Abstract
Background
Short-term recurrence of positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ribonucleic acid (RNA) polymerase chain reaction (PCR) in discharged coronavirus disease 2019 (COVID-19) patients attracts the public’s concern. This study aimed to determine the clinical and epidemiological results of such patients.
Methods
This retrospective study was conducted on 32 designated hospitals for COVID-19 patients discharged from January 14 to March 10, 2020. After 28-day followed-up, patients who tested positive again for SARS-CoV-2 RNA and confirmed by reverse-transcriptase polymerase chain reaction were re-admitted to hospital for further treatments. All of the close contacts of patients who tested positive again were asked to self-segregate for 14 days. Data of epidemiology, symptoms, laboratory tests, and treatments were analyzed in those patients, and their close contacts were investigated.
Results
Of 1282 discharged patients, 189 (14.74%) tested positive again for SARS-CoV-2 RNA during 28-day follow-up. The median time from discharge to the next positive test was 8 days (interquartile range [IQR], 5–13). Patients in the group that tested positive again were younger (34 vs 45 years, P < .001) with a higher proportion of moderate symptoms (95.77% vs 84.35%, P < .001) in the first hospitalization than in the negative group. During the second hospitalization, all patients who tested positive again showed normal peripheral white blood cells and lymphocytes and no new symptoms of COVID-19; 78.31% further improved on chest computed tomography scan compared with the first discharge, yet 25.93% accepted antiviral therapy. The median time of re-positive to negative test was 8 days (IQR, 4–15). None of the close contacts developed COVID-19.
Conclusions
Our data suggest that the short-term recurrence of positive SARS-CoV-2 RNA in discharged patients is not a relapse of COVID-19, and the risk of onward transmission is very low. This provides important information for managing COVID-19 patients.
Keywords: communicable, COVID-19, recurrence, relapse, retrospective
In this multicenter study involving 1282 discharged COVID-19 patients with 28-day follow-up, 14.74% tested positive again for SARS-CoV-2 RNA. However, our data suggest that re-positive is not a relapse of COVID-19, and the risk of onward transmission is very low.
Because the pandemic of coronavirus disease 2019 (COVID-19) has swept across the globe, attention has been drawn to the epidemiological and clinical features of patients [1–3], the therapeutic strategy [4, 5], and the ways to increase cure rate and decrease mortality in countries and regions [6]. However, the knowledge about the novel COVID-19 is still limited, particularly in the management of convalescence stage after discharge.
With increasing numbers of patients who were cured and discharged, we are facing serious problems with posthospital surveillance and follow-up of COVID-19 patients. Some sporadic case reports from many cities in China, such as Wuhan [7], Dongguan [8], Weihai [9], and Guangzhou [10], found that some patients had detectable recurrence of positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ribonucleic acid (RNA) polymerase chain reaction (PCR) in nasopharyngeal swab or anal swab 5–13 days after being discharged from the hospital. A recent report from Shenzhen city showed that the recurrence rate reached to 14.5% in 172 discharged patients [11]. Although these studies did not show the threat that was induced by the recurrence of positive SARS-CoV-2 RNA in patients, the findings aroused great concern for people because the evidence of highly human-to-human transmission was found in COVID-19 patients [12, 13]. Moreover, these archived studies were either case reports or small-sized and single-center studies with a short follow-up period; therefore, it was hard to ascertain the panorama of those populations with recurrence of viral RNA PCR positivity. In particular, the following questions were of great concern and have not been answered clearly. (1) What is the ratio of re-positive patients in all discharged COVID-19 patients in Guangdong Province? (2) Does the recurrence of positive SARS-CoV-2 RNA mean that COVID-19 would relapse like other infectious diseases such as typhoid fever [14, 15]? (3) Are re-positive patients transmissible to others? The answers to these questions can have a direct impact on a strategic formula for disease control and prevention. Considering the ongoing global pandemic of COVID-19, it is essential to carry out large-scale studies to better understand the issue of recurrence of positive SARS-CoV-2 RNA in COVID-19 patients. In response to these emerging concerns, we conducted a multicenter, retrospective, observational study of patients who tested positive again for COVID-19 and targeted 1282 patients from 32 designated hospitals in Guangdong Province, China.
METHODS
Study Design and Participants
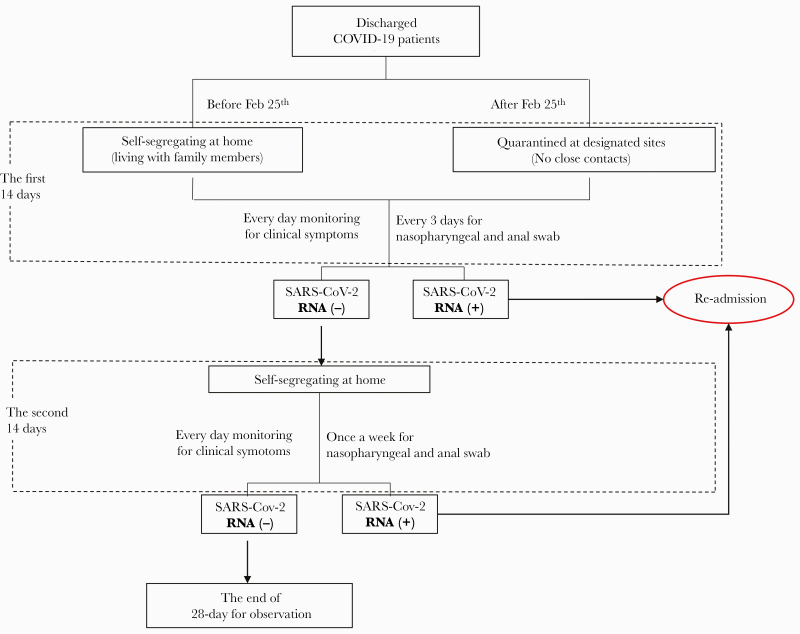
A multicenter, retrospective, observational study was conducted at 32 hospitals in Guangdong Province that were designated to treat COVID-19 patients [16]. All confirmed COVID-19 patients who were discharged from January 14 to March 10, 2020 were followed up for 28 days. According to the policy of Guangdong Province, the follow-up was divided into 2 stages. During the first stage, patients who were discharged before February 25, 2020 were required to self-segregate for 14 days (section A), whereas patients who were discharged after that time were mandatorily required to be quarantined in designated sites for 14 days (section B). Clinical symptoms (body temperature, cough, and other respiratory symptoms) of these patients were recorded every day, and SARS-CoV-2 RNA in nasopharyngeal and anal swabs specimens was tested every 3 days. During the second stage of the 14-day follow-up, patients were self-segregated at home (section C), and the community members tracked their symptoms by telephone and carried out viral RNA test once a week. See the protocol in Figure 1.
