Abstract
Background
Patients with inflammatory bowel disease [IBD] are considered immunosuppressed, but do not seem more vulnerable for COVID-19. Nevertheless, intestinal inflammation has shown to be an important risk factor for SARS-CoV-2 infection and prognosis. Therefore, we investigated the role of intestinal inflammation on the viral intestinal entry mechanisms, including ACE2, in IBD.
Methods
We collected inflamed and uninflamed mucosal biopsies from Crohn’s disease [CD] [n = 193] and ulcerative colitis [UC] [n = 158] patients, and from 51 matched non-IBD controls for RNA sequencing, differential gene expression, and co-expression analysis. Organoids from UC patients were subjected to an inflammatory mix and processed for RNA sequencing. Transmural ileal biopsies were processed for single-cell [sc] sequencing. Publicly available colonic sc-RNA sequencing data, and microarrays from tissue pre/post anti-tumour necrosis factor [TNF] therapy, were analysed.
Results
In inflamed CD ileum, ACE2 was significantly decreased compared with control ileum [p = 4.6E-07], whereas colonic ACE2 was higher in inflamed colon of CD/UC compared with control [p = 8.3E-03; p = 1.9E-03]. Sc-RNA sequencing confirmed this ACE2 dysregulation and exclusive epithelial ACE2 expression. Network analyses highlighted HNF4A as key regulator of ileal ACE2, and pro-inflammatory cytokines and interferon regulating factors regulated colonic ACE2. Inflammatory stimuli upregulated ACE2 in UC organoids [p = 1.7E-02], but not in non-IBD controls [p = 9.1E-01]. Anti-TNF therapy restored colonic ACE2 regulation in responders.
Conclusions
Intestinal inflammation alters SARS-CoV-2 coreceptors in the intestine, with opposing dysregulations in ileum and colon. HNF4A, an IBD susceptibility gene, seems an important upstream regulator of ACE2 in ileum, whereas interferon signalling might dominate in colon.
Keywords: COVID-19, ACE2, TMPRSS2, inflammatory bowel diseases, SARS-CoV-2, HNF4A, interferon, organoids, transcriptomics, single cell, intestinal inflammation
1. Introduction
Since the novel betacoronavirus SARS-CoV-2 was first reported in the province of Wuhan, China, at the end of 2019, the virus has spread worldwide. As of the 19 of August 2020, SARS-CoV-2 has caused more than 21.9 million infections, including 776 000 death globally.1 Despite being primarily a respiratory virus, coronavirus disease 2019 [COVID-19] can also present with non-respiratory signs, including digestive symptoms such as diarrhoea, nausea, and ageusia.2–4
Although it is thought that SARS-CoV-2 primarily infects the lungs with transmission via the respiratory route, the gastrointestinal tract may be an alternative viral target organ.3,5,6 Indeed, the SARS-CoV-2 receptor angiotensin converting enzyme 2 [ACE2] is highly expressed on differentiated enterocytes, with strong induction of generic viral response programmes upon viral binding.5–7 The cellular entry of coronaviruses depends on the binding of the spike [S] protein to a specific receptor, followed by an S protein priming by proteases, with key players ACE2 [receptor for the S protein] and TMPRSS2 [protease] in case of COVID-19.7–9 Furthermore, based on protein crystal structures, data predicted that the Middle East respiratory syndrome [MERS]-CoV receptor dipeptidyl peptidase 4 [DDP4] might act as a candidate binding target or co-receptor of SARS-CoV-2.10,11 In line, proteomic studies in COVID-19 patients suggested a prognostic role for DDP4.12 Upon cellular entry in nasal goblet secretory cells, lung type II pneumocytes, and ileal absorptive enterocytes, an interferon-driven mechanism is initiated, including the upregulation of ACE2 which further enhances infection.8
Why ACE2, the S protein receptor, is abundantly expressed on intestinal epithelium, is not entirely understood. Recent studies have addressed the homeostatic role of ACE2 on intestinal epithelial cells demonstrating defective intestinal amino acid absorption in ACE2-deficient mice.13 Mechanistically ACE2, independently of its role in the renin angiotensin system [RAS], is essential for regulating epithelial tryptophan absorption, expression of antimicrobial peptides, and consequently the ecology of the gut microbiome promoting homeostasis and preventing intestinal inflammation.14 Thus, ACE2 regulation could be linked to the pathogenesis of IBD, playing a role as modulator of epithelial immune homeostatic functions.
Individual susceptibility to COVID-19 may correlate with the expression of these designated [co]receptors. In this respect, studies investigating how inflammation affects ACE2, TMPRSS2, and/or DDP4 expression in ileum and colon, are limited and show conflicting data in inflammatory bowel disease [IBD].15,16 So far, data on COVID-19 in patients with IBD are rather limited,17–21 although they suggest that increasing age, a diagnosis of ulcerative colitis [UC] (as opposed to Crohn’s disease [CD]), and increasing disease activity are linked with a more severe course of COVID-19. In contrast, anti-inflammatory IBD therapy has not yet been associated with COVID-19 risk. Using a combination of bulk and single-cell transcriptomics and organoid cultures, we studied the intestinal expression of several SARS-CoV-2 co-receptors in the healthy gut and in IBD and investigated whether inflammation alters co-receptor expression.
2. Methods
2.1. Patients
This study was carried out at the University Hospitals Leuven [Leuven, Belgium]. All included patients had given written consent to participation in the Institutional Review Board approved IBD Biobank of University Hospitals Leuven, Belgium [B322201213950/S53684 and B322201110724/S52544]. Endoscopy-derived [un]inflamed mucosal biopsies were obtained cross-sectionally from IBD patients requiring colonoscopy during routine care [Supplementary Table S1, available as Supplementary data at ECCO-JCC online]. Samples from individuals undergoing colonoscopy for polyp detection were included as controls. Transmural ileal biopsies, derived during right hemicolectomy from CD patients and patients with colorectal cancer [CRC], were collected and stored in RPMI-1640 medium on ice until single cell isolation.
2.2. Organoids
Mucosal biopsies from both uninflamed and macroscopically inflamed colon segments [UC only] were processed as reported earlier.22–24 In short, crypts isolated as described before23 were embedded in Matrigel [phenol red free, growth factor reduced, Corning, NY, USA] diluted by 50% basal medium (DMEM:F12 supplemented with 1x GlutaMax, 10 mM HEPES and 100 U/ml penicillin, 100 µg/ml streptomycin [Gibco, Thermo Fisher Scientific, Waltham, Massachusetts, USA]). These organoids were then cultured in human expansion medium [basal medium supplemented with growth factors, as previously described24] for at least 4 weeks. Inflammation was then re-induced using an inflammatory mix (100 ng/ml tumour necrosis factor alpha [TNF-α], 20 ng/ml IL-1β, 1 μg/ml flagellin) over 24 h.22
2.3. Bulk transcriptomics
Inflamed biopsies were taken at the most affected site at the edge of an ulcerative surface, whereas uninflamed biopsies were taken randomly in macroscopically unaffected areas. All were stored in RNALater buffer [Ambion, Austin, TX, USA] and preserved at -80°C. As described previously,25 RNA from biopsies was isolated using the AllPrep DNA/RNA Mini kit [Qiagen, Hilden, Germany], and RNA libraries were prepared using the TruSeq Stranded mRNA protocol [Illumina, San Diego, USA]. RNA from organoids was extracted using the RNeasy Mini Kit [Qiagen] and libraries were constructed by the Lexogen QuantSeq 3’ mRNA-Seq Library Kit FWD [Lexogen, Vienna, Austria].22 All RNA libraries were sequenced by the Illumina HiSeq4000 [Illumina, San Diego, CA], with ~10‐20 M reads per biopsy RNA sample and ~3 M reads per organoid RNA sample. Raw sequencing data were aligned to the reference genome [GRCh37] using Hisat2 [version 2.1.0] 26 and absolute counts were generated using HTSeq.27 Counts were normalised for library size, and protein coding genes selected [Ensemble hg 19 reference build]28 using the DESeq2 package.29 A weighted gene co-expression network [WGCNA] was generated30 as described earlier.31,32 The module eigengene was defined as the first principal component summarising the expression patterns of all genes into a single expression profile within a given module. Genes showing the highest correlation with the module eigengene were referred to as hub genes. Pathway and upstream regulator analyses were performed using Ingenuity Pathway Analysis [IPA, QIAGEN, Aarhus, Denmark], with network visualisation via Cytoscape [v3.8.0].33 Publicly available microarray datasets of ileal and colonic biopsies [GEO GSE14580, GSE12251, GSE16879] were accessed to investigate the effect of anti-TNF therapy on genes of interest.34,35
2.4. Single-cell transcriptomics
Transmural ileal samples were treated with 1mM DTT and 1 mM EDTA in 1x Hank’s balanced salt solution [HBSS], and 1 mM EDTA in HBSS at 37°C for 30 min, respectively. Then tissue was transferred into a sterile gentleMACS C tube [Miltenyi Biotec], and digested with 5.4 U/mL collagenase D [Roche Applied Science], 100 U/mL DNase I [Sigma], and 39.6 U/mL dispase II [Gibco] with the gentleMACS™ Dissociator [program human_tumor_02.01]. Samples were incubated for 30 min at 37°C at 250 rpm. Dissociated samples were filtered with 70-µm cell strainers and treated with red blood cell lysis buffer [11814389001, Roche]. After centrifugation, single-cell suspensions were re-suspended in 0.4% BSA in PBS, and were immediately processed with 10 × 3’ v3 GEM kit, and loaded on a 10x chromium controller to create Single Cell Gel beads in Emulsion [GEM]. A cDNA library was created and assessed using a 10 × 3’ v3 library kit, and was then sequenced on a NovaSeq 6000 system [Illumina]. Pre-processing of the samples including alignment and counting was performed using Cell Ranger Software from 10x [Version: 3.0.2].
