Abstract
Background
Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in blood, also known as RNAemia, has been reported, but its prognostic implications are poorly understood. This study aimed to determine the frequency of SARS-CoV-2 RNA in plasma and its association with coronavirus disease 2019 (COVID-19) clinical severity.
Methods
An analytical cross-sectional study was performed in a single-center tertiary care institution and included consecutive inpatients and outpatients with confirmed COVID-19. The prevalence of SARS CoV-2 RNAemia and the strength of its association with clinical severity variables were examined and included intensive care unit (ICU) admission, invasive mechanical ventilation, and 30-day all-cause mortality.
Results
Paired nasopharyngeal and plasma samples were included from 85 patients. The median age was 55 years, and individuals with RNAemia were older than those with undetectable SARS-CoV-2 RNA in plasma (63 vs 50 years; P = .04). Comorbidities were frequent including obesity (37.6%), hypertension (30.6%), and diabetes mellitus (22.4%). RNAemia was detected in 28/85 (32.9%) of patients, including 22/28 (78.6%) who required hospitalization. In models adjusted for age, RNAemia was detected more frequently in individuals who developed severe disease including ICU admission (32.1 vs 14.0%; P = .04) and invasive mechanical ventilation (21.4% vs 3.5%; P = .02). All 4 deaths occurred in individuals with detectable RNAemia. An additional 121 plasma samples from 28 individuals with RNAemia were assessed longitudinally, and RNA was detected for a maximum duration of 10 days.
Conclusions
This study demonstrated a high proportion of SARS-CoV-2 RNAemia, and an association between RNAemia and clinical severity suggesting the potential utility of plasma viral testing as a prognostic indicator for COVID-19.
Keywords: SARS-CoV-2, COVID-19, RNAemia, blood, severity
This cross-sectional study assessed the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNAemia in 85 individuals, and association with clinical severity. Results demonstrate a high frequency of SARS-CoV-2 RNAemia, and an association between RNAemia and clinical severity variables including intensive care unit admission and mortality.
The coronavirus disease 2019 (COVID-19) is most frequently diagnosed from nasopharyngeal (NP) swab samples. Preliminary data suggest that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA may also be detected in plasma and serum [1, 2]. However, the frequency and prognostic implication of viral RNA detection in these sample types is not fully understood. Starting with the identification of individuals with detectable SARS-CoV-2 RNA in nasopharyngeal swab samples, we aimed to investigate the frequency of SARS-CoV-2 RNA in plasma and its association with COVID-19 clinical characteristics and severity.
METHODS
We performed a cross-sectional study at the Stanford Health Care (SHC) Clinical Virology Laboratory, which serves adult and pediatric tertiary care hospitals and affiliated primary care and specialty clinics in northern California. From the beginning of in-house SARS-CoV-2 testing on March 2, 2020, residual ethylenediaminetetraacetic acid (EDTA) whole blood was systematically collected from all SHC patients with a respiratory sample positive for SARS-CoV-2 RNA. NP samples collected with detectable SARS-CoV-2 RNA between March 2, 2020, and April 8, 2020, were identified and included in the study if residual EDTA whole blood had been collected within 72 hours before or after the NP collection. If available, residual whole blood samples from serial collections, with or without paired NP samples, were also included in the analysis. A volume of 400 µL of EDTA plasma from the residual whole blood was extracted and processed to assess for SARS-CoV-2 RNA. Testing was performed based on the adaptation of published real-time reverse transcription polymerase chain reaction (rRT-PCR) assays targeting the envelope (E) gene [3–5]. The standard cycle threshold (Ct) values of positive tests with this assay range from <20 to 45 cycles. Confirmatory testing of all plasma samples with a Ct value of 40 or greater was performed and resulted as positive if reproducible [4]. Given that viral culture was not performed to determine the viability of virus, the presence of viral RNA in plasma is referred to as RNAemia instead of viremia. Retrospective chart review was performed for all individuals in the study to identify basic demographic data, medical history, clinical presentation, and hospital admission. Outcome data collected included admission to an intensive care unit (ICU), need for invasive mechanical ventilation, and 30-day all-cause mortality. Furthermore, laboratory data were extracted for nasopharyngeal rRT-PCR results. Sample size was based on a convenience set of all available clinical samples without formal power calculation. Statistical analysis was completed using Stata v15.1. Univariable analysis was performed by χ 2 test or Fisher’s exact test when there were less than 5 datapoints per cell for categorical variables, and by Mann-Whitney U test for continuous variables. Multivariable analysis was performed by logistic regression including the top variable significantly associated with the outcome in univariable analysis. A 2-tailed P value of <.05 was considered significant. This study was approved by the Stanford Institutional Review Board (protocol #48973). Written individual consent was waived for this study.
