Abstract
The pandemic of coronavirus disease 2019 (COVID-19) urgently calls for more sensitive molecular diagnosis to improve sensitivity of current viral nuclear acid detection. We have developed an anchor primer (AP)-based assay to improve viral RNA stability by bioinformatics identification of RNase-binding site of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA and implementing AP dually targeting the N gene of SARS-CoV-2 RNA and RNase 1, 3, 6. The arbitrarily primed polymerase chain reaction (AP-PCR) improvement of viral RNA integrity was supported by (a) the AP increased resistance of the targeted gene (N gene) of SARS-CoV-2 RNA to RNase treatment; (b) the detection of SARS-CoV-2 RNA by AP-PCR with lower cycle threshold values (−2.7 cycles) compared to two commercially available assays; (c) improvement of the viral RNA stability of the ORF gene upon targeting of the N gene and RNase. Furthermore, the improved sensitivity by AP-PCR was demonstrated by detection of SARS-CoV-2 RNA in 70–80% of sputum, nasal, pharyngeal swabs and feces and 36% (4/11) of urine of the confirmed cases (n = 252), 7% convalescent cases (n = 54) and none of 300 negative cases. Lastly, AP-PCR analysis of 306 confirmed and convalescent cases revealed prolonged presence of viral loading for >20 days after the first positive diagnosis. Thus, the AP dually targeting SARS-CoV-2 RNA and RNase improves molecular detection by preserving SARS-CoV-2 RNA integrity and reveals the prolonged viral loading associated with older age and male gender in COVID-19 patients.
Keywords: SARS-CoV-2, COVID-19, RNA, anchor primer, bioinformatics analysis
INTRODUCTION
As of 6 May 2020, the pandemic of coronavirus disease 2019 (COVID-19) has reached 3.55 million cases with approximately 245 000 deaths (https://www.who.int), posing the greatest threat to global public health since the 1918 influenza pandemic. This unprecedented and ongoing worldwide outbreak calls for more sensitive molecular detection of the causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to limit human-to-human transmission and develop effective treatments. Many molecular detection methods for SARS-CoV-2 RNA were developed [1–5], including real-time polymerase chain reaction (RT-PCR) [1–4], multiplex nucleic acid amplification [6], metagenome sequencing [7, 8] and loop-mediated isothermal amplification assay (LAMP) [9]. Together with clinical and computed tomography imaging findings, nuclear acid-based detection of SARS-CoV-2 RNA is at the core of clinical diagnosis of COVID-19. However, computed tomography imaging cannot provide definite information on causative agents/virus [10, 11] and antibody-based diagnostic method, albeit faster (within 15 min), is limited by cross-reactivity with other coronaviruses and the belated appearance of antibody [12, 13], nuclear acid detection of SARS-CoV-2 RNA remains as the gold standard for the diagnosis of COVID-19. However, the sensitivity and reliability of nuclear acid detection have been questioned since clinical studies reported only 35–45% positive confirmation for total of >5000 specimens from clinically and epidemiologically suspected COVID-19 patients, China [10, 14, 15].
Most of the nuclear acid detection assays are largely focused on the RT-PCR amplification with the reported limit of detection (LOD) in the range of 200–500 copies/ml range [14, 16–18]. Mathematical modeling of RT-PCR amplification at 99% efficiency predicted to detect the N gene of SARS-CoV-2 RNA at 2.9 copies per reaction with the 95% hit rate [6, 19, 20]. Thus, further improvement in RT-PCR amplification is difficult since several nuclear acid detection assays have reportedly detected SARS-CoV-2 RNA at 5 copies/ml [16, 17]. By contrast, it remains untested if the efficiency of SARS-CoV-2 RNA detection can be improved by dampening the loss or degradation of virus RNA [21] during sampling process which contributes to underperformance of nuclear acid detection assays. The strategy to improve SARS-CoV-2 RNA stability is appealing for detecting SARS-CoV-2 RNA since coronaviruses stand out among RNA viruses for their unusually large genomes (∼30 kb) [22–24] and subject to a variety of viral RNA-specific and nonspecific degradation processes [21].
To overcome the bottleneck of nucleic acid detection of SARS-CoV-2 RNA, we have developed a novel assay with improved SARS-CoV-2 RNA stability by implementing an anchor primer (AP) dually targeting SARS-CoV-2 RNA (the N gene) and RNase 1, 3, 6. The improved SARS-CoV-2 RNA stability by arbitrarily primed polymerase chain reaction (AP-PCR) was confirmed by the resistance of the SARS-CoV-2 RNA:AP hetero-complex to RNase treatment of in sputum spiked with reference viral RNA, and by the AP-PCR detection of SARS-CoV-2 RNA with the reduced cycle threshold (CT) value in specimens from 252 confirmed COVID-19 cases compared to other assays. The improved sensitivity AP-PCR assay is supported by the detection of SARS-CoV-2 RNA in 70–80% of sputum, nasal swabs and pharyngeal swabs and 36% of urine specimens of 252 confirmed cases, four cases in 54 convalescent cases and none in 300 negative cases. Furthermore, our analysis of confirmed and convalescent cases revealed that COVID-19 patients could have prolonged viral load that was associated with older age and male gender.
METHODS
Ethical approval statement: This multicenter study was approved by the Institutional Ethical Review Committee of the School of Ophthalmology & Optometry, Wenzhou Medical University (WMU), China. The written informed consent was waived. The study was further reviewed and approved by the ethical committee of Zhejiang provincial center for disease control and prevention (CDC).
