Abstract
Background
From an isolated epidemic, coronavirus disease 2019 has now emerged as a global pandemic. The availability of genomes in the public domain after the epidemic provides a unique opportunity to understand the evolution and spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus across the globe.
Methods
We performed whole-genome sequencing of 303 Indian isolates, and we analyzed them in the context of publicly available data from India.
Results
We describe a distinct phylogenetic cluster (Clade I/A3i) of SARS-CoV-2 genomes from India, which encompasses 22% of all genomes deposited in the public domain from India. Globally, approximately 2% of genomes, which to date could not be mapped to any distinct known cluster, fall within this clade.
Conclusions
The cluster is characterized by a core set of 4 genetic variants and has a nucleotide substitution rate of 1.1 × 10–3 variants per site per year, which is lower than the prevalent A2a cluster. Epidemiological assessments suggest that the common ancestor emerged at the end of January 2020 and possibly resulted in an outbreak followed by countrywide spread. To the best of our knowledge, this is the first comprehensive study characterizing this cluster of SARS-CoV-2 in India.
Keywords: Clade I/A3i, COVID-19, phylogenomics, genetic epidemiology, India
Since the emergence of the outbreak in the Chinese city of Wuhan in late 2019, the novel coronavirus disease has spread widely to become a global pandemic, with approximately 25 million individuals infected worldwide and resulting in the death of >800 000 individuals [1]. The causative virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a member of the genus Betacoronavirus. During its transmission, the virus has differentiated into at least 10 clades globally and is continuously evolving [2]. This has implications in genetic epidemiology, surveillance, contact tracing, and the development of long-term strategies for mitigation of this disease [3].
The recent availability of whole-genome sequences of the SARS-CoV-2 from across the world deposited in public databases provides an unprecedented opportunity to understand the dynamics and evolution of the pathogen. The availability of genomic data in a public repository such as GISAID [4] also provides wider access to the resources and enables researchers across the globe to address pertinent hypotheses. Likewise, this gave us a unique scope to understand the introduction, evolution, and spread of the virus in India and understand it in the context of global clades circulating across the world.
In this manuscript, we report the sequences of SARS-CoV-2 isolates predominantly sampled from the states of Telangana and Tamil Nadu. Furthermore, we systematically analyzed the phylogenetic clusters of genomes from India and characterized a unique cluster of sequences (Clade I/A3i), which could not be classified into any of the previously annotated global clades. Isolates forming this cluster were predominant in several states and characterized by a shared set of 4 genetic variants. The cluster potentially arose from a single outbreak followed by a rapid spread across the country. To the best of our knowledge, this is the first comprehensive report of the novel and predominant cluster of sequences from India and suggests its distribution beyond India in many countries in South Asia, Oceania, and America.
MATERIALS AND METHODS
Patient Consent Statement
A written consent from the patients was obtained wherever applicable. The design and implementation of this work has been approved by a local ethical committee.
Sample Processing and Sequencing
Samples were collected and processed as per the guidelines of the Institutional Ethics Committee. Ribonucleic acid (RNA) was isolated from nasopharyngeal or oropharyngeal swabs collected in viral transport media as explained in Supplementary Methods. Purified RNA was sequenced using either a shotgun approach or the ARTIC v3 protocol, as detailed in Supplementary Methods [5].
Assembly of Sequencing Data
Quality control of the FASTQ files was performed using FastQC v0.11.7, and adaptors/poor quality bases were trimmed using Trimmomatic [6, 7]. Reads were aligned to the reference genome MN908947.3 using hisat2 [8]. Consensus sequence from the bam file was derived using seqtk and bcftools [9]. Samtools depth command was used to calculate the coverage across the genome [10]. The sequences were deposited in GISAID with accessions detailed in Supplementary Data 1.
Data Availability
All sequences generated in this study have been submitted to GISAID. The accession names of the samples and associated metadata are outlined in Supplementary Data 1.
Genomic Data Collection and Analysis
The datasets of Indian SARS-CoV-2 genomes deposited in GISAID (until August 7, 2020) were used for the analysis. Furthermore, 10 high-quality genomes from each of the 10 clades, respectively, as annotated by Nextstrain were retrieved from GISAID and used in the analysis. The datasets and acknowledgments are listed in Supplementary Data 2. We considered only high-quality genomes for evaluation of the nucleotide substitution rates, molecular clock, and phylogenetic clustering, because these would be sensitive to the quality of genomes. The criteria used for filtering low-quality genomes are outlined in Supplementary Methods.