Figure 1.
Flow chart for follow-up of coronavirus disease 2019 (COVID-19). As shown in the figure, during the first 14 days, patients discharged before February 25, 2020 in Guangdong Province were required to self-segregate for 14 days (A), whereas patients discharged after that time were mandatorily quarantined in designated sites for 14 days (B) and had no close contact since then. During the second 14 days, patients were self-segregated at home (C), and the community members followed up their symptoms by telephone and carried out viral ribonucleic acid (RNA) test once a week. Both A and C would generate close contacts. Once re-positive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA was tested, a second test would be carried out within 24 hours to determine the re-positive patient (see definition). Lung computed tomography was performed when positive SARS-CoV-2 RNA recurrence was confirmed. According to the results of SARS-CoV-2 RNA tests during follow-up, discharged patients were divided into a re-positive group and a negative group. Patients in the re-positive group were admitted to the designated hospital for further observation and treatment, and they were discharged again when they met the discharge criteria. Patients who had finished the processes above were included in our study.
Patient Consent Statement
This study was authorized by the Guangdong Provincial Health Commission and was approved by the Ethics Committee of the Guangdong Provincial People’s Hospital (No. GDREC2020028H). The whole course of treatment and follow-up for all COVID-19 patients were free. Data were collected by a public health surveillance system—the electronic medical information reporting system (E-System) built by the Guangdong Health Commission—and all personal data were deidentified before analysis. Written informed consent was waived due to the use of deidentified data for the purpose of public health surveillance.
Data Collection
To ensure the quality and homogeneity of medical treatment, the Guangdong COVID-19 Prevention and Control Headquarters was set up to direct and coordinate the management for COVID-19 patients across the province. An electronic medical information reporting system (E-System) was built by the Guangdong Health Commission for the entire provincial medical data collection, through which all data in this study were collected.
Information on demography, epidemiology, comorbidities, and clinical data of the first hospitalization was collected; clinical symptoms monitoring and SARS-CoV-2 RNA tests were made as described above. All patients who tested positive again (recurrence of SARS-CoV-2 RNA PCR positivity) were re-admitted to the hospital for observation or antiviral (arbidol, lopinavir, and ritonavir, etc) treatment. Daily clinical symptoms (temperature, cough, and other respiratory symptoms) and laboratory tests were recorded. All of the close contacts of patients who tested positive again were asked to self-segregate at home for 14 days. During the observation period, their clinical symptoms were monitored every day, and SARS-CoV-2 RNA in nasopharyngeal and anal swabs was checked every 3 days.
Specimen Collection
Swabs for detecting SARS-Cov-2 RNA were collected by qualified medical staff through standard procedures under level 3 biosafety protection according to the guidance of a previous study [17–19]. For nasopharyngeal specimens, a swab was inserted into the nostril parallel to the palate to a depth equal to the distance from the nostrils to the outer opening of the ear. The swab was left in place for several seconds to absorb secretions and then was slowly removed with rotation. For anal specimens, a swab was inserted into anus 2–3 cm, swiped the plica around it, and generally rotated for several circles, then removed it slowly. Specimens were immediately stored at 2–8°C and transported under the same conditions for viral RNA extraction and molecular testing.
Definitions
Recurrence of Positive Severe Acute Respiratory Syndrome Coronavirus 2 Ribonucleic Acid
In COVID-19 patients who were cured and left hospital after meeting the discharge criteria (see below), SARS-CoV-2 RNA tested by real-time reverse-transcriptase PCR (RT-PCR) was conducted on nasopharyngeal and anal swab and tested positive twice in each swab (24-hour sampling time interval) in the designated hospital [8, 10, 11].
Moderate Symptoms
Moderate symptoms consisted of fever and mild respiratory symptoms (cough, sore throat, runny nose, etc), multiple patchy shadowing and ground-glass opacity in lung computed tomography (CT), and normal range of vital signs [3, 16].
Severe Symptoms
Severe symptoms consisted of respiratory distress (respiratory rate ≥30 breaths/minute, SaO2 ≤93%, arterial oxygen tension/inspiratory oxygen fraction ≤300 mmHg under resting condition) (1 mmHg = 0.133 kPa). In addition, lung CT showed that the range of pulmonary lesions increased by more than 50% within 24–48 hours [4, 16].
Criteria for Discharge
Patients were discharged if they met all of the following criteria: (1) body temperature returned to normal for more than 3 days; (2) respiratory symptoms improved significantly; (3) substantial improvement in both lungs by CT scans; (4) SARS-CoV-2 assays in both nasopharyngeal and anal swabs specimens were negative for 2 consecutive times (24-hour interval) [5, 16].
Improvement of Chest Computed Tomography Images
Investigators noted improved CT images after comparing the last chest CT results and observing whether the size and density of pulmonary lesions were reduced; if the lesions were significantly reduced by more than 50%, it was considered to be a substantial improvement [14, 20].
Close Contacts
Close contacts are persons who have had close contact with re-positive patients without effective protection with masks, such as living and working together, which was confirmed by registered public health practitioners from the Center for Disease Control and Prevention (CDC) [21].
