5 (0.5) | 78 (4.7) | <.0001 |
| Respiratory diseaseb | 121 (4.5) | 11 (1.1) | 110 (6.6) | <.0001 |
| Kidney disease | 61 (2.3) | 8 (0.8) | 53 (3.2) | <.0001 |
| Human immunodeficiency virus infection | 5 (0.2) | 3 (0.3) | 2 (0.1) | .37 |
| Follow-up duration, d | 21 (14–28) | 21 (13–29) | 21 (15–28) | .62 |
| Laboratory results | ||||
| Creatinine, µmol/Lc | 77 (64–92) | 71 (60–84) | 80 (67–97) | <.0001 |
| Urea, mmol/L | 4.7 ± 4.0 | 4.1 ± 1.5 | 5.1 ± 4.9 | <.0001 |
| Sodium, mmol/L | 136.6 ± 4.2 | 138.8 ± 3.0 | 135.4 ± 4.2 | <.0001 |
| Potassium, mmol/L | 3.8 ± 0.5 | 3.9 ± 0.4 | 3.8 ± 0.6 | <.0001 |
| Albumin, g/L | 39.7 ± 5.4 | 41.5 ± 4.9 | 38.6 ± 5.4 | <.0001 |
| ALT, U/Lc | 22 (15–37) | 22 (15–34) | 23 (15–39) | .14 |
| ALP, U/Lc | 64 (52–83) | 62 (51–75) | 67 (53–90) | <.0001 |
| AST, U/Lc | 26 (20–38) | 26 (21–37) | 26 (19–39) | .37 |
| GGT, U/Lc | 46 (28–88) | 41 (26–62) | 58 (30–129) | .00033 |
| Total bilirubin, μmol/L | 8.9 ± 8.7 | 8.1 ± 4.7 | 9.4 ± 10.3 | <.0001 |
| Total protein, g/L | 73.7 ± 6.5 | 74.8 ± 5.5 | 73.0 ± 7.0 | <.0001 |
| Haptoglobin, g/L | 2.2 ± 1.3 | 2.6 ± 1.3 | 2.1 ± 1.2 | .14 |
| LDH, U/Lc | 233 (176–355) | 183 (157–225) | 300 (208–423) | <.0001 |
| CRP, mg/dL | 3.0 ± 5.2 | 1.4 ± 3.6 | 4.2 ± 5.8 | <.0001 |
| ESR, mm/h | 29.6 ± 29.1 | 25.2 ± 23.6 | 31.7 ± 31.3 | <.0001 |
| Prothrombin time, s | 11.9 ± 3.4 | 12.1 ± 1.1 | 11.9 ± 3.9 | .014 |
| International normalized ratio | 1.1 ± 0.3 | 1.1 ± 0.1 | 1.1 ± 0.3 | .80 |
| Hemoglobin, g/dL | 13.4 ± 1.8 | 13.9 ± 1.5 | 13.0 ± 1.8 | <.0001 |
| WCC, ×109/L | 6.4 ± 3.1 | 5.7 ± 2.0 | 6.7 ± 3.5 | <.0001 |
| WCC < 3.5 × 109/L, n (%) | 248 (9.4) | 88 (9.0) | 160 (9.6) | .63 |
| Lymphocyte, ×109/L | 1.2 ± 0.7 | 1.5 ± 0.7 | 1.0 ± 0.7 | <.0001 |
| Lymphocyte < 1 × 109/L, n (%) | 1146 (43.4) | 219 (22.6) | 927 (55.6) | <.0001 |
| Monocyte, ×109/L | 0.5 ± 0.3 | 0.5 ± 0.2 | 0.5 ± 0.3 | .21 |
| Neutrophil, ×109/L | 4.5 ± 2.9 | 3.7 ± 1.8 | 5.1 ± 3.2 | <.0001 |
| Eosinophil, ×109/L | 0.05 ± 0.1 | 0.07 ± 0.1 | 0.04 ± 0.2 | <.0001 |
| Platelet, ×109/L | 208.1 ± 82.0 | 228.3 ± 75.2 | 196.2 ± 83.5 | <.0001 |
| Platelet < 150 × 109/L, n (%) | 603 (22.9) | 114 (11.7) | 489 (29.4) | <.0001 |
| Fasting glucose, mmol/L | … | 5.8 ± 1.8 | … | - |
| Random glucose, mmol/L | … | 6.1 ± 2.4 | … | - |
| Ferritin, pmol/Lc | … | 658 (260–1528) | … | - |
| Procalcitonin, ng/mL | … | 0.2 ± 2.0 | … | - |
| Coinfections during follow-up, n (%) d | ||||
| Viral | 59 (2.2) | 22 (2.2) | 37 (2.2) | .94 |
| Bacterial | 139 (5.2) | 19 (1.9) | 120 (7.2) | <.0001 |
| Hypoxemia during follow-up, n (%) | 401 (14.9) | 53 (5.2) | 348 (20.8) | <.0001 |
| NSAID during follow-up, n (%) | 116 (4.3) | 38 (3.8) | 78 (4.7) | .26 |
| Antiviral treatment during follow-up, n (%) | ||||
| Ribavirin | 1942 (72.4) | 519 (51.2) | 1423 (85.2) | <.0001 |
| Lopinavir-ritonavir | 702 (26.2) | 592 (58.4) | 110 (6.6) | <.0001 |
| Interferon beta | 315 (11.7) | 315 (31.1) | 0 (0) | <.0001 |
| Oseltamivir | 201 (7.5) | 62 (6.1) | 139 (8.3) | .036 |
| Ganciclovir | 0 (0) | 0 (0) | 0 (0) | - |
| Acyclovir/famciclovir/valaciclovir | 24 (0.9) | 3 (0.3) | 21 (1.3) | .010 |
| Antibiotic treatment | 1926 (71.8) | 372 (36.7) | 1554 (93.1) | <.0001 |
| Antifungal treatment | 60 (2.2) | 0 (0) | 60 (3.6) | <.0001 |
| Corticosteroid | 1378 (51.4) | 42 (4.1) | 1336 (80.0) | <.0001 |
| Pulse methylprednisolone (≥250 mg daily) | 976 (36.4) | 4 (0.4) | 972 (58.2) | <.0001 |
| Peak daily dose (prednisolone equivalent, mg)d | 625 (100–625) | 37.5 (37.5–50) | 625 (625–625) | <.0001 |
| Intravenous immunoglobulin therapy | 76 (2.8) | 2 (0.2) | 74 (4.4) | <.0001 |
All comorbidities are represented as binary parameters. Categorical variables are presented as number (percentage). Alanine aminotransferase and follow-up duration are expressed in median (interquartile range), whereas other continuous variables were expressed in mean ± standard deviation. Qualitative and quantitative differences between subgroups were analyzed by χ 2 or Fisher’s exact tests for categorical parameters and Student t test or Mann-Whitney U test for continuous parameters, as appropriate.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; CRP, C-reactive protein; CoV, coronavirus; COVID-19, coronavirus disease 2019; ESR, erythrocyte sedimentation rate; ICD-9-CM, International Classification of Diseases, 9th edition, Clinical Modification; LDH, lactate dehydrogenase; SARS, severe acute respiratory syndrome; WCC, white cell count.
