Abstract
The technologies used for coronavirus testing consist of a pre-existing device developed to examine different pathologies, such as bacterial infections, or cancer biomarkers. However, for the 2019 pandemic, researchers knew that their technology could be modified to detect a low viral load at an early stage. Today, countries around the world are working to control the new coronavirus disease (n-SARS-CoV-2). From this perspective, laboratories, universities, and companies around the world have embarked on a race to develop and produce much-needed test kits. This review has been developed to provide an overview of current trends and strategies in n-SARS-CoV-2 diagnostics based on traditional and new emerging assessment technologies, to continuous innovation. It focuses on recent trends in biosensors to build a fast, reliable, more sensitive, accessible, user-friendly system and easily adaptable technology n-SARS-CoV-2 detection and monitoring. On the whole, we have addressed and identified research evidence supporting the use of biosensors on the premise that screening people for n-SARS-CoV-2 is the best way to contain its spread.
Keywords: COVID-19, n-SARS-CoV-2, Emerging technologies, Industry 4.0, Photothermal biosensor, Graphene-based biosensors, Smartphone sensors
1. Introduction
The current outbreak of new coronavirus 2019 (n-SARS-CoV-2), which was first reported in Wuhan, China, on 31 December 2019, has been declared by the World Health Organization (WHO) as an international health emergency because of the severity of the progression of the infection, which has resulted in the loss of more than 876, 616 people as of September 08, 2020 [1]. This n-SARS-CoV-2 epidemic is becoming more serious due to its continued global spread and the lack of appropriate treatment and diagnostic systems. In order to manage the n-CoV-2 SARS epidemic, international health agencies are making serious efforts to explore various aspects of therapy development and are paying particular attention to research on the need for intelligent diagnostic tools for rapid and selective detection of the n-SARS-CoV-2 protein.
The ability to diagnose the disease quickly and accurately has been one of the major failures of the health system in all countries. These failures are due to the limited number of test kits available, the limited number of certified testing facilities and the time required to obtain a result and providing information to the patient. In general, there are two types of techniques used to identify and quantify pathogens in practice: immunoassays and DNA-based tests [2]. A range of diagnostic methods to detect and identify infectious diseases are currently available, including direct microscopic examination, isolation of pathogens in culture, serological antibody response tests, and nucleic acid tests such as polymerase chain reaction (PCR) [3], [4], [5], [6], [7], [8], [9], [10], [11]. PCR has proven to be effective for the diagnosis of many microorganisms. Because of its incredible sensitivity, specificity, reproducibility, wide dynamic range and amplification speed, PCR has been advocated by infectious disease experts to identify organisms that cannot be cultured in vitro, or in cases where existing culture techniques are insensitive and/or require extended incubation times [12]. With traditional diagnostic methods, laboratory instruments are usually handled by qualified personnel in a centrally located laboratory facility as well as the time uncertainty caused by delays in testing, vacations, and other planned laboratory closures [13]. Taking into account these limitations and the need for real-time continuous monitoring capabilities among the various applications there is a need to examine other bioanalytical techniques [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]. For the design and development of miniaturized detection systems, it will be useful to introduce nanotechnologies and advanced system packaging, which are recommended for point-of-care (POC) diagnostics.
Overall, the n-SARS-CoV-2 epidemic can be managed through intelligent detection and diagnosis even in personalized medicine. For currently available diagnostic methods, biosensing technology offers several advantages, including the ability to make highly sensitive and instantaneous measurements using small amounts of analytes [24], [25], [26], [27], [28], [29].
In order to assist ongoing innovation, this article review has been developed to provide an overview of current n-SARS-CoV-2 diagnostic trends and strategies based on conventional and novel methodologies, including biosensing methodologies. It includes recent advancements in transduction system; nanotechnology and genetic engineering offer various strategies to improve the detection performance of biosensor as well as data on n-SARS-CoV-2 diagnostic trends.
2. Diagnostic approaches to n-SARS-CoV-2: Emerging techniques
The most commonly used coronavirus tests are the WHO-recommended nucleic acid amplification tests (NAAT) or real-time reverse transcription-polymerase chain reaction (RT-PCR), which detect the SARS-CoV-2 responsible for the n-SARS-CoV-2 disease. To identify viral nucleic acid, sequencing of the entire genome is therefore considered to be one of the most comprehensive approaches. In this method, the first reverse transcribes SARS-CoV-2 RNA into cDNA and then uses target-specific primers to amplify specific gene fragments in the cDNA [30]. While the technique was good and did not warrant any improvements, it was “just not available enough”. This can take up to 3–4 h, while results are available to patients within a few days in most countries. The fluorescence signal, which represents the copy number of the target sequence, can be easily detected during the amplification process. Five typical open reading frames (ORFs) have been identified in the SARS-CoV-2 genome [31], [32].
Another technique is the gene sequencing (GSN) method using the SmartXGene. Over the future, SmartXGene intends to introduce automated software to analyze the sequencing and impact of copy variants on a single 16S rRNA genome. In order to minimize the proportion of negative results, it is recommended to further test respiratory tract samples in highly suspect cases and to verify the quality of the sample. Many other options for diagnostic testing of n-SARS-CoV-2 are available; these are (i) Antigenic assays that allow the detection of specific proteins of SARS-CoV-2. These tests can be performed on nasopharyngeal swabs, lower respiratory tract samples. These tests allow the early diagnosis of the disease from the acute phase. However, due to their low performance, particularly in the case of low viral load, these antigenic tests are not currently recommended for clinical use in the context of n-SARS-CoV-2 [33]. (ii) Serological tests that allow the detection of specific antibodies (Ac) (immunoglobulin’s: Ig) produced by the organism and directed against SARS-CoV-2. These tests are performed on blood samples and may be useful in identifying patients who have developed immunity to SARS-CoV-2 whether they are symptomatic or not. As a corollary, serological testing could, under certain circumstances, identify patients who are or have been infected with SARS-CoV-2. Finally, these tests could also be useful in the collection of epidemiological data related to n-SARS-CoV-2 (patients actually infected, mortality rates, etc.). However, the relevance of the use of these tests in clinical practice depends on the prior availability of pathophysiological, technical, and clinical knowledge enabling their evaluation and validation. Some diagnostic technologies that have shown clinical feasibility in detecting SARS-CoV-2 are provided in Table 1 [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44].
Table 1.
Emerging diagnostics being developed for SARS-CoV-2.
| Technologies | Biomarker | Platform | Functioning | Types of clinical sample | References |
|---|---|---|---|---|---|
| RPA | Nucleic acid | CRISPR | PCR, perform CRISPR/Ca9-mediated lateral flow nucleic assay (CASLFA) | Serum | [34] |
| RT-RPA | Nucleic acid | CRISPR | RPA, SHERLOCK multiplexed signal detection via fluorescence | nasopharyngealswabs | [35] |
| LAMP | Nucleic acid | LAMP | isothermal DNA synthesis using self-recurring strand displacement reactions; positive detection leads to increased sample turbidity | throat swabs | [36] |
| RPA | Nucleic acid | RPA | forward and reverse primars blind to DNA and amplify strands at 37 °C | fecal and nasalswabs | [37] |
| RT-NASBA | Nucleic acid | NASBA | transcription-based amplification for RNA targets | nasal swabs | [38] |
| Rolling circle amplification | Nucleic acid | RCA | DNA polymerase used to extend a circular primer and repeatedly replicate the sequence | Serum | [39] |
| LAMP | Nucleic acid | RT-LAMP | reverse transcriptase LAMP reaction for RNA targets | nasopharyngealaspirates | [40] |
| ELISA | Protein | ELISA | enzymatic reaction to produce colored product in presence of target | Serum | [41] |
| Digital ELISA | Protein | SIMOA | digital readout of colored product by enzymatic reaction in presence of target | Serum | [42] |
| DNA-assisted immunoassay | Protein | Biobarcode assay | protein signal is indirectly detected by amplifying DNA conjugated to gold nanoparticle | Serum | [43] |
| Lateral flow | Protein | Rapid antigen test | gold-coated antibodies produce colorimetric signal on paper in presence of target | Serum | [44] |
Abbreviations:
RPA, Recombinase polymerase amplification; RT-RPA, reverse transcription recombinase polymerase amplification; LAMP, loop-mediated isothermal amplification; RT-LAMP, reverse transcription LAMP; ELISA, enzyme-linked immunosorbent assay; CRISPR, clustered regularly interspaced short palindromic repeats; RT-NASBA, reverse transcription-nucleic acid sequence-based amplification; SIMOA, Single molecule array.