Bioinformatics analysis of the secondary structure of the N gene of SARS-CoV-2 RNA and of RNase activity targeted by the AP
We first employed catRAPID [25] to identify ribonucleoproteins and RNA-binding region. In order to achieve better interaction region, we cut polypeptide and nucleotide sequences sliding windows into 500 nt fragments and used the step of 50 nt to scan through the entire 1630 bp of SARS-CoV-2 RNA to predict the interaction propensities. We then used two variants of RPISeq classifier (RPISeq-SVM, using Support Vector Machine, and RPIUSeq-RF, using Random Forest) for predicting whether or not the RNA-protein pair (SARS-CoV-2 RNA-RNase) interacts using only their linear sequence information [26]. Our choice of RF and Support Vector Machine (SVM) classifiers were also motivated by the demonstration that RPISeq achieved an AUC (Area under the Receiver Operating Characteristic curve) of 0.92–0.96 when analyzing two nonredundant benchmark datasets from the Protein-RNA interface Database (PRIDB) [27] and by several studies that have successfully used them on classification task of the RPI prediction [28]. Random Forest and SVM methods [26] for analysis of the binding property/affinity of the AP for RNase. Lastly, we used Minimum Free Energy algorithm to predict the secondary structure of the N gene of SARS-CoV-2 RNA by RNA structure software [26, 29] and analyzed the effect of the AP binding on the secondary structure of SARS-CoV-2 RNA (NC_004718). We further employed a local RNA structure alignment algorithm by RNAsmc [26] to compare structure similarity before and after the AP binding to assess the secondary structure changes of SARS-CoV-2.
AP-based RT-PCR (AP-PCR) detection of SARS-CoV-2 RNA
The AP was identified and designed to target the middle section of the N gene of SARS-CoV-2 RNA (GenBank: MN908947.3) to form an RNA:DNA hetero-complex which increase RNA stability and reduce its affinity for RNase. The AP was also designed to specifically bind to RNase 1, 3, 6 with high affinity to reduce RNase-mediated degradation of SARS-CoV-2 RNA [25, 26] and the AP sequence was 5’-CCTCTTCTCGTTCCTCATCACGTAGTCGCAACAGTTCAA-3′. Briefly, specimen’s lysates were prepared by thoroughly mixing ~300 μl specimen with 500 μl lysis buffer containing 1 mmol/L 2-Morpholinoethanesulfonic Acid, 100 mmol/L NaCl, 100 mmol/L KCl, 10 mmol/L Tris–HCl, 5 mol/L guanidium HCl, 1%Triton X-100, 0.1 mg/ml proteinase K, 0.1 mg/ml diatomite and 20 nM AP. Specimen lysates were incubated at 60°C for 10 min and then centrifuged for 30 s at 12 000 rpm. Upper aqueous phases were loaded onto the adsorbing column and allowed to elute by gravitation. The column was washed twice with washing buffer (600 μl, 400 μl) and centrifuged at 12 000 rpm for 30 s to discard the washing buffer. This was followed by eluting the column with prewarmed elution buffer (45 μl) and collected the elute by centrifugation at 12 000 rpm for 1 min.
AP-PCR detection of SARS-CoV-2-RNA was developed by adapting the method of the template-ready PCR [6] with modifications. The elution buffer (8 μl) were added to the reverse transcript reaction and PCR reaction mixture (22 μl) containing 50 mM Tris pH 9.0, 50 mM KCl, 1 mM DTT, 0.1 mg/ml BSA, 0.5 μM random primers, 0.5 μM oligo-T primers, 0.5 μM Gene-specific PCR primer, 0.2 mM dNTPs (Takara, China), 0.2 μM Probe WHN1-T1(Applied Biosystems, USA), 0.05 U/μl EX Taq DNA Polymerase (Takara, China) and 0.2 U/μl RT-enzymes (Takara, China). Gene-specific PCR primer sequences for the N gene assay were forward primer 5’-CAAGCCTCTTCTCGTTCCT-3′, reverse primer 5’-GCAGCAGATTTCTTAGTGACAG-3′ and the probe WHN1-T1 5’-FAM-ATTCAACTCCAGGCAGCAGTAG-MGB1–3′. The RT reaction mixture was applied to Applied Biosystem 7500 PCR machine (Applied Biosystems, USA) for amplification and detection using the program (42°C 10 min, 95°C 3 min, then 45 cycles of 95°C 10 s and 58°C 35 s). Participating laboratories used either Applied Biosystems QuantStudio™ 5 (Applied Biosystems, USA) or Bio-Rad CFX96 (Bio-Rad, USA), Roche Light Cycler 480II (Roche, Switzerland).
Clinical specimen collection and testing
The patients were consecutively enrolled from 23 January to 3 March 2020 [30, 31] in six medical centers and CDCs in Zhejiang province, China, including site 1 (Affiliated Taizhou Hospital of WMU), Site 2 (Wenzhou municipal CDC, WZ CDC), Site 3 (Zhejiang provincial CDC, ZJ CDC), Site 4 (1st affiliated hospital of WMU), Site 5 (2nd affiliated hospital of WMU) and Site 6 (Affiliated eye hospital of WMU). A total of 252 confirmed COVID-19, 54 COVID-19 convalescent and 300 COVID-19 negative, cases were included in the study cohort (see Table 2). All confirmed and convalescent patients were conferred to the criteria according to the Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment issued by the Chinese National Health Commission (6th edition). Most confirmed cases underwent multiple nucleic acid tests and sometimes using >2 different nucleic acid detection assays for SARS-CoV-2 RNA and combined with clinical and computer tomography analysis. (2) Convalescent COVID-19 patients: The patients referred to the confirmed COVID-19 patients who underwent treatment in Site 4 and Site 5 and had recovered and met the discharged criteria (i.e. two negative nuclear acid test results with >24 h interval). (3) SARS-CoV-2 RNA-negative controls: The subjects were the out-patients who underwent routine eye examination or regular operation unrelated to the COVID-19 but had similar demographic information as other groups at Site 5 and Site 6 and tested negative for SARS-CoV-2. The specimens collected from patients at five different sites included pharyngeal swabs, nasal swabs, sputum, feces and urine samples. The different types of the samples were distributed randomly throughout the entire disease courses (see Figure 4).
Table 2.