Confirmation for Recurrence of Positive Severe Acute Respiratory Syndrome Coronavirus 2 Ribonucleic Acid
Both nasopharynx swabs and anal swabs samples from each re-positive patient were collected for the RT-PCR test in a laboratory at the CDC in Guangdong Province. Once re-positive patients were reported by designated hospital, the specimens described above were collected by laboratory staff of the CDC. Total RNA was extracted using a prefilled viral total NA kit-Flex (catalog no. KFRPF-805296, Labserv; Fisher Scientific, https://www.fishersci.com) following manufacturer’s instructions. A commercial RT-PCR assay kit targeting the ORF1ab and N genes was used to detect SARS-CoV-2 RNA (catalog no. DA0931; DaAn Gene, Guangzhou, China). Amplification was performed on an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, https://www.thermofisher.com) as follows: 50°C for 15 minutes and then 95°C for 15 minutes, followed by 45 cycles of 94°C for 15 seconds and 55°C for 45 seconds. When both ORF1ab and N gene target amplification curves were generated within 40 cycles, the samples were considered to be positive for SARS-CoV-2 RNA.
Statistical Analysis
Continuous variables were expressed as median (interquartile range [IQR]) and compared by Mann-Whitney U test, except for continuous variables of cycle threshold (Ct) value of RT-PCR that conform to normal distribution, and were expressed as mean ± standard deviation and 95% confidence interval (CI). Categorical variables were expressed as number (%) and compared by χ 2 test or Fisher’s exact test as appropriate. A 2-sided α of less than 0.05 was considered statistically significant. Statistical analyses were done using the SPSS, version 25.0.
RESULTS
Features of Recurrence of Severe Acute Respiratory Syndrome Coronavirus 2 Ribonucleic Acid Polymerase Chain Reaction Positivity
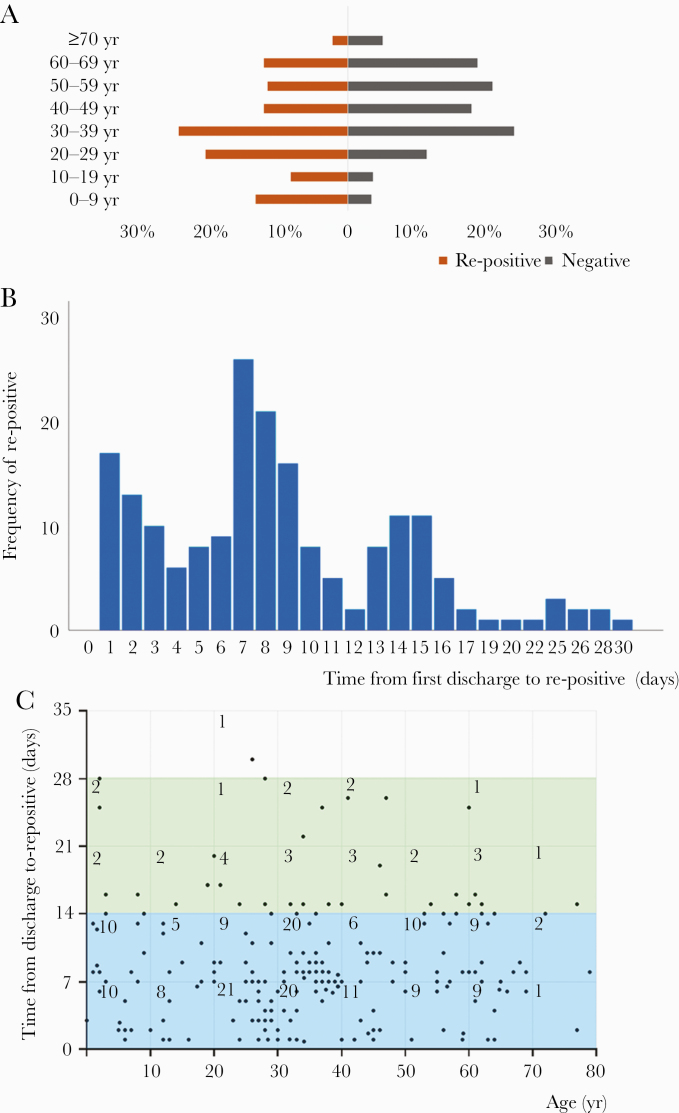
In the first 14 days of follow-up, 160 patients tested positive again for SARS-Cov-2 RNA, among them 111 re-positives were found from 809 discharged patients in section A and 49 re-positives from 473 cases in section B. The remaining 1122 cases were self-guarding for the next 14 days (section C), and 29 re-positives were found. Compared with the negative group, the re-positive group had a younger age (34 vs 45 years, P < .001) and more young components (63.49% vs 40.35% in 0–39 years range, P < .001) (Table 1, Figure 2A).
Table 1.