3. Industry 4.0
Fourth technologies revolution also known as industry 4.0 consists of advanced manufacturing and information technologies, in order to meet the specific needs of different areas of the human being in a shorter period of time. They provide effective means to deal with the speed, scale, and impact of the n-SARS-CoV-2 pandemic [45], [46], [47]. However, the global distribution of these technologies is far from uniform, posing a significant disadvantage to lagging countries and their vulnerable populations in their ability to reduce risk and slow disease transmission. To facilitate contact tracing, verification of symptoms, and prediction of epidemics and vulnerabilities, artificial intelligence (AI) and mobile technology offer data collection tools [48], [49]. To identify patients with severe symptoms of the disease, HealthCare collaborators, for example, created an n-SARS-CoV-2 screening tool based on AI, and the Canadian Blue Dot model issued an early warning on December 31, 2019, indicating that the new coronavirus would spread around the world [50], [51].
The use of AI-based computer vision cameras is used to assess whether social distance is maintained in public places, and alarmingly, AI-based thermal cameras have been used to scan public spaces and identify potentially sick people. According to accounts, Blue Dot, a group of researchers has drawn up a list of the 20 main destination cities where Wuhan passengers would arrive in the wake of the epidemic. They warned that these cities could be at the forefront of the global spread of the disease. Furthermore, several important technologies in Industry 4.0 that can help in cases of n-SARS-CoV-2 epidemics including IOT, Big Data, virtual reality, holography, cloud computing, autonomous robot, 3D scanning [52], [53], [54], [55], [56], [57], [58].
4. Nanoscale virus biosensors
In the field of virus biosensing, new developments have been made, including fluorescence [59], light scattering [60], [61], surface-enhanced raman scattering (SERS) [62], [63], electrochemistry [64], [65], microcantilevers [66], and surface plasmon resonance (SPR) [67], [68]. Biosensor refers to an analytical device that produces a quantifiable signal proportional to the concentration of an analyte. They consist of a transducer and biologically active elements or materials, such as nucleic acids, enzymes, and antibodies, which can detect analytes by specific interactions [69]. By using the appropriate combination of biometric system and transducer, the biosensor reacts with the analyte present in the sample, thus indicating the concentration of the analyte in the electronic signal. With the advancement in scientific and technological progress, these devices will play highly important roles in all sectors of human Endeavour. In particular, biosensors will provide the basis for cheap, simple, sensitive, specific, and time-consuming equipment that can be used to find chemical information, the presence of analytes, and provide sophisticated analytical techniques for non-professionals and the public. It is important to quickly develop new market opportunities in this sector.
The detection of viral diseases by biosensors must be rapid, highly accurate, and sensitive [70]. A higher affinity, selectivity, and specificity can determine the success or failure of all detection technologies, which is essential for the design and operation of biosensors. This makes it difficult to estimate which biorecognition is used by a given target pathogen [70], [71]. The two main approaches to biorecognition are the detection of viral nucleic acid (NA) sequences [64], [70], [72] and the detection of specific viral biomolecules, such as protein/surface antigen [73], [74], [75]. The specificity and sensitivity of nanotechnology-based biosensors are high after being labeled with NA probes, antibodies, or other specific molecules with affinity to the target structures [76].
Biosensors based on NA have always been a hot topic and have provided broad perspectives for clinical diagnosis [77]. Overall, NA-based tests are more specific and sensitive than immunological tests, which are faster and more reliable [78]. The signal can be generated or amplified using various labeling molecules, such as newly used electroactive substances, fluorophores, radioisotopes, enzymes, or haptens (antibodies can be used) [70]. The development of the market for antibody-based diagnostic methods requires a new, rapid, and accurate immunodiagnostic method. Reagent-free biosensor“ strategies based on antibodies and natural or artificial binding proteins have been described until now [79]. The nucleic acid test is usually performed by detecting the presence of the virus in the patient's sputum (or saliva) or nasal secretions (glanders) [80]. Alternatively, in the antibody test strip, blood samples are collected from patients that contain antibodies to the virus [81]. These antibodies can be detected in patient plasma, serum, or whole blood.
Real-time biosensors detect analytes of interest almost continuously and play an important role in the efficient generation and processing of data, supporting real-time decision making and rapid operation [82]. Therefore, much current research in this field focuses on integrating real-time measurement with on-chip sensing functions to build multifunctional nanosensors [83].
4.1. Optical biosensors
Virus detection is essential for the pharmaceutical industry, disease prognosis, and surveillance. Developments in biosensor technologies can provide POC diagnostics of various detection strategies, including optical biosensors [84]. Optical techniques are highly sensitive and can detect even a single molecule, but require the fluorophore molecule to be attached to the target [85], [86]. Various modes of optical measurement exist (i.e., absorption; reflection; fluorescence; chemiluminescence; and phosphorescence) [87]. In the field of virus detection, biosensors-based SPR and fluorescence are the most common and promising methods. Similarly, the latest developments in fiber optic technology indicate that in the near future, optical biosensors could become powerful tools for real-time and remote virus detection [88], [89], [90]. SPR is a form of reflectance spectroscopy and has been widely used in the development of biosensors. It has been shown to play an important role in immunogenicity, food analysis, proteomics, drug discovery, and DNA analysis [91], [92], [93]. This method is particularly attractive for direct label-free detection.
Using nanotechnology, a variety of useful methods have been developed to improve the performance of SPR biosensing tests. For example, by using metallic nanoparticles, the sensitivity of the SPR can be greatly improved, thereby improving the plasmon field and increasing the contrast of the index [94]. The term “Localized SPR (LSPR)” is technically used in the context of nanoscale structures. According to Mie theory, electrons cannot move within the internal framework of nano-sized metal particles, and this oscillation of the collective charge density is called LSPR [95].
More recently, intending to detect the n-SARS-CoV-2 in the air, scientists at the Swiss Federal Laboratory have developed a sensor for fast and reliable monitoring [19]. For clinical diagnosis, these sensors can be used as an alternative method for real-time measurement of virus concentration in the air. Using plasma photothermal effect (PPT) and local surface plasmon resonance (LSPR) using two-dimensional gold nano-islands (AuNI), they can functionalize complementary DNA receptors, which can treat SARS-CoV-2. The receptors on the sensor are complementary sequences to the coronaviruses’ unique RNA sequences to help reliably identify the virus. Researchers plan to further develop the sensor to measure the concentration of coronavirus in the air. So far, that system the researchers plan to develop involves sucking in air, concentrating aerosols and releasing RNA from viruses [19]. Although the sensor is faster and more accurate than RT-PCR, it cannot eliminate barriers to exposure to viral particles, and if the viral strain mutates and changes morphology, it lacks long-term specificity. In another work, Yu et al. reported a label-free LSPR detection system based on nanoisland to detect adenovirus [96]. The nanoisland platform can greatly improve LD and sensitivity and can locate and excite SP and control the associated evanescent near field. However, nano-island synthesis has produced random and uncontrollable patterns, of which only certain models can produce better results [96]. Protein genetic fusion with metal-binding properties offers an alternative approach for the direct interaction between the protein and the gold surface, in addition to the classical gold-thiol method.
One group recently integrated a digital amplification process into a biosensor based on a sample-response chip to quantify nucleic acids, further simplifying the entire nucleic acid analysis process [97]. After adding the sample, the cells are lysed and the nucleic acid is bound to the magnetic beads. Once the washing and elution steps are complete, the vacuum self-priming method is used for isothermal amplification and then the fluorescence imaging is passively drawn into the digital RPA region. Since the fluorescence probe is labeled with UV-detectable carboxyfluorescein (FAM), the intensity of the fluorescence signal is proportional to the concentration of amplicons. Within 45 min, the biosensor could perform three main nucleic acid testing steps without the need for complex instrumentation and control systems. It holds great promise for the rapid and accurate detection of n-SARS-CoV-2 in patient samples.