Demography of 252 confirmed COVID-19 cases from three sites in the study
| Site 1 | Site 2 | Site 3 | |
|---|---|---|---|
| Total cases | 57 | 115 | 75 |
| Age, mean, year | 53 (4–79) | 45 (1–93) | 40 (1–85) |
| Male, No. (%) | 31 (54.39) | 58 (50.43) | 38 (50.66) |
| Specimen type, No. (%) | Sputum 37 (64.91) Nasal swab 3 (5.26) Pharyngeal swab 10 (17.54) Feces 7 (12.28) |
Sputum 38 (33.04) Nasal swab 18 (15.65) pharyngeal swab /Sputum 44(38.26) Feces 15 (13.04) |
Sputum 11 (14.67) Nasal swab 22 (29.33) Pharyngeal swab 41 (54.67) Unknown 1 (1.33) |
| Detection rate (%) | 70.18 | 80.87 | 78.67 |
| Mean CT value | 31.72 | 27.75 | 32.27 |
Figure 4.
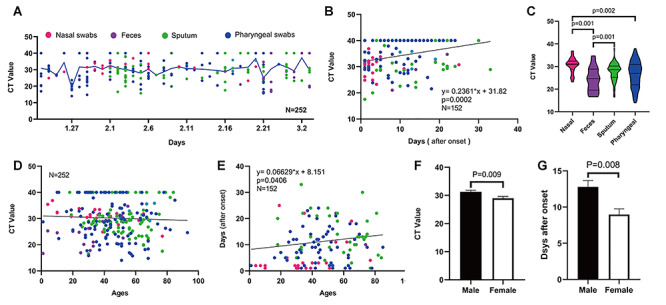
Prolonged SARS-CoV-2 RNA loading and its associated factors of age and sex as revealed by the AP-PCR. A. The chronological changes of SARS-CoV-2 RNA loading during epidemics in Zhejiang province from 23 January to 3 March 2020. The viral RNA loading from different collecting sites was evenly distributed throughout the epidemic period (color symbol = specimen type). B. There was a positive linear correlation between the days after the first positive diagnosis and the CT Value (r = 0.091, P = 0.0002). C. The biodistribution of SARS-CoV-2 RNA loading among four different collecting sites: Analysis of the 252 confirmed patients showed the highest positive rates in Nasal swabs (96%), followed by sputum (76.7%), pharyngeal swabs (74.6%), feces (69.6%). D. There was no correlation between the CT value and age (r = 0.0021, P = 0.463). E. Older age was associated with a longer duration of the SARS-CoV-2 RNA loading (r = 0.002159, P = 0.0406). F. Male patients have lower SARS-CoV-2 RNA loadings than female, as evidenced by higher Ct value in male (n = 132, 31.3 ± 0.57, mean ±SE) than female (n = 120, 29±0.66 mean ± SE) in specimens from 252 confirmed cases (P = 0.0086, t-test). G, Male COVID-19 patients showed the longer duration of the viral RNA loading by 4 days when compared to female patients after the first positive test (P = 0.0077, t-test).
RT-PCR test for SARS-CoV-2 RNA was conducted at the certified diagnostic laboratories at Sites 1–6.
RESULTS
The AP bound to the N gene of SARS-CoV-2 RNA and RNase 1, 3, 6 to improve viral RNA stability as revealed by bioinformatics analysis
To date, many molecular detection methods for SARS-CoV-2 RNA were developed with their advantages and disadvantages (see the major methods [1, 4, 9, 32–35] listed in the Table 1). By calculating the affinity beyond the interactive threshold, catRAPID analysis coupled with Random Forest and SVM predicted that the AP has significant binding affinity for RNase. We found that the highest interaction propensity in the region of SARS-CoV-2 RNA (N gene) from 550–750 (AP sequences selected 545–583: CCTCTTCTCGTTCCTCATCACGTAGTCGCAACAGTTCAA). RIPSeq-RF and RIPSeq-SVM analysis suggested that RNase 1, 3 and 6 were the potential targets of the AP (Figure 1C). The analysis of the effect of the AP binding on overall secondary structure of SARS-CoV-2 RNA revealed that the score for overall secondary structure similarly (with or without the AP binding) was 7.70 (with 10 being maximal similarity), suggesting that the AP binding did not affect the overall secondary structure of SARS-CoV-2 RNA. However, RNAsmc analysis indicated that the AP binding altered the local secondary structure of segments of the N gene of SARS-CoV-2 RNA (Figure 1B), leading to the reduced affinity of SARS-CoV-2 RNA for RNase.
Table 1.
Advantages and disadvantages of current nuclear acid detection methods for SARS-CoV-2 RNA
| Methods | Advantages | Disadvantage | References |
|---|---|---|---|
| RT- qPCR | • Quantitative real-time PCR. • High throughput,• Relatively sensitive,• Specificity,• Multiplex specimens. | • Extra RNA extraction.• Time-consuming (2- 4 h).• False-negative | Zhu N, et al. 2020. [1] Corman VM, et al. 2020. [4] |
| RT- LAMP | • An exponential amplification at a constant temperature.• Easy-to-operate,• Time-efficient (30 min). | • Extra RNA extraction.• False-positive/negative• Inappropriate for complex specimen. | Park GS, et al. 2020. [9] |
| COVID-19 Penn-RAMP | • Two-stage isothermal amplification based on recombinase polymerase amplification.• High sensitivity. | • Extra RNA extraction.• False negatives/positives.• Time-consuming (50–80 min).• Inappropriate for complex specimen. | Mohamed, et al., (2020). [32] |
| COVID-19-RdRp/Hel assay | • A novel RT-qPCR assay targeting RdRp/Hel,• High sensitivity,• Multiplex specimens. | • Extra RNA extraction.• False-negative• Time-consuming (≥ 2 h). | Chan, et al., (2020). [33] |
| Droplet digital PCR (ddPCR) | • Based on the principles of limited dilution, end-point PCR, and Poisson statistics.• Absolute quantification for viral loading,• Highest sensitivity of all molecular detections,• Specificity. | • Extra RNA extraction.• Time-consuming (≥ 2 h).• False positives,• Complex operation. | Suo, et al., (2020). [34] |
| CRISPR methods | • CRISPR-based nucleic acid detection.• High sensitivity,• Multiplex specimens. | • Extra RNA extraction.• False negatives/positives.• Time-consuming (60 min). | Zhang, Abudayyeh, et al., (2020). [35] |
| AP-PCR | • AP dually targeting SARS-CoV-2 RNA and RNase.• Increased RNA stability• High sensitivity,• Multiplex specimens. | • Time-consuming (< 70 min).• False-negative |
Figure 1.