aRespiratory system disease was defined by ICD-9-CM diagnosis codes for pneumonia other than SARS-related pneumonia (ICD-9-CM codes: 480–487.0) in previous 3 months, chronic obstructive pulmonary disease and allied conditions (ICD-9-CM codes: 490–496), pneumoconioses and other lung diseases due to external agents (ICD-9-CM codes: 500–508) in previous 3 months, and other diseases of respiratory system (ICD-9-CM codes: 510–519) in previous 3 months.
bOther nervous system disease was defined by ICD-9-CM diagnosis codes for inflammatory diseases of the central nervous system (ICD-9-CM codes: 320–327), hereditary and degenerative diseases of the central nervous system (ICD-9-CM codes: 330–337), and other disorders of the central nervous system (ICD-9-CM codes: 340–345).
cPresented in median (interquartile range).
dBased on nasopharyngeal-aspirate reverse transcription-polymerase chain reaction and/or serology, sputum culture, and blood culture.
Hematologic and Biochemical Findings
The initial blood count showed similar prevalence of leukopenia in in 88 (9.0%) of COVID-19 and in 160 (9.6%) of SARS patients, whereas fewer COVID-19 patients had moderate lymphopenia (22.6% vs 55.6%) and thrombocytopenia (11.7% vs 29.4%) on presentation. Prothrombin time remained normal in most cases (Table 1 and Supplementary Table 5).
Serum chemical values were normal in the majority of COVID-19 patients. They had lower serum creatinine level, lower LDH level, and lower C-reactive protein level than SARS patients. The results of laboratory tests performed on presentation are listed in Table 1. The results in a single imputation data set are shown in Supplementary Table 6.
Microbiologic and Virologic Findings
The prevalence of viral coinfections was similar in COVID-19 and SARS patients, whereas bacterial coinfections were less common among COVID-19 patients (Table 1). COVID-19 patients had higher prevalence of rhinovirus/enterovirus, but lower prevalence of influenza A and B. The most common bacterial pathogens isolated from respiratory tract of COVID-19 patients were Staphylococcus aureus and Hemophilus influenzae (Table 2).
Table 2.
Nasopharyngeal-Aspirate (NPA), Sputum, and Blood Sample Results
| COVID-19 N = 1013 | SARS N = 1670 | P Value | |
|---|---|---|---|
| Nasopharyngeal-aspirate RT-PCR and/or serology | |||
| Influenza A | 0 (0%) | 20 (1.2%) | .00047 |
| Influenza B | 0 (0%) | 10 (0.6%) | .017 |
| Parainfluenza viruses 1–3 | 5 (0.5%) | 5 (0.3%) | .52 |
| Respiratory syncytial virus | 4 (0.4%) | 1 (0.1%) | .071 |
| Rotavirus | 0 (0%) | 0 (0%) | - |
| Adenovirus | 6 (0.6%) | 4 (0.2%) | .19 |
| Rhinovirus/enterovirus | 13 (1.3%) | 0 (0%) | <.0001 |
| Metapneumovirus | 1 (0.1%) | 0 (0%) | .38 |
| Sputum culture | |||
| Haemophilus influenzae | 5 (0.5%) | 24 (1.4%) | .022 |
| Haemophilus parainfluenzae | 0 (0%) | 2 (0.1%) | .53 |
| Klebsiella pneumoniae | 1 (0.1%) | 8 (0.5%) | .17 |
| Klebsiella species | 0 (0%) | 17 (1.0%) | .0013 |
| Streptococcus pneumoniae | 0 (0%) | 10 (0.6%) | .017 |
| Streptococcus—Group G | 0 (0%) | 1 (0.1%) | 1.00 |
| Methicillin-resistant Staphylococcus aureus (MRSA) | 0 (0%) | 40 (2.4%) | <.0001 |
| Staphylococcus aureus | 6 (0.6%) | 10 (0.6%) | .98 |
| Coagulase −ve staphylococci | 0 (0%) | 2 (0.1%) | .53 |
| Blood culture | |||
| Klebsiella pneumoniae | 0 (0%) | 1 (0.1%) | 1.00 |
| Staphylococcus aureus | 0 (0%) | 1 (0.1%) | 1.00 |
| Staphylococcus albus | 0 (0%) | 6 (0.4%) | .089 |
| Staphylococcus epidermidis | 0 (0%) | 2 (0.1%) | .53 |
| Staphylococcus haemolyticus | 0 (0%) | 1 (0.1%) | 1.00 |
| Staphylococcus hominis | 1 (0.1%) | 0 (0%) | .38 |
| Coagulase −ve Staphylococcus | 1 (0.1%) | 0 (0%) | .38 |
Qualitative and quantitative differences between subgroups were analyzed by χ 2 or Fisher’s exact tests for categorical parameters, as appropriate.