As a new alternative, based on a colorimetric detection method using a paper-based 3D microfluidic biosensor, the application of fuchsin in the detection of DNA amplicon was studied [98]. Without DNA amplicons, the addition of the sodium sulfite molecule and fushsine produces the colorless fuchsine leucosulphonic acid or leucofushsine. Furthermore, in the presence of DNA amplicons, a bond between the aldehyde groups of the DNA and the sulfonates groups is disrupted and the bond between the hydrogen sulfite and the central C atom is produced, producing fuchsin with a chromophoric structure, which appears to be violet signals [99] . The colorimetric signal generated by this biosensor is simple and can be detected with the naked eye without the need for an external reader, showing hope for rapid diagnosis of n-SRAS CoV-2 infections on the POC.
In the modern era, rapid development of DNA technology has provided a feasible route to creating nanoscale materials. The field of DNA-based photonic structures is in its initial discovery stage and this means that its potential is still wide open. Researchers continue to focus primarily on demonstrating proof-of-principle research, followed by the formulation of optimized design principles and methods. As a result, it is not clear which of the above areas of application will be truly expanded and developed or will have even greater academic interest. From a material point of view, the main challenge comes from the nature of the DNA structure itself and all the optically active elements that need to be assembled on or around the DNA structure. A fundamental understanding of these optical processes will help drive development of next-generation photonic nanomaterials.
More recently, Jiao et al., reported a DNA-nanoscaffold hybrid chain reaction-based nucleic acid as simple and specific for the assay of SARS-CoV-2 RNA [27]. The nanoscaffold was constructed by hybridizing the DNA hairpin probes (H1) with long strand. Then, the SARS will initiate the hybridization oh H1 and free H2 DNA probes along the nanoscaffold and illuminated DNA nanostring in instantly obtained. With the help of nanoscaffold, the fluorescence intensity is much stronger in the presence of free H1 and H2. The developed methodology present low reaction time to without 10 min and high signal in wide temperature range of 15–35 °C. However, the output of the fluorescent signal requires the assistance of the equipment. So further effort need to be made in point of care testing [27].
4.2. Graphene/Black phosphorus based biosensors
Since mid-2004, graphene has been one of the most attractive materials [100]. From band structure standpoint, graphene is a zero band gap material. The electrons and the holes it can easily generate have a positive and negative electric field (i.e. electric doping) respectively. This is called an ambipolar characteristic, in which both the positive and negative gate voltages generate a drain current. The ambipolar behavior can also be generated by a change in solution tension through the top gate. Graphene is considered as a material for the next generation of semiconductor devices for its special properties, linked in particular to its high electron/hole mobility, its transparency and mechanical resistance, etc. The development of new multi-purpose textile fabrics has attracted a great deal of interest in recent years. In polymers or textiles, incorporated graphene or graphene derivatives can improve the properties of fabrics for specific applications.
Graphene could be used to make better masks, gloves and gowns for medical teams. The face mask sector is hot right now. The new high-tech masks designed to kill the virus, thanks to antiviral nanoparticles embedded in the protective material, are even more effective than standard masks [101]. To meet the demand, ZEN Graphene Solutions Co, Ltd. (TSXV: ZEN) and Graphene Composites Ltd. (GC) have combined their expertise to produce new high-tech masks and other protective garments that can damage viruses through the development of virucidal graphene-based composite ink [102]. They reported that the face masks not only block the virus but can kill it.
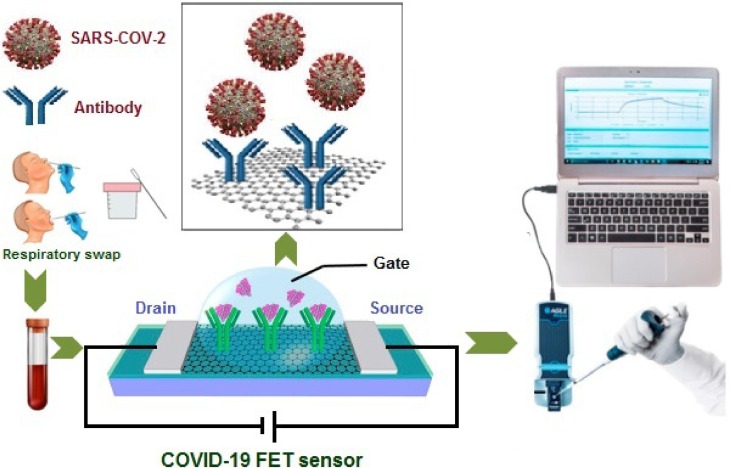
In biosensors, the graphene can be used as a means of identifying and capturing biomarkers: biomolecules commonly found in serum, saliva, and tissue can provide information on disease conditions and stages [103], [104]. Extensive research has been carried out on graphene field-effect transistors (gFETs) for biosensor applications, using different geometries and triggering methods. Biosensors with gFETs integrated between bioreceptors and ion-sensitive FETs have emerged as the most developed candidates. Indeed, about half a century after the invention of the first generation of FETs by Bergveld in 1970, these transducers have evolved into many biosensor applications [105]. In a typical FET system, due to the high specificity and binding affinity, the sensing element is attached to the sensing channel (semiconductor path), which is connected to the source (S) and drain (D) electrodes to capture the target [106]. Edmond Changkyun Park, Seung Il Kim and their colleagues developed a gFET biosensor that detects SARS-CoV-2 in nasopharyngeal swabs from patients with n-SARS-CoV-2 in less than a minute [17]. This sensor was produced by coating the gate of the transistor made up of graphene sheets, with an antibody that was specific against SARS-CoV-2 spike protein (see Fig. 1 ).
Fig. 1.
Schematic representation of the operating procedure of the n-SARS-CoV-2 FET sensor.
The assays were performed on a FET based- graphene sheet with high electronic conductivity. The attachment to the antibody results in a change in electrical current when they add either the purified spike protein or the cultured SARS-CoV-2 virus to the sensor. The FET sensor clearly distinguishes between patient and normal specimens and the assay is approximately 2 to 4 times less sensitive than RT-PCR. The device showed excellent semiconductor characteristics and could be used to detect SARS-CoV-2 down to 1 fg/mL in phosphate-buffered saline and 100 fg/mL clinical transport medium. The experimental results demonstrated that the FET biosensor could be a candidate for rapidly screening n-SARS-CoV-2 patients in early stages of the disease, with extra benefits of low-cost and ease of use. However, it lacks the specificity in longer terms if viral strain gets mutated and changes its morphology [17].
The assistance of nanotechnology (such as graphene) and organic materials in FET biosensors can reduce the cost factor, and facilitate the production of commercial miniaturized devices. Point-of-care diagnostics can soon create a revolution in healthcare if solutions related to the commercial cost and reliability factor of FET biosensors are adopted in a practical and widespread manner.
Black phosphorus (BP) or phosphorus-based biosensors have also been widely explored for medical diagnosis, as has graphene [107], [108], [109]. BP has a pleated lattice structure along the armchair direction and a double-layer structure in the zigzag direction. It is covalently bound in the plane and the weak van der Waals interactions between the two layers. Due to the unique structure and in-plane anisotropy of the orthorhombic folds, it exhibits a higher surface-to-volume ratio, extremely high hole mobility and higher molecular adsorption energy than other 2D materials [110], [111]. The properties of BP and photoelectric applications have been extensively studied, but little attention has been paid to its potential biomedical applications. This may be mainly due to the lack of biocompatibility of BP, i.e. when exposed to a biological environment containing small biomolecules and low concentrations [112], [113], [114], [115].