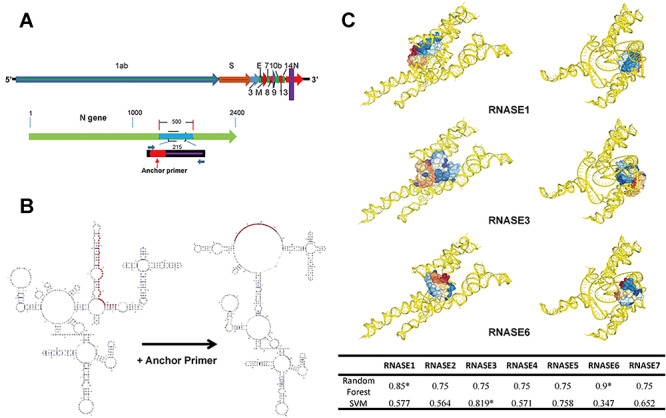
Design of the anchor primer dually targeting the N gene of SARS-CoV-2 RNA and RNase 1,3,6. A. The schematic diagram of the designed AP (the purple stripe) targeting the middle portion of the N gene regions of SARS-CoV-2 RNA (the red stripe). B. The alteration of the secondary structural of SARS-CoV-2RNA before and after AP binding. The AP binding altered the secondary structure of the N gene of SARSCoV-2 RNA, resulting in reduced affinity of SARS-CoV-2 RNA for RNase 1,3,6. C. The Random Forest and SVM analyses indicate that the AP has high binding affinity for RNase 1, 3 and 6 beyond the interactive threshold.
To determine the specificity of the AP-RNase interaction, we also performed analysis of the cDNA sequence randomization of AP while keeping the ATGC composition rate and sequence length constant to construct the ‘randomized’ control sets. RIPSeq-RF and RIPSeq-SVM algorithms were used to predict the binding state of the ‘randomized’ control sequence to RNases (with the composition rate of the ATGC and the length of the sequence being constant). The results showed that the ‘randomized’ control sequences were more inclined to be unbound than the AP sequence by SVM algorithm (data not shown).
AP-PCR detected SARS-CoV-2 RNA at an LOD of 20 copies/ml and 100% specificity using sputum spiked with SARS-CoV-2 RNA
We determined the effect of the AP on the SARS-CoV-2 RNA stability and RNase activity using sputum from healthy volunteers spiked with series of 10-fold dilutions of SARS-CoV-2 viral RNA (Figure 2A). We found that the AP presence in the lysis buffer indeed increased the resistance to RNase treatment of the DNA:RNA hetero-complex and enhanced PCR amplification of SARS-CoV-2 RNA as evident by the increased PCR amplification plot and appearance of PCR products by gel analysis (Figure 2B). We also determined the effects of varying concentration of AP (with fixed PCR primers) and varying concentrations of PCR primers (with fixed AP) on the performance of AP-PCR assay using sputum spiked with SARS-CoV-2 RNA (2095 copies/ml). We found that altered ratio of the AP versus PCR primers did not affect the overall performance (Figure 2C).
Figure 2.
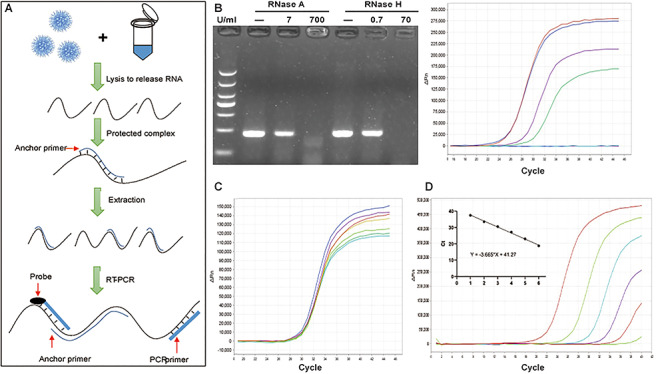
The detection of SARS-CoV-2 RNA by AP-PCR in sputum spiked with SARS-CoV-2 RNA. A. Schematic illustration of the AP-PCR procedure. Mixing the sputum specimens (spiked with SARS-CoV-2RNA) with lysis buffer containing AP led to the formation of DNA:RNA hetero-complexes. This was followed by RT-PCR amplification of 215 bp fragment of the N gene of SARS-CoV-2 RNA. B. In the sputum from healthy volunteers spiked with SARS-CoV-2 RNA, AP improved RT-PCR amplification after the RNase R treatment. C. In sputum spiked with SARS-CoV-2 RNA, we determined the effect of varying concentrations of AP (with fixed PCR primers) and varying PCR primers (with the fixed AP) on AP-PCR amplification efficiency. Varying concentrations of AP or PCR primers did not affect AP-PCR amplification efficiency (P = 0.52). D. The determination of the LOD of the AP-PCR assay using sputum spiked with series of 10-folddilution of SARS-CoV-2 RNA (20-2095 copies/ml). The negative linear correlation was established with β as -3.665 (r = 0.995, P < 0.0001).
We determined the analytic sensitivity (LOD) of the AP-PCR detection of SARS-CoV-2 RNA using sputum spiked with series of 10-fold dilution of SARS-CoV-2 viral RNA (20–2095 copies/ml). The results showed that the CT values were 20.02 ± 0.23, 23.34 ± 0.22, 26.47 ± 0.33 (Mean ± SEM), from the sputum spiked with SARS-CoV-2 RNA at 29.04 ± 0.48, 31.51 ± 0.75, 33.79 ± 1.32, (Mean ± SEM), respectively. This led to the establishment of the liner standard curve for the conversion of the CT value to the copies/ml (Figure 2D). We also demonstrated a good reproducibility of SARS-CoV-2 RNA detection at concentrations of 20 copies/ml of SARS-CoV-2 RNA for 10 repeated tests. ROC analysis indicated that the threshold CT value at 40 yielded 95% agreement of the SARS-CoV-2 RNA reference material and 100% agreement of the negative control.
