Abstract
Background
Nucleic acid amplification testing is a critical tool for addressing the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. Specimen pooling can increase throughput and conserve testing resources but requires validation to ensure that reduced sensitivity does not increase the false-negative rate. We evaluated the performance of a real-time reverse transcription polymerase chain reaction (RT-PCR) test authorized by the US Food and Drug Administration (FDA) for emergency use for pooled testing of upper respiratory specimens.
Methods
Positive specimens were selected from 3 prevalence groups, 1%–3%, >3%–6%, and >6%–10%. Positive percent agreement (PPA) was assessed by pooling single-positive specimens with 3 negative specimens; performance was assessed using Passing-Bablok regression. Additionally, we assessed the distributions of RT-PCR cycle threshold (Ct) values for 3091 positive specimens.
Results
PPA was 100% for the 101 pooled specimens. There was a linear relationship between Ct values for pooled and single-tested specimens (r = 0.96–0.99; slope ≈ 1). The mean pooled Ct shifts at 40 cycles were 2.38 and 1.90, respectively, for the N1 and N3 targets. The median Cts for 3091 positive specimens were 25.9 (N1) and 24.7 (N3). The percentage of positive specimens with Cts between 40 and the shifted Ct was 1.42% (N1) and 0.0% (N3).
Conclusions
Pooled and individual testing of specimens positive for SARS-CoV-2 demonstrated 100% agreement, which demonstrates the viability of pooled specimens for SARS-COV-2 testing using a dual-target RT-PCR system. Pooled specimen testing can help increase testing capacity for SARS-CoV-2 with a low risk of false-negative results.
Keywords: pooled testing, positive percent agreement, RT-PCR, SARS-CoV-2
The ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has resulted in an unprecedented worldwide demand for laboratory testing. As of July 19, 2020, more than 3.7 million people in the United States and over 14 million worldwide have been diagnosed with coronavirus disease 2019 (COVID-19; https://coronavirus.jhu.edu/map.html). Timely access to SARS-CoV-2 nucleic acid testing is a critical tool for patient management, controlling the spread of the epidemic and informing public health efforts, highlighting the key role of the clinical laboratory in the health care system. The average number of daily tests for SARS-CoV-2 performed in the United States has reached >700 000 (https://covidtracking.com/data/us-daily); however, this increased demand for testing has put pressure on the laboratory supply chain, resulting in shortages of testing materials and instrumentation, leading to delays in obtaining test results.
Pooled diagnostic testing offers a means to reduce utilization of testing supplies and reagents while increasing laboratory testing throughput. In a simple Dorfman [1, 2] pooled testing scheme, a number of individually collected specimens are combined in a single well or tube and tested together. If the pooled test result is negative, results for all individual specimens may be immediately reported as negative. If a pool is positive, then each specimen in the pool must be tested individually before the patient results can be reported. The optimal number of specimens that can be included in a pool to maximize efficiency is determined by the prevalence of positive specimens [1, 3, 4] in the population being tested and is further constrained by the sensitivity of the test to reliably detect a positive signal in a diluted negative specimen pool. It is critical, therefore, to validate pooling strategies for diagnostics tests to ensure that the false-negative rate remains below an acceptable threshold.
Pooling specimens to increase testing efficiency and conserve testing resources has been used for HIV and hepatitis B virus and hepatitis C virus screening [5, 6]. Pooling has also been evaluated for the detection of influenza virus and bacterial pathogens from nasopharyngeal or throat swab specimens [7, 8] and for SARS-CoV-2 testing using pool sizes of between 4 and 64 specimens [3, 4, 9–11]. Here we describe the performance of pooled testing of upper respiratory specimens for a US FDA Emergency Use Authorization (EUA) real-time reverse transcription PCR (RT-PCR) test performed in low-prevalence populations for high-throughput testing. This study was performed as part of an FDA EUA application to authorize the use of pooled testing for detecting SARS-CoV-2 RNA.