Characteristics of Patients in Re-Positive and Negative Groups at the First Hospitalizationa
| Characteristics | Total (n = 1282) | Re-positive Group (n = 189) | Negative Group (n = 1093) | P Value |
|---|---|---|---|---|
| Sex (male), n (%) | 626 (48.83%) | 103 (54.50%) | 523 (47.85%) | .091 |
| Age (years), median (IQR) | 43.00 (32.00–57.00) | 34.00 (24.00–49.00) | 45.00 (33.00–58.00) | <.001 |
| <40 | 561 (43.76%) | 120 (63.49%) | 441 (40.35%) | <.001 |
| Incubation (days), median (IQR) | 8.00 (5.00–13.00) | 9.00 (6.00–13.00) | 8.00 (5.00–13.00) | .236 |
| Comorbidities, n (%) | 269 (20.98%) | 21 (11.11%) | 248 (22.69%) | <.001 |
| Diabetes | 59 (4.60%) | 7 (3.70%) | 52 (4.76%) | .523 |
| Hypertension | 124 (9.67%) | 10 (5.29%) | 114 (10.43%) | .027 |
| Cardiovascular disease | 34 (2.65%) | 2 (1.06%) | 32 (2.93%) | .140 |
| Cerebrovascular disease | 11 (0.86%) | 1 (0.53%) | 10 (0.91%) | NA |
| Chronic kidney disease | 9 (0.70%) | 0 | 9 (0.82%) | NA |
| Chronic lung disease | 34 (2.65%) | 4 (2.12%) | 30 (2.74%) | .620 |
| Chronic liver disease | 40 (3.12%) | 3 (1.59%) | 37 (3.39%) | .189 |
| History of cancer | 14 (1.09%) | 0 | 14 (1.28%) | NA |
| Symptoms, n (%) | ||||
| Fever | 862 (67.24%) | 115 (60.85%) | 747 (68.34%) | .043 |
| Cough | 616 (48.05%) | 72 (38.10%) | 544 (49.77%) | .003 |
| Fatigue | 149 (11.62%) | 16 (8.47%) | 133 (12.17%) | .142 |
| Myalgia | 106 (8.27%) | 17 (8.99%) | 89 (8.14%) | .695 |
| Diarrhea | 33 (2.57%) | 3 (1.59%) | 30 (2.74%) | .461 |
| Dyspnea | 91 (7.10%) | 8 (4.23%) | 83 (7.59%) | .097 |
| PE, Median (IQR) | ||||
| Respiratory rate | 20.00 (19.00–20.00) | 20.00 (19.00–20.00) | 20.00 (19.00–20.00) | .733 |
| Heart rate | 83.00 (77.00–92.00) | 85.00 (77.00–95.75) | 82.00 (77.00–91.00) | .059 |
| Mean arterial pressure | 93.00 (86.00–100.33) | 91.67 (84.67–99.33) | 93.33 (86.00–100.33) | .118 |
| Oxygen saturation (SaO2) | 98.00 (97.70–99.00) | 98.00 (97.80–99.00) | 98.00 (97.70–99.00) | .224 |
| Laboratory Test, Median (IQR) | ||||
| PCT | 0.06 (0.03–0.18) | 0.06 (0.02–0.18) | 0.06 (0.03–0.18) | .544 |
| WBC | 5.46 (4.34–6.70) | 5.40 (4.30–6.68) | 5.46 (4.35–6.71) | .620 |
| Lymphocyte | 1.48 (1.10–1.94) | 1.59 (1.17–2.05) | 1.46 (1.10–1.91) | .039 |
| AST | 21.00 (16.60–28.40) | 22.00 (17.00–30.50) | 21.00 (16.40–28.00) | .196 |
| ALT | 21.90 (14.00–34.00) | 19.00 (13.00–31.70) | 22.00 (14.00–34.10) | .071 |
| CK | 54.00 (37.00–80.00) | 57.50 (38.33–76.00) | 53.50 (37.00–80.00) | .475 |
| Moderate symptoms, n (%) | 1103 (86.04%) | 181 (95.77%) | 922 (84.35%) | <.001 |
| Severe symptoms, n (%) | 179 (13.96%) | 8 (4.23%) | 171 (15.65%) | <.001 |
| Length of hospitalization (days), median (IQR) | 18.00 (14.00–24.00) | 17.00 (13.00–23.00) | 19.00 (14.00–25.00) | .013 |
| 0–9 years | 14.00 (12.00–22.00) | 14.00 (12.25–18.75) | 17.00 (12.00–25.00) | NA |
| 10–19 years | 15.00 (11.50–21.00) | 13.5 (9.00–18.75) | 16.00 (12.50–22.00) | NA |
| 20–29 years | 16.50 (13.00–22.00) | 15.00 (13.00–21.00) | 17.00 (13.00–23.00) | NA |
| 30–39 years | 16.00 (13.00–22.00) | 17.00 (13.5–23.5) | 16.00 (13.00–22.00) | NA |
| 40–49 years | 19.00 (15.00–24.00) | 17.50 (15.00–21.00) | 19.00 (15.00–24.00) | NA |
| 50–59 years | 20.00 (15.00–25.00) | 21.00 (17.00–30.50) | 20.00 (15.00–25.00) | NA |
| 60–69 years | 22.00 (16.00–28.25) | 18.00 (14.50–26.50) | 22.00 (16.00–29.00) | NA |
| ≥70 years | 21.00 (15.50–27.00) | 23.00 (19.75–27.00) | 20.00 (15.00–27.00) | NA |
| Treatment, n, (%) | ||||
| Antiviral | 1093 (85.26%) | 160 (84.66%) | 933 (85.36%) | .801 |
| Antibiotics therapy | 289 (22.54%) | 49 (25.93%) | 240 (21.96%) | .228 |
| Oxygen therapy | 1211 (94.46%) | 175 (92.59%) | 1036 (94.78%) | .224 |
| Corticosteroids therapy | 162 (12.64%) | 22 (11.64%) | 140 (12.81%) | .655 |
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; CK, creatine kinase; IQR, interquartile range; NA, not applicable; PCT, procalcitonin; PE, physical examination; WBC, white blood cells.
aData are presented as median (IQR) or n (%). P values are calculated by χ 2 test or Mann-Whitney U test between re-positive patients and negative patients.
NOTE. Missing Data: In Table 1, 477 patients lack data of incubation days for vague, ambiguous exposure history. Twelve patients lack data of respiratory rate, 10 patients lack data of heart rate, 36 patients lack mean arterial pressure (MAP), and 106 patients lack SaO2 at the first-time admission due to incomplete admission records. Two hundred thirty-five patients lack data of PCT, 100 patients lack WBC count, 107 patients lack lymphocyte count, 119 patients lack AST and ALT, and 350 patients lack data of CK due to lacking of inspection or incomplete laboratory test records. In the re-positive group, 2 patients lack data of respiratory rate, 3 patients lack data of heart rate, 16 patients lack MAP, and 20 patients lack oxygen saturation at the first-time admission. Five patients lack PCT data, 28 patients lack WBC and lymphocyte counts, 37 patients lack AST, 38 patients lack ALT, and 78 patients lack CK data.