RESULTS
A total of 7240 samples from SHC patients were tested for SARS-CoV-2 corresponding to 4485 outpatients, 2452 emergency department patients, and 303 inpatients; 453 (6.3%) were positive. Of these, 85 individuals with paired NP and plasma samples were included for analysis (Table 1). The overall median age was 55 years (interquartile range [IQR], 40–69) and only 2 pediatric cases met inclusion criteria, aged 9 and 17 years. One individual with multiple concurrent medical conditions in this cohort acquired SARS-CoV-2 infection in the hospital, with favorable clinical outcome. Individuals with detectable RNAemia were significantly older than those without RNAemia (63 vs 50 years; P = .04). Comorbidities were prevalent, including obesity in 32/85 (37.6%), hypertension in 26/85 (30.6%), and diabetes mellitus in 19/85 (22.4%); no association was detected between each comorbidity and RNAemia. The overall median number of days of symptoms at the time of NP testing was 7 days (IQR, 4–11) and was similar in individuals with and without RNAemia. The most common symptoms were cough (87.1%) and fever (69.8%). RNAemia was detected in a total of 28/85 (32.9%) individuals, and 17/85 (20.0%) were admitted to the ICU. For patients admitted to the ICU, acute respiratory distress syndrome (ARDS) was present in 9/17 (52.9%), and non-ARDS respiratory failure in 6/17 (35.3%). RNAemia was detected in 7/9 (77.8%) individuals with ARDS and in 2/6 (33.3%) of those with non-ARDS respiratory failure.
Table 1.
Demographic, Clinical, and Laboratory Characteristics of 85 Patients With Paired Nasopharyngeal and Plasma Samples Tested for SARS-CoV-2
| Overall Patients (n = 85) | Detectable Plasma RNA (n = 28) | Nondetectable Plasma RNA (n = 57) | Unadjusted P Valuea | ||
|---|---|---|---|---|---|
| Age, median (IQR) | 55 (40–69) | 63 (47.5–71) | 50 (37–67) | .04 | |
| Sex, no. (%) | M | 40 (47.1) | 14 (50.0) | 26 (45.6) | .7 |
| F | 45 (52.9) | 14 (50.0) | 31 (54.4) | ||
| Location at presentation, no. (%) | ED | 79 (92.9) | 26 (92.9) | 53 (93.0) | 1 |
| Outpatient | 5 (5.9) | 2 (7.1) | 3 (5.3) | ||
| Inpatient | 1 (1.2) | 0 | 1 (1.7) | ||
| Immunocompromised, no. (%) | No | 78 (91.8) | 25 (89.3) | 53 (93.0) | .7 |
| Yes | 7 (8.2) | 3 (10.7) | 4 (7.0) | ||
| Comorbidities, no. (%) | Obesity (BMI ≥30) | 32 (37.6) | 10 (35.7) | 22 (38.6) | .9 |
| (Obesity status unknown) | 7 (8.2) | 3 (10.7) | 4 (7.0) | ||
| Diabetes mellitus | 19 (22.4) | 7 (25.0) | 12 (21.1) | .7 | |
| HTN | 26 (30.6) | 9 (32.1) | 17 (29.8) | .8 | |
| COPD/asthma | 12 (14.1) | 3 (10.7) | 9 (15.8) | .7 | |
| Days of symptoms at time of NP swab collection, median (IQR) | 7 (4–11) | 7 (4–14) | 7 (4–11) | .7 | |
| Symptoms, no. (%) | Fever | 60 (70.6) | 20 (71.4) | 40 (70.2) | .9 |
| Cough | 74 (87.1) | 24 (85.7) | 50 (87.7) | .8 | |
| Headache | 21 (24.7) | 4 (14.3) | 17 (29.8) | .3 | |
| (Headache unknown) | 3 (3.5) | 1 (3.6) | 2 (3.5) | ||
| Fatigue | 35 (41.2) | 16 (57.1) | 19 (33.3) | .1 | |