METHODS
Clinical Specimens
Deidentified specimens collected between May 2020 and early July 2020 that had been previously tested using the Quest Diagnostics SARS-CoV-2 RNA Qualitative Real-Time RT-PCR EUA were recovered from frozen (–10°C to –30°C) storage. Specimen types included upper respiratory specimens (nasopharyngeal, midturbinate, nasal swabs) collected in viral transport media (UTM, UTM-RT), phosphate-buffered saline (PBS), or normal saline. Swabs were limited to those with a synthetic tip such as Dacron, Flocked, or Nylon and an aluminum or plastic shaft as specified in the FDA EUA instructions for use (https://www.fda.gov/media/136231/download). Sequentially tested single-positive and single-negative specimens were selected.
Pooled Testing
A pool size of 4 specimens was selected based on the estimated prevalence of SARS-CoV-2-positive specimens submitted for testing (7%–11%) and the corresponding recommended pool size for maximal pooling efficiency found in the FDA guidance document Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-public-health-emergency-revised). In the Quest Diagnostics SARS-CoV-2 RNA Qualitative Real-Time RT-PCR EUA (comparator) method (https://www.fda.gov/media/136231/download), 200 µL of patient specimen is combined with 250 μL of lysis buffer in the first step of the nucleic acid isolation procedure. In the pooling method, 50 μL from each of 4 patient specimens is combined with 250 µL of lysis buffer in the first step of the nucleic acid isolation procedure. The remaining steps in the nucleic acid isolation and real-time RT-PCR procedures are unchanged. Briefly, the assay utilizes a 1-step reverse transcription and PCR amplification with SARS-CoV-2-specific primers and real-time detection with SARS-CoV-2-specific probes for the N1 and N3 targets of the virus nucleocapsid gene. Following 50 cycles of RT-PCR, a specimen is deemed positive for SARS-CoV-2 RNA when the Ct values for both targets are <40 cycles. A specimen is deemed negative when the Ct values for both targets are ≥40 cycles and the internal amplification control is valid. If the Ct value for a single detector is <40 cycles while the Ct for the second detector is ≥40 cycles, the test result is deemed inconclusive and the specimen is retested. Pooled specimens with negative results and valid internal control results are individually defined as negative. Pooled specimens with positive or inconclusive results or invalid internal control results are repeated as individual specimens to determine the final result for each.
Study Design
The Quest Diagnostics Informatics database was utilized to geographically stratify specimens according to SARS-CoV-2 test positivity rate into 3 groups: group 1: positivity rate of 1%–3%; group 2: positivity rate of 3%–6%; and group 3: positivity rate of 6%–10%. Each prevalence group included specimen remnants selected from at least 2 separate geographic locations. We performed sensitivity studies in all 3 prevalence groups by combining 1 positive sample with 3 negative samples. All single-positive specimens were repeated at the same time as the single-positive pools to ensure that there was no degradation of the archived specimens. The linearity and the shift in the Ct values between pooled and singlicate results were assessed using Passing-Bablok regression [12, 13]. The shifted upper range for the Ct values for the N1 and N3 targets was defined as (40 – y intercept)/slope. Specificity studies were performed in all 3 prevalence groups by combining 4 negative samples per pool.
The informatics database was further used to select 3091 deidentified positive test results from the 3 defined prevalence groups selected from US counties for in silico analyses. The percentage of positive test results in the range between the upper limit of a positive result for the singlicate assay (40 cycles) and the upper limit minus the Ct shift seen in the pools (eg, 37.0 for the N1 detector in prevalence group 1) was then calculated to predict the potential number of high Ct value false negatives in a pooled testing design.
Statistical Analysis
Statistical analysis was performed using Analyze-it for Microsoft Excel, version 5.65.3.
Human Subjects
This study utilized deindentified specimen remnants and retrospectively collected deindentified data from previously tested specimens. No human subjects were utilized in this study, and thus patient consent was not applicable.
RESULTS
Sensitivity of Pooled Testing
Three groups of single-positive pooled specimens were prepared using sequentially tested single-positive and single-negative specimen remnants from 3 prevalence populations as described in the “Methods” section. Of the single-positive samples, 30 were included from group 1, 36 were included from group 2, and 35 were included from group 3 (Supplementary Table 1). Overall, 44.6% of the positive specimens were from women and 52.5% were from men, and the median patient age was 38 years (Supplementary Table 1). We obtained 100% percent positive agreement (PPA) between the pooled and singlicate tests (Table 1). There were 2 inconclusive pools (Ct values of only 1 of the 2 detectors <40) in prevalence group 3, which were positive for SARS-CoV-2 based on singlicate retesting.