Phylogenetic Analysis and Divergence Estimation
Phylogenetic analysis of the samples was performed as detailed previously following the standard protocol for analysis of SARS-CoV-2 genomes provided by Nextstrain [11, 12]. BEAST v1.10.4 was used for the analysis of nucleotide substitution rates and the estimation of times to the most recent common ancestor. The detailed methodology for phylogenetic tree construction and dating analysis is provided in Supplementary Methods. The resulting tree was used to infer mutations and identify clades. The values used for each parameter in the protocol are given in Supplementary Data 4.
Functional Evaluation of Variants
Wuhan-Hu-1 genome (NC_045512) was used as reference wherever applicable. The variants were also evaluated for the functional consequences using SIFT [13]. A SIFT score of 0.0 to 0.05 was interpreted to have a deleterious effect. The functional effects of protein variants identified in the clades were assessed using the PROVEAN web server, using a default threshold value of −2.5 [14]. In addition, PhyloP conservation scores and base-wise GERP rejected substitutions scores for the variants were computed [15, 16]. Sites having positive PhyloP scores were predicted to be conserved, whereas positive GERP scores were considered indicative of a site under evolutionary constraint. The variants were also checked for overlaps with immune epitope predictions as given on UCSC Genome Browser for SARS-CoV-2.
RESULTS
Demographics and Quality of Viral Genomes
The samples sequenced encompass 303 genomes in total, majorly collected from the states of Telangana and Tamil Nadu. The age of the patients ranged from 1.5 to 80 years, with >80% (275 of 303) within the age bracket of 20–60 years (Supplementary Figure S1A). A total of 294 samples were sequenced using an amplicon-based approach with a target of ~2 million paired-end reads per sample. We could achieve an average coverage of >1000× in all cases, with a uniform representation from all amplicons (Supplementary Figure S1B, top, and S1C). Three samples were sequenced using a shotgun sequencing approach and had an average coverage of approximately 100×. The coverage across the genome was uniform (Supplementary Figure S1B, bottom). The samples and metadata for the isolates sequenced and deposited in the public domain are summarized in Supplementary Data 1.
Phylogenetic Clusters
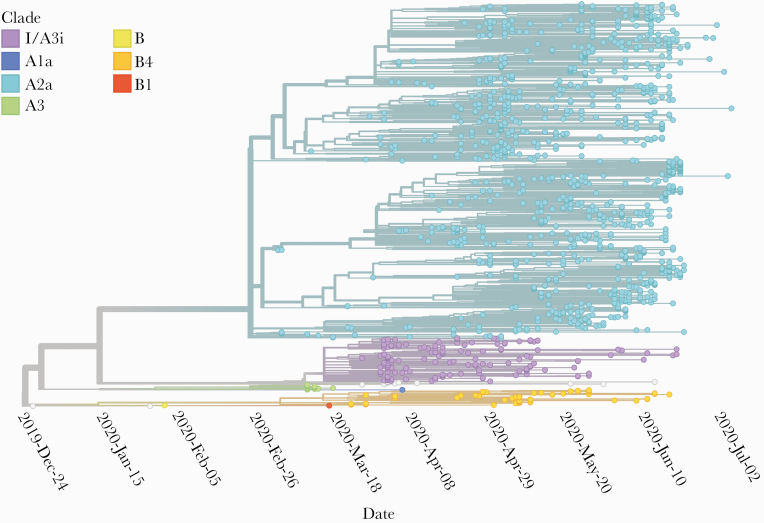
A total of 2212 genome sequences of the SARS-CoV-2 were available for analysis as of August 7, 2020 from India including the genomes sequenced by our group. After removal of low-quality sequences, the dataset resulted in a total of 1377 genomes submitted from 16 institutions (including 275 of our 303 genomes) (Supplementary Data 2). The genomes isolated from India were found to be classified under 7 clusters (Figure 1). Six of these clusters are known clades identified by Nextstrain: A1a, A2a, A3, B, B1, and B4 [17]. The first and the major cluster encompassed 1143 (83%) of genomes, which fell into the A2a clade. The clade was represented by samples derived from multiple states across the country including Gujarat, Maharashtra, Telangana, West Bengal, Odisha, Karnataka, Uttarakhand, Tamil Nadu, and Haryana.