Publicly available colonic single-cell RNA sequencing data [sc-RNA seq] [Single Cell Portal, SCP 259] were downloaded and visualised using the SCP data browser.36 For colonic epithelial single-cell data, tSNE coordinates and publicly available annotation with the data were used for visualisation and analysis.
Annotation of the ileal data was performed using SingleR R package, with inbuilt Human Cell Atlas data as reference. Quality control, clustering, and dimensionality reduction of sc-RNA seq data was performed using Seurat R package [Version 3.1.5].37,38 Data from each 10x run were integrated after performing SCTransform on each dataset, with percentage of mitochondrial genes set as a parameter to be regressed. Single Cell Network Inference [SCENIC] analysis was performed using a python implementation of the SCENIC pipeline [PySCENIC] [version 0.9.19].39
2.5. Immunofluorescence staining
Transmural ileal biopsies, obtained during abdominal surgery in patients with IBD and CRC, were fixed in 4% formalin and embedded in paraffin, and sections of 5 µm were cut [Translational Cell & Tissue Research Laboratory, University Hospitals Leuven, and at VIB & KU Leuven Center for Brain & Disease Research]. After deparaffinisation, antigen retrieval was done in Tris-EDTA buffer [10 mM Tris base, 1 mM EDTA solution, 0.05% Tween 20, pH 9.0] at 95°C for 30 min; 1% BSA in PBST [0.1% Tween-20 and 0.5% sodium azide] was used to block non-specific binding of detection antibodies and gently permeabilise before ACE2 and Cytokeratin AE1/AE3 staining. In brief, ACE2 [Polyclonal, Cell Signaling Technology] and cytokeratin [IgG1-kappa, clone AE1/AE3, Dako] were applied in 1% BSA, followed by donkey anti-rabbit Cy3 [Jackson Immuno Research] and donkey anti-mouse Alexa fluor 488 [Invitrogen]. Slides were mounted in SlowFade™ Diamond Antifade Mountant [Invitrogen], and stored at 4 °C before imaging. Images were acquired using a Zeiss LSM 780 at the Cell and Tissue Imaging Cluster [CIC] at KU Leuven.
2.6. Genetics
All samples were genotyped using the Illumina GSA array. All single nucleotide polymorphisms [SNPs] and samples with more than 10% missingness rate were removed, as were SNPs with minor allele frequency [MAF] <0].001. Genotypes for rs6017342 [HNF4A] were extracted. All steps were performed using PLINK [v1.90b4.9].40
2.7. Statistical analysis
Statistical analysis was performed using R 3.6.2 [R foundation, Vienna, Austria]. Pearson correlation coefficients were computed to assess the correlation between individual genes. Multivariate regression analysis was performed using the R package ‘lm.beta’. Continuous variables on graphs were expressed as median and interquartile range [IQR]. ACE2, DPP4, and TMPRSS2 comparisons were done using two-sample t tests or Wilcoxon tests, as appropriate; and multiple testing correction was applied (adjusted p [adj. p], Benjamini‐Hochberg method).
3. Results
3.1. Intestinal ACE2, TMPRSS2, and DPP4 expression in IBD patients versus non-IBD controls
First, we studied the expression patterns of ACE2, DPP4, and TMPRSS2 in ileum and colon biopsies from 351 IBD patients [193 CD, 158 UC] and 51 non-IBD controls, based on bulk RNA sequencing.
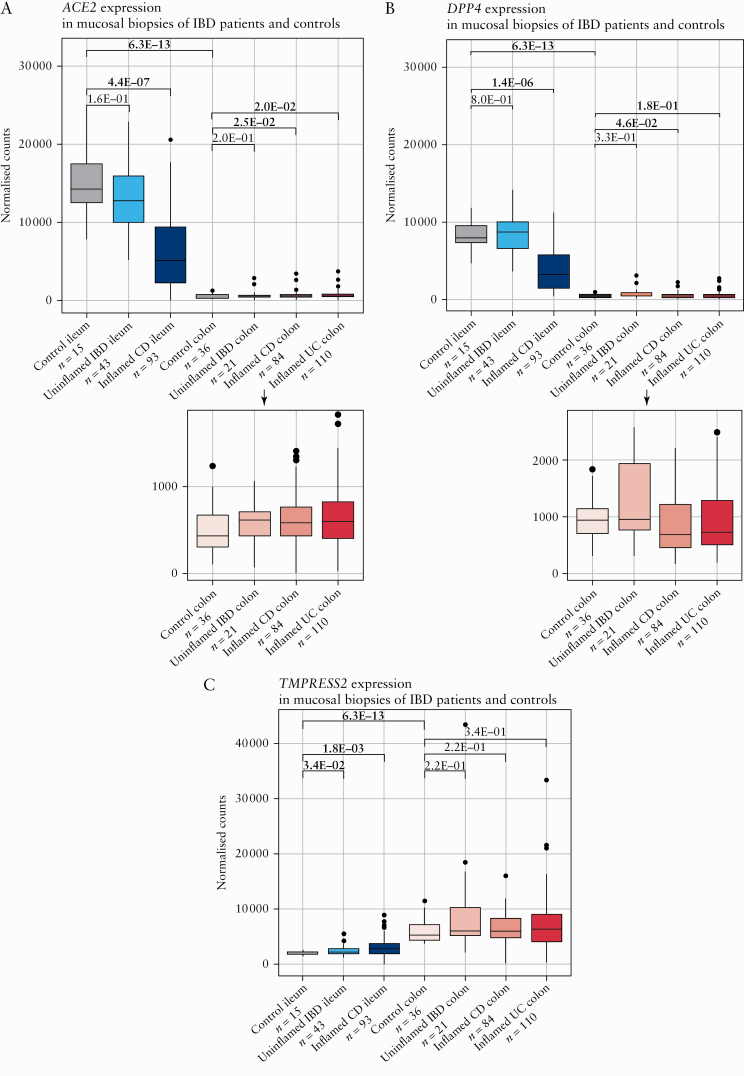
In non-IBD controls, ACE2 and DPP4 expression levels were strongly increased in ileum compared with colon 9fold change ([FC] = 32.0, p = 6.3E-13, adj. p = 1.9E-12; FC = 16.5, p = 6.3E-13, adj. p = 1.9E-12) [Figure 1A, B]. In contrast, ileal TMPRSS2 was lower compared with colon [FC = -2.9, p = 6.3E-13, adj. p = 1.9E-12] [Figure 1C].