Abbreviations: COVID-19, coronavirus disease 2019; RT-PCR, reverse transcription-polymerase chain reaction; SARS, severe acute respiratory syndrome.
Pharmacological Treatment for COVID-19 and SARS Patients
Of the 1013 COVID-19 patients, 372 (36.7%) had received antibiotics, 592 (58.4%) lopinavir-ritonavir, 519 (51.2%) ribavirin, 315 (31.1%) interferon beta, 42 (4.1%) corticosteroid therapy (4 received pulse methylprednisolone), and 2 (0.2%) IVIG. Of the 1670 SARS patients, 1554 (93.1%) had received antibiotics, 1423 (85.2%) ribavirin, 110 (6.6%) lopinavir-ritonavir, 1336 (80.0%) corticosteroid therapy (972 received pulse methylprednisolone), and 74 (4.4%) IVIG (Table 1).
Clinical Outcomes
Fifty-five (5.4%) COVID-19 patients and 432 (25.9%) SARS patients had reached the primary endpoint in 30 days, respectively. Among the 1013 COVID-19 patients, 53 (5.2%) were admitted to the ICU, all because of respiratory failure. Mechanical ventilatory support was required in 22 patients (2.2%). The clinical characteristics of these patients are summarized in Supplementary Table 7. By 7 June 2020, 4 patients had died (CFR, 0.4%); a total of 1006 (99.3%) patients had been discharged [18]. By 7 June 2020, among the 53 patients admitted to the ICU, 1 (1.9%) remained in the ICU, 2 (3.8%) had been discharged from the ICU but remained in the hospital, 48 (90.6%) had been discharged from the hospital, and 2 (3.8%) had died.
Of the 1670 SARS patients, 333 (19.9%) were admitted to the ICU within 30 days. Invasive mechanical ventilatory support was required in 61 (3.7%) patients, whereas 43 (2.6%) required noninvasive ventilator support. By 30 June 2003, 286 patients had died (CFR, 17.1%). Among those admitted to the ICU, 136 (40.8%) patients had died. Supplementary Table 8 shows the comparisons between COVID-19 and SARS patients who developed the primary endpoint.
Factors Predictive of ICU Admission, Invasive Mechanical Ventilation, and Death in COVID-19 and SARS Patients
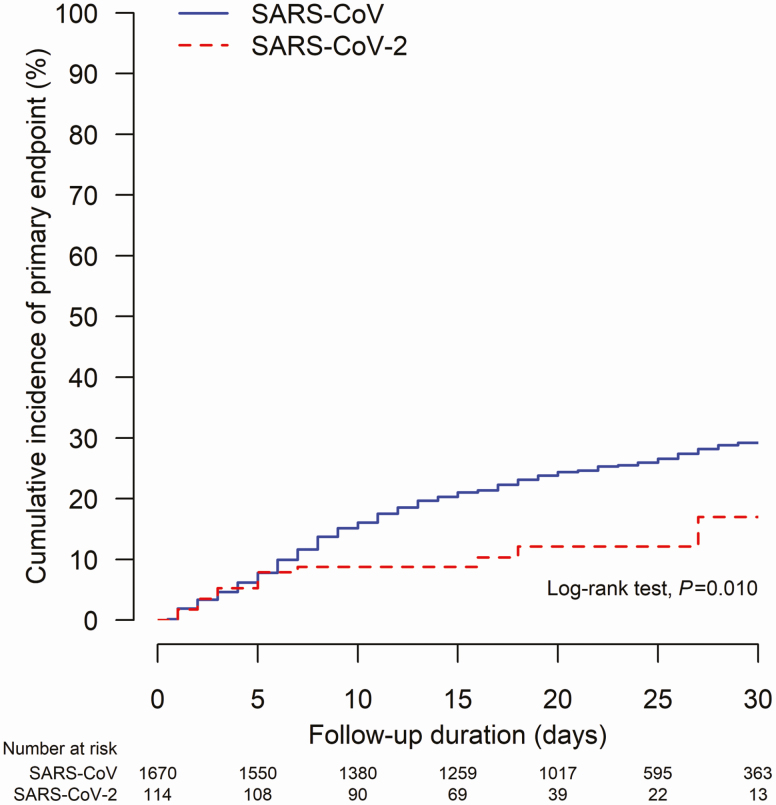
Supplementary Table 9 shows the comparisons between patients who developed and did not develop the primary endpoint. Univariate analysis showed that SARS, advanced age, male gender, comorbidities, laboratory parameters, coinfections, and treatment used for the viral infections were significant predictive factors for the primary endpoint (Table 3). On multivariable analysis, COVID-19 was associated with 71% lower risk of primary endpoint compared with SARS (aHR 0.29; 95% CI, .21–.40; P < .0001; Figure 1). Other factors that were predictive of an adverse outcome included advanced age (aHR per year 1.01; 95% CI, 1.01–1.02; P = .00015), male (aHR 1.24; 95% CI, 1.02–1.50; P = .027), diabetes mellitus (aHR 2.14; 95% CI, 1.74–2.63; P < .0001), and bacterial or viral coinfection (aHR 1.74; 95% CI, 1.36–2.22; P < .0001) (Table 3).