Table 1.
Sensitivity of Pooled SARS-CoV-2 RT-PCR Testing
| Prevalence Groupa | No. | Negative Pools | Inconclusive Poolsb | Positive Pools | Invalid Poolsc | PPA, % |
|---|---|---|---|---|---|---|
| 1 | 30 | 0 | 0 | 30 | 0 | 100 |
| 2 | 36 | 0 | 0 | 36 | 0 | 100 |
| 3 | 35 | 0 | 2 | 33 | 0 | 100 |
Abbreviations: Ct, cycle threshold; PPA, positive percent agreement, RT-PCR, reverse transcription polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
aSpecimens were drawn from 3 geographic prevalence groups, as described in the “Methods” section. For pooled testing, 1 positive sample with 3 negative samples.
bAn inconclusive result occurs when 1 of the Ct values for the N1 and N3 targets is positive (<40 Cts) and the other is negative (≥40 Cts). Inconclusive pools are resolved through singlicate retesting.
cAn invalid result occurs when the internal positive control does not yield an acceptable Ct value. Invalid pools are then tested in singlicate.
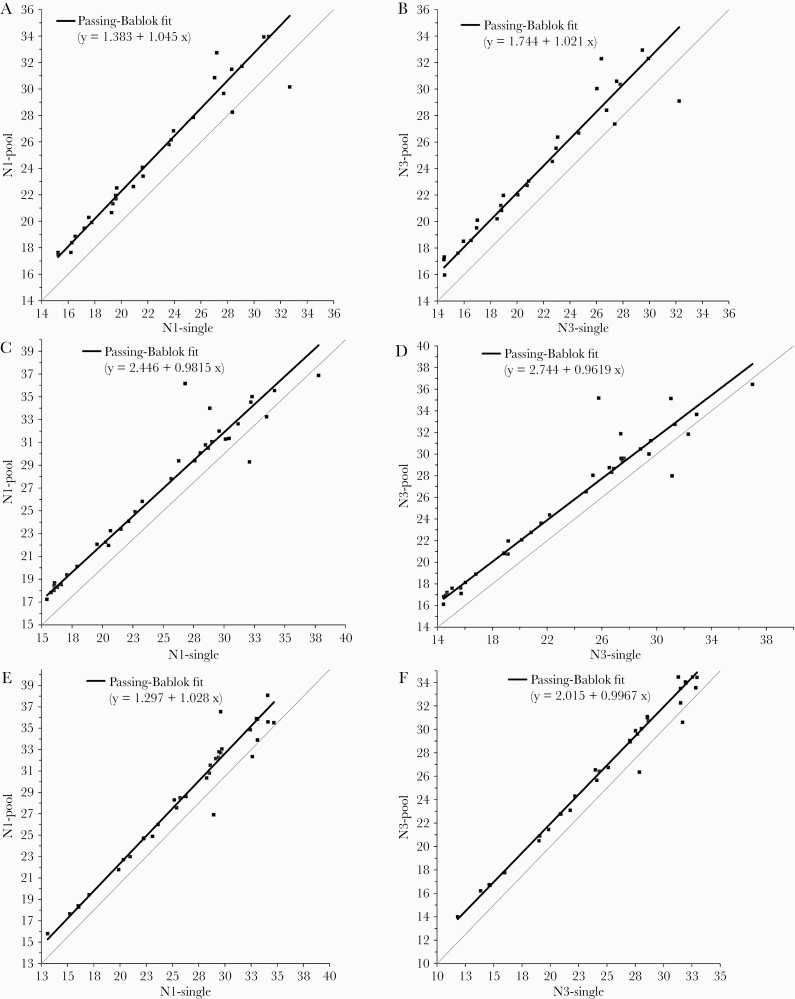
We performed Passing-Bablok regression for the N1 and N3 detector Ct values in each of the 3 groups to assess the linearity between the pooled and singlicate Ct values and to determine the shift in Ct as a measure of the reduction in sensitivity resulting from the dilution of positive samples in a negative pool (Figure 1A–F). The average regression correlation coefficients ± SD for the N1 and N3 targets in the 3 groups were 0.97 ± 0.007 and 0.97 ± 0.016, respectively, indicating good linearity. The average slopes were 1.02 ± 0.033 for the N1 target and 0.993 ± 0.030 for the N3 target, indicative of minimal proportional bias (Table 2). The average reduction in sensitivity ± SD for the pooled tests as measured by the shift in Ct values was 1.71 ± 0.64 for N1 and 2.17 ± 0.52 for N3 (Table 2). These shifts are close to the shift of 2 Cts, which would be the expected estimate for a 1:4 dilution of a positive specimen.