The development of an electrochemical biosensor with an aptamer-functionalized BP nanostructured electrode has been reported in a recent study [114]. BP is coated with poly-L-lysine which allows the functionalization of BP with anti-Ab-aptamers. A coulombic interaction between the aptamers and the PLL allows immobilization of the aptamers on the nanosheets. The presence of target antigens causes the direct oxidation of iron (ii) to iron (iii) at the electrode surface by the electron transfer mechanism. BP-based biosensors show a higher detection sensitivity and specificity, achieving the detection limit down to pg level. The proposed biosensor technology has the potential for highly sensitive detection against coronaviruses in patient blood samples.
4.3. Smartphone-embedded sensors
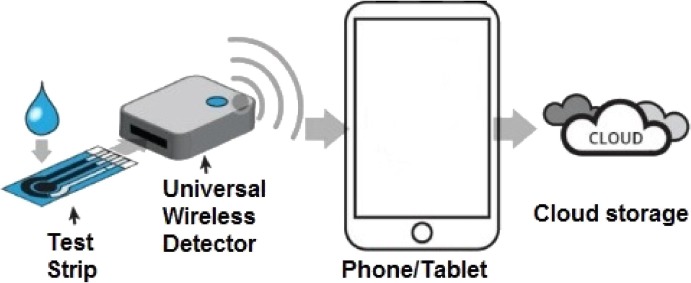
Modern smartphones incorporating multitasking operating systems, high-performance microprocessors, a rich set of sensors, and wireless and wired communication technologies are very popular and widely distributed around the world. For the development of low-cost detection systems, these are ideal devices, especially for low-income developing countries and rural areas that do not have access to diagnostic laboratories and expensive instruments. Various elements need to be considered when designing a smartphone-based detection system, such as sensor performance, acquisition rate, and privacy strategies when personal data is to be shared in the cloud (Fig. 2 ).
Fig. 2.
Peripheral biosensors that share analysis in the cloud.
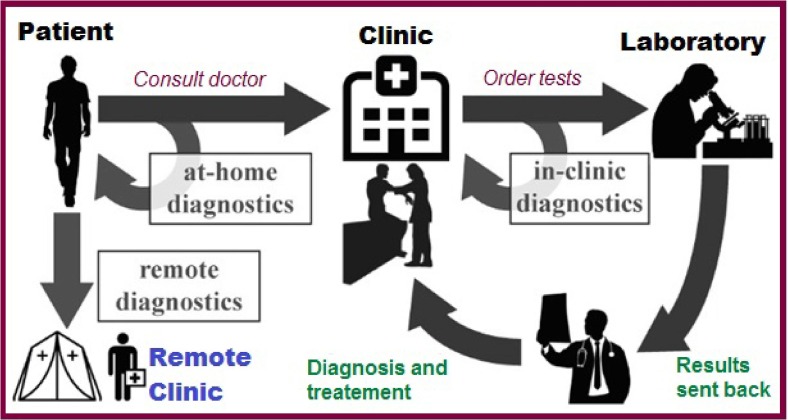
Smartphones are increasingly being used to detect clinically relevant analytes [116]. In particular, they are used to detect human pathogenic cells for the diagnosis of cancer and tuberculosis [117]. Recently, the development of POC instruments has made it possible to perform many tests at the point of need, outside the laboratory. However, the exponentially increasing performance of complementary metal-oxidesemiconductor photocameras may make smartphones more suitable for use as biosensors and portable analysis devices [118]. In most cases, smartphones do not work alone as laboratory instruments. They are complemented by other accessories. These improved devices have great potential as platforms for healthcare in the workplace [119], [120] bringing diagnostic tools closer to the patient and providing faster and more frequent feedback loops [121], [122] (Fig. 3 ).
Fig. 3.
Proposal of a modern health system concept complemented by POC biosensors.
Throughout the n-SARS-CoV-2 crisis, smartphones and smart devices have already played an important role. To assess the possibility of n-SARS-CoV-2 infection, researchers and governments have used them variously: by tracking people's movements (to find out who they have come into contact with), by measuring blood pressure, or even by helping doctors identify potential n-SARS-CoV-2 patients through voice recognition. Some require adding sensors not incorporated into the device, others require environments previously prepared with different equipment types, while others only use the device’s embedded hardware, but the results do not have the required precision.
Recently, Sun et al. developed a smartphone-based system for the rapid, multiplexed detection of specific nucleic acids in five pathogens that can cause equine respiratory infectious diseases [13]. For the detection of pathogenic DNAs, the system has been used and can be easily adapted for the detection of RNA viruses using an isothermal, one-step, RT-loop-mediated amplification (LAMP) protocol that adds reverse transcriptase to the LAMP reaction mixture without changing the buffer or reaction conditions. Coinfection diagnosis is one of the most advantages of this sensor, is able to detect one or more specific targets simultaneously. For high concentrations, LAMP reactions take less than 30 min and the entire detection process can be completed within one hour using inexpensive and portable equipment, allowing veterinarians or physicians to diagnose infections at the point of care and report outbreaks remotely for effective outbreak surveillance. The sensor works by detecting viruses on surfaces using a swab test. Another work by Tabib-Azar and Subhashish Dolai reported a self-test for n-SARS-CoV-2 via smartphone sensor by using swab [123]. The main idea was to enable people to have their sensors to detect Zika in places that they travel and are now being reinvented to test for n-SARS-CoV-2 instead. This alternative method would enable quick and cheap testing, while also providing to be significantly more comfortable. It's about an inch wide and connects to a host device via Bluetooth and draws power from a smartphone charging port. There is also a mobile application that asks you to deposit a saliva sample to read it. The results would then be displayed on a cell phone within 60 s. The sensor is reusable. By producing a small electric current, it is possible to destroy the previous sample by heating it and removing or disintegrating the virus.
Overall, most diagnostic methods require blood or plasma, and even a simple lancet device for blood or plasma collection is invasive and not entirely suitable. This limits the spread of these biosensor formats. Some analytes have been measured in saliva and sweat, as the collection of these body fluids is easier and less stressful for patients. However, there are disadvantages to using these fluids. In fact, the concentration of several analytes in saliva is lower than the concentration in blood (at least 10 to 100) [124]. Therefore, the analytical method used must be highly sensitive. It should also be noted that salivary concentrations of many analytes do not reflect plasma concentrations, making it difficult to correlate analytical data with relevant diagnostic information. Different methods have been used to study biosensors based on smartphones. The ideal biosensor format fully integrates the biometric process with the sensor (smartphone), providing a stand-alone biosensor. This may be the best solution. However, for obvious technical reasons, less exploration has been done.
4.4. Other biosensors
Viral Imprint Technology (VIT) represents another viable option, which involves the preparation of a synthetic polymer matrix to capture viral particles via Molecular Imprint Polymers (MIPs). Depending on the target, the impression process is performed in the presence of the target in the form of a whole viral particle, a single protein, or a single epitope, to generate a specific receptor/binding site with a high affinity for the target virus [125], [126], [127]. For the electrochemical detection of Zika virus, VIT has been used using surface-printed polymers and graphene oxide composites [128]. It has also been used for the detection of human adenovirus (AdV), considered as a model virus facilitating the development and application of rapid quantification of the virus [129]. Moreover, Through the use of VIT, it was possible to differentiate between the different subtypes of human seasonal influenza (influenza A virus) by integrating MIPs on a quartz microbalance transducer (QCM).
In another work, a colloidal gold-immunochromatographic assay (GICA) was developed against SARS-CoV-2 by Li et al. [130]. Nevertheless, since this test is based on GICA technology, it is very fast, easy to use and easy to read. Such a fast and simple point-of-care lateral flow immunoassay has been developed to detect human IgM and IgG antibodies to the SARS-CoV-2 virus in the blood. The detection of color was performed with a mixture of gold nanoparticles (AuNP) conjugated to recombinant S-protein antigen and AuNP-rabbit-IgG. In addition to its use as a diagnostic tool, the GICA test could also permit low-cost monitoring of populations. In fact, this technique was used on SARS-CoV-2 patients, symptomatic or asymptomatic cases, and the observed sensitivity and specificity were 88.66% and 90.63%, respectively. The results of this serological test can then be combined with RT-PCR to improve diagnostic efficiency, even if the detection limit of the test has not yet been estimated [130].