Table 3.
Univariate and Multivariable Analysis With Cox Proportional Hazard Model on Factors Associated with Primary Endpoint (A Composite Endpoint of Intensive Care Unit Admission, Use of Invasive Mechanical Ventilation, and Death) Among COVID-19 and SARS Patients After Multiple Imputation
| Parameters | Univariate Analysis | Multivariable Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P Value | aHR (95% CI) | P Value | |
| COVID-19 (vs SARS) | 0.21 (.16–.28) | <.0001 | 0.32 (.23–.44) | <.0001 |
| Age, y | 1.03 (1.03–1.04) | <.0001 | 1.01 (1.00–1.02) | .00022 |
| Male | 1.38 (1.15–1.64) | .00047 | 1.23 (1.02–1.49) | .030 |
| Hemoglobin | 0.85 (.81–.88) | <.0001 | … | |
| White blood cell | 1.10 (1.08–1.11) | <.0001 | … | |
| Platelet | 0.997 (.996–.998) | <.0001 | 0.997 (.996–.998) | <.0001 |
| Alanine aminotransferase | 1.002 (1.001–1.003) | <.0001 | … | |
| Albumin | 0.90 (.89–.91) | <.0001 | 0.97 (.95–.99) | .0011 |
| Total bilirubin | 1.01 (1.01–1.02) | .00014 | 0.99 (.98–1.00) | .029 |
| International normalized ratio | 1.56 (1.29–1.90) | <.0001 | … | |
| Creatinine | 1.002 (1.001–1.002) | <.0001 | … | |
| C-reactive protein | 1.08 (1.07–1.08) | <.0001 | 1.04 (1.03–1.06) | <.0001 |
| Lactate dehydrogenase (per 100 U/L) | 1.21 (1.19–1.24) | <.0001 | 1.11 (1.08–1.15) | <.0001 |
| Neutrophil-to-lymphocyte ratio | 1.06 (1.06–1.07) | <.0001 | 1.02 (1.01–1.03) | .0032 |
| Circulatory system disease | 3.55 (2.97–4.24) | <.0001 | … | |
| Digestive system disease | 1.89 (1.49–2.38) | <.0001 | … | |
| Diabetes mellitus | 4.30 (3.58–5.18) | <.0001 | 2.13 (1.74–2.61) | <.0001 |
| Malignant tumor | 2.83 (2.02–3.97) | <.0001 | … | |
| Nervous system disease | 1.91 (1.41–2.60) | <.0001 | 0.48 (.34–.68) | <.0001 |
| Respiratory disease | 2.33 (1.73–3.13) | <.0001 | … | |
| Chronic kidney disease | 3.31 (2.33–4.72) | <.0001 | … | |
| Bacterial or viral coinfection | 3.17 (2.51–4.00) | <.0001 | 1.71 (1.34–2.20) | <.0001 |
| Hypoxia during follow-up | 2.52 (2.08–3.06) | <.0001 | … | |
| Lopinavir-ritonavir during follow-up | 0.33 (.25–.44) | <.0001 | … | |
| Ribavirin during follow-up | 0.78 (.64–.95) | .013 | 0.44 (.35–.56) | <.0001 |
| Steroid during follow-up | 1.45 (1.21–1.74) | <.0001 | … | |
Abbreviations: aHR, adjusted hazard ratio; CI, confidence interval; COVID-19, coronavirus disease 2019; HR, hazard ratio; SARS, severe acute respiratory syndrome.
Figure 1.
Cumulative incidence of primary endpoint (a composite endpoint of death, intensive care unit admission, and use of invasive mechanical ventilation) in patients with SARS-CoV-2/COVID-19 versus SARS-CoV infection/SARS. Abbreviations: COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Factors Predictive of ICU Admission, Mechanical Ventilation, and Death in COVID-19 Patients
Five predictive factors were identified for adverse outcome on multivariable analysis: age (aHR 1.02; 95% CI, 1.00–1.04; P = .040), diabetes mellitus (aHR 3.21; 95% CI, 1.72–6.00; P = .00026), baseline LDH level (aHR per 100 U/L 1.48; 95% CI, 1.15–1.91; P = .0027), C-reactive protein (aHR 1.07; 95% CI, 1.02–1.12; P = .0065), and platelet count (aHR 0.995; 95% CI, .991–1.000; P = .031) (Table 4).
Table 4.