Figure 1.
Mucosal ACE2, DPP4, and TMPRSS2 in IBD patients and controls. [A] Boxplots of mucosal ACE2 as measured by RNA sequencing [normalised counts]. [B] Boxplots of mucosal DPP4 as measured by RNA sequencing [normalised counts]. [C] Boxplots of mucosal TMPRSS2 as measured by RNA sequencing [normalised counts]. Significant comparisons [nominal p-values] are highlighted in bold. CD, Crohn’s disease; control, non-IBD controls; IBD, inflammatory bowel disease; UC, ulcerative colitis.
When turning to tissue from IBD patients, ACE2 and DPP4 levels in uninflamed IBD ileum were similar to those observed in matched control ileum [p = 1.6E-01, adj. p = 2.4E-01; p = 8.0E-01, adj. p = 8.0E-01] [Figure 1A, B]. TMPRSS2 however, was upregulated compared with control ileum [FC = 1.2, p = 3.4E-02, adj. p = 1.0E-01] [Figure 1C]. In uninflamed IBD colon, expression levels of ACE2, DDP4, and TMPRSS2 did not differ from control colon [p = 2.0E-01, adj. p = 6.0E-01; p = 3.3E-01, adj. p = 3.3E-01; p = 2.2E-01, adj. p = 3.3E-01] [Figure 1A‐C].
In inflamed CD ileum, ACE2 and DPP4 expression was significantly decreased compared with control ileum [FC = -2.8, p = 4.4E-07, adj. p = 1.3E-06; FC = -2.5, p = 1.4E-06, adj. p = 2.1E-06] [Figure 1A, B]. TMPRSS2 behaved conversely, with a significant upregulation in inflamed ileum versus control ileum [FC = 1.4, p = 1.8E-03, adj. p = 1.8E-03] [Figure 1C]. At colonic level, ACE2 expression was higher in inflamed CD and UC colon than in control colon [FC = 1.4, p = 2.5E-02, adj. p = 7.5E-02; FC = 1.4, p = 2.0E-02, adj. p = 6.0E-02, respectively] [Figure 1A]. Except for a decrease in DPP4 expression in inflamed CD colon versus control colon [FC = 1.3, p = 4.6E-02, adj. p = 6.9E-02], no dysregulations were observed for colonic DPP4 and TMPRSS2 [p ≤3.4E-01, adj. p ≤3.4E-01] [Figure 1B, C].
Despite ACE2 being X-linked, multivariate analysis did not reveal any contribution of sex to mucosal ACE2 expression [p = 5.1E-01], nor of age [p = 1.4E-01], diagnosis [p = 5.6E-01], or disease duration [p = 5.2E-01]. Intestinal ACE2 expression was significantly affected by biopsy location [p = 2.5E-34] and inflammatory state [p = 4.2E-12] [Supplementary Table S2, available as Supplementary data at ECCO-JCC online].
3.2. Gene co-expression analysis of the ACE2-, DPP4-, and TMPRSS2-related networks
To get a better understanding of the biological network of ACE2, DPP4, and TMPRSS2, we performed WGCNA on all mucosal biopsies.
At ileal level, we identified 18 co-expression modules [clusters] ranging in size from 106 to 1465 genes [Supplementary Figure S1A, available as Supplementary data at ECCO-JCC online]. One module contained both ACE2 and DPP4 [module ‘blue’; 1134 genes] [Supplementary Table S3, available as Supplementary data at ECCO-JCC online]. The strongest correlation with the eigengene [ie, the principal component] of this ACE2/DPP4-module was found for hub genes MMP5 [r = 0.94, p = 8.6E-74], ZNF664 [r = 0.94, p = 3.7E-71] and DPP4 [r = 0.93, p = 1.2E-68] [Supplementary Figure S1A]. Moreover, ACE2 also seemed to have a central role in this co-expression network with a correlation value of r = 0.86 [p = 4.6E-45] [Supplementary Figure S1A]. Pathway analysis of the ACE2/DPP4-module found enrichment for epithelium-related metabolic pathways such as xenobiotic metabolism signalling, nicotine degradation ii, and melatonin degradation [p < 1.0E-08]. Predicted upstream analysis (using curated datasets in ingenuity pathway analysis [IPA]) highlighted the transcription regulator HNF4A, an IBD susceptibility gene, as the most likely upstream regulator of the ACE2/DPP4-module [p = 1.2E-11].
TMPRSS2 belonged to a separate module ‘yellow’ [1126 genes] with hub gene COA3 [r = 0.92, p = 4.7E-61] [Supplementary Figure S1A, Supplementary Table S4, available as Supplementary data at ECCO-JCC online]. Genes within this module were mainly related to mitochondrial functions [eg. oxidative phosphorylation, mitochondrial dysfunction and sirtuin signalling, p < 1.6E-29], and their top upstream regulator was again HNF4A [p = 1.5E-27].
At colonic level, 24 co-expression modules were present ranging in size from 128 to 2267 genes [Supplementary Figure S1B]. In contrast to the ileum, colonic ACE2 and DPP4 were not co-expressed [Supplementary Table S3], with ACE2 being part of module ‘green’ [797 genes]. Here, ACE2 co-clustered with TMPRSS2. The ACE2-module with top hub gene TMEM63B [r = 0.89, p = 5.8E-81] did not show significant enrichment for specific pathways. Upstream analysis of this module ranked TNF and again HNF4A as the top regulators [p = 7.7E-06; p = 9.4E-03].
Last, we studied the relationship between mucosal ACE2 and HNF4A expression levels. Ileal ACE2 expression strongly correlated with ileal HNF4A expression [r = 0.69, p <2.2E-16], whereas colonic levels showed limited correlation [r = 0.2, p = 1.3E-03] [Supplementary Figure S2, available as Supplementary data at ECCO-JCC online].
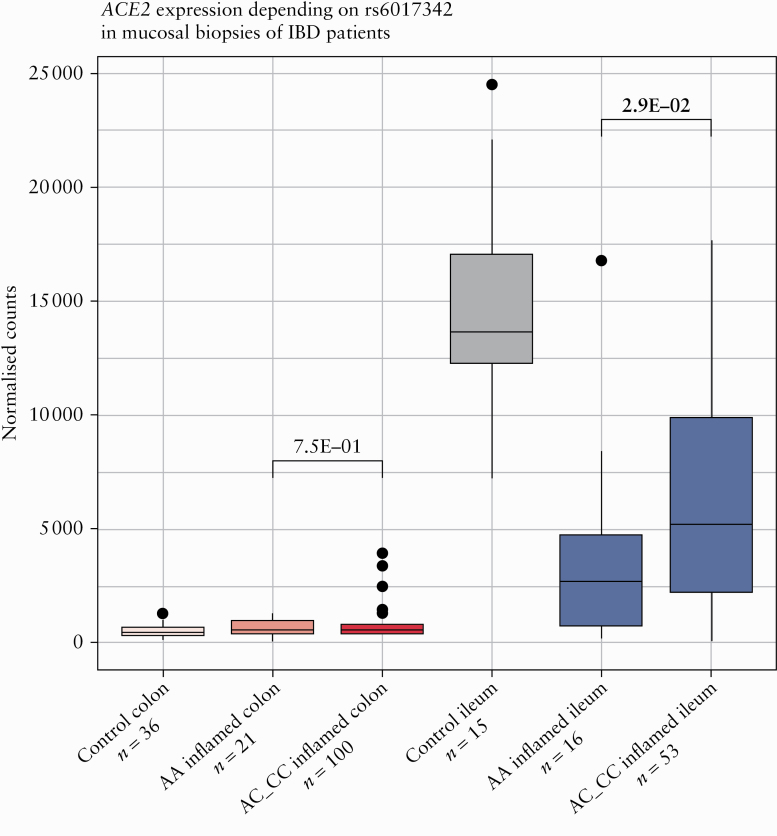
3.3. Single nucleotide polymorphisms in HNF4A linked to ACE2 expression in ileum but not in colon
As the expression of ACE2-modules was found to be driven by the IBD susceptibility locus, HNF4A, we next studied the genetic variability in rs6017342 [ie, the causal IBD variant in this locus41], and its relationship with ACE2 and HNF4A expression, both in inflamed ileum and in colon. Ileal ACE2 levels were lower in patients carrying the HNF4A-AA genotype, compared with patients carrying the C-allele, ie, HNF4A-AC or HNF4A-CC genotypes [p = 2.8E-02] [Figure 2]. Colonic ACE2 expression was independent of the HNF4A genotype [p = 6.7E-01].