Figure 1.
Phylogenetic clusters and clades as generated by Nextstrain for the dataset of 1377 high-quality Indian severe acute respiratory syndrome coronavirus 2 genomes. Indian genomes were found to fall under 7 clusters with the majority of the genomes falling under clade A2a. The second largest cluster in India (purple) has been designated as clade I/A3i.
The second largest cluster consisted of 160 genomes (11.6%). This cluster of sequences could not be classified into any of the 10 clade sequences defined by Nextstrain, and it did not share the nucleotide compositions that define any of the 10 clades [17]. This cluster was found to have diverged from the A1a and A3 clades, and most, but not all of the sequences, shared a variant (L3606F in ORF1a) with members of the A3 and A1a clades. We call this the A3i clade in cognizance of this fact. Therefore, to avoid potential conflict with the nomenclature followed by Nextstrain, we define this cluster of sequences as Clade I/A3i, for the unique occurrence as a dominant cluster among SARS-CoV-2 genome sequences from India, and also because this clade is largely formed by sequences from India (Supplementary Figures S2 and S3). The other clusters encompassed the B4, A3, A1a, B, and B1 clades with 52 and 17 genomes falling into the clusters A3 and B4, respectively, and clades A1a, B, and B1 having 1 genome each.
Molecular Definition of the Cluster
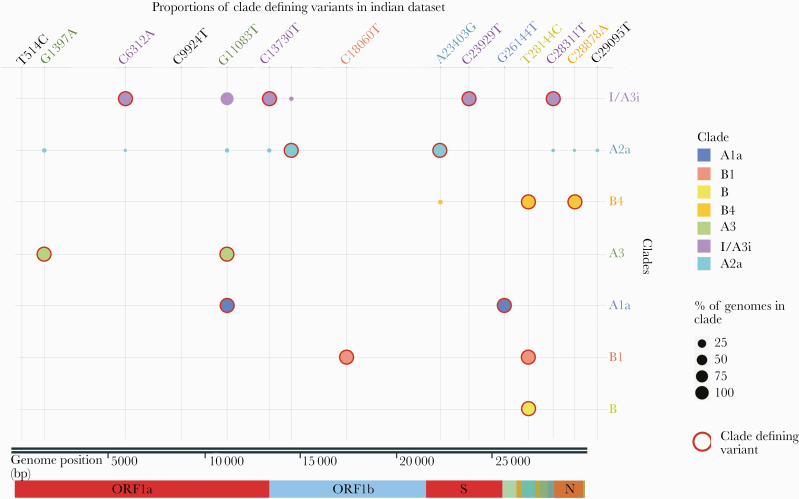
A discriminant analysis was performed for all variants in any genome defined by the cluster of sequences. Systematic analysis of members of the cluster revealed that a set of 4 variants (C6312A, C13730T, C23929T, and C28311T) was shared by a majority of members of the cluster (Figure 2). A total of 149 genomes of the 160 genomes (93%) in the cluster shared the combination of variants. This unique combination of variants was shared by none of the other genomes that were assigned to any other clade.
Figure 2.
Shared variants among the variants that define the clusters of the Indian severe acute respiratory syndrome coronavirus 2 genomes. The size of the circle represents the allele frequencies of the respective variants, and clade-defining variants are marked with a red circle.
We further analyzed the global datasets for identifying the genomes that displayed matches for all 4 variants that defined the Clade I/A3i. Our prospective search retrieved a total of 362 high-quality genomes (Supplementary Data 5). Of the retrieved genomes, the largest number of genomes originated from Singapore, which had 219 genomes and constituted 53% of the high-quality genomes from Singapore. The other genomes originated from several countries including Malaysia, Australia, United States, Canada, Taiwan, Japan, Thailand, Philippines, Oman, Guam, and Saudi Arabia. However, the members in the clade contributed to a much smaller proportion of the clades/clusters identified in the respective countries. Of these, 23 were sampled from a date earlier than the earliest sample of this cluster from India and were from the United States, Canada, Australia, Thailand, Saudi Arabia, Taiwan, Singapore, Malaysia, Japan, and Brazil (Figure 3B).