| (Fatigue unknown) | 3 (3.5) | 1 (3.6) | 2 (3.5) | ||
| GI | 35 (41.2) | 12 (42.9) | 23 (40.4) | .8 | |
| (GI unknown) | 2 (2.4) | 1 (3.6) | 1 (1.8) | ||
| Lymphopenia at presentation, no. (%) | No | 37 (43.5) | 11 (39.3) | 26 (45.6) | .6 |
| Yes | 48 (56.5) | 17 (60.7) | 31 (54.4) | ||
| NP Ct value, no. (%) | Ct <25 | 23 (27.1) | 11 (39.3) | 12 (21.1) | .1 |
| Ct 25–35 | 37 (43.5) | 12 (42.9) | 25 (43.9) | ||
| Ct >35 | 25 (29.4) | 5 (17.9) | 20 (35.1) | ||
| Median NP Ct (IQR) | 30.1 (24.0–35.7) | 27.1 (21.1–34.4) | 31.6 (25.9–36.2) | .05 | |
| Median plasma Ct (IQR) | … | 37.5 (35.1–39.6) | … | ... |
aχ 2, Fisher’s exact test, or Mann-Whitney U test.
Abbreviations: BMI, body mass index; COPD, chronic obstructive pulmonary disease; Ct, cycle threshold; ED, emergency department; F, female; GI, gastrointestinal; HTN, hypertension; ICU, intensive care unit; IQR, interquartile range; M, male; NP, nasopharyngeal.
In the unadjusted analysis, ICU admission occurred significantly more frequently in individuals with RNAemia compared with those with undetectable SARS-CoV-2 RNA in plasma (32.1% vs 14.0%; P = .05), as did invasive mechanical ventilation (21.4% vs 3.5%; P = .01) and mortality (14.3% vs 0%; P = .01) (Table 2). In the age-adjusted analysis, the association was maintained between RNAemia and ICU admission (P = .04), and between RNAemia and invasive mechanical ventilation (P = .02). All 4 deaths occurred in individuals with RNAemia, and adjusted analysis could not be performed for mortality. Although an initial association was detected between the median viral load in the nasopharynx as estimated by rRT-PCR Ct value and detection of RNAemia, this association did not persist with age adjustment. Furthermore, plasma E gene Ct values were high, which is consistent with low viral burden, with a median of 37.5 (IQR, 35.1–39.6). A total of 15/17 (88.2%) plasma samples in severely ill patients were collected before or on the same day as their ICU admission. For the 16 individuals with suspected community-acquired COVID-19 admitted to the ICU, time from hospital admission to the ICU was short, with 13/16 (81.3%) transferred on the same day or within 1 day of admission.
Table 2.
COVID-19 Clinical Severity Outcomes and Association With Detectable RNAemia in 85 Patients
| Overall Patients (n = 85) | Detectable Plasma RNA (n = 28) | Nondetectable Plasma RNA (n = 57) | Unadjusted P Valuea | Adjusted P Valueb | |
|---|---|---|---|---|---|
| Hospital admission, no. (%) | 59 (69.4) | 22 (78.6) | 37 (64.9) | .2 | .3 |
| ICU admission, no. (%) | 17 (20.0) | 9 (32.1) | 8 (14.0) | .05 | .04 |
| Invasive mechanical ventilation, no. (%) | 8 (9.4) | 6 (21.4) | 2 (3.5) | .01 | .02 |
| 30-day all-cause mortality, no. (%) | 4 (4.7) | 4 (14.3) | 0 | .01 | ...c |
aχ 2 or Fisher’s exact test.
bLogistic regression; model adjusted for age.
cCould not be calculated given no mortality events in the nondetectable plasma RNA group.