Figure 1.
Passing-Bablok regression analysis of N1 and N3 cycle threshold values for pooled and single-tested positive specimens. A: group 1, N1 target. B: group 1, N3 target. C: group 2, N1 target. D: group 2, N3 target. E: group 3, N1 target. F: group 3, N3 target.
Table 2. .
Passing-Bablok Regression Analysesa
| N1 Target | N3 Target | |||||
|---|---|---|---|---|---|---|
| Prevalence Group | Ct Shiftb | Slope | Correlation Coefficient | Ct Shiftb | Slope | Correlation Coefficient |
| 1 | 3.05 | 1.05 | 0.970 | 2.53 | 1.02 | 0.962 |
| 2 | 1.74 | 0.980 | 0.963 | 1.27 | 0.960 | 0.961 |
| 3 | 2.35 | 1.03 | 0.976 | 1.89 | 1.00 | 0.989 |
| Mean (SD) | 2.38 ± 0.65 | 1.01 ± 0.03 | 0.970 ± 0.007 | 1.90 ± 0.63 | 0.993 ± 0.03 | 0.971 ± 0.016 |
Abbreviation: Ct, cycle threshold.
aCt values from positive specimens tested in pools and in singlicate were analyzed by Passing-Bablok regression analysis, as shown in Figure 1A–F.
bThe Ct shift at 40 cycles was defined as 40 – (40 – intercept)/slope.
Specificity of Pooled Testing
Pools were prepared from negative specimens using sequentially tested single-negative specimen remnants from 3 prevalence populations as described in the “Methods” section (Supplementary Table 3). The negative percent agreement (NPA) for pooled sample testing was 99% for group 1 (102/103) with 1 inconclusive pool and 100% for groups 2 and 3 (Supplementary Table 4). The overall NPA across all 3 groups was 99.6%. A single pool yielded an inconclusive result in group 1, with an N1 Ct value of 39.89, just below the positive Ct cutoff of 40. The initial Ct values of the 4 specimens comprising this pool were verified to be undetectable (>50 cycles); further, single-sample retesting was not performed.
Distribution of Ct Values in Positive Samples
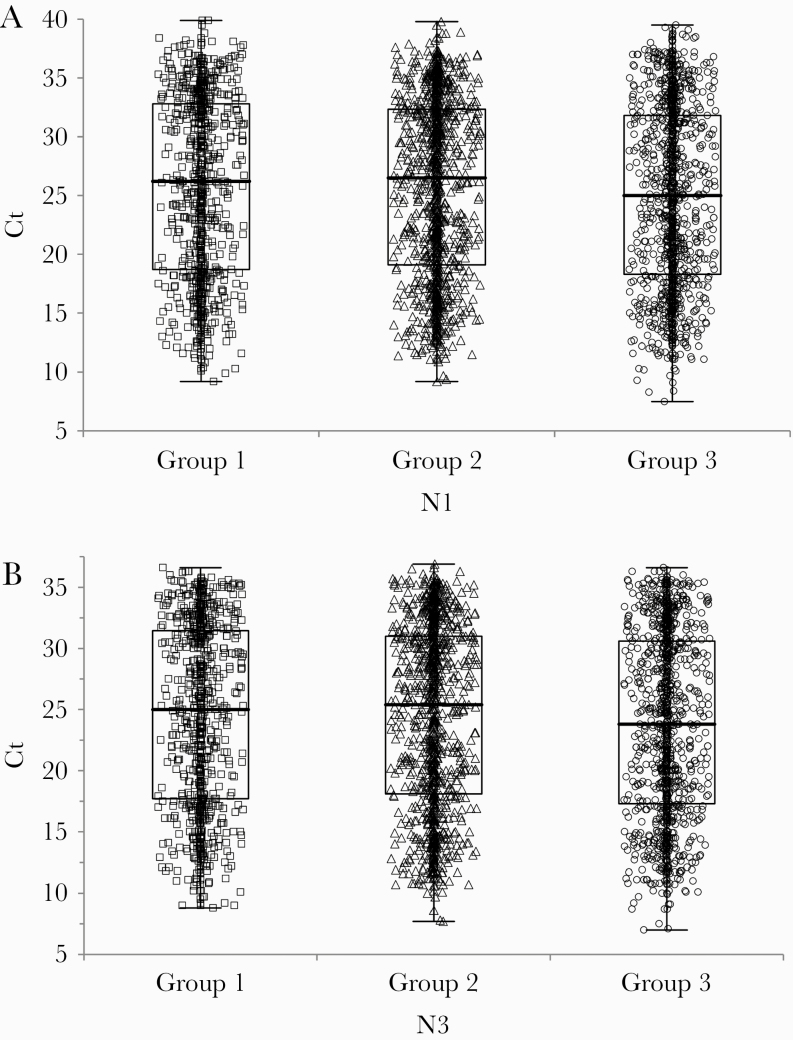
Three groups of single-positive specimens were selected from 3 prevalence populations. Each prevalence group included positive specimens selected from multiple geographic locations (Supplementary Table 5): 820 positives were included from group 1; 1113 from group 2, and 1158 from group 3. Patient sex and age distributions are shown in Supplementary Table 6. The median group 3 Ct values for N1 were slightly lower than for groups 1 and 2 by 1.2 and 1.5 Cts, respectively (P = .0303) (Figure 2A), and the median group 3 Ct values for N3 were 1.2 and 1.6 Cts lower than those for groups 1 and 2 (P = .0271) (Figure 2B). The upper 95th percentiles for the Ct value distributions were 36.4, 36.1, and 36.5 for the 3 groups for the N1 target and 35.0, 34.8, and 34.6 for the N3 target.