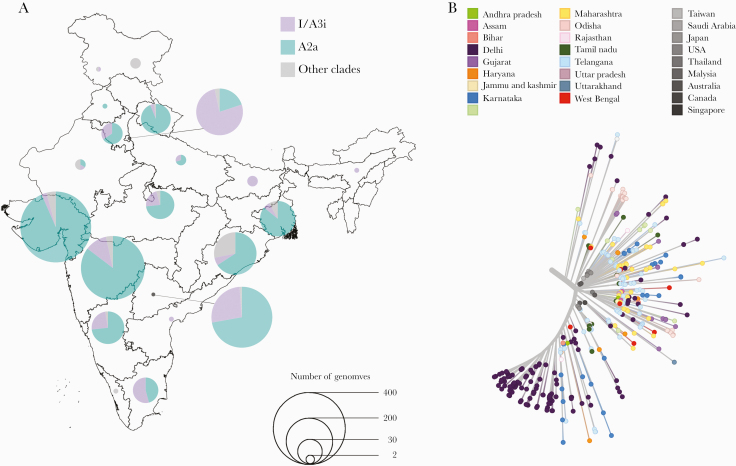
Figure 3.
(A) Proportion of the I/A3i clade (purple) and A2a (teal) in the genomes sequenced from different states of India. The proportion of the A2a clade (teal) is also shown for comparison, whereas all other clades are shaded gray. (B) The short tree of the I/A3i clade diverging from a central point suggests a single point of introduction and spread across the different states. The 23 global genomes that were sampled before the first Indian genome from this cluster are highlighted in gray.
Nucleotide Substitution Rates
Mutation rates were calculated for the Indian sequences using BEAST, with the WH1 genome as the root. Our analysis suggests that the substitution rate is 1.76 × 10–3 (95% highest posterior density [HPD] 1.57 × 10–3–1.99 × 10–3) per site per year for the entire Indian SARS-CoV-2 genomes put together. This also confirms the estimates previously made [18].
The substitution rate was also computed for the individual clades. The gene-wise substitution rates were also similarly calculated for the major clusters. The analysis suggests that the I/A3i clade has a nucleotide substitution rate of 1.1 × 10–3 variants per site per year compared with the estimate of 1.73 × 10–3 variants per site per year for the prevalent A2a clade and 1.76 × 10–3 variants per site per year for all the high-quality genomes from India analyzed. The nucleotide substitution rate suggests that the evolution of the I/A3i clade is largely determined by changes in the structural proteins—Nucleocapsid (N) and Membrane (M) genes, compared with the A2a, the globally predominant clade, which is determined by changes in the Spike (S) genes (Table 1).
Table 1.
Nucleotide Substitution Rates of the Different Structural Protein Genes and Genome-Wide Across the Different Clusters and Clades in Indiaa
| Clade/ Cluster | S Gene | E Gene | M Gene | N Gene | Genome |
|---|---|---|---|---|---|
| All (N = 1376) | 3.55 × 10–3 | 4.57 × 10–3 | 4.69 × 10–3 | 6.94 × 10–3 | 1.76 × 10–3 |
| A2a (N = 1143) | 3.49 × 10–3 | 5.42 × 10–3 | 3.67 × 10–3 | 5.85 × 10–3 | 1.73 × 10–3 |
| I/A3i (N = 149) | 0.94 × 10–3 | 3.5 × 10–3 | 2.26 × 10–3 | 1.54 × 10–3 | 1.1 × 10–3 |
| B4 (N = 52) | 1.18 × 10–3 | 3.9 × 10–3 | 1.15 × 10–3 | 3.33 × 10–3 | 1.12 × 10–3 |
| A3 (N = 17) | 1.36 × 10–3 | 1.89 × 10–3 | 1.28 × 10–3 | 7.23 × 10–3 | 1.85 × 10–3 |
aThe estimates for A1a, B, and B1 were not computed because the clades encompassed very few genomes from India.