Alternatively, textile-based biosensors such as thread- based, cloth-based, or fabric-based biosensors have been developed with simple manufacturing processes and improved test performance. For example, work has been done to incorporate hydrophobically adjustable polysiloxanes into tissue-based biosensors to delay fluid flow in lateral flow tests and improve detection sensitivity [131]. Similar to conventional lateral flow test strip, the fluidic delay in thread increases interactions between gold nanoparticles-antibodies (AuNP-Ab) and targets. The increased number of interactions, under optimal conditions, produces more AuNP-Abtarget complexes, showing ten times greater sensitivity than unmodified biosensors. This biosensor is simple to manufacture and highly sensitive, revealing immense potential for the detection of IgG and IgM in patients treated with n-SARS-CoV-2 for appropriate health monitoring.
Trinh et al. developed a fully integrated foldable biosensor encapsulated in agarose for long-term reagent storage and multiple fluorescence detection [132]. To prevent evaporation of the sample, the sample is placed in the reaction area, sealed with an adhesive sealing film and placed on a portable heater for amplification. The amplification process is followed by the removal of the sealing film and the detection zone, followed by folding and dipping in the reaction zone. UV light is used to visualize the reaction between the amplicons and silver ions. The test zone became browner in color as the concentration of amplicons increased. The biosensor is simple and user-friendly and should detect n-SARS-CoV-2 nucleic acids in patient samples.
5. Conclusion
The detection and reporting of infectious pathogens in a rapid, sensitive, and specific manner is important for patient management and outbreak surveillance. With their ability to diagnose in real-time with high specificity from a low-concentration sample, biosensors are much more reliable than the rapid test for the detection of coronaviruses. The use of nanobiosensors has been regarded as the more advantageous approach for detecting new coronavirus disease n-SARS-CoV-2. The cost-effectiveness, simplicity, rapidity, and portability of these biosensors play a crucial role in POC applications.
Future work should include specificity improvement or combination with other tests such as rapid nucleic acid tests to further confirm the test result. Meanwhile, current work has also attempted to improve the detection sensitivity, simplicity, and performance of biosensors. Enhanced enzyme signaling, sample concentration, or much simpler fluid control strategies can be used to improve test sensitivity. The use of portable power sources, such as batteries in biosensors, is an important way to improve their functionality, especially in rural areas where electricity supply is limited.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest to this work.
References
- 1.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200907-weekly-epi-update-4.pdf?sfvrsn=f5f607ee_2.
- 2.Cesewski E., Johnson B.N. Electrochemical biosensors for pathogen detection. Biosens Bioelectron. 2020;159 doi: 10.1016/j.bios.2020.112214. 112214-112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kradin R.L. Online and Print. Elsevier Health Sciences; 2010. Diagnostic Pathology of Infectious Disease E-Book: Expert Consult. [Google Scholar]
- 4.Sellon D.C., Long M. Elsevier Health Sciences; 2013. Equine Infectious Diseases E-Book. [Google Scholar]
- 5.Yang S., Rothman R.E. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. LancetInfect. Dis. 2004;4(6):337–348. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morel A.S., Dubourg G., Prudent E., Edouard S., Gouriet F., Casalta J.P.…Raoult D. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34(3):561–570. doi: 10.1007/s10096-014-2263-z. [DOI] [PubMed] [Google Scholar]
- 7.Amar C.F.L., East C.L., Gray J., Iturriza-Gomara M., Maclure E.A., McLauchlin J. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993–1996) Eur. J. Clin. Microbiol. Infect. Dis. 2007;26(5):311–323. doi: 10.1007/s10096-007-0290-8. [DOI] [PubMed] [Google Scholar]
- 8.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.…Mulders D.G. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sea-Liang N., Sereemaspun A., Patarakul K., Gaywee J., Rodkvamtook W., Srisawat N.…Hemachudha T. Development of multiplex PCR for neglected infectious diseases. PLoS Negl.Trop. Dis. 2019;13(7) doi: 10.1371/journal.pntd.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas E.J., Leber A., Ardura M.I. Broad-range PCR application in a large academic pediatric center: clinical value and challenges in diagnosis of infectious diseases. Pediatr. Infect. Dis. J. 2019;38(8):786–790. doi: 10.1097/INF.0000000000002308. [DOI] [PubMed] [Google Scholar]
- 11.Shinohara K., Kutsuna S., Takasaki T., Moi M.L., Ikeda M., Kotaki A.…Hayakawa K. Zika fever imported from Thailand to Japan, and diagnosed by PCR in the urine. J. Travel Med. 2016;23(1):tav011. doi: 10.1093/jtm/tav011. [DOI] [PubMed] [Google Scholar]
- 12.Louie M., Louie L., Simor A.E. The role of DNA amplification technology in the diagnosis of infectious diseases. CMAJ. 2000;163(3):301–309. doi: 10.1016/s1381-1169(00)00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun F., Ganguli A., Nguyen J., Brisbin R., Shanmugam K., Hirschberg D.L.…Cunningham B.T. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip. 2020;20(9):1621–1627. doi: 10.1039/d0lc00304b. [DOI] [PubMed] [Google Scholar]
- 14.Palmieri V., Papi M. Can graphene take part in the fight against COVID-19? Nano Today. 2020;33 doi: 10.1016/j.nantod.2020.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maghdid H.S., Zrar Ghafoor K., Safaa Sadiq A., Curran K., Rabie K. A novel AI-enabled framework to diagnose coronavirus COVID 19 using smartphone embedded sensors: design study. arXiv. 2020 arXiv-2003. [Google Scholar]
- 16.Mahmoudi M. Emerging biomolecular testing to assess risk of mortality from COVID-19 infection. Mol. Pharm. 2020 doi: 10.1021/acs.molpharmaceut.0c00371. [DOI] [PubMed] [Google Scholar]
- 17.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B.…Kim S.J. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 18.Kim H., Park M., Hwang J., Kim J.H., Chung D.R., Lee K.S., Kang M. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019;4(5):1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- 19.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 20.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal. Chem. 2017;89(10):5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pejcic B., De Marco R., Parkinson G. The role of biosensors in the detection of emerging infectious diseases. Analyst. 2006;131(10):1079–1090. doi: 10.1039/b603402k. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H.…Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92(10):7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 23.Yang N., Tanner J.A., Wang Z., Huang J.D., Zheng B.J., Zhu N., Sun H. Inhibition of SARS coronavirus helicase by bismuth complexes. Chem. Commun. 2007;42:4413–4415. doi: 10.1039/b709515e. [DOI] [PubMed] [Google Scholar]
- 24.Janissen R., Sahoo P.K., Santos C.A., Da Silva A.M., Von Zuben A.A., Souto D.E.…Oliveira D.S. InP nanowire biosensor with tailored biofunctionalization: ultrasensitive and highly selective disease biomarker detection. Nano Lett. 2017;17(10):5938–5949. doi: 10.1021/acs.nanolett.7b01803. [DOI] [PubMed] [Google Scholar]
- 25.Liu J., Chen X., Wang Q., Xiao M., Zhong D., Sun W.…Zhang Z. Ultrasensitive monolayer MoS2 field-effect transistor based DNA sensors for screening of down syndrome. Nano Lett. 2019;19(3):1437–1444. doi: 10.1021/acs.nanolett.8b03818. [DOI] [PubMed] [Google Scholar]
- 26.Liu C., Mao B., Martinez V., Chen X., Li Y., He L.…Shen H. A facile assay for rapid detection of COVID-19 antibodies. RSC Adv. 2020;10(47):28041–28048. doi: 10.1039/d0ra04107f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao J., Duan C., Xue L., Liu Y., Sun W., Xiang Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan B., Broza Y.Y., Li W., Wang Y., Wu S., Liu Z.…Liu W. Multiplexed nanomaterial-based sensor array for detection of COVID-19 in exhaled breath. ACS Nano. 2020 doi: 10.1021/acsnano.0c05657. [DOI] [PubMed] [Google Scholar]
- 29.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J.…Zorn K. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J.…Jervey S.R. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6(5):591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J.…Tsoi H.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L., Liu W., Zhang Q., Xu K., Ye G., Wu W.…Mei Y. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerging Microbes Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19--8-april-2020.