Figure 2.
Mucosal ACE2 and HNF4A in IBD patients and controls depending on rs6017342 genotype. Boxplots of mucosal ACE2 as measured by RNA sequencing [normalised counts]. Significant comparisons are highlighted in bold. IBD, inflammatory bowel disease; controls, non-IBD controls.
3.4. Decrease of ACE2/TMPRSS2 double-positive cells in inflamed ileum, but not in colon
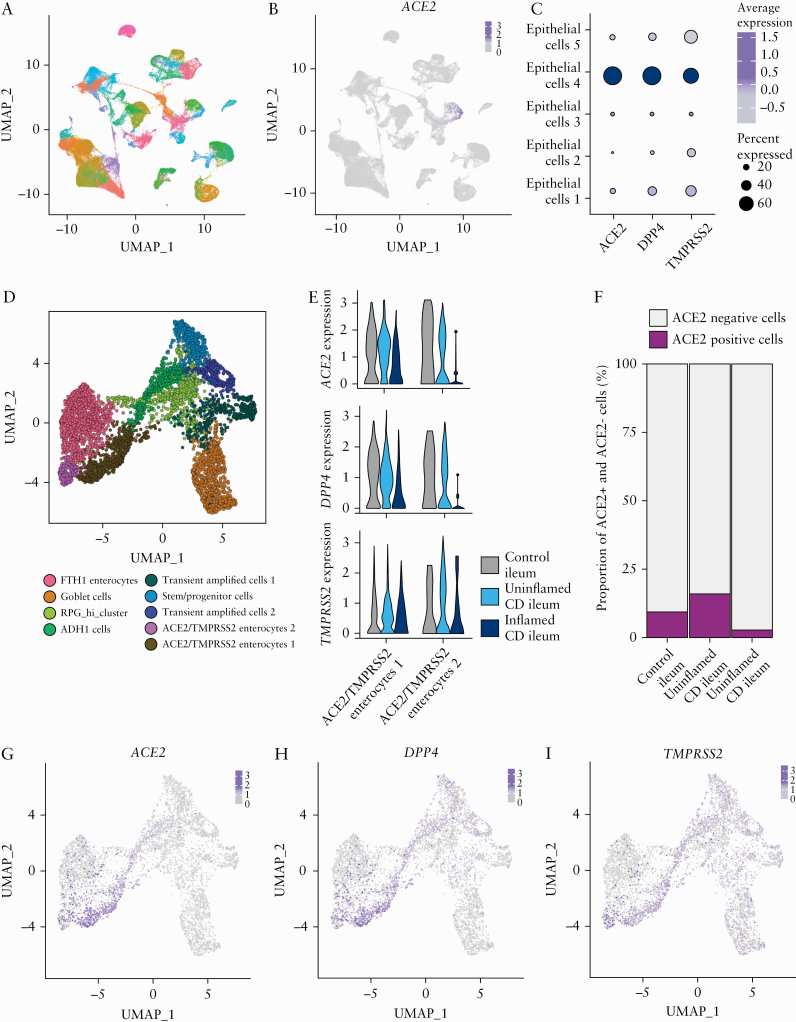
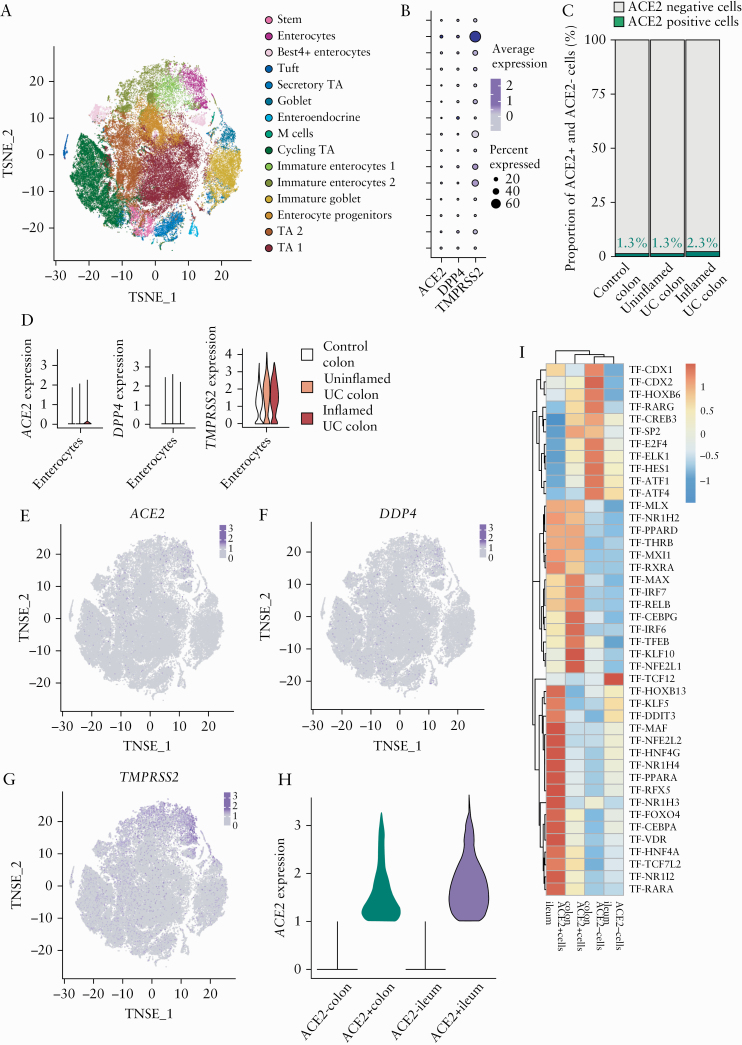
ACE2 expression in the gastrointestinal tract is primarily found in absorptive enterocytes,8,42 which could indirectly be confirmed through the significant correlation [p < 2.2E-16] between mucosal ACE2 and several epithelial marker genes [APOA1, SI, FABP6, ENPEP] [Supplementary Figure S3, available as Supplementary data at ECCO-JCC online]. To further examine the expression of genes associated with risk of SARS-CoV-2 infection in IBD patients, we employed sc-RNA seq to profile transmural biopsies of [un]inflamed regions of resected tissue from six CD patients undergoing ileocaecal resection. Unaffected ileal tissue from five patients with CRC undergoing right hemicolectomy was used as control. A total of 78 722 cells were used for downstream analyses containing a similar number of cells from each type of tissue [inflamed CD, uninflamed CD, and healthy tissue] [Supplementary Figure S4B, available as Supplementary data at ECCO-JCC online]; 61 cell clusters belonging to epithelial, immune, and stromal cells were obtained using unsupervised clustering [Figure 3A; Supplementary Figure S4A]. Cell clusters were annotated by correlating the cluster gene expression profiles with Human Cell Atlas using SingleR, as previously described.43ACE2 expression was found exclusively in epithelial cell clusters [Figure 3B, C], which could also be confirmed using immunofluorescence staining [Figure 4]. To define the epithelial cell subtypes expressing ACE2 at deeper resolution, clusters annotated as epithelial cells by SingleR were extracted and re-clustered [Figure 3D]. The re-clustered epithelial cell subtypes were annotated using a marker panel designed based on previous reports [Supplementary Figure S4C].44 Three enterocyte clusters were identified, out of which two clusters co-expressed ACE2, TMPRSS2, and DPP4. Most prominent ACE2 expression was observed in the ACE2/TMPRSS2 Enterocytes 1 cluster [Figure 3G‐I; Supplementary Figure S4D, available as Supplementary data at ECCO-JCC online].