Estimating Time to Most Recent Common Ancestor and Age of the Cluster
The date of the most recent ancestor for the dataset of all Indian SARS-CoV-2 genomes, with WH1 genome sequence included, was computed using BEAST. The median time to most recent common ancestor (tMRCA) was December 10, 2019 (95% HPD November 24 to December 24), confirming the previous estimates of the origin of the epidemic in Wuhan city of China [19]. The tMRCA for the I/A3i clade, as well as the A2a clade, which constituted the majority of samples, was also computed. Clade A2a, which is the predominant clade in India, had a tMRCA of January 15, 2020 (95% HPD interval December 25, 2019–February 2, 2020), whereas clade I/A3i had a tMRCA of January 26, 2020 (95% HPD interval January 1, 2020–February 15, 2020).
Functional Consequences of the Variants
The majority of the variants that defined other clades were predicted to be neutral by PROVEAN, with the exception of G251V, which defines the A1a clade. Three variants that define Clade I/A3i (C6312A, C13730T, and C28311T) resulted in amino acid changes with potentially deleterious functional consequences, as predicted by SIFT, and mapped to conserved genomic loci in the SARS-CoV-2 genome (Table 2). One of these variants, A97V in the RDRP protein (corresponds to A88V in ORF1b), is located in its NiRAN domain, which is suggested to be important in RNA binding and nucleotidylation activity [20]. Both SIFT and PROVEAN analyses suggest that the effect of this mutation is deleterious in nature; however, because both alanine and valine are hydrophobic amino acids, the exact effect of the mutation needs to be experimentally validated. Of notable significance is the P13L variant (C28311T) in the Nucleocapsid protein, which is required for the viral entry into the cells. The variant maps to the intrinsically disordered region (IDR) domain of the N protein and SIFT predicts the variant to be deleterious, although the PROVEAN analysis categorized it as a neutral mutation.
Table 2.
Functional Characteristics of the Four Variants That Define Clade I/A3i and Other Clades Across the Worlda
| Clade | Gene | Site | Mutation | PROVEAN Score/Prediction | SIFT Score/Prediction | Conservation Scores |
|---|---|---|---|---|---|---|
| A1a | ORF3a | G26144T | G251V | −8.581 | 0 | PhyloP: 4.256 |
| Deleterious | Deleterious | GERP: 1.65 | ||||
| A1a, A3, I/A3i | ORF1a | G11083T | L3606F | −1.4 | 0.01 | PhyloP: −1.32286 |
| Neutral | Deleterious | GERP: −3.3 | ||||
| A2 | S | A23403G | D614G | 0.598 | 0.3 | PhyloP: 2.25839 |
| Neutral | Tolerated | GERP: 1.65 | ||||
| A2a | ORF1b | C14408T | P314L | −0.914 | 0.31 | PhyloP: 3.30748 |
| Neutral | Tolerated | GERP: 1.65 | ||||
| A3 | ORF1a | G1397A | V378I | −0.199 | 0.62 | PhyloP: 0.227575 |
| Neutral | Tolerated | GERP: −1.81 | ||||
| A7 | ORF1a | C9924T | A3220V | −2.049 | 0.04 | PhyloP: 3.30935 |
| Neutral | Deleterious | GERP: 1.65 | ||||
| B, B1, B2, B4 | ORF8 | T28144C | L84S | 2.333 | 0.37 | PhyloP: −1.52089 |
| Neutral | Tolerated | GERP: −0.206 | ||||
| B4 | N | G28878A | S202N | −0.404 | 0 | PhyloP: 4.256 |
| Neutral | Deleterious | GERP: 1.65 | ||||
| I/A3i | N | C28311T | P13L | −1.23 | 0 | PhyloP: 3.27687 |
| Neutral | Deleterious | GERP: 1.65 | ||||
| I/A3i | RDRP/ORF1b | C13730T | A97V (A88V in ORF1b) | −3.611 | 0 | PhyloP: 3.31844 |
| Deleterious | Deleterious | GERP: 1.65 | ||||
| I/A3i | ORF1a | C6312A | T2016K | −0.352 | 0.03 | PhyloP: 3.29661 |
| Neutral | Deleterious | GERP: 1.61 |
aPROVEAN scores of less than −2.5 are considered deleterious in nature. Similarly, SIFT scores of 0 to 0.05 are considered deleterious.