Figure 2.
Distribution of N1 and N3 cycle threshold (Ct) values for 3091 positive specimens in groups 1, 2, and 3. A: N1 target. B: N3 target. Median Ct differences between groups were assessed using the Kruskal-Wallis test. N1: P = .0303. N3: P = .0271.
In Silico Sensitivity Analysis
In order to estimate the fraction of positive specimens that could potentially yield false-negative results in pooled testing, we subtracted the Ct value shifts estimated in the Passing-Bablock regression analyses (Figure 1 and Table 2) from 40, the upper Ct limit defining a single-positive result. We then tabulated the number of positive results in prevalence groups 1, 2, and 3 that fall in the shifted range. We found that 1.5% (46/3091) positive specimens had N1 Ct values in the range of 40 – shiftN1, but no specimens had N3 Ct values in the range of 40 – shiftN3 (Table 3). Thus, we would predict that in pooled specimen testing 0.61% of pools would yield inconclusive results to be resolved by singlicate testing, while no pools would yield false-negative results due to the shift in Ct values.
Table 3. .
Percentage of Single-Positive Specimens With Ct Values Within the Shifted Rangea
| Prevalence Groupb | No. | Shifted Range (N1) | Shifted Totals (N1) | % Shifted (N1)c | Shifted Range (N3) | Shifted Totals (N3) | % Shifted (N3)c |
|---|---|---|---|---|---|---|---|
| 1 | 820 | 37.0–40 | 22 | 2.7 [1.8–4.0] | 37.5–40 | 0 | 0.0 [0.0–0.69] |
| 2 | 1113 | 38.3–40 | 7 | 0.63 [0.3–1.3] | 38.7–40 | 0 | 0.0 [0.0–0.51] |
| 3 | 1158 | 37.6–40 | 17 | 1.5 [0.9–2.3] | 38.1–40 | 0 | 0.0 [0.0–0.49] |
| All groups combined | 3091 | 46 | 1.5 [1.1–2.0] | 0 | 0.0 [0.0–0.18] |
Abbreviation: Ct, cycle threshold.
aThe shift in N1 and N3 Ct values for pooled testing was calculated from Passing-Bablok regression analysis of pooled and singlicate positive specimens. Shift = (40 – intercept)/slope. The number of single-positive specimens with Ct values in the range of 40 – shiftN1 and 40 – shiftN3 was then tabulated.
bSpecimens were drawn from 3 geographic prevalence groups, as described in the “Methods” section.
cShown as value [95% CI].