- 34.Wang X., Xiong E., Tian T., Cheng M., Lin W., Wang H.…Zhou X. Clustered regularly interspaced short palindromic repeats/cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14(2):2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 35.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai M., Ninomiya A., Minekawa H., Notomi T., Ishizaki T., Van Tu P.…Odagiri T. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J. Virol. Methods. 2007;141(2):173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Amer H.M., El Wahed A.A., Shalaby M.A., Almajhdi F.N., Hufert F.T., Weidmann M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J. Virol. Methods. 2013;193(2):337–340. doi: 10.1016/j.jviromet.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wat D., Gelder C., Hibbitts S., Cafferty F., Bowler I., Pierrepoint M.…Doull I. The role of respiratory viruses in cystic fibrosis. J. Cyst. Fibros. 2008;7(4):320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martel N., Gomes S.A., Chemin I., Trépo C., Kay A. Improved rolling circle amplification (RCA) of hepatitis B virus (HBV) relaxed-circular serum DNA (RC-DNA) J. Virol. Methods. 2013;193(2):653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 40.Shirato K., Nishimura H., Saijo M., Okamoto M., Noda M., Tashiro M., Taguchi F. Diagnosis of human respiratory syncytial virus infection using reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2007;139(1):78–84. doi: 10.1016/j.jviromet.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowe T., Abernathy R.A., Hu-Primmer J., Thompson W.W., Lu X., Lim W.…Katz J.M. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 1999;37(4):937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L.…Ferrell E.P. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010;28(6):595. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thaxton C.S., Elghanian R., Thomas A.D., Stoeva S.I., Lee J.S., Smith N.D.…Mirkin C.A. Nanoparticle-based bio-barcode assay redefines “undetectable” PSA and biochemical recurrence after radical prostatectomy. Proc. Natl. Acad. Sci. 2009;106(44):18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosch I., De Puig H., Hiley M., Carré-Camps M., Perdomo-Celis F., Narváez C.F.…Durbin A. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017;9(409):eaan1589. doi: 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng G.J., Liu L.T., Qiang X.J., Liu Y. 2016 International Conference on Information System and Artificial Intelligence (ISAI) IEEE; 2016. Industry 4.0 development and application of intelligent manufacturing; pp. 407–410. [Google Scholar]
- 46.Grasselli G., Pesenti A., Cecconi M. Critical care utilization for the COVID-19 outbreak in Lombardy, Italy: early experience and forecast during an emergency response. JAMA. 2020;323(16):1545–1546. doi: 10.1001/jama.2020.4031. [DOI] [PubMed] [Google Scholar]
- 47.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Bernard (2020). How AI could help in the fight against COVID-19. https://www.techrepublic.com/article/how-ai-could-help-in-the-fight-against-covid-19/. (Accessed on May 19, 2020).
- 49.Naudé W. Institute of Labor Economics (IZA); 2020. Artificial Intelligence Against COVID-19: An Early Review (No. 13110) [Google Scholar]
- 50.K. Kreuzhuber. How AI, Big Data and Machine Learning can be used against the Corona virus. ARS Electronica Blog, 19 March (2020).
- 51.Bogoch I., Watts A., Thomas-Bachli A., Huber C., Kraemer M., Khan K. Pneumonia of unknown aaetiology in Wuhan, China: potential for international spread via commercial air travel. J. Travel Med. 2020;27(2):1–3. doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.N. Izadp, & W. Naudé. DISCUSSION PAPER SERIES artificial intelligence against COVID-19. An Early REv, 13110 (2020).
- 53.Petropoulos Georgios. Artificial intelligence in the fight against COVID-19 [Internet]. Available from: https://www.bruegel.org/2020/03/artificialintelligence- in-the-fight-against-covid-19/.
- 54.Bean Randy. Big data in the time of coronavirus (COVID-19). CIO Netw [Internet]. Available from: https://www.forbes.com/sites/ciocentral/2020/03/30/big-data-in-the-time-of-coronavirus-covid-19/#161ff87558fc.
- 55.He S. Using the Internet of Things to fight virus outbreaks [Internet]. Available from: https://www.technologynetworks.com/immunology/articles/usingthe-internet-of-things-to-fight-virus-outbreaks-331992.
- 56.P. Dialani. How virtual reality is helping to deal with COVID-19 [internet]. 2020. Available from: https://www.analyticsinsight.net/virtualreality-helping-deal-covid-19/.
- 57.C. Irvine. New holographic virtual events will reach millions amid coronavirus (COVID-19) crisis [internet]. DVE holographics. Available from: https://www.prnewswire.com/news-releases/new-holographic-virtual-events-will reachmillions-amid-coronavirus-covid-19-crisis-301025496.html.
- 58.C. Lawrence. Is cloud computing the superhero of covid-19? Dev Hub [Internet]. 2020 Mar; Available from: https://www.codemotion.com/magazine/dev-hub/cloud-manager/cloud-computing-covid-19/.
- 59.Mukundan H., Anderson A.S., Grace W.K., Grace K.M., Hartman N., Martinez J.S., Swanson B.I. Waveguide-based biosensors for pathogen detection. Sensors. 2009;9(7):5783–5809. doi: 10.3390/s90705783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X., Dong X., Zhang K., Han X., Fang X., Zhang Y. A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst. 2013;138(2):642–650. doi: 10.1039/c2an36099c. [DOI] [PubMed] [Google Scholar]
- 61.Wang X., Li Y., Wang H., Fu Q., Peng J., Wang Y.…Zhan L. Gold nanorod-based localized surface plasmon resonance biosensor for sensitive detection of hepatitis B virus in buffer, blood serum and plasma. Biosens. Bioelectron. 2010;26(2):404–410. doi: 10.1016/j.bios.2010.07.121. [DOI] [PubMed] [Google Scholar]
- 62.Tripp R.A., Dluhy R.A., Zhao Y. Novel nanostructures for SERS biosensing. Nano Today. 2008;3(3–4):31–37. [Google Scholar]
- 63.Li M., Cushing S.K., Liang H., Suri S., Ma D., Wu N. Plasmonic nanorice antenna on triangle nanoarray for surface-enhanced Raman scattering detection of hepatitis B virus DNA. Anal. Chem. 2013;85(4):2072–2078. doi: 10.1021/ac303387a. [DOI] [PubMed] [Google Scholar]
- 64.Lazerges M., Bedioui F. Analysis of the evolution of the detection limits of electrochemical DNA biosensors. Anal. Bioanal. Chem. 2013;405(11):3705–3714. doi: 10.1007/s00216-012-6672-5. [DOI] [PubMed] [Google Scholar]
- 65.Iost R.M., Madurro J.M., Brito-Madurro A.G., Nantes I.L., Caseli L., Crespilho F.N. Strategies of nano-manipulation for application in electrochemical biosensors. Int. J. Electrochem. Sci. 2011;6(7):2965–2997. [Google Scholar]
- 66.Timurdogan E., Alaca B.E., Kavakli I.H., Urey H. MEMS biosensor for detection of Hepatitis A and C viruses in serum. Biosens. Bioelectron. 2011;28(1):189–194. doi: 10.1016/j.bios.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 67.Riedel T., Rodriguez-Emmenegger C., de los Santos Pereira A., Bědajánková A., Jinoch P., Boltovets P.M., Brynda E. Diagnosis of Epstein-Barr virus infection in clinical serum samples by an SPR biosensor assay. Biosens. Bioelectron. 2014;55:278–284. doi: 10.1016/j.bios.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 68.Inci F., Tokel O., Wang S., Gurkan U.A., Tasoglu S., Kuritzkes D.R., Demirci U. Nanoplasmonic quantitative detection of intact viruses from unprocessed whole blood. ACS Nano. 2013;7(6):4733–4745. doi: 10.1021/nn3036232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Songa E.A., Somerset V.S., Waryo T., Baker P.G., Iwuoha E.I. Amperometric nanobiosensor for quantitative determination of glyphosate and glufosinate residues in corn samples. Pure Appl. Chem. 2009;81(1):123–139. [Google Scholar]