Univariate and Multivariable Analysis with Cox Proportional Hazard Model on Factors Associated with Primary Endpoint (A Composite Endpoint of Intensive Care Unit Admission, Use of Invasive Mechanical Ventilation, and Death) Among COVID-19 Patients After Multiple Imputation
| Parameters | Univariate Analysis | Multivariable Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P Value | aHR (95% CI) | P Value | |
| Age, y | 1.05 (1.04–1.07) | <.0001 | 1.02 (1.00–1.04) | .040 |
| Male | 1.63 (.94–2.85) | .084 | … | |
| Hemoglobin | 0.88 (.74–1.03) | .12 | … | |
| White blood cell | 1.14 (1.02–1.27) | .023 | … | |
| Platelet | 0.99 (.99–1.00) | .0065 | 0.995 (.991–1.000) | .031 |
| Alanine aminotransferase | 1.01 (1.00–1.02) | .0030 | … | |
| Albumin | 0.87 (.83–.90) | <.0001 | … | |
| Total bilirubin | 1.02 (.98–1.07) | .31 | … | |
| International normalized ratio | 2.14 (.46–9.97) | .33 | … | |
| Creatinine | 1.02 (1.01–1.03) | .00020 | … | |
| C-reactive protein | 1.14 (1.11–1.17) | <.0001 | 1.07 (1.02–1.12) | .0065 |
| Lactate dehydrogenase (per 100 U/L) | 2.17 (1.34–3.52) | .0031 | 1.48 (1.15–1.91) | .0027 |
| Neutrophil-to-lymphocyte ratio | 1.24 (1.18–1.30) | <.0001 | … | |
| Circulatory system disease | 5.62 (3.31–9.54) | <.0001 | … | |
| Digestive system disease | 3.48 (1.58–7.70) | .0020 | … | |
| Diabetes mellitus | 10.06 (5.90–17.15) | <.0001 | 3.21 (1.72–6.00) | .00026 |
| Malignant tumor | 2.88 (.70–11.83) | .14 | … | |
| Nervous system disease | 1.41 (.20–10.19) | .73 | … | |
| Respiratory disease | 5.90 (1.84–18.89) | .0028 | … | |
| Chronic kidney disease | 5.47 (1.33–22.46) | .018 | … | |
| Bacterial or viral coinfection | 1.41 (.44–4.52) | .56 | … | |
| Hypoxia during follow-up | 2.22 (.95–5.17) | .066 | … | |
| Lopinavir-ritonavir during follow-up | 0.95 (.56–1.62) | .85 | … | |
| Ribavirin during follow-up | 1.11 (.65–1.88) | .71 | … | |
| Interferon beta during follow-up | 1.14 (.66–1.99) | .64 | … | |
| Steroid during follow-up | 4.02 (1.90–8.51) | .0003 | … | |
Abbreviations: aHR, adjusted hazard ratio; CI, confidence interval; COVID-19, coronavirus disease 2019; HR, hazard ratio.
DISCUSSION
This is the first report to compare the clinical outcomes of COVID-19 patients and SARS patients with detailed patient-level data. Among our cohort of the first 1013 COVID-19 patients in Hong Kong, the CFR was 0.4%, and 5% had ICU admission or death within 30 days of hospital admission. Among patients admitted to the ICU, 4% died. Age, diabetes, and biochemical laboratory parameters were predictive of adverse outcomes, whereas none of the antiviral treatments were associated with clinical outcomes. COVID-19 was associated with 71% lower risk of adverse outcomes compared with SARS.
In our territory-wide cohort of COVID-19 patients in Hong Kong, 5% required ICU care and CFR was 0.4%. At the time of our study, testing for COVID-19 involved a wide range of patients, from those who were asymptomatic to those who were critically ill. Since the early weeks of 2020, testing for COVID-19 was done for hospitalized patients with pneumonia irrespective of contact and travel history and for patients with influenza-like illness and pneumonia presenting to private and public outpatient clinics. Later, testing was expanded to include asymptomatic inbound travellers and close contacts of confirmed cases [19]. Testing of ~800 serum samples taken from the general population of Hong Kong after the pandemic did not reveal any seropositive individuals [20]. All patients diagnosed with COVID-19 were hospitalized in Hong Kong for quarantine purposes, irrespective of disease severity. At the time of our analysis, 99.3% of our patients had already been discharged from the hospital or had died. Therefore, our observed CFR should be close to the true CFR of COVID-19 patients, with minimal risk of biases from preferential selection of severe cases and delayed reporting of deaths, as in studies performed in the early phase of novel disease epidemics [21].
Observed and estimated CFRs of COVID-19 across different populations around the world have varied greatly. Among the first 82 719 laboratory-confirmed cases in China, the overall CFR was 5.65% [22]. However, the CFR was much higher in Hubei province (5.9%–7.7%) than in other provinces outside Hubei (.86%–.98%) [22, 23]. The average CFR in Italy until March 2020 was 7.5%, with CFRs in different regions ranging from 3.1% to 16.7% [24]. Until early April 2020, the CFR in the United States was 3.2%, ranging from 0.7% to 5.7% among different states [25].
There are several reasons for the large differences between CFRs observed in different countries or cities. Accessibility to medical care and national strategies for testing and case identification are possibly the major factors causing differences in reported CFRs. Countries with scarce resources in proactive contact tracing and identification of milder or asymptomatic cases would inevitably see a falsely low denominator in estimating the CFR, leading to a falsely high CFR [26, 27]. As explained previously, our observed CFR likely reflected closely the true CFR of all symptomatic and asymptomatic patients with COVID-19. Another situation in which a complete dataset was available for all diagnosed patients was from the Diamond Princess cruise ship, in which an age-adjusted CFR of 0.5% was observed, which was very similar to our observation [26].
Another likely reason for the much lower CFR in Hong Kong than that in many other countries was the lower incidence of COVID-19 and the lower burden on surge capacity of our healthcare system. Italy, for example, reported a linear negative correlation between CFR and ICU admission rate among different provinces, indicating higher mortality being associated with absence of ICU care because of the operational capacity of ICUs being exceeded [24]. Mortality among patients admitted to ICU in other cities ranged from 26% to 62% early in the pandemic [28–30]. In our cohort, among those admitted to ICU, only 42% received mechanical ventilation and the CFR was only 4%, implying that patients who were less critically ill were admitted to the ICU for close monitoring. These findings highlight the importance of public health measures to “flatten the curve” in preventing depletion of hospital resources and direct impact on patient mortality [19, 31].