Figure 3.
Decrease of ACE2/TMPRSS2 double-positive cells in inflamed ileum in CD patients. [A] Uniform manifold approximation and projection [UMAP] plot showing unsupervised clustering of integrated single-cell RNA sequencing data from control, uninflamed and inflamed ileal tissue. [B] Expression of ACE2 overlaid on the UMAP plot as in A. [C] Expression of ACE2, DPP4, and TMPRSS2 in ileal epithelial cells. [D] UMAP showing epithelial sub-clusters obtained upon re-clustering only the epithelial cells in ileum. [E] Expression and distribution of ACE2, DPP4, and TMPRSS2 in the two enterocyte clusters co expressing ACE2 and TMPRSS2, split between control, uninflamed, and inflamed samples. [F] Proportion of ACE2+ and ACE2- cells in control, uninflamed, and inflamed samples in the ileal epithelial cells. [G‐I] Gene expression overlaid on the UMAP Plot as in panel D of ACE2, DPP4, and TMPRSS2, respectively. CD, Crohn’s disease; control, non-IBD controls.
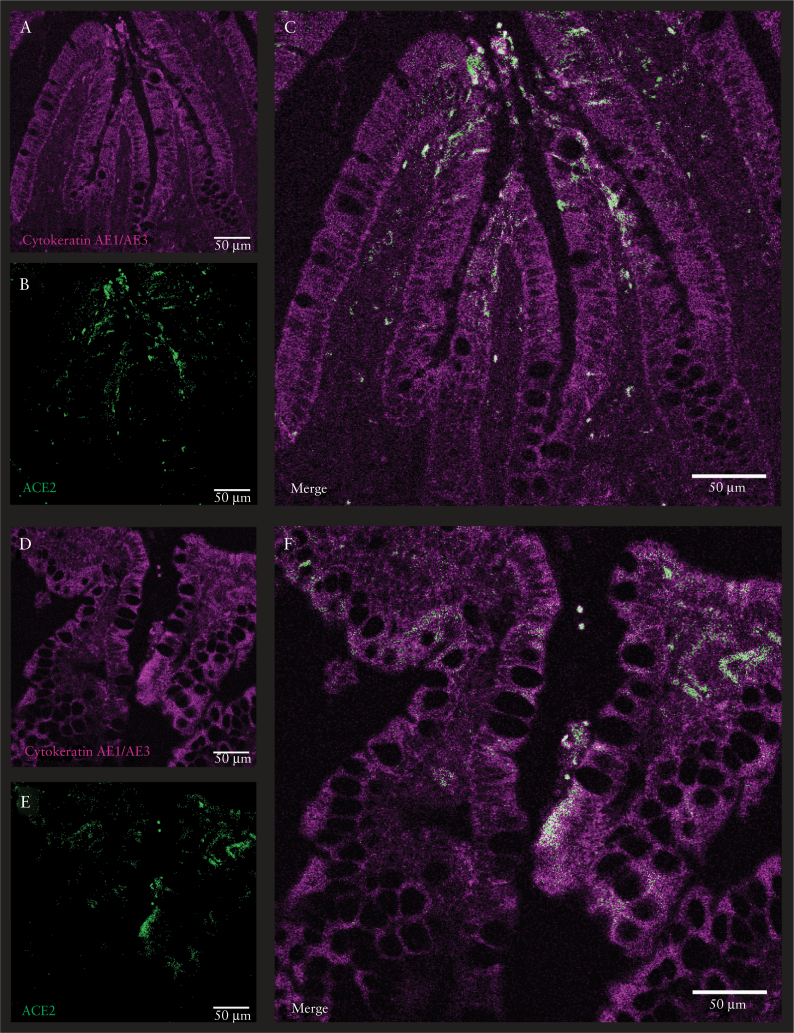
Figure 4.
Cytokeratin AE1/AE3 and ACE2 expression in human gut. Confocal microscopy images of human gut in which ACE2-positive epithelial cells were stained with cytokeratin AE1/AE3 [magenta] and ACE2 [green]. The scale bar in the immunofluorescent image represents 50 μm. Normal ileum from patient with colorectal cancer [A‐C]; inflamed ileum from patient with Crohn’s disease [D‐F].
Next, we asked whether ACE2 expression varied across ileal tissue in an inflammatory state, as observed in our bulk transcriptomic data [Figure 1A]. ACE2 expression and frequency of ACE2-positive cells were clearly reduced in ileum of patients with active CD, compared with uninflamed or healthy tissue [Figure 3E, F; Supplementary Figure S4E]. A similar reduction of DPP4 expression was observed in the inflamed samples in the ACE2/TMPRSS2 Enterocytes 1 and ACE2/TMPRSS2 Enterocytes 2 clusters [Figure 4E]. In linenwith this, reduction of ACE2 expression in inflamed ileum compared with healthy tissue was also confirmed with confocal imaging [Figure 4].
To define ACE2 expression in healthy and inflamed colon, we visualised publicly available colonic sc-RNA seq data containing 366 650 cells from colonic mucosa obtained in 18 [in]active UC patients and 12 healthy individuals [Single Cell Portal, SCP 259] [Supplementary Figure S5A‐C, available as Supplementary data at ECCO-JCC online].36 As for the ileum, ACE2 was solely expressed in colonic epithelium, mainly in a subset of enterocytes [Figure 5A, B; Supplementary Figure S5D]. As in the ileum, the ACE2-positive colonic enterocyte cluster co-expressed TMPRSS2 and DPP4 [Figure 5B, E‐G]. However, in contrast to ileum, colonic ACE2 expression was mainly restricted to enterocytes isolated from patients with active UC, while undetectable in colonic enterocytes isolated from the mucosa of healthy subjects [Figure 5C, D; Supplementary Figure S5E].
Figure 5.
Increased colonic ACE2 expression in the epithelial cells of patient with active UC. [A] t-distributed stochastic neighbour embedding [tSNE] plot showing the clustering and annotation of epithelial cells from the colon as reported in Smillie et al.36 [B] Expression of ACE2, DPP4, and TMPRSS2 in epithelial cell clusters of the colon. [C] Proportion of ACE2+ colonic epithelial cells in control, uninflamed, and inflamed samples. [D] Expression of ACE2, DPP4, and TMPRSS2 in colonic enterocytes from control, uninflamed, and inflamed samples. [E‐G] Expression of ACE2, DPP4, and TMPRSS2 overlaid on tSNE shown in panel A. [H] Expression level of ACE2 in ACE2+ cells of colon and ileum. [I] Heatmap showing scaled area under the curve [AUC] values of top 15 specific and highly enriched regulons [average AUC >0.1] in ACE2+ or ACE2- compartments in integrated data of ileal and colonic single-cell data identified by SCENIC analysis; control, non-IBD controls; UC, ulcerative colitis.
To compare expression and regulation of ACE2 between colon and ileum, we performed an integrated analysis of epithelial cells from colon and ileum [Supplementary Figure S6A, B, available as Supplementary data at ECCO-JCC online]. In colonic ACE2-positive epithelial cells, ACE2 expression was lower compared with levels in ileal ACE2-positive epithelial cells [Figure 5H]. Furthermore, using SCENIC we performed genomic regulatory networks analysis of the epithelial cells to identify specific transcription programmes in ACE2-expressing enterocytes, both in ileum and colon. As demonstrated using bulk RNA analysis, we found a relatively higher HNF4A regulon activation in ileal ACE2-positive cells, compared with colonic ACE2 enterocytes [Figure 5I]. Differently, colonic ACE2-expressing enterocytes were found to have increased regulon activity of interferon-responsive factors, such as IRF6 and IRF7, when compared with ileum [Figure 5I].