Abbreviations: COVID-19, coronavirus disease 2019; ICU, intensive care unit.
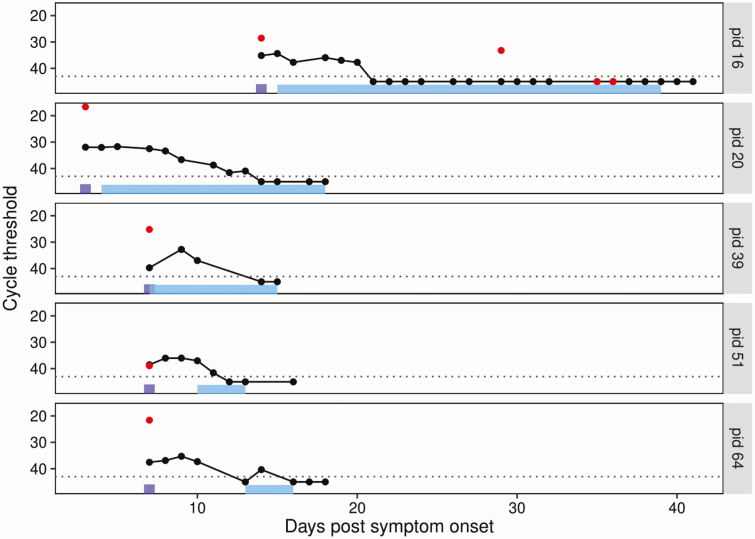
An additional 121 plasma samples from the 28 individuals with RNAemia detected on the paired plasma sample were tested to assess longitudinal trends. Each individual underwent a median of 2 (IQR, 0–7) repeat SARS-CoV-2 plasma tests, with a range of 1 to 27 days between tests. The longest period of RNAemia was 10 days from the time of the first detectable RNAemia result; this occurred in an ICU-admitted patient. SARS-CoV-2 rRT-PCR Ct values were comparable to those seen in the initial paired samples with a median of 37.2 (IQR, 34.8–39.2). Detailed paired NP and plasma values are presented in Figure 1 for the individuals with 5 or more longitudinal samples, including 1 sample collected before ICU admission.
Figure 1.
Longitudinal SARS-CoV-2 nasopharyngeal and plasma data for individuals with 5 or more samples including at least 1 sample collected before ICU admission. The dotted background line represents the real-time reverse transcription polymerase chain reaction threshold. Black circles represent the cycle threshold result of plasma samples tested for SARS-CoV-2. Red dots represent the cycle threshold result of nasopharyngeal samples tested for SARS-CoV-2. The purple rectangles represent the time of hospital admission. The light blue rectangles represent the timeline of ICU stay. Patient identification numbers appear on the right. Abbreviations: ICU, intensive care unit; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2.
DISCUSSION
This analytical cross-sectional study assessed 85 individuals with a nasopharyngeal swab positive for SARS-CoV-2 RNA for detection of RNAemia in paired plasma samples. These findings show that RNAemia was present in almost one-third of samples tested. This proportion is higher than previously reported from a cohort of COVID-19-positive individuals in China, where only 3/307 (1%) of plasma samples tested for SARS-CoV-2 were detected [1]. Other small studies have reported varying rates of RNAemia from plasma (0%–20%), whole blood (8%–40%), and serum samples (0%–17%) [2, 6–11]. However, direct comparison of these results is challenging because of the use of different testing protocols, including differences in specimen extraction volumes (ranging from 80 to 200 μL), sample types examined (plasma, serum, and whole blood), viral genomic targets tested (N, S, E genes or ORF1ab region), variable timing of blood collection relative to symptom chronology, and different patient populations. For example, using the most similar plasma rRT-PCR method to this study as a comparator, the higher prevalence of RNAemia may be explained by the higher proportion of individuals with severe disease in this cohort (ICU admission 20.0% vs 11.1%) [8]. By comparison, clinical illness with SARS-CoV-1 was almost always severe, and RNAemia was reported to occur in up to 75% of plasma samples between 5 and 7 days of illness, and up to 78% of serum samples on admission [12–14].