Two of the variants, C6312A in ORF1a and C13730T in ORF1b, also mapped to immune epitope predictions (HLA-A0201 binding peptides) from NetMHC 4.0, available on UCSC Genome Browser and as listed on UCSC Genome Browser for SARS-CoV-2 [21]. The potential consequences of the variants in the immune response could not be ascertained.
Defining the Origin and Spread From the Cluster
The presence of a short tree of Clade I/A3i with divergence from a single point suggests a single point of introduction [22]. The single point of divergence also suggests that the origin and spread of the cluster were possibly from a single outbreak (Figure 3). The clustering of samples around January 2020 suggests a rapid spread spanning multiple regions across the country. The first sequence from the cluster in India was GMC-KN443/2020 (Accession ID EPI_ISL_431103, deposited by Department of Microbiology, Gandhi Medical College and Hospital, Hyderabad, India) sampled on March 16, 2020 from an Indonesian traveler from the state of Telangana. Of the 14 states from which the data for high-quality genomes were made available, the I/A3i clade was represented in 11 of the states.
Considering all the genomic data available from India, the I/A3i clade is represented in 446 genomes (22%) and represented from 17 of the 20 states from which the genomes originated. The geographical distribution and the proportion of the Clade I/A3i isolates are depicted in Figure 3. The states of Delhi, Telangana, Maharashtra, Karnataka, and Tamil Nadu have the highest proportions of this clade, followed by Haryana, Madhya Pradesh, West Bengal, Odisha, Uttar Pradesh, and Bihar (Supplementary Data 6).
Temporal Shifts in the Prevalent Clades
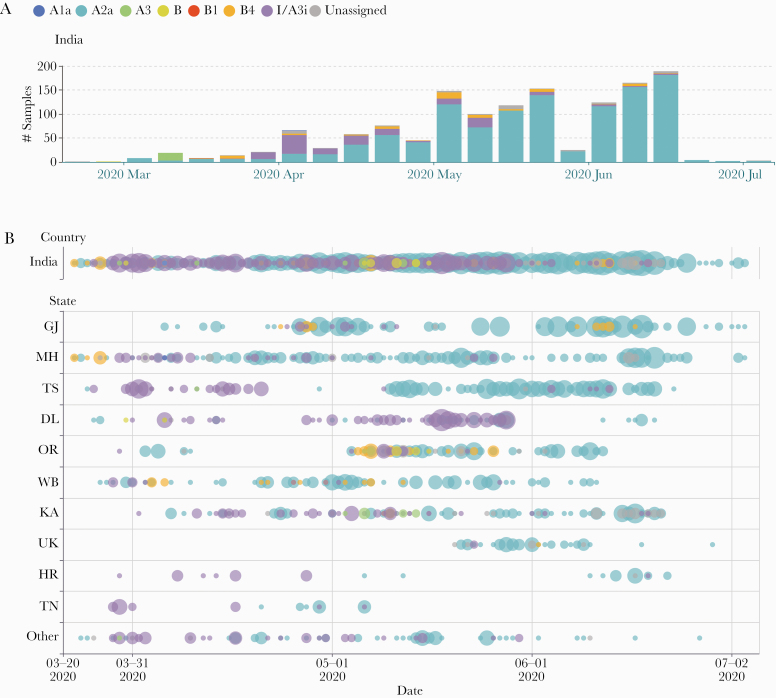
After the initial outbreak of coronavirus disease 2019 (COVID-19) in India, most of the samples collected in the months of March and April belonged to the I/A3i clade (Figure 4A). In fact, it was the predominant clade in almost all of the states where data were collected in March and April, with the exception of West Bengal and Gujarat. However, by late April and early May, a shift in the prevalent clade was observed. All states, except Delhi, showed an increased representation of the A2a clade (Figure 4B). It is interesting to note that in Gujarat, the most prominent clade remained A2a throughout the period of April to July, with meager representation of I/A3i and B4 clades. Odisha was a mixed bag during the month of May, with almost equal representation of I/A3i, A2a, and B4 clades. However, recent samples collected from Odisha in the month of June all belonged to A2a clade. Further sample collection and sequencing is needed to assess this shift in the predominance of clades reliably.
Figure 4.