- 70.Dai Tran L., Nguyen B.H., Van Hieu N., Tran H.V., Le Nguyen H., Nguyen P.X. Electrochemical detection of short HIV sequences on chitosan/Fe3O4 nanoparticle based screen printed electrodes. Mater. Sci. Eng., C. 2011;31(2):477–485. [Google Scholar]
- 71.Esfandyarpour R., Esfandyarpour H., Harris J.S., Davis R.W. Simulation and fabrication of a new novel 3D injectable biosensor for high throughput genomics and proteomics in a lab-on-a-chip device. Nanotechnology. 2013;24(46) doi: 10.1088/0957-4484/24/46/465301. 465301-465301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Solanki P.R., Patel M.K., Kaushik A., Pandey M.K., Kotnala R.K., Malhotra B.D. Sol-gel derived nanostructured metal oxide platform for bacterial detection. Electroanalysis. 2011;23(11):2699–2708. [Google Scholar]
- 73.Krejcova L., Nejdl L., Hynek D., Krizkova S., Kopel P., Adam V., Kizek R. Beads-based electrochemical assay for the detection of influenza hemagglutinin labeled with CdTe quantum dots. Molecules. 2013;18(12):15573–15586. doi: 10.3390/molecules181215573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Esseghaier C., Ng A., Zourob M. A novel and rapid assay for HIV-1 protease detection using magnetic bead mediation. Biosens. Bioelectron. 2013;41:335–341. doi: 10.1016/j.bios.2012.08.049. [DOI] [PubMed] [Google Scholar]
- 75.Lin Y.Y., Wang J., Liu G., Wu H., Wai C.M., Lin Y. A nanoparticle label/immunochromatographic electrochemical biosensor for rapid and sensitive detection of prostate-specific antigen. Biosens. Bioelectron. 2008;23(11):1659–1665. doi: 10.1016/j.bios.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 76.Yao C.Y., Fu W.L. Biosensors for hepatitis B virus detection. World J. Gastroenterol. 2014;20(35):12485–12492. doi: 10.3748/wjg.v20.i35.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao W., Zhang A., Chen Y., Chen Z., Chen Y., Lu F., Chen Z. A novel probe density controllable electrochemiluminescence biosensor for ultra-sensitive detection of Hg2+ based on DNA hybridization optimization with gold nanoparticles array patterned self-assembly platform. Biosens. Bioelectron. 2013;49:139–145. doi: 10.1016/j.bios.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 78.Qasim M., Lim D.J., Park H., Na D. Nanotechnology for diagnosis and treatment of infectious diseases. J. Nanosci. Nanotechnol. 2014;14(10):7374–7387. doi: 10.1166/jnn.2014.9578. [DOI] [PubMed] [Google Scholar]
- 79.Ueda H., Dong J. From fluorescence polarization to Quenchbody: Recent progress in fluorescent reagentless biosensors based on antibody and other binding proteins. Biochim. Biophys. Acta (BBA)-Proteins Proteomics. 2014;1844(11):1951–1959. doi: 10.1016/j.bbapap.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 80.Zhifeng J., Feng A., Li T. Consistency analysis of COVID-19 nucleic acid tests and the changes of lung CT. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104359. 104359-104359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S.…Zhang Y. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020 doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yogeswaran U., Chen S.M. A review on the electrochemical sensors and biosensors composed of nanowires as sensing material. Sensors. 2008;8(1):290–313. doi: 10.3390/s8010290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reddy R.K. Portland State University; OR: 2007. Nanomonitors: Electrical Immunoassay for Clinical Diagnostics Implementation of a Microfabricated Biosensor for the Detection of Proteins. [Google Scholar]
- 84.Sin M.L., Mach K.E., Wong P.K., Liao J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagnostics. 2014;14(2):225–244. doi: 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobayashi M., Kikuchi N., Sato A. Optical tomography of fluorophores in dense scattering media based on ultrasound-enhanced chemiluminescence. Appl. Phys. Lett. 2015;106(2) 021103-021103. [Google Scholar]
- 86.Tansi F.L., Rüger R., Rabenhold M., Steiniger F., Fahr A., Hilger I. Fluorescence-quenching of a liposomal-encapsulated near-infrared fluorophore as a tool for in vivo optical imaging. JoVE (Journal of Visualized Experiments) 2015;95 doi: 10.3791/52136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Collings A., Caruso F. Biosensors: recent advances. Rep. Prog. Phys. 1997;60(11):1397–1445. [Google Scholar]
- 88.Kerslake E.D.S., Wilson C.G. Pharmaceutical and biomedical applications of fiber optic biosensors based on infra red technology. Adv. Drug Deliv. Rev. 1996;21(3):205–213. [Google Scholar]
- 89.Uttamchandani D., McCulloch S. Optical nanosensors—towards the development of intracellular monitoring. Adv. Drug Deliv. Rev. 1996;21(3):239–247. [Google Scholar]
- 90.Monk D.J., Walt D.R. Optical fiber-based biosensors. Anal. Bioanal. Chem. 2004;379(7–8):931–945. doi: 10.1007/s00216-004-2650-x. [DOI] [PubMed] [Google Scholar]
- 91.Karlsson R. SPR for molecular interaction analysis: a review of emerging application areas. J. Mol. Recognit. 2004;17(3):151–161. doi: 10.1002/jmr.660. [DOI] [PubMed] [Google Scholar]
- 92.Bianchi N., Rutigliano C., Tomassetti M., Feriotto G., Zorzato F., Gambari R. Biosensor technology and surface plasmon resonance for real-time detection of HIV-1 genomic sequences amplified by polymerase chain reaction. Clin. Diagnostic Virol. 1997;8(3):199–208. doi: 10.1016/s0928-0197(97)00025-1. [DOI] [PubMed] [Google Scholar]
- 93.Kukanskis K., Elkind J., Melendez J., Murphy T., Miller G., Garner H. Detection of DNA hybridization using the TISPR-1 surface plasmon resonance biosensor. Anal. Biochem. 1999;274(1):7–17. doi: 10.1006/abio.1999.4241. [DOI] [PubMed] [Google Scholar]
- 94.Moon S., Kim Y., Oh Y., Lee H., Kim H.C., Lee K., Kim D. Grating-based surface plasmon resonance detection of core-shell nanoparticle mediated DNA hybridization. Biosens. Bioelectron. 2012;32(1):141–147. doi: 10.1016/j.bios.2011.11.047. [DOI] [PubMed] [Google Scholar]
- 95.Barnes W.L., Dereux A., Ebbesen T.W. Surface plasmon subwavelength optics. Nature. 2003;424(6950):824–830. doi: 10.1038/nature01937. [DOI] [PubMed] [Google Scholar]
- 96.Yu H., Kim K., Ma K., Lee W., Choi J.W., Yun C.O., Kim D. Enhanced detection of virus particles by nanoisland-based localized surface plasmon resonance. Biosens. Bioelectron. 2013;41:249–255. doi: 10.1016/j.bios.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 97.Yin J., Zou Z., Hu Z., Zhang S., Zhang F., Wang B.…Mu Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab Chip. 2020;20(5):979–986. doi: 10.1039/c9lc01143a. [DOI] [PubMed] [Google Scholar]
- 98.Trinh T.N.D., Lee N.Y. A foldable isothermal amplification microdevice for fuchsin-based colorimetric detection of multiple foodborne pathogens. Lab Chip. 2019;19(8):1397–1405. doi: 10.1039/c8lc01389f. [DOI] [PubMed] [Google Scholar]
- 99.Mello M.L.S., de Campos Vidal B. The Feulgen reaction: a brief review and new perspectives. Acta Histochem. 2017;119(6):603–609. doi: 10.1016/j.acthis.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 100.Novoselov K.S., Geim A.K., Morozov S.V., Jiang D., Zhang Y., Dubonos S.V.…Firsov A.A. Electric field effect in atomically thin carbon films. Science. 2004;306(5696):666–669. doi: 10.1126/science.1102896. [DOI] [PubMed] [Google Scholar]
- 101.Bhattacharjee S., Joshi R., Chughtai A.A., Macintyre C.R. Graphene modified multifunctional personal protective clothing. Adv. Mater. Interfaces. 2019;6(21):1900622. doi: 10.1002/admi.201900622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.https://www.zengraphene.com/.