Figure 2.
Age distributions and the features of re-positive ribonucleic acid (RNA) in re-positive patients. As shown in A, age pyramid count graph, patients in the re-positive group account for 12.70% in patients aged 0–9 years, 7.94% in patients aged 10–19 years, 19.58% in patients aged 20–29 years, 23.28% in patients aged 30–39 years, 11.64% in patients aged 40–49 years, 11.11% in patients aged 50–59 years, 11.64% in patients aged 60–69 years, and 2.12% in patients aged ≥70 years, whereas patients in the negative group account for 3.20% in patients aged 0–9 years, 3.39% in patients aged 10–19 years, 10.89% in patients aged 20–29 years, 22.87% in patients aged 30–39 years, 17.02% in patients aged 40–49 years, 19.85% in patients aged 50–59 years, 17.93% in patients aged 60–69 years, and 4.85% in patients aged ≥70 years. As shown in B, bars represent frequency of patients tested re-positive from first discharge to re-positive at different re-positive time. Taller bars illustrate 1–15 days, presented with 3 peaks at the 1st, 7th, and 14th day. Eighteen cases (9.5%) exceeded 15 days for severe acute respiratory syndrome coronavirus 2 RNA re-positive. As shown in C, time from first discharge to re-positive was divided by per 7 days and matched with age group divided by per 10 years. The figures in the boxes represent the number of cases. The area of 0–21 days in the y axis, especially of 0–14 days, contains the largest number of dots, which represent patients. The number of patient days from first discharge to re-positive are as follows: 89 patients, ≤7 days; <71 patients, 7 days; <20 patients, 14 days, ≤14 days; <8 patients, 21 days; <1 patient, ≤28 days, 28 days . In addition, in the x-axis, the density of dots in the area of 20–40 years is the highest; 81 dots are located in this area.
The median re-positive time from the first discharge was 8 days (IQR, 5–13), which was mainly concentrated within 15 days (90.48%), and presented with 3 peaks at the 1st, 7th, and 14th day, respectively (Figure 2B, Table 2). Most of the recurrence for positive SARS-Cov-2 RNA (84.66%) occurred within 2 weeks, among which 54.50% were in the range of 0–39 years old (Figure 2C).
Table 2.
Clinical Features of Re-positive Patients at the Second Hospitalizationa
| Clinical Features | Total (n = 189) |
|---|---|
| Days from last negative to re-positive, median (IQR) | 10.00 (7.00–15.00) |
| Days from first discharge to re-positive, median (IQR) | 8.00 (5.00–13.00) |
| New symptoms of COVID-19 | None |
| Clinical Symptoms | |
| Fever | 0 |
| Cough | 30 (15.87%) |
| Fatigue | 2 (1.06%) |
| Myalgia | 2 (1.06%) |
| Diarrhea | 1 (0.53%) |
| Dyspnea | 7 (3.70%) |
| None | 152 (80.42%) |
| Length of second hospitalization (days), median (IQR) | 10.00 (6.00–17.00) |
| Chest CT Scan Alteration Compared With the First Discharge, n (%) | |
| Improvement | 140 (74.07%) |
| Healing | 8 (4.23%) |
| No alteration | 36 (19.05%) |
| No check | 5 (2.65%) |
| Antiviral therapy | 49 (25.93%) |
| No antiviral therapy | 140 (74.07%) |
Abbreviations: CT, computed tomography; COVID-19, coronavirus disease 2019; IQR, interquartile range.
aData are presented as median (IQR) or n (%).
NOTE. Missing Data: In Table 2, 27 patients lack data of white blood cell (WBC) and lymphocyte count at the second-time admission and 37 patients lack WBC and lymphocyte count at the second-time discharge due to lack of inspection or incomplete laboratory test records. Two patients lack data regarding length of second-time hospitalization because they had not been discharged from hospital by April 7, 2020 (the closing day of our study), 1 of whom also lacked data of time from re-positive to negative because it had not turn to severe acute respiratory syndrome coronavirus 2 ribonucleic acid negative again.
Characteristics of Re-Positive Patients at the First Hospitalization
In clinics, re-positive patients showed a larger proportion of moderate symptoms (95.8% vs 84.4%, P < .001) at the first admission but a lesser proportion of comorbidities (11.11% vs 22.69%, P < .001) and a lower incidence of fever (60.85% vs 68.34%, P = .043) and cough (38.10% vs 49.77%, P = .003) than negative patients. The clinical treatment protocol for re-positive patients was not different from treatment for negative ones, including antivirals (84.66%), antibiotics (25.93%), corticosteroids (11.64%), and oxygen therapy (92.59%). The median length of the first hospitalization in re-positive patients was shorter than the negative ones (17 days vs 19 days, P = .013). The median time from the last negative to re-positive of viral RNA was 10 days (IQR, 7–15) (Table 1).