(A) A week-wise stacked bar displaying the proportion of clades across India, starting from the week of March 1, 2020. (B) A bubble plot depicting the change in the predominant clades with time in various states. X-axis indicates the date on which the sample was collected, and color indicates the clade. Only those states with collection data across at least 2 months are plotted.
Demographics of the Patients
Of the members in Clade I/A3i, 112 were male (70%) whereas 44 were female (27.5%). The mean age was 35.7 years (confidence interval [CI], 33.2–38.2 years). For A2a cluster, 761 were male (66.6%) and 353 were female (30.9%), whereas the mean age was 40.8 years (CI, 39.8–41.8 years). Although age and clinical outcomes were found to be significantly different between Clade I/A3i and other clades (P = .00042 and P = .000075, respectively), sex was not found to be significantly different between Clade I/A3i and the other clades (χ 2 = 0.68, P > .05). Patient details for the Indian samples as provided by GISAID are available in Supplementary Data 7.
DISCUSSION
Genomic evolution coupled with the appropriate tools such as genome sequencing provides a unique opportunity to understand the spread and evolution of pathogens [23, 24]. The emergence of COVID-19 as a global pandemic and the availability of the Open Data for SARS-CoV-2 genomes from across the globe facilitated by genomic databases such as GenBank and GISAID has truly opened up new opportunities to understand the pathogen and its spread and evolution at an unprecedented rate [4, 25]. Whole-genome sequencing of SARS-CoV-2 has also been extensively used in understanding epidemics at a macro- as well as microlevels, at hospitals [26].
In this report, we describe a distinct cluster of sequences from genomes of SARS-CoV-2 sequenced and deposited from multiple laboratories across India, which we classify as the I/A3i clade. This distinct cluster could not be classified into any of the 10 clade annotations as described by Nextstrain, and it was characterized by a unique combination of 4 variants that was shared by more than 95% of the isolates falling in the cluster. The cluster was predominantly found in genomes from India; although additional members could also be found from genomes deposited in other countries, they form a minor proportion of the genomes from the respective countries. As per Nextstrain, the Indian genomes constituted more than 30% of the global genomes for this cluster.
In-depth analysis of the genome cluster suggests a comparable rate of nucleotide substitutions with other predominant clades, although a gene-wise estimate of substitution suggests a distinct mode of evolution, driven by the Nucleocapsid (N) and Membrane (M) genes, and sparing of the Spike (S) gene in contrast to predominant diversity in the Spike (S) gene in A2a clade, the globally predominant clade [27]. However, it has not escaped our attention that host genetic factors could modulate the evolution of the virus genome, and without large-scale host genomic studies, the causal relationships cannot be conclusively established.
The cluster suggests a potential single introduction around February, followed by a countrywide spread, mostly affecting the South Indian states as evidenced by the tMRCA as well as the short cluster. Our analysis suggests that the Clade I/A3i was represented in almost all states from which genomes are available. Members of the Clade I/A3i formed the predominant class of isolates from the states of Delhi, Telangana, Maharashtra, Karnataka, and Tamil Nadu and the second largest in membership in Haryana, Madhya Pradesh, West Bengal, Odisha, Uttar Pradesh, and Bihar.
CONCLUSIONS
Put together, the cluster of genomes (Clade I/A3i) forms a distinct cluster, predominantly found amongst Indian SARS-CoV-2 genomes, with limited representation outside the region. To the best of our knowledge, this is the first comprehensive study characterizing the distinct and predominant cluster of SARS-CoV-2 in India. This report also exemplifies the fact that timely and open access to genomic data can provide unique insights into the genetic epidemiology of pathogens.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Data 1. Metadata for the genome sequences submitted by our group.
Supplementary Data 2. List of Indian and global high-quality GISAID submissions used in this study, and acknowledgments of the contributing authors.
Supplementary Data 3. List of sites masked before variant analysis.
Supplementary Data 4. Parameter values used for phylogenetic tree construction and the prior values used in the analysis of nucleotide substitution rates using BEAST.
Supplementary Data 5. List of 362 high-quality global GISAID submissions falling under Clade I/A3i.
Supplementary Data 6. Statewise proportions of genomes belonging to the I/A3i clade.
Supplementary Data 7. Metadata of clinically relevant information associated with the genomes deposited in GISAID from India.