The detection of SARS-CoV-2 RNA with lower CT values upon AP-PCR indicates improved viral RNA integrity
We further analyzed the CT values from the same specimens by AP-PCR assay and compared them with the CT values obtained by commercial SARS-CoV-2 RNA tests during initial positive diagnosis. Consistent with the improved SARS-CoV-2 RNA stability, we found that CT values of AP-PCR assay were lower than the commercial kit A by 2.27 cycles for the specimens from Site 1 (Figure 3A n = 33, P = 0.0001, paired t-test), and by 3.74 cycles for the specimens from Site 2 (Figure 3B n = 59, P = 0.0005, paired t-test). Similarly, CT values by our AP-PCR assay were lower than the commercial kit B by 3.79 cycle (n = 54, P < 0.0001, paired t-test) in Site 3 (Figure 3C). After converting to the RNA copies/ml, we found that AP-PCR detected more SARS-CoV-2 RNA (1668 copies/ml) than the commercially available tests.
Figure 3.
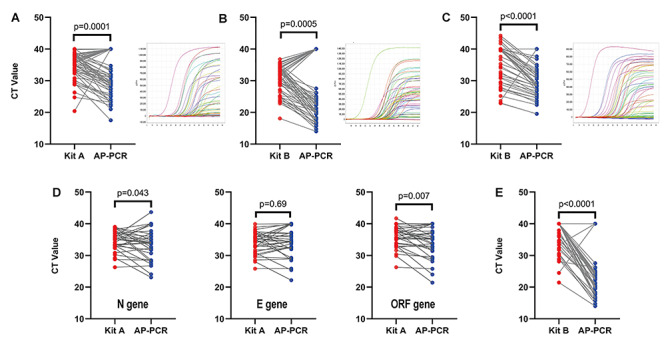
Detection of SARS-CoV-2 RNA with lower CT value by AP-PCR in specimens from 252 confirmed cases. The CT value obtained with the AP-PCR assay was lower than that obtained with the commercial kit A by 2.27 cycles in analyzing the specimens from Site 1. (A) (n = 33, P = 0.0001, paired t-test) and by 3.79 cycles for the specimens from Site 3. (C) (n = 54, P < 0.0001, paired t-test). B. The CT values obtained with the AP-PCR assay was lower than that obtained with the commercial kit B by 3.74 cycles for the specimens from Site 2 (n = 59, P < 0.0001, paired t-test). D. In specimens from Site 1, AP-based isolation yielded lower Ct value targeting the N (by 1.42 cycles, n = 33, P = 0.043 paired t-test) and ORF (by 1.71 cycle, n = 33, P = 0.007, paired Student’s t-test) gene than that of the commercial RNA isolation. E. In specimen from Site 2, the AP-based RNA isolation also improved SARS-CoV-2 RNA stability (by 2.29 cycles, n = 59, P < 0.0001 paired Student’s t-test).
To dissect out the specific step conferring this improved RNA stability, we performed a parallel analysis of the same specimens by two different types of RNA isolation procedures (AP-PCR and commercial kit) followed by the same PCR amplification. Using the specimens from Site 1, we found that the CT values (for the N gene of SARS-CoV-2 RNA) were significantly lower using AP-based isolation system as compared with the commercial RNA isolation (Figure 3D n = 57, P = 0.043). Similarly, AP-based RNA isolation yielded lower CT values in detecting SARS-CoV-2 RNA compared to the commercial RNA isolation for the specimens from Site 2 (Figure 3E n = 59, P < 0.0001). Thus, the AP-based RNA isolation was the critical step conferring RNA protection by AP-PCR assay. Furthermore, we assessed whether the AP-PCR improved the RNA integrity of the E and ORF genes (possibly by inhibiting RNase) (Figure 3D). AP-PCR detected the ORF1ab gene of SARS-CoV-2 RNA with lower CT value than the commercial assay (by 1.71 cycles, n = 33, P = 0.0074, paired t-test) but not for the E gene of SARS-CoV-2 RNA (n = 33, P = 0.6894) (Figure 3). This was corresponding to improved detection of SARS-CoV-2 RNA by ~10 fold. Thus, the AP dually targeting N gene and RNase effectively improved the integrity of the SARS-CoV-2 RNA beyond the N gene.
The AP-PCR detected SARS-CoV-2 viral RNA in 70–80% of specimens from 252 confirmed COVID-19 cases
To validate AP-PCR detection sensitivity and specificity in clinical specimens, we exploited 252 cases of confirmed COVID-19 patients with mixed patient population of severe and mild cases (see Table 2), 54 cases of convalescent COVID-19 patients and 300 negative controls. (1) COVID-19 confirmed cases: the demography of the patient’s age and sex were comparable to that of the previous reports [3, 36]. These specimens were not specifically aggregated but rather mostly evenly distributed through the entire epidemic course (from 23 January to 3 March). Different types of specimens were also largely evenly distributed throughout the epidemic course. We found that AP-PCR detected SARS-CoV-2 RNA in 70–80% of the confirmed cases, with the detection rate of 70.2% at Site 1, 80.9% Site 2 and 78.7% Site 3, respectively. The mean CT value of all specimen types by AP-PCR was 30.2 cycles, corresponding to 2095 copies/ml.
(2) COVID-19 convalescent cases: We also examined 54 convalescent cases (range 18–81 years and 66.7% male and 33.3% female) and confirmed negative for 50 convalescent cases. However, we detected SARS-CoV-2 RNA in the specimen from four convalescent cases which was deemed negative by the commercial tests, indicating the improved detection sensitivity of SARS-CoV-2 RNA by AP-PCR.
(3) COVID-19-ngegative controls: We also tested a total of 300 clinical samples from the out-patients at Site 6 and found that none of these patients were tested positive, confirming the specificity of our AP-PCR assay.
Molecular detection of SARS-CoV-2 viral loading dynamics and its associated factors
We employed the AP-PCR to investigate SARS-CoV-2 viral loading in terms of duration, biodistribution and its associated risk factors in the specimens from 252 confirmed cases and 54 convalescent cases.