Features of Negative Conversion in Re-Positive Patients
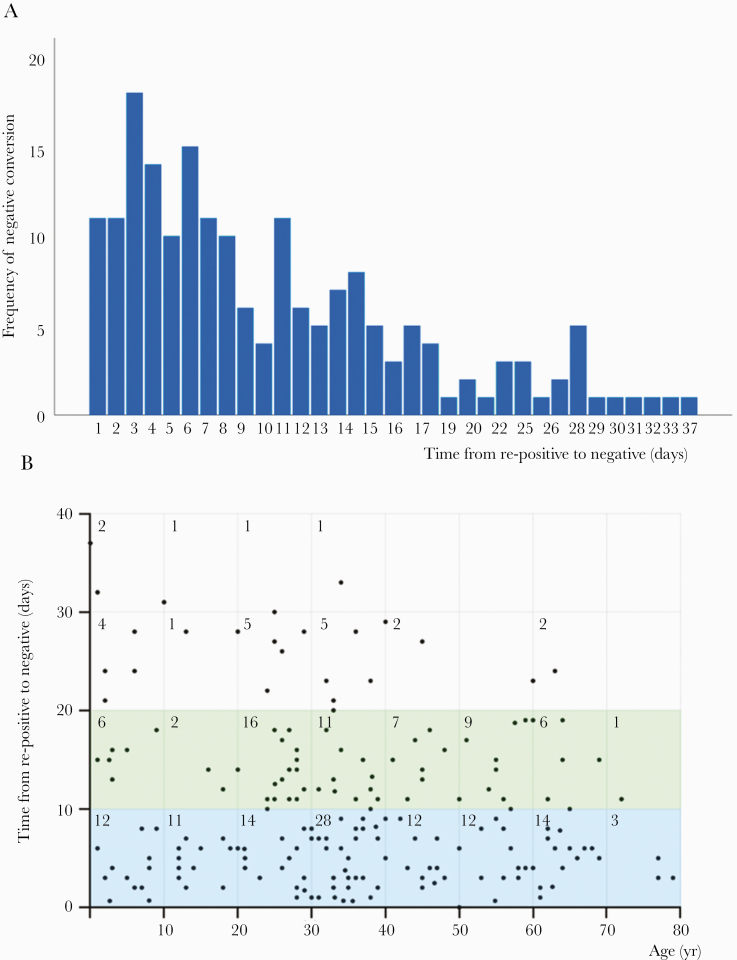
In total, 188 of 189 re-positive patients turned negative eventually during the second hospitalization in 1 to 37 days by April 7, 2020. The median conversion time from re-positivity was 8 days (IQR, 4–15) (Table 2). The majority (87.77%) of them converted within 20 days; however, 12.23% cases needed 21–37 days to recover (Figure 3A). A total of 46.28% of re-negatives were 0–39 years old and converted within 2 weeks (Figure 3B). The remaining patient did not test negative until May 3, 2020.
Figure 3.
Features of negative conversion of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) ribonucleic acid (RNA) in re-positive patients. As shown in A, re-positive patients turned negative eventually at the second hospitalization with conversion time ranging from 1 to 37 days. Taller bars illustrate 1–19 days with peak at 3 days. A total of 87.23% patients turned to negative from re-positive within 19 days, 78.19% in 15 days, and 12.77% (n = 24) needed a longer time of 20–37 days to convert. As shown in B, the area of 0–20 days in the y-axis, especially of 0–10 days, contains the largest number of dots. One hundred six patients spent <10 days from re-positive to negative again, while fifty eight patients spent 10-19 days, nineteen patients spent 20-29 days, five patients spent 30-39 days. The figures in the boxes represent the number of case. Data are missing for 1 patient because the patient was not SARS-Cov-2 RNA negative again after being re-positive by April 7, 2020, which was the closing day of our study.
Characteristics of Re-Positive Patients at the Second Hospitalization
The median length of the second hospitalization for patients who tested positive again was 10 days (IQR, 6–17). Several patients still had lingering symptoms from the last disease course of COVID-19—30 (15.87%) patients cough at second admission, 2 (1.06%) had fatigue, 2 (1.06%) had myalgia, 1 (0.53%) had diarrhea, and 7 (3.70%) had dyspnea—and 152 (80.42%) patients had relief from symptoms, but were only re-admitted for re-positive SARS-Cov-2 RNA. Nevertheless, none of the 189 patients had newly developed symptoms of COVID-19 during the second hospitalization. White blood cell and lymphocyte counts were within normal range either in the second admission (5.70 × 109/L and 1.82 × 109/L, respectively) or at discharge (6.01 × 109/L and 1.99 × 109/L, respectively) from hospitalization. Chest CT scans also showed no aggravations (140 improved, 8 cured, and 36 no remarkable changes) compared with the first discharge. Only 49 cases (25.93%) accepted antiviral therapy (Table 2).
The Cycle Threshold Value of Real-Time Reverse-Transcriptase Polymerase Chain Reaction for Recurrence of Positive Severe Acute Respiratory Syndrome Coronavirus 2 Ribonucleic Acid
After our best efforts, 121 samples from 61 re-positive patients, including 61 nasopharynx swabs and 60 anal swabs, were collected and tested by RT-PCR. The Ct values of ORF1ab gene and N gene in nasopharynx specimen were 37.55 ± 1.32 (95% CI, 36.88–38.22; n = 61) and 36.56 ± 1.39 (95% CI, 36.07–37.05; n = 61) and 37.05 ± 2.13 (95% CI, 36.06–38.05; n = 60) and 36.00 ± 2.25 (95% CI, 35.17–36.80; n = 60) in anal specimen, respectively.
Clinical Outcomes of Close Contacts
After strict investigations during the 28-day follow-up period, only the patients in section A (Figure 1) generated close contacts. Of a total of 111 re-positives in section A, 69 generated 209 close contacts. The median age of the contacts was 46 years old (IQR, 37–65), and 26.80% were male. None of them reported a travel history to the COVID-19 epidemic area or had ever been infected by SARS-CoV-2. During the 14 days of quarantine, none of them developed clinical symptoms of COVID-19, eg, fever, cough, or dyspnea. The tests for SARS-CoV-2 RNA in nasopharyngeal or anal swab specimens were all negative.
DISCUSSIONS
Coronavirus disease 2019 has infected more than 6 million people [22]. Recurrence of positive SARS-CoV-2 RNA after discharge is a serious public health concern for the management of discharged COVID-19 patients and prevention of further spread of COVID-19. This retrospective study aimed to (1) analyze clinical and epidemiological information of 1282 patients discharged from 32 designated hospitals in Guangdong Province and (2) reveal the features of short-term recurrence of positive SARS-CoV-2 RNA in those populations.