3.5. Ileum and colon: different key regulators in ACE2-positive cells
We then asked whether particular expression patterns within ACE2-positive cells depend on the tissue and/or inflammatory state, and studied which upstream regulators were linked to these changes. When comparing expression profiles of ACE2-positive cells from inflamed CD ileum with control ileum, we found 56 differentially expressed genes [adj. p <0.05, FC >2.0]. Predicted upstream regulators of these genes were HNF4A [inhibited, p = 2.3E-04] and IFNγ [activated, p = 5.2E-05]. At the colonic level, we identified 54 differentially expressed genes in ACE2-positive cells from inflamed colon, as compared with control tissue. TNF, lipopolysaccharides, IFNγ, and IL-1β were predicted as top-ranked upstream regulators [activated, p ≤1.9E-15].
3.6. Inflammatory stimuli result in upregulation of ACE2 and TMPRSS2 in organoids from IBD patients but not from healthy individuals
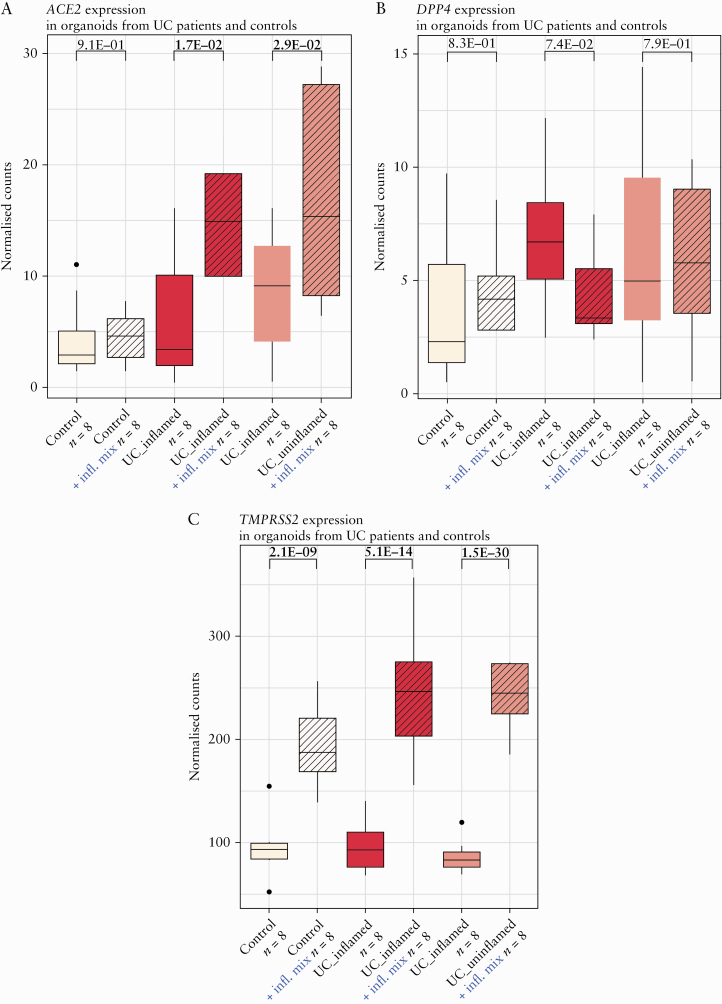
Because of the clear upregulation of ACE2 in inflamed colonic mucosa [Figure 1A] and the prediction of TNF as key regulator in ACE2-positive cells, we investigated the effect of an inflammatory stimulus on ACE2 expression in an ex vivo organoid model. In organoids derived from controls, inflammatory stimuli did not affect ACE2 expression [p = 9.1E-01, adj. p = 9.1E-01] [Figure 6A]. Strikingly, in organoids derived from inflamed or uninflamed colonic biopsies from UC patients, addition of an inflammatory stimulus did significantly upregulate ACE2 [FC = 2.4, p = 1.7E-02, adj. p = 2.6E-02; FC = 2.0, p = 2.9E-02, adj. p = 4.4E-02] [Figure 6A]. No significant effect on DPP4 expression could be observed [p = 7.4E-02, adj. p = 1.0E + 00; p = 7.9E-01, adj. p = 1.0E-00], whereas TMPRSS2 was significantly upregulated after inflammatory stimulation [FC = 2.6, p = 5.1E-14, adj. p = 1.5E-13; FC = 2.8, p = 1.5E-30, adj. p = 4.4E-30] [Figure 6B, C].
Figure 6.
Organoid ACE2, DPP4, and TMPRSS2 in UC patients and controls with and without addition of an inflammatory mix. [A] Boxplots of organoid ACE2 as measured by RNA sequencing [normalised counts]. [B] Boxplots of organoid DPP4 as measured by RNA sequencing [normalised counts]. [C] Boxplots of organoid TMPRSS2 as measured by RNA sequencing [normalised counts]. Significant comparisons [nominal p-values] are highlighted in bold; control, non-IBD controls; UC, ulcerative colitis.
3.7. Anti-TNF therapy restores colonic, but not ileal, epithelial ACE2 regulation in anti-TNF responders
Given that the ex vivo model clearly confirmed the effect of a pro-inflammatory mix, including TNF, on epithelial ACE2 expression, we subsequently studied the effect of neutralising TNF [through administration of infliximab] on intestinal ACE2 expression in IBD patients with active endoscopic disease. Paired transcriptomic data, generated before first infliximab administration and 4–6 weeks after treatment initiation, confirmed a significant downregulation of colonic ACE2 in endoscopic remitters, but not in non-remitters [p = 1.8E-04, p = 6.5E-01, respectively] [Supplementary Figure S7], available as Supplementary data at ECCO-JCC online. In contrast, infliximab therapy did not significantly affect ileal ACE2 expression in remitters and non-remitters [p = 7.8E-02, p = 2.25E-01, respectively].
4. Discussion
Many patients with IBD have long-term exposure to corticosteroids, thiopurines, methotrexate, small molecules, and/or biologic agents, classifying them as high-risk patients because of their immunosuppression. In addition, intestinal inflammation has shown to be an important risk factor for SARS-CoV-2 infection and prognosis in IBD.17–21 However, emerging evidence now suggests that IBD patients do not seem more vulnerable for COVID-19. To reconcile these observations, we investigated the role of intestinal inflammation on the potential viral intestinal entry mechanisms, through bulk and single-cell transcriptomics, immunofluorescence, and ex vivo organoid cultures in patients with IBD.
In contrast to previous bulk data,15 we observed significant alterations in intestinal ACE2 expression depending on the location and inflammatory state, at both tissue and single-cell mRNA levels, as at protein level. ACE2 expression was limited exclusively to epithelial cells, in both ileum and colon. Hence, ACE2 dysregulation in bulk transcriptomics, as a result of massive influx of immunocytes at the site of inflammation, could be excluded.
It is suggested that SARS-CoV-2 infects epithelial cells, causing cytokine and chemokine release, resulting in acute intestinal inflammation characterised by infiltration of neutrophils, macrophages, and T cells,45 with associated shedding of faecal calprotectin and increased systemic IL-6 response46 and IFN signallng.8 Similar to recent data,16,47,48 we found a significant downregulation of ACE2 in inflamed ileum and a significant ACE2 upregulation in inflamed colon. This opposing effect of inflammation on intestinal ACE2 expression in small and large intestine was striking, which could be attributed—based on sc-RNA data—to different key transcription factors active between ileal and colonic ACE2-positive cells.
Being an IBD susceptibility locus,49 epithelial HNF4A plays a protective role in IBD by consolidating the epithelial barrier,50 especially in small intestine.51 HNF4A has also been found as a transcriptional sensor of inflammation,52 plays a key role as transcription factor in the regulation of angiotensinogen metabolism,53 and has recently been predicted to regulate intestinal ACE2 expression.42 The decrease in ACE2 in inflamed ileum does therefore not come as a surprise. In individuals carrying the minor AA genotype at the IBD HNF4A susceptibility locus, ileal ACE2 expression was even further downregulated, without any effect on colonic ACE2. Of note, our sc-RNAseq data showing ACE2 downregulation in enterocytes from inflamed CD ileum further suggest an intrinsic regulation of ACE2. In addition, as we observed significant correlations between enterocyte markers and ileal ACE2 as well as an overall decrease in number of cells expressing ACE2 in inflamed CD ileum, a loss of enterocytes might also explain lower ACE2 levels.