(1) Duration of the SARS-CoV-2 viral loading: The virus load was fluctuated largely around the average level throughout the entire epidemic period [30, 31] without a clear peak (Figure 4A). This indicated that the virus load was probably not a contributing factor to the dynamic of epidemic. Furthermore, we analyzed the viral loading dynamic after the first positive diagnosis by plotting the viral loading against the days after the diagnostic date (i.e. the days between the first positive-testing date and the current sampling/testing day). We found that viral loading remained at high level over the first two-weeks after the first positive test. This pattern apparently applied to four different specimen types.
(2) Biodistribution of SARS-CoV-2 viral load: We found significant difference in the CT values among four different specimen types, with highest from nasal swabs (96%, 24/25), followed by sputum (76.5%, 65/85), pharyngeal swabs (74.57%, 85/114), feces (69.6%, 16/23) (Figure 4C). Notably, we found SARS-CoV-2 positive in 4 out of 11 urine specimens examined, suggesting that the kidney might be a putative target of the virus [37, 38].
(3) Correlation with age: we analyzed the CT values from four types of specimens against age and found that there was no correlation between the CT value and age (Figure 4D). However, when we analyzed the correlation between age and the days after the first positive diagnosis, we found that older age was associated with the longer duration of the SARS-CoV-2 viral loading (Figure 4B n = 152, R2 = 0.0912, P = 0.0002). Thus, SARS-CoV-2 viral loading was relatively longer in the older population.
(4) Correlation with sex: Our analysis of 252 confirmed cases showed that CT value in male (mean ± SE, n = 132, 31.3 ± 0.57,) was lower than female (n = 120, 29 ± 0.66 mean ± SE) by 2.3 cycles (Figure 4F P = 0.0086, t-test). Moreover, male patients were associated with the longer duration of the SARS-CoV-2 viral loading after the first diagnosis (Figure 4E n = 152, R2 = 0.02783, P = 0.04). Thus, male was an independent factor for the prolonged SARS-CoV-2 viral loading.
DISCUSSION
APs dually targeting SARS-CoV-2 RNA and RNase improves the viral RNA stability and detection of viral loading from 252 confirmed and 54 convalescent COVID-19 patients
We proposed and validated a novel strategy improve SARS-CoV-2 RNA integrity by implementing the AP dully targeting the N gene of SARS-CoV-2 RNA and RNase 1,3,6. Our bioinformatics analysis, ex vivo experimental findings and clinical investigation support the validity of the dual actions of AP-PCR: (i) the 39 bp of the AP sequence in the middle regions of the N gene containing beta-sheet structures [22–24] has high affinity for RNase 1, 3 and 6. Thus, AP can not only bind directly to RNase, to competitively inhibit RNase activity but also to the N gene of SARS-CoV-2 RNA to form an RNA:DNA hetero-complex, resulting in altered secondary structure and reduced affinity for RNase. (ii) In sputum from healthy volunteer spiked with SARS-CoV-2 RNA the presence of AP in the lysis buffer inhibited RNase activity, resulting in the enhanced PCR amplification of SARS-CoV-2 RNA. (iii) clinical studies with four types of specimens from 252 confirmed COVID-19 cases demonstrated that the CT value by AP-PCR assay was lower than that obtained with two commercially available assays by ~2.7 cycles (or by 10 copies /ml). (iv) The AP against the N gene stabilizes not only the N gene but also ORF1ab gene, indicating the improved stability of entire SARS-CoV-2 RNA.
The improved SARS-CoV-2 RNA stability by AP-PCR is important for overcoming the current bottleneck in the detection of SARS-CoV-2 RNA in clinical specimens. The rapid nature of epidemic in China prevented us from performing parallel comparison of the detection of the SARS-CoV-2 RNA by the AP-PCR assay with other assays in the clinically and epidemiologically suspected COVID-19 cases amid the epidemic in China. Instead, we performed parallel assessments of the AP-PCR and commercially available assays on the duplicated specimens from the confirmed case, (i) AP-PCR detected 70–80% of the 252 confirmed COVID-19 cases using one reaction targeting one gene, probably outperforming the commercially available assays for confirming suspected cases with only 30–50% detection rate after often multiple and repeated testing [10, 14, 15]. (ii) The AP-PCR assay was able to detect SARS-CoV-2 RNA in 70% of the 144 confirmed case compared to 45–50% using the commercially available tests. (iii) AP-PCR positively detected SARS-CoV-2 RNA in four convalescent cases that were deemed as ‘negative’ by the commercial assays. (iv) AP-PCR detected SARS-CoV-2 RNA in 36% (4/11) urine samples, in comparison with none of the 72 and 12 urine specimens tested positive in two previous reports [39, 40]. Collectively, these findings support the hypothesis that AP-PCR assay improved RNA stability and detection sensitivity of SARS-CoV-2 virus.
The improved SARS-CoV-2 RNA stability and enhanced detection sensitivity by this AP-PCR method can be further improved by identifying and employing multiple sequence targets of the SARS-CoV-2 RNA genome with the dual actions, and by coupling with other parallel RNA degradation protection strategies by AP targeting mRNA decay, decapping enzymes, RNA-binding protein and cellular endonuclease (RNase L) [21, 41].
Prolonged viral loading and associated factors in COVID-19 patients
SARS-CoV-2 viral loading, its biodistribution and dynamics are a determinant of clinical outcomes of COVID-19 patients. Early studies on the viral loading were often carried out with a limited number of specimens but usually with multiple samples to uncover the SARS-CoV-2 viral loading pattern. As a complementary analysis, this study collected and analyzed each sample from different individual, collected at different times after onset of symptoms and from four different sites, and thus provided a broad populational view on the dynamic changes of viral loading and its putative contribution to the disease.
Consistent with earlier reported studies [42, 43], AP-PCR analysis revealed prolonged detection of SARS-CoV-2 RNA for >20 days in 152 confirmed COVID-19 cases. Prolonged detection of viral RNA of 20 days or longer has also been reported for patients with MERS-CoV or SARS-CoV infections [44, 45]. Notably, the prolonged SARS-CoV-2 viral loading may not be the same as viral shedding, as after one week of the disease onset the viral cultures from sputum samples were largely unsuccessful despite of high level of SARS-CoV-2 viral loading [46].