In this study, we found that 14.74% (n = 189) of 1282 discharged patients in Guangdong Province had recurrence of positive SARS-CoV-2 RNA during 28 days of follow-up, and the median time of re-positive RNA distributed from 7 to 11 days at different age range. Lapse time from the first discharge was mainly within 15 days (90.48%).
Based on the strict process of prevention and control policy in Guangdong Province, all positive results of RNA must be rechecked with the same source of samples within 24 hours and issued by the CDC of Guangdong Province before they were diagnosed as re-positives. We believe that the recurrence of positive viral RNA in all COVID-19 cases is universal. A recent small-sized study in the neighboring city of Guangdong showed similar data to ours [11].
Furthermore, we showed that these re-positive of viral RNA means the continuation of the first infection, but not a re-infection of virus particles.
Epidemiological Evidence
The median length of the first hospitalization of re-positive cases was 17 days (IQR, 13–23). It was reported that the specific antibody of immunoglobulin (Ig)G can be detected on the 4th day after symptom onset, and 85% of the patients could produce IgG on the 10th–13th day and 100% on the 17th day [23], which supported that most of the patients in our study had produced specific antibodies at the time of discharge. Meanwhile, we combed the close contacts of the re-positive patients and found that they had not been to the epidemic area nor were they exposed to COVID-19 patients. Even though an asymptomatic case of COVID-19 re-infection confirmed by whole-genome sequencing was reported [24], we noted that this case was diagnosed 142 days after the first symptomatic episode when the patient traveled to Spain and the United Kingdom and had a lot access to infected people. In this study, 49 discharged patients were tested for recurrence of positive SARS-CoV-2 RNA during the mandatory quarantine period without contacting anyone but medical staff (section B of Figure 1), which was the solid evidence to rule out re-infection. Therefore, the possibility of re-infection in this group is extremely low.
Clinical Evidence
In this study, although a small group of patients still had remaining symptoms from last disease course of COVID-19—such as cough, fatigue, and dyspnea—all 189 re-positive patients showed neither new symptoms of COVID-19 nor aggravation on chest CT images during follow-up. Most of chest CT images for them further improved when compared with their first discharge. In addition, white blood cell and lymphocyte counts were within the normal range. All the above data indicated that the immunocompetence of re-positive patients was recovering.
Virological Evidence
The Ct value of specific ORF1ab gene in anal and nasopharynx specimens derived from re-positive cases ranged from 36 to 40, which were close to the detection limit of quantitative PCR. This suggested that the SARS-CoV-2 RNA content in re-positive cases was too low to provoke an onset of disease. Moreover, re-positive patients turned SARS-CoV-2 RNA negative again within 8 days (IQR, 4–15), and 74.07% of them did not accept antiviral therapy at their second admission, indicating clearance of virus achieved automatically. Therefore, our study suggests that short-term recurrence for positive SARS-CoV-2 RNA in discharged patients did not represent relapse of disease but rather a phenomenon in a stage of the viral clearance process.
Our data also show that there was no communicability of re-positive patients. None of the 209 close contacts of 69 patients developed clinical symptoms of COVID-19 (eg, fever or cough), and the RT-PCR also tested negative in both nasopharyngeal and anal swab samples. Therefore, it is possible that re-positive patients were barely communicable. Additional evidence suggests that the viral load of SARS-CoV-2 in nasopharyngeal swab reached extremely low limits of detection until the 21st day [25]. The live virus can only be isolated in nasopharyngeal swabs within 8 days after symptom onset with a viral load more than 106 copies/mL [26]. In the median 18 days of hospitalization at the first admission, we determined that the possibility of excreting live virus was extremely low.
Finally, we explain why the recurrence of positive viral RNA frequently occurred in younger populations. Previous studies reported that the time of virus shedding in COVID-19 survivals was 17–24 days, and it even reached 49 days in individual cases [27, 28]. In our data, however, the median hospitalization time for negative group of patients was significantly longer than the re-positive group, especially in ages <30 years. It is possible that the insufficient length of hospital stay may be one of the explanations why the SARS-CoV-2 RNA was tested re-positive after discharge. In our study, patients in the re-positive group had younger age, lower incidence of fever and cough at admission with less proportion of severe symptoms, but higher lymphocyte counts. Although the younger patients and patients with mild symptoms could recover within a shorter period of time after hospitalization, meeting the discharge criteria did not mean that the virus was completely cleared. Thus, the larger proportion of recurrence for positive SARS-CoV-2 RNA in younger patients may be related to the shorter time of hospitalization.
There are limitations in our study. First, due to the complexity of the epidemic situation, we cannot achieve a cohort-based design and implementation in a short period of time, nor can we identify the virus genotypes. Second, serum antibody test and live virus isolation of re-positive patients were not conducted, and less than half of re-positive patients were drawn for viral RNA confirmation because total samples could not be obtained. Third, only 209 close contacts were generated, which might not be enough to fully assess the risk for communicability.
CONCLUSIONS
In conclusion, in this multicenter, retrospective, observational study with the large samples and 28-day follow-up among the discharged COVID-19 patients in Guangdong Province, China, we found that (1) the younger and more physically active patients accounted for a large proportion of recurrence for positive viral RNA after discharge, (2) the recurrence is not a relapse of COVID-19, and (3) the re-positive patients are not communicable. This may be a common process in convalescence of disease, with low risk of onward transmission of the virus. Our findings have potentially important information for clinicians currently managing COVID-19 patients. However, due to the limited knowledge regarding SARS-CoV-2 at this time, close monitoring and followed quarantine after discharge from hospital are still needed.