The association between RNAemia, ICU admission, invasive mechanical ventilation, and mortality observed in this study suggests the potential utility of plasma SARS-CoV-2 RNA testing as a prognostic indicator. This is further supported by the high proportion of ICU admissions occurring for COVID-19-related respiratory complications. Although this study was not performed in a controlled setting, most plasma samples with presence of RNAemia in severely ill patients were collected before or on the same day as their ICU admission, supporting a possible role for the early detection of at-risk individuals. Preliminary data from a limited number of patients have shown an association between RNAemia and severe COVID-19 and support the presence of RNA in extrapulmonary sites [11]. A similar association between detection of SARS-CoV-1 RNA in serum and clinical complications including oxygen desaturation, mechanical ventilation, and mortality was noted in 2004 [15]. Thus, in the absence of a robust COVID-19 clinical scoring system, plasma may be considered as a complementary modality for the early identification of individuals likely to develop severe COVID-19.
This study has several limitations. First, it is limited by its single-center experience and use of a convenience set of samples for SARS-CoV-2 testing that favored overall selection of individuals with more severe disease given the need for having had a blood sample collection. However, that a signal for ICU admission was detected despite this patient selection lends further support to the clinical importance of RNAemia. Second, the median time from symptom onset to first plasma sample collection was 7 days; collection of plasma samples earlier in the disease course may have been advantageous to inform prognosis. However, given that individuals with COVID-19 symptoms often seek care only several days after illness onset, this represents a pragmatic approach. Third, in the absence of viral culture data, it cannot be determined at present whether detectable RNA represents intact infectious virus or inactive, nonreplicating nucleic acid. A recent study attempted to address this question but failed to detect SARS-CoV-2 rRT-PCR in all serum samples; as a result, culture could not be pursued [9]. Further work should thus be performed to investigate this question given the potential important biosafety and blood product transfusion implications, though nonseverely ill individuals are less likely to be RNAemic [16]. Finally, given most plasma viral load results were below the level of quantification of the assay, Ct values were used as surrogate measures of quantitative viral burden. Although viral load values may have been preferable for analysis, Ct values remain a reliable means to assess the association between nasopharyngeal results and RNAemia and have been used in other similar studies [1, 6, 8].
In summary, this study demonstrated a high overall proportion of detectable SARS-CoV-2 RNAemia, an association between plasma viral detection and older age, and severe clinical disease including need for ICU admission, invasive mechanical ventilation, and 30-day all-cause mortality. Further studies are required to investigate the prognostic potential of RNAemia to facilitate early therapeutic and supportive interventions before the development of severe clinical manifestations of COVID-19.
Notes
Acknowledgments. The authors thank Stephen Nash for helpful discussion on statistical analysis and critical review of the manuscript.
Financial support. None.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 2020; 9:386–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hogan CA, Sahoo MK, Pinsky BA. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 2020; 323:1967–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. U.S. Food and Drug Administration. Stanford health care clinical virology laboratory SARS-CoV-2 test EUA summary Available at: https://www.fda.gov/media/136818/download. Accessed 7 May 2020.
- 6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 2020; 323:1488–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 10. Lescure FX, Bouadma L, Nguyen D, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis 2020; 20:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen W, Lan Y, Yuan X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect 2020; 9:469–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen W, Xu Z, Mu J, et al. Antibody response and viraemia during the course of severe acute respiratory syndrome (SARS)-associated coronavirus infection. J Med Microbiol 2004; 53:435–8. [DOI] [PubMed] [Google Scholar]
- 13. Wang WK, Fang CT, Chen HL, et al. ; Members of the SARS Research Group of National Taiwan University College of Medicine-National Taiwan University Hospital Detection of severe acute respiratory syndrome coronavirus RNA in plasma during the course of infection. J Clin Microbiol 2005; 43:962–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng LF, Wong M, Koh S, et al. Detection of severe acute respiratory syndrome coronavirus in blood of infected patients. J Clin Microbiol 2004; 42:347–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hung IF, Cheng VC, Wu AK, et al. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis 2004; 10:1550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang L, Yan Y, Wang L. Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev 2020; 34:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]