Clinical studies demonstrated that COVID-19 disproportionally and more severely affected aged population [47] with a higher incidence of multi-organ involvement and a higher mortality rate [48]. Our analysis of four different specimens from 252 confirmed cases showed that the older age was correlated with longer duration of the viral loading. This collaborates with two earlier studies [42, 43] to support the observation of age as an independent factor for prolonged viral loading. This is expected as aging is associated with impairment of innate and adaptive immune response [49].
Our analysis showed that mean CT value in specimens from 132 male patients was lower than that from 120 female patients. Moreover, the median duration after the first positive testing was 13 days in male and 9 days for female, indicating that male gender may be an independent risk factor for prolonged SARS-CoV-2 viral loading. Gender differences in immune responses [49], and in expression and modulation of human angiotensin-converting enzyme 2 (ACE2) [50] may contribute to the increased viral load and prolonged duration of SARS-CoV-2 viral loading in male patients.
We found that SARSS-CoV-2RNA was detected in all four specimen types, indicating that this virus may wide spread throughout multiple systems and organs in the body, pointing to other potential transmission routes (other than respiratory droplets). In contrast to the report of lacking positive detection from 72 urine samples [40], we found positive SARS-CoV-2 viral RNA in 4 out of 11 specimens examined, implying that the kidney is putative target of SARS-CoV-2 infection as suggested by histological evidence of viral protein in the postmortem renal epithelial cells [37].
There are several limitations of the interpretation of the data collected in this study, including lack of multiple samples from the same individual, lack of quantitative analysis of viral loads (such as digital RT-PCR) and lack of concurrent viral culture to validate the viral loading and infectivity. Nonetheless, this novel AP-PCR assay with the improved RNA stability has revealed prolonged SARS-CoV-2 viral loading for >20 days, and older age and male sex are independent risk factors for prolonged SARS-CoV-2 viral loading.
Key points
Bioinformatics analyses identified the anchor primer (AP) dually targeting the N gene of SARS-CoV-2 RNA and RNase 1,3,6 to increase SARS-CoV-2 RNA stability by directly binding AP to RNase and indirectly via forming a SARS-CoV-2 RNA:RNase hetero-complex.
The AP-PCR improved viral RNA integrity with increased resistance of the N gene and ORF gene of SARS-CoV-2 RNA to RNase treatment and with the detection of SARS-CoV-2 RNA at lower (−2.7 cycles) cycle threshold values.
The AP-PCR improved molecular detection with the detection rate of 70–80% multiple complex samples and 36% of urine from 252 confirmed cases, and 7% of 54 convalescent cases.
The AP-PCR revealed prolonged SARS-CoV-2 viral loading and its association with older age and male sex in COVID-19 patients.
Acknowledgments
We first and foremost thank the medical staff and the patients and their families at four medical centers (the Affiliated First Hospital, Affiliated Second hospital and Affiliated Eye hospital of Wenzhou Medical University and Taizhou Eng Zhe Medical Center) as well as two CDC (Wenzhou and Zhejiang CDC at Hangzhou) for their dedication and support for the project while coping with the current global pandemic. We greatly appreciate Bingyu Yin, Yongping Cheng, Yinghai Ye, Yang Yang, Yunfeng Gu, Xiangtian Zhou for providing critical assistance in collecting and preserving clinical specimens. We sincerely appreciate the discussion and critical feedback from Dr. Yuanguo Zhou with invaluable insight during the project. We thank Drs. Guoping Cai and Jing Lin for reading and revision of the manuscript, Liping Zhang, Wei Guo and Wu Zheng for data analysis and discussion, Lifang Zhang bioinformatics analysis of SARS-CoV-2 virus structure. We also appreciate colleagues at the Eye hospital of WMU who contributed their precious research reagents for this project and provided assistance for transportation of the testing reagents during this emergency time (February 2020). Lastly, we thank the current and formal members of the Molecular Neuropharmacology Lab who provide spiritual support as well as daily assistance for the project during this difficult time.
Tao Xu is an instructor at School of Optometry and Ophthalmology of Wenzhou Medical University (WMU) with research in computational neuroscience.
Jingu Wang is a scientist at Hangzhou Baisaisi Biotechnology Co., Ltd, with research in clinical diagnostic.
Bingjie Hu is a research assistant at Taizhou Hospital of WMU with research in laboratory medicine.
Guosi Zhang is a research assistant at School of Optometry and Ophthalmology of WMU with research in bioinformatics.
Wu Zhou is an associate Professor at the First Affiliated Hospital of WMU with research in laboratory medicine.
Meiqin Zheng is a professor at School of Optometry and Ophthalmology and Eye Hospital, WMU, with research interest in laboratory medicine.
Bo Shen is a professor at Taizhou Hospital of WMU with research in laboratory medicine.
Baochang Sun is an associate professor at Wenzhou Municipal Center for Disease Control and Prevention, with research in laboratory medicine.
Yanjun Zhang is a professor at Zhejiang Provincial Center for Disease Control and Prevention, with research in microbiology.
Yin Chen is an associate professor at Zhejiang Provincial Center for Disease Control and Prevention, with research in laboratory medicine.
Jian Yu is a research assistant at the Second Affiliated Hospital & Yuying Children’s Hospital of WMU, with research in laboratory medicine.
Min Liang is a lecturer of the First Affiliated Hospital of WMU with research in laboratory medicine.
Jingye Pan is a professor at the First Affiliated Hospital of WMU with research in critical care medicine.
Chengshui Chen is a professor at the First Affiliated Hospital of WMU with research in respiratory & critical care medicine.
Haixiao Chen is a professor and medical director of Taizhou Hospital of WMU.
Minghua Jiang is a professor at the Second Affiliated Hospital & Yuying Children's Hospital of WMU, with research in laboratory medicine.
Liangde Xu is a professor at School of Optometry and Ophthalmology of WMU with research in bioinformatics.