We next sought to determine the potential number of all SARS-CoV-2 single positive results between late March and mid-July 2020 tested at Quest Diagnostics Infectious Disease (San Juan Capistrano, CA, USA) that could have yielded a false-negative result in pooled testing. We found only a single positive test result (N1 Ct = 38.3, N3 Ct = 39.3) out of 44 217 positives that would have been reported as negative had pool testing been used, while 613/44 217 (1.4%) positives would have yielded an inconclusive result, necessitating singlicate testing. This finding represents a false-negative rate of 0.002%.
DISCUSSION
We have demonstrated that upper respiratory specimens collected in viral transport media (UTM, UTM-RT), PBS, or saline from low-prevalence populations can be tested in pools of 4 specimens, with comparable results to single-specimen testing. Agreement between pooled and singlicate SARS-CoV-2-positive tests was demonstrated at 100% for 101 positive samples spanning a range of Ct values. Regression analysis showed a strong linear relationship between the Cts for pooled and single positive specimens, with a slope near 1.0, indicating a lack of proportional bias resulting from pooled testing. The decrease in Ct values stemming from pooled testing was near the theoretical estimate (log2(n) where n = pool size) of 2 Cts for a pool size of 4 and did not result in any false-negative results. Pooled testing of 4 previously tested negative specimens demonstrated excellent specificity. Only a single pool out of 247 yielded a positive result for the N1 target near the upper Ct limit of the assay, while the result for the N3 target was negative. Testing of the 247 negative pools would have equated to the reporting of 988 negative results that would not have required individual testing.
Further analysis of >3000 positive specimens demonstrated that, in a dual target RT-PCR system, no positive specimens would have been miscalled due to the use of pooled testing. This finding was also confirmed in a larger data set of >44 000 positive results in which only a single specimen had both N1 and N3 Ct values in the shifted Ct range, corresponding to a false-negative rate of 0.002%. We found that the N3 RT-PCR target provides better sensitivity than the N1 target, with Ct values of 1.1–1.4 units lower on average at the 95th percentile of the Ct value distributions. While <1.5% of the specimens had an N1 Ct value in the shifted Ct range, none of the Ct values for the N3 target were in the shifted Ct range. Interestingly, specimens from the highest-prevalence group (6%–10%) in this population had a slightly lower median Ct for both the N1 and N3 targets than for the 2 lower-prevalence groups. Although this difference was statistically significant, the difference was small, and the Ct distributions largely overlapped between the 3 groups. It is therefore difficult to ascribe any biological or clinical significance to these differences, which may be related to geography or other factors, rather than to prevalence.
Several recent studies have evaluated specimen pooling strategies for SARS-CoV-2 RT-PCR testing. Yelin et al. [10] evaluated pools of 16–64 specimens and found an average increase of 1.24 Cts for each 2-fold dilution. As the Ct values for the positive samples used in the study averaged 24–25, the false-negative rate was low but estimated to be 10% at the highest fold pooling. Ben-Ami et al. [3] evaluated 184 SARS-CoV-2 specimens tested by RT-PCR in singlicate and in pools of 8 specimens and found no loss in diagnostic accuracy. They then applied 8 sample pools to screen >26 000 specimens from asymptomatic health care workers for SARS-CoV-2 and identified 31 positives (0.12%). In this low-prevalence population, a large gain in testing efficiency was achieved. Abdalhamid et al. [4] utilized a pool size of 5 to assess 60 randomly selected specimens drawn from a population with an estimated prevalence of 5%. They identified 2 positive pools that were subsequently tested individually, resulting in a savings of 38 extractions and RT-PCR reactions compared with single-sample testing.