Differences in host determinants of cellular entry and other molecular mechanisms of pathogenesis may result in variations in genetic susceptibility among different ethnic groups to this infection [32], although large variations in CFRs observed within the same country could not be explained by genetic factors alone [22, 24].
Practice in prescribing medications with presumed antiviral activity varies greatly among different countries. However, to date, although antivirals like remdesivir or interferon beta, have been shown to reduce time to recovery and viral clearance [33, 34], there is no antiviral agent proven to reduce mortality in randomized clinical trials. In our own cohort, use of lopinavir-ritonavir, ribavirin, and interferon therapies was not associated with a reduced risk of adverse clinical outcomes both in univariate and multivariable analyses. Therefore, the use or nonuse of various antiviral agents is unlikely to cause significant differences in CFRs among different countries.
Patients with COVID-19 had a 71% lower risk of adverse clinical outcomes than patients with SARS in 2003 in Hong Kong. Differences in host characteristics partly accounted for this difference because patients with COVID-19 were generally younger and had fewer comorbidities. However, because the association of SARS with adverse clinical outcomes persisted after adjustment of host characteristics and use of antiviral and steroid treatment, higher virulence of SARS-CoV than SARS-CoV-2 is the most possible reason for the lower CFR observed in COVID-19. Although 1.2% of COVID-19 patients were asymptomatic and 81% had mild infections [35], asymptomatic or subclinical infections were rare in SARS, as shown in seroprevalence studies [36], Genomic differences, particularly amino acid substitutions concentrated in 2 nonstructural proteins and spike protein, might explain the differences in pathogenicity between the 2 viruses [37, 38], Moreover, Hong Kong had been the major epicenter of the SARS outbreak in 2003 [1]. This explained the similar number of COVID-19 and SARS patients in our cohort, despite the global number of COVID-19 patients far exceeding that of SARS. The huge burden on hospital, and particularly ICU, resources thus partly explains the higher CFR of SARS patients in Hong Kong (17.0%), compared with the global CFR (9.5%) in 2003 [27].
We have identified older age, diabetes, higher LDH, higher C reactive protein, and lower platelet count as independent predictors of adverse outcomes among COVID-19 patients. All of these variables have been identified as risk factors for mortality or adverse outcomes in other cohorts [39–42]. Nevertheless, different host characteristics and laboratory parameters were identified from various cohorts around the world as predictors of adverse outcomes. These differences may possibly be due to variations in disease severity and ethnic differences. For example, although all confirmed patients were hospitalized in Hong Kong, fewer than 40% of all confirmed patients were hospitalized in a state in United States, and disease severity varies between ethnic groups [40].
In particular, diabetes carried a 3-fold higher risk of adverse clinical outcomes in COVID-19 in our cohort and was an independent predictor of adverse outcomes in both COVID-19 and SARS. Among 18 571 COVID-19 patients in the United States, diabetes was more prevalent in those requiring ICU admission (32%) and hospitalization (24%) than those managed as outpatients (6%) [43]. In a cohort of 1099 COVID-19 patients in China, diabetes was also more prevalent in those with ICU admission, mechanical ventilation, or death (27% vs 6%) [44]. Glycosylated hemoglobin had a linear correlation with inflammation, hypercoagulability, and hypoxia in COVID-19 patients [45]. Diabetes is associated with impaired innate immunity, and pneumonia is an increasingly important cause of mortality in patients with diabetes [46]. Diabetes predisposed patients to cardiac and renal injuries, which were common complications in 45%–71% and 57%–88% of patients with severe or fatal disease, respectively [47, 48]. Diabetes also induces expression of angiotensin-converting enzymes in lung, liver, and heart tissues. Activation of angiotensin 1 and 2 receptors in diabetes may enhance pro-inflammatory cytokine responses, thereby increasing the risk of acute respiratory distress syndrome in COVID-19. Effective control of glucose and blood pressure in COVID-19 patients may help to reduce local inflammatory response and dampen the acute effects of viral infection [49].
Our study has provided important outcome data that facilitate the clinical management, quarantine arrangement, and the resource allocation amid the COVID-19 outbreak [50]. The established risk factors, namely advanced age and presence of comorbidities, facilitate early identification of population at risk, so that such people should strictly adopt social distancing or even staying at home [51, 52]. We are still in the middle of an ongoing outbreak worldwide and we have to identify hospitalized patients at risk of deterioration as soon as possible based on these risk factors [39].
The strength of our study includes a territory-wide cohort that covers about 90% of the inpatient service and essentially all the SARS and COVID-19 cases in Hong Kong. Data from real-life cohorts represent a wider spectrum of patients such that the findings from real-life cohorts are thus more readily applicable to routine clinical practice. Our study has a few limitations. First, the mean age of COVID-19 patients in our cohort was generally younger than other reported hospitalized cohorts [39, 40]. This was largely because of the large proportion of imported cases, who were younger patients involved in international travelling. Therefore, our results may not be extrapolated to regions with predominantly local transmission. Second, we missed 85 of 1755 (4.8%) of SARS patients in 2003 because of diagnosis coding. Nonetheless, we believe missing less than 5% of the patients does not have major impact on the findings because the proportion of deaths in our cohort (286/1670; 17.1%) was consistent with what was reported officially in 2003 (299/1755; 17.0%). Third, COVID-19 and SARS patients might have been different in terms of the baseline clinical characteristics (eg, age, gender, comorbidities) such that our study might be subjected to confounding as in other observational studies. Therefore, we applied multivariable adjustment on important baseline characteristics. Fourth, missing data on laboratory measurements might lead to biases as in other retrospective studies, though these biases can partially be compensated for by our respectable cohort size. Missing data were rare for common laboratory parameters because they are regularly checked in our routine clinical practice. Yet, some less common laboratory parameter, such as troponin, ferritin, or procalcitonin, might not be checked for every single patient because of minor variations of clinical practice in different hospitals. Multiple imputation with 20 imputed data sets was used to reduce the possible selection bias resulting from missing data [53]. Fifth, ascertainment bias may affect the reliability of the study because of inaccurate entry of certain diagnosis codes for comorbidities, namely diabetes mellitus and cardiovascular disease. We minimized this bias by including laboratory as well as medication data for certain diagnoses (diabetes mellitus, hypertension).