Supplementary Data 8. Online resource. An online and updated resource for SARS-CoV-2 genomes from India, their clade assignments and distribution across the country is available at http://clingen.igib.res.in/genepi/phylovis/.
Acknowledgments
We acknowledge the GISAID database and the contributors of genomic data, without which this analysis was not possible. We thank the COVID-19 volunteer team of Centre for Cellular and Molecular Biology (CCMB) for help in initial sample processing. The full acknowledgements of data and contributors are available in Supplementary Data 2. We also acknowledge funding from the Council of Scientific and Industrial Research (CSIR India).
Author contributions. R. K. M., D. T. S., and V. S. conceptualized and designed the study. S. B. and P. M. processed the sequencing data. S. B., P. M., and P. S. performed sequence data analysis and visualization. B. J. performed the analysis for phylogenetic clustering, molecular characterization, and quantification of relatedness. S. K., L. Z., S. Sh., and N. G. conducted the experiments. S. R. , K. K., S. Sr., D. R. G., A. B. S., K. B. T., and K. T. supervised processing and provision of samples and data. D. T. S., V. S., and B. J. prepared the manuscript with inputs from R. K. M. All authors read and approved the manuscript.
Disclaimer. The funders had no role in the design of the experiments, preparation of the manuscript or decision to publish.
Financial support. B. J. is a recipient of the Junior Research Fellowship from CSIR India.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. World Health Organization. Coronavirus disease (COVID-19) Weekly Epidemiological Update. Available at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200831-weekly-epi-update-3.pdf?sfvrsn=d7032a2a_4. Accessed 31 August 2020.
- 2. Sun J, He WT, Wang L, et al. COVID-19: Epidemiology, Evolution, and Cross-Disciplinary Perspectives. Trends Mol Med 2020; 26: 483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications [published online ahead of print April 27, 2020]. Genomics 2020; doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data–from vision to reality. Euro Surveill 2017; doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quick J. NCoV-2019 Sequencing Protocol V2. Protocols.io. 2020; published online March 14 Available at: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v2-bdp7i5rn. Accessed 30 August 2020.
- 6. Andrews S. Babraham bioinformatics-FastQC a quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 30 August 2020.
- 7. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim D, Paggi JM, Park C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019; 37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011; 27:2987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H, Handsaker B, Wysoker A, et al. ; 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jolly B, Scaria V. Computational analysis and phylogenetic clustering of SARS-nCov-2 genomes. Available at: https://docs.google.com/document/d/1B5NxWFwsRz_vD5Y6EwjKxkRamsPLVfs1MjVoziU1Zq0/edit?usp=sharing&usp=embed_facebook. Accessed 31 August 2020. [DOI] [PMC free article] [PubMed]
- 12. Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34:4121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vaser R, Adusumalli S, Leng SN, et al. SIFT missense predictions for genomes. Nat Protoc 2016; 11:1–9. [DOI] [PubMed] [Google Scholar]
- 14. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015; 31:2745–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 2010; 20:110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cooper GM, Stone EA, Asimenos G, et al. ; NISC Comparative Sequencing Program Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005; 15:901–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bedford T, Neher R, Hadfield J, et al. Nextstrain. Nextstrain/ncov. GitHub. Available at: https://github.com/nextstrain/ncov. Accessed 30 August 2020.
- 18. Duchene S, Featherstone L, Haritopoulou-Sinanidou M, et al. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evolution 2020; doi: 10.1093/ve/veaa061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao Y, Yan L, Huang Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020; 368:779–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jurtz V, Paul S, Andreatta M, et al. NetMHCpan-4.0: improved peptide-MHC Class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 2017; 199:3360–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grubaugh ND, Ladner JT, Lemey P, et al. Tracking virus outbreaks in the twenty-first century. Nat Microbiol 2019; 4:10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang C, Liu Z, Chen Z, et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol 2020; doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang X, Wu C, Li X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev 2020; doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res 2013; 41:D36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meredith LW, Hamilton WL, Warne B, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koyama T, Weeraratne D, Snowdon JL, Parida L. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens 2020; 9:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences generated in this study have been submitted to GISAID. The accession names of the samples and associated metadata are outlined in Supplementary Data 1.