However, as the prevalence of SARS-CoV-2 increases in the tested population, the efficiency of pooled testing will decrease. The current positivity rate for SARS-CoV-2 specimens submitted for laboratory testing as of July 2020 is much higher than that for low-prevalence screening or asymptomatic populations, and the national testing positivity rate is estimated at 7%–8% as of early July 2020 (https://covidtracking.com/). At prevalences between 1% and 3%, an 8-specimen pool will yield greater testing efficiencies than a 4-specimen pool: For example, at 3% prevalence, a 4-specimen pool would require 36 tests per 100 specimens, whereas an 8-specimen pool would require only 34 tests (https://bilder.shinyapps.io/PooledTesting/) [4]. In contrast, at 8% prevalence, the 8-specimen pool would require 61 tests per 100 specimens compared with only 53 tests per 100 specimens for the 4-specimen pool. At a 10% prevalence, at least 40% fewer tests would be run with a 4-specimen pool strategy compared with single-specimen testing (https://bilder.shinyapps.io/PooledTesting/) [4]. The balance between pool size and sensitivity is also a key consideration for pooling strategies. For a pool size of 4, the dilution factor would be 1:4, compared with 1:8 for a pool size of 8. The stated limit of detection (LOD) in the instructions for use (IFU) for the Quest Diagnostics RT-PCR test is viral copies 136 copies/mL. Using a 1:4 pooling dilution, the LOD would then be 544 copies/mL, whereas a 1:8 pooling dilution would increase the LOD to 1088 cp/mL. Given these considerations, a 4-specimen pool is appropriate to maximize testing efficiency while minimizing the loss of sensitivity, in accordance with the recommendations in the FDA guidance document Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/policy-coronavirus-disease-2019-tests-during-public-health-emergency-revised).
Our study had several limitations. First, this study utilized previously tested samples that had been stored frozen and then thawed for retesting in the study. In a previous study in our laboratory [14], we demonstrated that SARS-CoV-2 specimens in a variety of transport media remain stable while stored under refrigerated or frozen conditions. However, in the current study, specimens were stored frozen for longer periods than previously assessed. The greatest impact would have been to increase Ct values for high-Ct positive specimens. Nevertheless, we had no false-negative results in this study. As pooled testing is intended for use on freshly obtained specimens under the same conditions used for single-specimen testing, retesting of frozen specimens is unlikely to affect pooled testing.
Second, not all transport media or specimen types were evaluated in this pooling study. For example, specimens collected in COPAN Eswab were excluded from the pooling studies. Internal stability studies with specimens spiked in COPAN Eswab have demonstrated reduced stability (loss of >3 Cts) following increased freeze-thaw cycles (≥2 times; data not shown). Given that this study utilized archived specimens, the optimal conditions for such media may not have been utilized. A variety of collection media, including ESwab, are approved for CoV testing using commercially available assays. Pooling of specimens in the study was not limited to the use of like media for a pool of 4 specimens; however, the media pooled were limited to UTM, PBS, and saline. As PBS and saline are commonly used as a diluent, the impact of the pooling of the different transport media was likely minimal. Further work is needed with prospective specimens before additional media types can be included in a pooling protocol.
Third, no clinical information was available for the patient specimens that were utilized in this study. Thus we were not able to correlate the SARS-CoV-2 test result with clinical diagnoses of COVID-19, stage of disease, or presence of symptoms. However, given the results we have presented here for pooled testing as well as our retrospective analysis of patient Ct values, pooling of 4 samples is unlikely to have a significant impact on clinical management. Patients undergoing testing soon after exposure, asymptomatic patients being screened for COVID-19, and positive patients who never go on to develop symptoms may have lower SARS-CoV-2 viral loads that could theoretically impact the clinical accuracy of pooling. In a follow-up retrospective analysis (manuscript in preparation), we found that although SARS-CoV-2-positive patients in some of these categories do have higher Ct values, only a very small fraction (<0.15%) of these Cts would fall above the shifted Ct value for the more sensitive N3 RT-PCR target.
Finally, it is important to stress that the implementation of pooled testing in a high-throughput laboratory adds additional operational challenges and complexity to the testing process. It requires the validation of automated liquid handling processes to pool the specimens for testing as well as robust support from one’s information technology department to enable laboratory information system (LIS) reporting of pooled results to the LIS. These considerations need to be addressed before implementing a pooled testing approach in the laboratory.