In conclusion, CFR of COVID-19 observed in Hong Kong was 0.4%. COVID-19 was associated with an ~71% lower risk of adverse clinical outcomes compared with SARS. Patients with risk factors, namely advanced age and presence of diabetes, would be at much higher risk of death and ICU admission. In view of the ongoing outbreak worldwide, we have to identify patients at risk of deterioration as soon as possible based on these risk factors. Health authorities should allocate adequate resources, in particular intensive care facilities, based on the trajectories of the numbers of confirmed cases and well ahead to avoid collapse of the healthcare systems.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. All authors were responsible for the study concept and design. G. W., T. Y., Y.-K. T., and G. L. were responsible for the acquisition and analysis of data, had full access to all of the data in the study, and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors were responsible for the interpretation of data, the drafting, and critical revision of the manuscript for important intellectual content.
Potential conflicts of interest. G. L. has served as an advisory committee member for Gilead, Merck, and GSK; speaker for Merck and Gilead; and received research grant from Gilead, Merck, and GSK. T. Y. has served as an advisory committee member and a speaker for Gilead Sciences. V. W. has served as an advisory committee member for 3V-BIO, AbbVie, Allergan, Boehringer Ingelheim, Echosens, Gilead Sciences, Intercept, Janssen, Novartis, Novo Nordisk, Perspectum Diagnostics, Pfizer, TARGET-NASH, and Terns, and a speaker for Bristol-Myers Squibb, Echosens, Gilead Sciences, and Merck; he has also received a research grant from Gilead Sciences. H. C. is an advisor for AbbVie, Aptorum, Arbutus, Hepion, Intellia, Janssen, Gilead, GSK, GRAIL, Medimmune, Merck, Roche, Vaccitech, VenatoRx, and Vir Biotechnology, and a speaker for Mylan, Gilead, and Roche. D. H. has served as an advisory committee member for Roche. G. W. has served as an advisory committee member for Gilead Sciences; as a speaker for Abbott, Abbvie, Bristol-Myers Squibb, Echosens, Furui, Gilead Sciences, Janssen, and Roche; and received research grant from Gilead Sciences. All other authors declare that they have no competing interests. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hui DSC, Zumla A. Severe acute respiratory syndrome: historical, epidemiologic, and clinical features. Infect Dis Clin North Am 2019; 33:869–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Oragnization. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. Available at: https://www.who.int/csr/sars/country/table2004_04_21/en/. Accessed 22 June 2020.
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Oragnization. Coronavirus disease 2019 (COVID-19) Situation Report—154 on 22 June 2020. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Accessed 23 June 2020.
- 5. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheung NT, Fung V, Chow YY, Tung Y. Structured data entry of clinical information for documentation and data collection. Stud Health Technol Inform 2001; 84:609–13. [PubMed] [Google Scholar]
- 7. The Hospital Authority. Hospital authority statistical report 2018–2019. Available at: https://www3.ha.org.hk/data/HAStatistics/StatisticalReport/2018–2019. Accessed 30 July 2020.
- 8. Lai JC, Wong GL, Yip TC, et al. Chronic hepatitis B increases liver-related mortality of patients with acute hepatitis E: a territorywide cohort study from 2000 to 2016. Clin Infect Dis 2018; 67:1278–84. [DOI] [PubMed] [Google Scholar]
- 9. Lui GCY, Wong NS, Wong RYK, et al. Antiviral therapy for hepatitis B prevents liver injury in patients with tuberculosis and hepatitis B coinfection. Clin Infect Dis 2020; 70:660–6. [DOI] [PubMed] [Google Scholar]
- 10. Yip TC, Wong VW, Chan HL, Tse YK, Lui GC, Wong GL. Tenofovir is associated with lower risk of hepatocellular carcinoma than entecavir in patients with chronic HBV infection in China. Gastroenterology 2020; 158:215–25 e6. [DOI] [PubMed] [Google Scholar]
- 11. Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003; 348:1986–94. [DOI] [PubMed] [Google Scholar]
- 12. Sung JJ, Wu A, Joynt GM, et al. Severe acute respiratory syndrome: report of treatment and outcome after a major outbreak. Thorax 2004; 59:414–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. American Diabetes A. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2018. Diabetes Care 2018; 41:S13–27. [DOI] [PubMed] [Google Scholar]
- 14. Little RJ, D’Agostino R, Cohen ML, et al. The prevention and treatment of missing data in clinical trials. N Engl J Med 2012; 367:1355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. White IR, Royston P. Imputing missing covariate values for the Cox model. Stat Med 2009; 28:1982–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rubin DB, Schenker N. Multiple imputation in health-care databases: an overview and some applications. Stat Med 1991; 10:585–98. [DOI] [PubMed] [Google Scholar]
- 17. Wood AM, White IR, Royston P. How should variable selection be performed with multiply imputed data? Stat Med 2008; 27:3227–46. [DOI] [PubMed] [Google Scholar]
- 18. Protection CfH. Latest situation of novel coronavirus infection in Hong Kong. Available at: https://chp-dashboard.geodata.gov.hk/nia/zh.html. Accessed 22 June 2020.