Jia Qu is a professor and director at School of Optometry and Ophthalmology and Eye Hospital, WMU, with research in ophthalmology.
Jiang-Fan Chen is a professor at School of Optometry and Ophthalmology, WMU, with research in neuroscience.
Funding
This research was supported by the COVID-19 emergency fund from Zhejiang Provincial Science and Technology Bureau (2020C03128), Scientific Innovation Fund for COVID-19 from the Wenzhou Municipal City (ZY202001–4) and Scientific Innovation Fund for COVID-19 from the Wenzhou Municipal City (ZY202001–7).
References
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020;382:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shen M, Zhou Y, Ye J, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal 2020;10:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li C, Debruyne D, Spencer J, et al. High sensitivity detection of SARS-CoV-2 using multiplex PCR and a multiplex-PCR-based metagenomic method. bioRxiv 2020. doi: 10.1101/2020.03.12.988246. [DOI] [Google Scholar]
- 7. Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect 2020;9:313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Houldcroft CJ, Beale MA, Breuer J. Clinical and biological insights from viral genome sequencing. Nat Rev Microbiol 2017;15:183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park GS, Ku K, Baek SH, et al. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) assays targeting SARS-CoV-2. J Mol Diagn 2020;22:729–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology 2020;296:E32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y, Xia L. Coronavirus disease 2019 (COVID-19): Role of chest CT in diagnosis and management. AJR Am J Roentgenol 2020;214:1280–6. [DOI] [PubMed] [Google Scholar]
- 12. Pan Y, Li X, Yang G, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect 2020;81:e28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol 2020. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang X, Yao H, Xu X, et al. Limits of detection of six approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2). Clin Chem 2020;66:977–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y, Yao L, Li J, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol 2020;92:903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan JF, Yip CC, To KK, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J Clin Microbiol 2020;58:e00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nalla AK, Casto AM, Huang MW, et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer/probe sets and one assay kit. J Clin Microbiol 2020;58:e00557–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfefferle S, Reucher S, Norz D, et al. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill 2020;25:2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shirato K, Nao N, Katano H, et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis 2020;73:304–7. [DOI] [PubMed] [Google Scholar]
- 20. Yu F, Yan L, Wang N, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis 2020;71:793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abernathy E, Glaunsinger B. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 2015;479-480:600–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci U S A 2018;115:E162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat Med 2020;26:450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Forster P, Forster L, Renfrew C, et al. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A 2020;117:9241–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Livi CM, Klus P, Delli Ponti R, et al. catRAPID signature: identification of ribonucleoproteins and RNA-binding regions. Bioinformatics 2016;32:773–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang H, Lu X, Chen F, et al. Landscape of SNPs-mediated lncRNA structural variations and their implication in human complex diseases. Brief Bioinform 2018:1–11. [DOI] [PubMed] [Google Scholar]
- 27. Lewis BA, Walia RR, Terribilini M. PRIDB: a Protein-RNA interface database. Nucleic Acids Res 2011;39:D277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muppirala UK, Honavar VG, Dobbs D. Predicting RNA-Protein interactions using only sequence information. Bmc Bioinformatics 2011;12:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu ZZ, Mathews DH. Experiment-assisted secondary structure prediction with RNAstructure. Methods Mol Biol 2016;1490:163–76. [DOI] [PubMed] [Google Scholar]
- 30. Ruan L, Wen M, Zeng Q, et al. New measures for COVID-19 response: a lesson from the Wenzhou experience. Clin Infect Dis 2020;71:866–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang W, Cao Q, Qin L. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):A multi-center study in Wenzhou city, Zhejiang, China. J Infect 2020;80:388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohamed E-T-B, Haim H, Jinzhao S. A single and two stage, closed-tube, molecular test for the 2019 novel coronavirus (COVID-19) at home, clinic, and points of entry. ChemRxiv 2020. doi: 10.26434/chemrxiv.11860137. [DOI] [Google Scholar]
- 33. Chan JFW, Yip CCY, To KKW, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol 2020;58:e00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suo T, Liu X, Feng J, et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect 2020;9:1259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang F. AOO, & Gootenberg J. S.: A protocol for detection of COVID-19 using CRISPR diagnostics. 2020. https://broad.io/sherlockprotocol.
- 36. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Diao B, Wang C, Wang R, et al. Human Kidney is a Target for Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wadman M. How does coronavirus kill? Clinicians trace a ferocious rampagethrough the body, from brain to toes. Science 2020. doi: 10.1126/science.abc3208. [DOI] [Google Scholar]
- 39. Team C-I. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 2020;26:861–8. [DOI] [PubMed] [Google Scholar]
- 40. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Molleston JM, Cherry S. Attacked from all sides: RNA decay in antiviral defense. Viruses 2017;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu K, Chen Y, Yuan J, et al. Factors associated with prolonged viral RNA shedding in patients with COVID-19. Clin Infect Dis 2020;71:799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020;20:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 2003;361:1767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oh MD, Park WB, Choe PG, et al. Viral load kinetics of MERS coronavirus infection. N Engl J Med 2016;375:1303–5. [DOI] [PubMed] [Google Scholar]
- 46. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. [DOI] [PubMed] [Google Scholar]
- 47. Lauc G, Sinclair D. Biomarkers of biological age as predictors of COVID-19 disease severity. Aging (Albany NY) 2020;12:6490–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen T, Dai Z, Mo P, et al. Clinical characteristics and outcomes of older patients with coronavirus disease 2019 (COVID-19) in Wuhan, China (2019): a single-centered, retrospective study. J Gerontol A Biol Sci Med Sci 2020;glaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Giefing-Kroll C, Berger P, Lepperdinger G, et al. How sex and age affect immune responses, susceptibility to infections, and response to vaccination. Aging Cell 2015;14:309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Clotet-Freixas S, Soler MJ, Palau V, et al. Sex dimorphism in ANGII-mediated crosstalk between ACE2 and ACE in diabetic nephropathy. Lab Invest 2018;98:1237–49. [DOI] [PubMed] [Google Scholar]