Acknowledgments
We thank all designated hospitals in no particular order: Guangdong Provincial People’s Hospital (Guangdong Academy of Medical Sciences), Guangdong Second Provincial General Hospital, The Third Affiliated Hospital of Sun Yat-sen University, Nanfang Hospital of Southern Medical University, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou Eighth People’s Hospital, Guangzhou Women and Children’s Medical Center, The Third People’s Hospital of Shenzhen, The Fifth Affiliated Hospital Sun Yat-sen University, Shantou Central Hospital, The First Affiliated Hospital of Shantou University Medical College, The First People’s Hospital, Yuebei Second People’s Hospital of Foshan, Heyuan People’s Hospital, Meizhou People’s Hospital, Huizhou Municipal Central Hospital, People’s Hospital of Shanwei, Dongguan People’s Hospital, Zhongshan Second People’s Hospital, Jiangmen Central Hospital, Central People’s Hospital of Zhanjiang, Affiliated Hospital of Guangdong Medical University, Maoming People’s Hospital, The First People’s Hospital of Zhaoqing, Qingyuan People’s Hospital, Chaozhou Central Hospital, Jieyang People’s Hospital, The Ninth People’s Hospital of Dongguan, The Fourth People’s Hospital of Foshan, Yangjiang Hospital, The Third Affiliated Hospital of Sun Yat-Sen University, Yuedong Hospital, Jiangmen Wuyi Hospital of Traditional Chinese Medicine. We also thank the following: all healthcare workers involved in the diagnosis, treatment, and collection of data for patients that was used in our study; Yu-feiDuan, Guan-xian Liu, Li Chen, Fa-bin Zhang, Hong-xiang Feng, Jie-wu Guo, Yong-jia Chen, Rui-jie Yao, Ming Li, and Yong-ping Du (Health Commission of Guangdong Province) for facilitating the collection of patients’ data; and statistical team members Xiao-qing Lin, Min-Qing Li, Hai-dong Chen, Shu-gen Luo, and Jin-hong Wang (the Rescue Team of Guangdong Provincial COVID-19 Prevention and Control Headquarters) for their dedication to data collection and verification for COVID-19 patients.
Author contributions. M. F. and S.-L. C. conceived of the idea and designed the study. S.-L. C., H. X., H.-Y. F., J.-F. S., and X. L. had full access to all data in the study and took responsibility for the integrity of the data and data analysis. S.-S. H., H. X., J.-L. H., W.-L. S., and R.-J. W. were responsible for data collection. M. F., S.-L. C., H.-Y. F., J.-F. S., H. X., Y.-Y. D., and L. Z. contributed to writing of the report. All authors contributed to data acquisition, data interpretation, and reviewed and approved the final version.
Financial support. This study is supported by the Health Commission of Guangdong Province and the Department of Science and Technology of Guangdong Province. This study is also supported by Special Project on Emergency Response to Control of Novel Coronavirus Infection of Guangdong Province. This work was funded by the Special Project on Emergency Response to Control of COVID-19 Infection of Guangdong Province (No. 2020B1111330003) and the National Natural Science Foundation of China (No. 81701875).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu K, Fang YY, Deng Y, et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl) 2020; 133:1025–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guan WJ, Ni ZY, Hu Y, et al. ; China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zumla A, Hui DS, Azhar EI, et al. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet 2020; 395:e35–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arabi YM, Mandourah Y, Al-Hameed F, et al. ; Saudi Critical Care Trial Group Corticosteroid therapy for critically ill patients with Middle East respiratory syndrome. Am J Respir Crit Care Med 2018; 197:757–67. [DOI] [PubMed] [Google Scholar]
- 6. Li LQ, Huang T, Wang YQ, et al. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol 2020; 92:577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lan L, Xu D, Ye G, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA 2020; 323:1502–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang B, Liu S, Dong Y, et al. Positive rectal swabs in young patients recovered from coronavirus disease 2019 (COVID-19). J Infect 2020; 81:e49–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qu YM, Kang EM, Cong HY. Positive result of Sars-Cov-2 in sputum from a cured patient with COVID-19. Travel Med Infect Dis 2020; 34:101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen D, Xu W, Lei Z, et al. Recurrence of positive SARS-CoV-2 RNA in COVID-19: a case report. Int J Infect Dis 2020; 93:297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuan J, Kou S, Liang Y, et al. PCR assays turned positive in 25 discharged COVID-19 patients. Clin Infect Dis 2020; ciaa398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu XW, Wu XX, Jiang XG, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ 2020; 368:m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kucharski AJ, Russell TW, Diamond C, et al. ; Centre for Mathematical Modelling of Infectious Diseases COVID-19 working group Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis 2020; 20:553–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waddington CS, Darton TC, Woodward WE, et al. Advancing the management and control of typhoid fever: a review of the historical role of human challenge studies. J Infect 2014; 68:405–18. [DOI] [PubMed] [Google Scholar]
- 15. Greisman SE, Woodward TE, Hornick RB, et al. Typhoid fever: a study of pathogenesis and physiologic abnormalities. Trans Am Clin Climatol Assoc 1961; 73:146–61. [PMC free article] [PubMed] [Google Scholar]
- 16. National Health Commission, National Administration of Traditional Chinese Medicine. Translation: diagnosis and treatment protocol for novel Coronavirus pneumonia (Trial Version 7) Infectious Microbes & Diseases 2020; 2:48–54. [Google Scholar]
- 17. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020; 382:1177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun J, Xiao J, Sun R, et al. Prolonged persistence of SARS-CoV-2 RNA in body fluids. Emerg Infect Dis 2020; 26:1834–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus-infected pneumonia in Wuhan, China. JAMA 2020; 323:1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ding J, Tuan WJ, Temte JL. Managing close contacts of COVID-19 confirmed cases in Metropolitan areas in China. J Public Health Manag Pract 2020; 26:345–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. World Health Organization. Coronavirus disease (COVID-19) pandemic. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 1 June 2020.
- 23. Long Q, Liu B, Deng H, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 24. To KK, Hung IF, Ip JD, et al. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis 2020; doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020; 26:672–5. [DOI] [PubMed] [Google Scholar]
- 26. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 27. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395:1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tan L, Kang X, Zhang B, et al. A special case of COVID-19 with long duration of viral shedding for 49 days. medRxiv. doi: 10.1101/2020.03.22.20040071. [DOI]