- 19. Cowling BJ, Ali ST, Ng TWY, et al. Impact assessment of non-pharmaceutical interventions against coronavirus disease 2019 and influenza in Hong Kong: an observational study. Lancet Public Health 2020; 5:e279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. To KK, Cheng VC, Cai J, et al. Seroprevalence of SARS-CoV-2 in Hong Kong and in residents evacuated from Hubei province, China: a multicohort study. Lancet Microbe 2020; 1:e111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lipsitch M, Donnelly CA, Fraser C, et al. Potential biases in estimating absolute and relative case-fatality risks during outbreaks. PLoS Negl Trop Dis 2015; 9:e0003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deng X, Yang J, Wang W, et al. Case fatality risk of the first pandemic wave of novel coronavirus disease 2019 (COVID-19) in China. Clin Infect Dis 2020 May 15; ciaa578.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leung K, Wu JT, Liu D, Leung GM. First-wave COVID-19 transmissibility and severity in China outside Hubei after control measures, and second-wave scenario planning: a modelling impact assessment. Lancet 2020; 395:1382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Immovilli P, Morelli N, Antonucci E, Radaelli G, Barbera M, Guidetti D. COVID-19 mortality and ICU admission: the Italian experience. Crit Care 2020; 24:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Team CC-R. Geographic differences in COVID-19 cases, deaths, and incidence–United States, February 12-April 7, 2020. MMWR Morb Mortal Wkly Rep 2020; 69:465–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Faust JS, Del Rio C. Assessment of deaths from COVID-19 and from seasonal influenza. JAMA Intern Med 2020; 180:1045–6. [DOI] [PubMed] [Google Scholar]
- 27. Rajgor DD, Lee MH, Archuleta S, Bagdasarian N, Quek SC. The many estimates of the COVID-19 case fatality rate. Lancet Infect Dis 2020; 20:776–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhatraju PK, Ghassemieh BJ, Nichols M, et al. Covid-19 in critically ill patients in the seattle region–case series. N Engl J Med 2020; 382:2012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grasselli G, Zangrillo A, Zanella A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA 2020; 323:1574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020; 8:475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barrett K, Khan YA, Mac S, Ximenes R, Naimark DMJ, Sander B. Estimation of COVID-19-induced depletion of hospital resources in Ontario, Canada. CMAJ 2020; 192:E640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paniri A, Hosseini MM, Akhavan-Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J Biomol Struct Dyn 2020:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid-19–preliminary report. N Engl J Med 2020 . May 22:NEJMoa2007764. [DOI] [PubMed] [Google Scholar]
- 34. Hung IF, Lung KC, Tso EY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 2020; 395:1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu Z, McGoogan JM. Characteristics of and important lessons from the Coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA 2020; 323:1239–42. [DOI] [PubMed] [Google Scholar]
- 36. Leung GM, Lim WW, Ho LM, et al. Seroprevalence of IgG antibodies to SARS-coronavirus in asymptomatic or subclinical population groups. Epidemiol Infect 2006; 134:211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect 2020; 26:729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020; 27:325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395:1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Price-Haywood EG, Burton J, Fort D, Seoane L. Hospitalization and mortality among black patients and white patients with Covid-19. N Engl J Med 2020; 382:2534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 2020; 58:1021–8. [DOI] [PubMed] [Google Scholar]
- 42. Bello-Chavolla OY, Bahena-Lopez JP, Antonio-Villa NE, et al. Predicting mortality due to SARS-CoV-2: a mechanistic score relating obesity and diabetes to COVID-19 outcomes in Mexico. J Clin Endocrinol Metab 2020; 105:dgaa346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prevention CfDCa. Interim U.S. guidance for risk assessment and public health management of healthcare personnel with potential exposure in a healthcare setting to patients with 2019 novel Coronavirus (2019-nCoV). Available at: https://www.cdc.gov/coronavirus/2019-ncov/hcp/guidance-risk-assesment-hcp.html. Accessed 22 June 2020.
- 44. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Z, Du Z, Zhu F. Glycosylated hemoglobin is associated with systemic inflammation, hypercoagulability, and prognosis of COVID-19 patients. Diabetes Res Clin Pract 2020; 164:108214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma RCW, Holt RIG. COVID-19 and diabetes. Diabet Med 2020; 37:723–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao J, Tu WJ, Cheng W, et al. Clinical features and short-term outcomes of 102 patients with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis 2020; 71:748–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Du Y, Tu L, Zhu P, et al. Clinical features of 85 fatal cases of COVID-19 from Wuhan. A retrospective observational study. Am J Respir Crit Care Med 2020; 201:1372–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bornstein SR, Dalan R, Hopkins D, Mingrone G, Boehm BO. Endocrine and metabolic link to coronavirus infection. Nat Rev Endocrinol 2020; 16:297–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Emanuel EJ, Persad G, Upshur R, et al. Fair allocation of scarce medical resources in the time of Covid-19. N Engl J Med 2020; 382:2049–55. [DOI] [PubMed] [Google Scholar]
- 51. Lagier JC, Colson P, Tissot Dupont H, et al. Testing the repatriated for SARS-Cov2: should laboratory-based quarantine replace traditional quarantine? Travel Med Infect Dis 2020; 34:101624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stein RA. COVID-19 and rationally layered social distancing. Int J Clin Pract 2020; 74:e13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sterne JA, White IR, Carlin JB, et al. Multiple imputation for missing data in epidemiological and clinical research: potential and pitfalls. BMJ 2009; 338:b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.