In summary, we have demonstrated that pooled testing of 4 upper respiratory specimens from populations with a prevalence ≤10% with a sensitive dual-target RT-PCR test for SARS-CoV-2 is highly correlated to single-specimen testing and does not generate false-negative test results. This pooling strategy can improve testing capacity while reducing reagent and supply utilization and therefore afford better access and a more rapid turnaround time for patients in need of testing and help combat the SAR-CoV-2 pandemic.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Author contributions. G.A.B.: data curation, formal analysis, investigation, resources, validation, writing—review & editing. R.M.K.: conceptualization, data duration, formal analysis, investigation, methodology, project administration, resources, software, validation, visualization, writing—original draft, review & editing. R.E.B.: data curation, formal analysis, resources, writing—review & editing. B.M.F.: data curation, software, writing—review & editing. L.U.: investigation, resources, validation, writing—review & editing. H.-R.L.: investigation, resources, validation, writing—review & editing. H.W.K.: conceptualization, data curation, writing—review & editing. N.J.K.: conceptualization, formal analysis, funding acquisition, methodology, supervision, writing—review & editing. E.M.M.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, supervision, validation, visualization, writing—original draft, review & editing.
Financial support. This work was supported by Quest Diagnostics Incorporated (Secaucus, NJ, USA).
Potential conflicts of interest. All authors of this manuscript are employees of Quest Diagnostics Incorporated (Secaucus, NJ, USA), a diagnostics laboratory that provides SARS-CoV-2 testing. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Dorfman R. The detection of defective members of large populations. Ann Math Stat 1943; 14:436–40. [Google Scholar]
- 2. Bilder CR, Tebbs JM. Pooled-testing procedures for screening high volume clinical specimens in heterogeneous populations. Stat Med 2012; 31:3261–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ben-Ami R, Klochendler A, Seidel M, et al. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin Microbiol Infect 2020; 26:1248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abdalhamid B, Bilder CR, McCutchen EL, et al. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol 2020; 153:715–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Emmanuel JC, Bassett MT, Smith HJ, Jacobs JA. Pooling of sera for human immunodeficiency virus (HIV) testing: an economical method for use in developing countries. J Clin Pathol 1988; 41:582–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mine H, Emura H, Miyamoto M, et al. ; Japanese Red Cross NAT Research Group High throughput screening of 16 million serologically negative blood donors for hepatitis B virus, hepatitis C virus and human immunodeficiency virus type-1 by nucleic acid amplification testing with specific and sensitive multiplex reagent in Japan. J Virol Methods 2003; 112:145–51. [DOI] [PubMed] [Google Scholar]
- 7. Edouard S, Prudent E, Gautret P, et al. Cost-effective pooling of DNA from nasopharyngeal swab samples for large-scale detection of bacteria by real-time PCR. J Clin Microbiol 2015; 53:1002–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van TT, Miller J, Warshauer DM, et al. Pooling nasopharyngeal/throat swab specimens to increase testing capacity for influenza viruses by PCR. J Clin Microbiol 2012; 50:891–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lohse S, Pfuhl T, Berkó-Göttel B, et al. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yelin I, Aharony N, Shaer Tamar E, et al. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta E, Padhi A, Khodare A, et al. Pooled RNA sample reverse transcriptase real time PCR assay for SARS CoV-2 infection: a reliable, faster and economical method. PLoS One 2020; 15:e0236859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Passing H, Bablok W. A new biometrical procedure for testing the equality of measurements from two different analytical methods. Application of linear regression procedures for method comparison studies in clinical chemistry, part I. J Clin Chem Clin Biochem 1983; 21:709–20. [DOI] [PubMed] [Google Scholar]
- 13. Passing H, Bablok W. Comparison of several regression procedures for method comparison studies and determination of sample sizes. Application of linear regression procedures for method comparison studies in clinical chemistry, part II. J Clin Chem Clin Biochem 1984; 22:431–45. [DOI] [PubMed] [Google Scholar]
- 14. Rogers AA, Baumann RE, Borillo GA, et al. Evaluation of transport media and specimen transport conditions for the detection of SARS-CoV-2 using real time reverse transcription PCR. J Clin Microbiol 2020; doi: 10.1128/jcm.00708-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.