- 103.Zhou L., Mao H., Wu C., Tang L., Wu Z., Sun H.…Chen X. Label-free graphene biosensor targeting cancer molecules based on non-covalent modification. Biosens. Bioelectron. 2017;87:701–707. doi: 10.1016/j.bios.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 104.Kim D.J., Sohn I.Y., Jung J.H., Yoon O.J., Lee N.E., Park J.S. Reduced graphene oxide field-effect transistor for label-free femtomolar protein detection. Biosens. Bioelectron. 2013;41:621–626. doi: 10.1016/j.bios.2012.09.040. [DOI] [PubMed] [Google Scholar]
- 105.Bergveld P. Development of an ion-sensitive solid-state device for neurophysiological measurements. IEEE Trans. Biomed. Eng. 1970;1:70–71. doi: 10.1109/tbme.1970.4502688. [DOI] [PubMed] [Google Scholar]
- 106.Vu C.A., Chen W.Y. Field-effect transistor biosensors for biomedical applications: recent advances and future prospects. Sensors. 2019;19(19):4214. doi: 10.3390/s19194214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qian X., Gu Z., Chen Y. Two-dimensional black phosphorus nanosheets for theranostic nanomedicine. Mater. Horiz. 2017;4(5):800–816. [Google Scholar]
- 108.Ge X., Xia Z., Guo S. Recent advances on black phosphorus for biomedicine and biosensing. Adv. Funct. Mater. 2019;29(29):1900318. [Google Scholar]
- 109.Luo M., Fan T., Zhou Y., Zhang H., Mei L. 2D black phosphorus–based biomedical applications. Adv. Funct. Mater. 2019;29(13):1808306. [Google Scholar]
- 110.Li L., Yu Y., Ye G.J., Ge Q., Ou X., Wu H.…Zhang Y. Black phosphorus field-effect transistors. Nat. Nanotechnol. 2014;9(5):372–377. doi: 10.1038/nnano.2014.35. [DOI] [PubMed] [Google Scholar]
- 111.Reich E.S. Phosphorene excites materials scientists. Nature. 2014;506(7486):19. doi: 10.1038/506019a. [DOI] [PubMed] [Google Scholar]
- 112.Li P., Zhang D., Liu J., Chang H., Sun Y.E., Yin N. Air-stable black phosphorus devices for ion sensing. ACS Appl. Mater. Interfaces. 2015;7(44):24396–24402. doi: 10.1021/acsami.5b07712. [DOI] [PubMed] [Google Scholar]
- 113.Hanlon D., Backes C., Doherty E., Cucinotta C.S., Berner N.C., Boland C.…Zhang S. Liquid exfoliation of solvent-stabilized few-layer black phosphorus for applications beyond electronics. Nat. Commun. 2015;6(1):1–11. doi: 10.1038/ncomms9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kumar V., Brent J.R., Shorie M., Kaur H., Chadha G., Thomas A.G.…Burke M.G. Nanostructured aptamer-functionalized black phosphorus sensing platform for label-free detection of myoglobin, a cardiovascular disease biomarker. ACS Appl. Mater. Interfaces. 2016;8(35):22860–22868. doi: 10.1021/acsami.6b06488. [DOI] [PubMed] [Google Scholar]
- 115.Shao J., Xie H., Huang H., Li Z., Sun Z., Xu Y.…Wang H. Biodegradable black phosphorus-based nanospheres for in vivo photothermal cancer therapy. Nat. Commun. 2016;7(1):1–13. doi: 10.1038/ncomms12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee D., Chou W.P., Yeh S.H., Chen P.J., Chen P.H. DNA detection using commercial mobile phones. Biosens. Bioelectron. 2011;26(11):4349–4354. doi: 10.1016/j.bios.2011.04.036. [DOI] [PubMed] [Google Scholar]
- 117.Gopinath S.C., Tang T.H., Chen Y., Citartan M., Lakshmipriya T. Bacterial detection: From microscope to smartphone. Biosens. Bioelectron. 2014;60:332–342. doi: 10.1016/j.bios.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 118.Smith Z.J., Chu K., Espenson A.R., Rahimzadeh M., Gryshuk A., Molinaro M.…Wachsmann-Hogiu S. Cell-phone-based platform for biomedical device development and education applications. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wac K. Smartphone as a personal, pervasive health informatics services platform: literature review. Yearbook Med. Inf. 2012;21(01):83–93. [PubMed] [Google Scholar]
- 120.Kenyon J.I., Poropatich R., Holtel M.R. Cell phones in telehealth and otolaryngology. Otolaryngol. Clin. North Am. 2011;44(6):1351–1358. doi: 10.1016/j.otc.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 121.Petryayeva E., Algar W.R. Toward point-of-care diagnostics with consumer electronic devices: the expanding role of nanoparticles. RSC Adv. 2015;5(28):22256–22282. [Google Scholar]
- 122.Bueno L., de Araujo W.R.T.R., Paixão L.C. Woodhead Publishing; 2017. Medical Biosensors for Point of Care (POC) Applications in Med. Biosens. Point Care POC Appl. pp. 183–201. [Google Scholar]
- 123.Dolai S., Tabib-Azar M. Microfabricated Nano-Gap Tunneling Current Zika Virus Sensors with Single Virus Detection Capabilities. IEEE Sens. J. 2020;20(15):8597–8603. [Google Scholar]
- 124.Roda A., Girotti S., Lodi S., Preti S. Development of a sensitive enzyme immunoassay for plasma and salivary steroids. Talanta. 1984;31(10):895–900. doi: 10.1016/0039-9140(84)80218-0. [DOI] [PubMed] [Google Scholar]
- 125.Tai D.F., Lin C.Y., Wu T.Z., Chen L.K. Recognition of dengue virus protein using epitope-mediated molecularly imprinted film. Anal. Chem. 2005;77(16):5140–5143. doi: 10.1021/ac0504060. [DOI] [PubMed] [Google Scholar]
- 126.Lu C.H., Zhang Y., Tang S.F., Fang Z.B., Yang H.H., Chen X., Chen G.N. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012;31(1):439–444. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 127.Hussein H.A., Hassan R.Y., El Nashar R.M., Khalil S.A., Salem S.A., El-Sherbiny I.M. Designing and fabrication of new VIP biosensor for the rapid and selective detection of foot-and-mouth disease virus (FMDV) Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111467. [DOI] [PubMed] [Google Scholar]
- 128.Tancharoen C., Sukjee W., Thepparit C., Jaimipuk T., Auewarakul P., Thitithanyanont A., Sangma C. Electrochemical biosensor based on surface imprinting for zika virus detection in serum. ACS Sens. 2018;4(1):69–75. doi: 10.1021/acssensors.8b00885. [DOI] [PubMed] [Google Scholar]
- 129.Gast M., Kühner S., Sobek H., Mizaikoff B. Understanding the viral load during the synthesis and after rebinding of virus imprinted particles via real-time quantitative PCR. Analyst. 2018;143(11):2616–2622. doi: 10.1039/c8an00300a. [DOI] [PubMed] [Google Scholar]
- 130.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S.…Zhang Y. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Choi J.R., Nilghaz A., Chen L., Chou K.C., Lu X. Modification of thread-based microfluidic device with polysiloxanes for the development of a sensitive and selective immunoassay. Sens. Actuators, B. 2018;260:1043–1051. [Google Scholar]
- 132.Trinh T.N.D., La H.C., Lee N.Y. Fully integrated and foldable microdevice encapsulated with agarose for long-term storage potential for point-of-care testing of multiplex foodborne pathogens. ACS Sens. 2019;4(10):2754–2762. doi: 10.1021/acssensors.9b01299. [DOI] [PubMed] [Google Scholar]