Remarkably, a very recent genome-wide association study [GWAS] identified 3p21.31 as a genetic locus associated with COVID-19–induced respiratory failure.54 This locus covers a cluster of six genes [SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1], with the identified risk allele [ie, worse COVID-19 outcome] being associated with increased SCL6A20 expression. Strikingly, SCL6A20 is known to be regulated by HNF4A.55
Although the colonic ACE2 co-expression cluster in bulk tissue was also enriched for HNF4A as upstream transcriptional regulator, single-cell data revealed that colonic ACE2 expression seems primarily driven by interferon regulator factors. Upstream regulating analysis further supported that pro-inflammatory cytokines, including TNF, IFNγ, and IL-1β, contribute to colonic ACE2 upregulation. Hypothetically, elevated colonic ACE2 levels in patients with active inflammation might promote viral entry and, in theory, could promote COVID-19 disease severity. However, functional data are currently lacking to prove this hypothesis. Furthermore one could question this hypothesis, as downregulated ACE2 in inflamed ileum remains much higher than in normal and IBD colon. However, ACE2 expression is the most abundant in the small intestine, followed by the large intestine, whereas its expression is limited in the respiratory system.56–58 Moreover, a recent study in human small intestinal organoids observed similar SARS-CoV-2 infection rates between enterocyte precursors and enterocytes, whereas ACE2 expression was ~1000-fold higher in differentiating organoids as compared with proliferating organoids. This suggests that lower levels of ACE2—as observed in the colon—may be sufficient for viral entry.5
Although there is yet no direct evidence that altered expression of intestinal ACE2 directly affects SARS-CoV-2 intestinal entry and tropisms to different intestinal sites,59 using ex vivo organoid models we confirmed that pro-inflammatory cytokines can upregulate colonic epithelial ACE2 expression in IBD patients, but not in healthy individuals. Different genetic susceptibility and/or microbial composition may be responsible for the difference in response to inflammatory stimuli observed in controls and in IBD. Indeed, it has already been demonstrated that organoids from UC patients maintain some inherent differences as compared with non-IBD tissue,22,60 presumably reflecting inherent genetic factors which could result in a more sensitive epithelium.
Being the key example of a complex immune-mediated entity where environmental and microbial factors modulate the immune response in a genetically susceptible host,61 the differences in ACE2 expression upon inflammatory stimuli between colon and ileum in patients with IBD may also be attributed to differences in the intestinal microbiome. Lipopolysaccharides, comprising the wall of Gram-negative bacteria, was indeed identified as one of the key drivers of the ACE2 gene cluster in colon, but not in ileum. However, blind use of antibiotics or probiotics for COVID-19 is not recommended until a better understanding of the effect of SARS-CoV-2 on gut microbiota is obtained.62
National and international registries suggest active IBD as a risk factor for [complicated] COVID-19.17–21 Adequate disease management, by appropriate dampening of intestinal inflammation, therefore seems key in protecting IBD patients from COVID-19. The ACE2 upregulation in inflamed colon could potentially affect viral cell entry in active UC patients and/or CD patients with colonic involvement, although functional data are currently lacking. So far, international registries have not yet reported any COVID-19 outcome data in IBD patients by disease location.
Of note, several key cytokines implicated in IBD pathogenesis,61,63 and also key drivers of ACE2 colonic expression in this study, are currently under investigation as potential therapeutic targets for COVID-19, including TNF, IFNγ, IL-1β, and IL-6.64 Although further evidence is warranted if these anticytokine therapies can dampen the observed cytokine storm in COVID-19, we demonstrated that anti-TNF therapy does restore intestinal ACE2 dysregulation in a subset of IBD patients.
In this study, we acknowledge the lack of data on SARS-CoV-2 infected patients, a sequencing depth not enabling a search for HNF4A alternative splicing and isoforms with pro- and anti-inflammatory effects,65 and the lack of additional functional validation experiments [eg., intestinal HNF4A regulation of ACE2, the role of intestinal ACE2 in SARS-CoV-2 entry]. Despite these limitations, the replication of our findings on several levels [tissue and single-cell gene expression, protein expression, and ex vivo models] highlights the robustness of our observations. Current guidelines do not promote stopping immunosuppressive and biologic drugs in IBD patients without symptoms suggestive of COVID-19. On the contrary, immunosuppressive and biologic drugs may protect against the development of severe forms of COVID-19 infection.66
In conclusion, using bulk and single-cell transcriptomic datasets as well as ex vivo organoid cultures, we demonstrated that intestinal inflammation could alter the expression of SARS-CoV-2 entry mechanisms in the intestinal epithelium, with opposing dysregulations seen in ileum and colon. HNF4A, an IBD susceptibility gene and transcriptional regulator of one of the key Covid-19 GWAS loci, seems an important upstream regulator of ACE2 expression in ileal tissue. In contrast, colonic ACE2 expression seems to depend on interferon-regulating factors and pro-inflammatory cytokines.
Supplementary Material
Funding
KA is a doctoral fellow and SV and MF are Senior Clinical Investigators of the Research Foundation Flanders [FWO], Belgium. GM’s laboratory is supported by an FWO grant [G.0D83.17N], a grant from the International Organization for the Study of Inflammatory Bowel Diseases [IOIBD], a grant from the European Crohn´s and Colitis Organisation [ECCO], and grants from the KU Leuven Internal Funds [C12/15/016 and C14/17/097]. SV and GM are funded by a Strategic Basic Research FWO grant [S008419N].
Conflict of Interest
BV reports financial support for research from Pfizer; lecture fees from Abbvie, Ferring, Takeda Pharmaceuticals, Janssen, and R Biopharm; consultancy fees from Janssen and Sandoz. JS reports lecture fees from Abbvie, Takeda, Janssen, and Nestle Health Sciences. MF reports financial support for: research from Amgen, Biogen, Janssen, Pfizer, Takeda; consultancy from Abbvie, Boehringer-Ingelheim, MSD, Pfizer, Sandoz, Takeda, and Thermo Fisher; speaking from Abbvie, Amgen, Biogen, Boehringer-Ingelheim, Falk, Ferring, Janssen, Lamepro, MSD, Mylan, Pfizer, Sandoz, and Takeda. GM received financial support for research from DSM Nutritional Products, Karyopharm Therapeutics, and Janssen. SV reports financial support for: research from MSD, AbbVie, Takeda, Pfizer, J&J; lectures from MSD, AbbVie, Takeda, Ferring, Centocor, Hospira, Pfizer, J&J, Genentech/Roche; consultancy from MSD, AbbVie, Takeda, Ferring, Centocor, Hospira, Pfizer, J&J, Genentech/Roche, Celgene, Mundipharma, Celltrion, SecondGenome, Prometheus, Shire, Prodigest, Gilead, Galapagos.
Author Contributions
BV: study design, data acquisition and interpretation, statistical analysis, and drafting of the manuscript. SaV: study design, data acquisition and interpretation, statistical analysis, and drafting of the manuscript. SAR: data acquisition and interpretation [single-cell RNA], statistical analysis, and critical revision of the manuscript. BJK: data acquisition and interpretation [single-cell RNA and immunostainings] and critical revision of the manuscript. KA: data acquisition and interpretation [organoid data], statistical analysis, and critical revision of the manuscript. IC: data acquisition [genetics] and critical revision of the manuscript. JS: data interpretation and critical revision of the manuscript. MF: data interpretation and critical revision of the manuscript. GM: supervision, data acquisition, and interpretation, critical revision of the manuscript. SV: study design, supervision, data interpretation and critical revision of the manuscript. All authors agreed on the final manuscript.
References
- 1. World Health Organization. Coronavirus Disease [Covid-2019] Situation Reports. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Accessed May12, 2020.
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020;158:1831–3.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mao R, Qiu Y, He JS, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2020;5:667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamers MM, Beumer J, van der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020;369:50–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stanifer ML, Kee C, Cortese M, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep 2020;32:107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sungnak W, Huang N, Bécavin C, et al. ; HCA Lung Biological Network . SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 2020;26:681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ziegler CGK, Allon SJ, Nyquist SK, et al. ; HCA Lung Biological Network. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020;181:1016–35.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li Y, Zhang Z, Yang L, et al. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. iScience 2020;23:101160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou J, Li C, Zhao G, et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv 2017;3:eaao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van de Veerdonk FL, Janssen NAF, Grondman I, et al. A systems approach to inflammation identifies therapeutic targets in SARS-COV-2 infection. medRxiv 2020:2020.05.23.20110916. [Google Scholar]
- 13. Singer D, Camargo SM, Ramadan T, et al. Defective intestinal amino acid absorption in Ace2 null mice. Am J Physiol Gastrointest Liver Physiol 2012;303:G686–95. [DOI] [PubMed] [Google Scholar]
- 14. Hashimoto T, Perlot T, Rehman A, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012;487:477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Burgueño JF, Reich A, Hazime H, et al. Expression of SARS-CoV-2 entry molecules ACE2 and TMPRSS2 in the gut of patients with IBD. Inflamm Bowel Dis 2020;26:797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krzysztof NJ, Christoffer LJ, Rahul K, et al. Age, inflammation and disease location are critical determinants of intestinal expression of sars-cov-2 receptor ace2 and tmprss2 in inflammatory bowel disease. Gastroenterology 2020;159:1151–1154.e2. doi: 10.1053/j.gastro.2020.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bezzio C, Saibeni S, Variola A, et al. ; Italian Group for the Study of Inflammatory Bowel Disease [IG-IBD] . Outcomes of COVID-19 in 79 patients with IBD in Italy: an IG-IBD study. Gut 2020;69:1213–7. [DOI] [PubMed] [Google Scholar]
- 18. Brenner E, Ungaro R, Colombel J, Kappelman M. Secure-IBD Database Public Data Update. 2020. covidibd.org. Accessed 12 May, 2020.
- 19. Taxonera C, Sagastagoitia I, Alba C, et al. 2019 novel coronavirus disease [covid-19] in patients with inflammatory bowel diseases. Aliment Pharmacol Ther. 2020;52:276–283. [Epub ahead of print 7 June 2020] doi: 10.1111/apt.15804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lukin DJ, Kumar A, Hajifathalian K, et al. Baseline disease activity and steroid therapy stratify risk of covid-19 in patients with inflammatory bowel disease. Gastroenterology 2020:S0016-5085(20)34738-7. doi: 10.1053/j.gastro.2020.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khan N, Patel D, Xie D, et al. Impact of anti-TNF and thiopurines medications on the development of covid-19 in patients with inflammatory bowel disease: a nationwide VA cohort study. Gastroenterology 2020:S0016-5085(20)34737-5. doi: 10.1053/j.gastro.2020.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnauts K, Verstockt B, Santo Ramalho A, et al. Ex vivo mimicking of inflammation in organoids derived from patients with ulcerative colitis. Gastroenterology 2020:S0016-5085(20)34736-3. doi: 10.1053/j.gastro.2020.05.064. [DOI] [PubMed] [Google Scholar]
- 23. Vanhove W, Nys K, Arijs I, et al. Biopsy-derived intestinal epithelial cell cultures for pathway-based stratification of patients with inflammatory bowel disease. J Crohns Colitis 2018;12:178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vancamelbeke M, Laeremans T, Vanhove W, et al. Butyrate does not protect against inflammation-induced loss of epithelial barrier function and cytokine production in primary cell monolayers from patients with ulcerative colitis. J Crohns Colitis 2019;13:1351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verstockt B, Verstockt S, Veny M, et al. Expression levels of 4 genes in colon tissue might be used to predict which patients will enter endoscopic remission after vedolizumab therapy for inflammatory bowel diseases. Clin Gastroenterol Hepatol 2020;18:1142–51.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anders S, Pyl PT, Huber W. HTSeq – a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yates A, Akanni W, Amode MR, et al. Ensembl 2016. Nucleic Acids Res 2016;44:D710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verstockt B, Verstockt S, Creyns B, et al. Mucosal IL13RA2 expression predicts nonresponse to anti-TNF therapy in Crohn’s disease. Aliment Pharmacol Ther 2019;49:572–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verstockt S, De Hertogh G, Van der Goten J, et al. Gene and miRNA regulatory networks during different stages of Crohn’s disease. J Crohns Colitis 2019;13:916–30. [DOI] [PubMed] [Google Scholar]
- 33. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009;58:1612–9. [DOI] [PubMed] [Google Scholar]
- 35. Arijs I, Quintens R, Van Lommel L, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis 2010;16:2090–8. [DOI] [PubMed] [Google Scholar]
- 36. Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178:714–30.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang H, Fang M, Jostins L, et al. ; International Inflammatory Bowel Disease Genetics Consortium . Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barker H, Parkkila S. Bioinformatic characterization of angiotensin-converting enzyme 2, the entry receptor for SARS-COV-2. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y, Song W, Wang J, et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med 2020;217:e20191130. doi: 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vabret N, Britton GJ, Gruber C, et al. ; Sinai Immunology Review Project . Immunology of COVID-19: current state of the science. Immunity 2020;52:910–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Effenberger M, Grabherr F, Mayr L, et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut 2020;69:1543–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Potdar AA, Dube S, Naito T, et al. Reduced expression of COVID-19 host receptor, ACE2 is associated with small bowel inflammation, more severe disease, and response to anti-TNF therapy in Crohn’s disease. medRxiv 2020. [Google Scholar]
- 48. Suárez-Fariñas M, Tokuyama M, Wei G, et al. Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-COV-2 related disease. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mirkov MU, Verstockt B, Cleynen I. Genetics of inflammatory bowel disease: beyond NOD2. Lancet Gastroenterol Hepatol 2017;2:224–34. [DOI] [PubMed] [Google Scholar]
- 50. Ahn SH, Shah YM, Inoue J, et al. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis 2008;14:908–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Montenegro-Miranda PS, van der Meer JHM, Jones C, et al. A novel organoid model of damage and repair identifies HNF4α as a critical regulator of intestinal epithelial regeneration. Cell Mol Gastroenterol Hepatol 2020;10:209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Babeu JP, Boudreau F. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J Gastroenterol 2014;20:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yanai K, Hirota K, Taniguchi-Yanai K, et al. Regulated expression of human angiotensinogen gene by hepatocyte nuclear factor 4 and chicken ovalbumin upstream promoter-transcription factor. J Biol Chem 1999;274:34605–12. [DOI] [PubMed] [Google Scholar]
- 54. Ellinghaus D, Degenhardt F, Bujanda L, et al. Genomewide association study of severe covid-19 with respiratory failure. N Engl J Med 2020;NEJMoa2020283. doi:10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fishilevich S, Nudel R, Rappaport N, et al. Genehancer: genome-wide integration of enhancers and target genes in genecards. Database [Oxford] 2017;2017:bax028. doi:10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 57. Hikmet F, Méar L, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci 2020;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Neurath MF. COVID-19 and immunomodulation in IBD. Gut 2020;69:1335–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dotti I, Mora-Buch R, Ferrer-Picón E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut 2017;66:2069–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol 2016;13:13–27. [DOI] [PubMed] [Google Scholar]
- 62. Mak JWY, Chan FKL, Ng SC. Probiotics and COVID-19 ‐ authors’ reply. Lancet Gastroenterol Hepatol 2020;5:722–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Friedrich M, Pohin M, Powrie F. Cytokine networks in the pathophysiology of inflammatory bowel disease. Immunity 2019;50: 992–1006. [DOI] [PubMed] [Google Scholar]
- 64. Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol 2020;20:355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chellappa K, Deol P, Evans JR, et al. Opposing roles of nuclear receptor HNF4alpha isoforms in colitis and colitis-associated colon cancer. Elife 2016;5:e10903. doi: 10.7554/eLife.10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. D’Amico F, Danese S, Peyrin-Biroulet L; ECCO COVID taskforce . Inflammatory bowel disease management during the coronavirus-19 outbreak: a survey from the European Crohn’s and Colitis Organisation. Gastroenterology 2020;159:14–9.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.