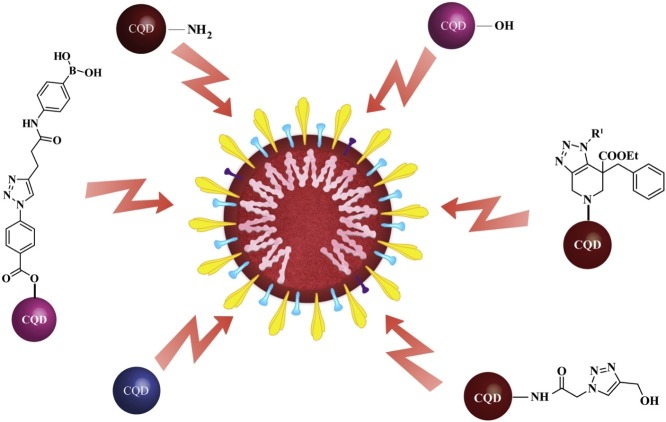
Graphical abstract
Keywords: Carbon quantum dots, Heteroatom doped, Surface functionalization, SARS-CoV-2, Human coronavirus, Click chemistry
Abstract
Preventing the trajectory of human coronaviruses including the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic could rely on the sprint to design a rational roadmap using breakneck strategies to counter its prime challenges. Recently, carbon quantum dots (CQDs), zero-dimensional (0D) carbon-based nanomaterials, have emerged as a fresh antiviral agent owing to their unique physicochemical properties. Additionally, doping instils beneficial properties in CQDs, augmenting their antiviral potential. The antiviral properties of CQDs can be reinforced by heteroatom doping. Bestowed with multifaceted features, functionalized CQDs can interact with the spike protein of the human coronaviruses and perturb the virus-host cell recognition. Recently, triazole derivatives have been explored as potent inhibitors of human coronaviruses by blocking the viral enzymes such as 3-chymotrypsin-like protease (3CLpro) and helicase, important for viral replication. Moreover, they offer a better aromatic heterocyclic core for therapeutics owing to their higher thermodynamic stability. To curb the current outbreak, triazole functionalized heteroatom co-doped carbon quantum dots (TFH-CQDs) interacting with viral cells spanning the gamut of complexity can be utilized for deciphering the mystery of its inhibitory mechanism against human coronaviruses. In this quest to unlock the potential of antiviral carbon-based nanomaterials, CQDs and triazole conjugated CQDs template comprising a series of bioisosteres, CQDs-1 to CQDs-9, can extend the arsenal of functional antiviral materials at the forefront of the war against human coronaviruses.
Introduction
The world is facing the potential catastrophe of a global pandemic due to the rapid surge in the number of people being infected with the novel human coronavirus i.e. SARS-CoV-2, leaving the world in dire need of possible cure regimens and effective therapies [1,2]. In the pursuit of finding effective therapeutics, viral entry, mode of attachment, and replication play a pivotal role in intervention strategies. Due to their unique physicochemical properties and inexpensive nature, CQDs have emerged as a rising star in the arena of antiviral materials. The cousin members in the family of fluorescent carbon nanomaterials, i.e. CQDs and graphene quantum dots (GQDs) are bestowed with superior properties such as excellent biocompatibility, resistance to photobleaching, solubility, and chemical inertness [3,4]. CQDs and GQDs are 0D materials possessing sp2 clusters, predominant quantum confinement, and small size. They share several common properties such as excellent photoluminescence, fluorescence, solubility in water [5]; whereas they can sometimes be distinguished in other properties such as surface to volume ratio, hydrodynamic diameter, crystalline structure, and their excitation/emission wavelengths. The PL bandwidth of CQDs is much wider than that of GQDs. Intriguing, GQDs are mainly composed of sp2 hybridized carbon while CQDs possess both sp3 and sp2 hybridized carbon. GQDs are usually found in crystalline nature whereas CQDs can be crystalline/semi-crystalline/amorphous in nature [3,6]. The graphene lattices inside the GQDs are produced from graphene-based starting materials or the rigid synthetic chemistry of graphene-like smaller polycyclic aromatic hydrocarbon molecules [5]. Unlike the discrete and quasi-spherical CQDs (sizes below 10 nm), GQDs are disks with a diameter across less than 20 nm as graphene sheets with lateral dimensions less than 100 nm in single, double and few (3–10) layers [[7], [8], [9]]. Generally, surface modifications are different in both cases. The antiviral properties of CQDs can be reinforced by heteroatom doping [10]. The properties arising from co-doping of CQDs [11] can be used in synergy with surface-functionalized triazole derivatives for generating potential antiviral candidates that may act either via signal transduction or cytosolic protein interaction. Taking into account the promising properties of CQDs, they can be further investigated to combat human coronaviruses (e.g. SARS-CoV-2). Understanding the receptor recognition mechanism of human coronaviruses and their binding with the substrate pocket of the host cell is imperative to elucidate the inhibitory mechanism of TFH-CQDs, which is a key factor to control their infection. Herein, cracking the antiviral potential of CQDs can extend the armament of these materials against human coronaviruses.
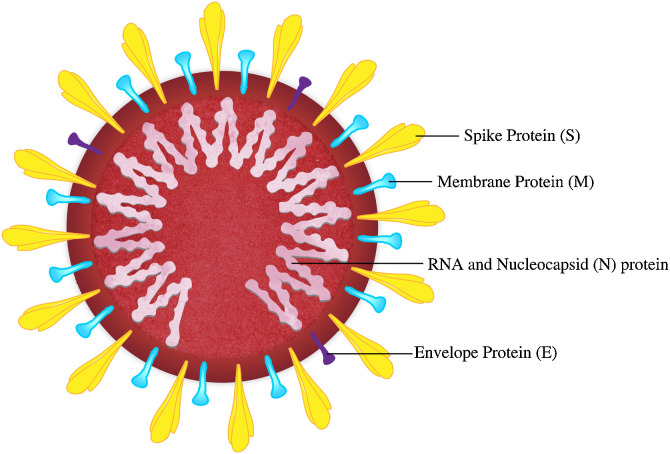
Human coronaviruses are positive-sense, single-stranded, enveloped RNA viruses, armed with four structural proteins, namely the heavily glycosylated spike (S), envelope (E), nucleocapsid (N), and membrane (M) proteins (Fig. 1 ), wherein the transmembrane S and M proteins are responsible for the viral association during replication [12,13]. They bind to cell surface proteins on target cells, followed by proteinase priming of S proteins on the viral surface leading to viral internalization either via endosomal fractions, which are subsequently acidified, or accumulated through a non-endosomal route. The S protein of the recent human coronavirus e.g. SARS-CoV-2 encompasses two subunits (S1 and S2) wherein receptor-binding-domain (RBD) within S1 subunit joins to the human angiotensin-converting enzyme 2 (hACE2) receptor (dimer) [14]. SARS-CoV and SARS-CoV-2 identify the same receptor hACE2. Addressing the increased pathogenicity and high transmission efficiency of the recent SARS-CoV-2, the following factors are taken into account: (i) Several residue changes in the SARS-CoV-2 RBD are responsible for stabilizing two virus-binding hotspots at the RBD–hACE2 interface. This, in conjunction with the more compact conformation of the hACE2-binding ridge, is responsible for enhancing the SARS-CoV-2 RBD’s hACE2 binding affinity. The modifications in the amino acid (Aa) residues of the SARS-CoV-2 RBD can lead to enhanced hydrophobic interactions and salt bridge formations, contributing to increased pathogenicity in contrast to SARS-CoV [15]. (ii) In contrast to SARS-CoV, a specific furin-like protease recognition pattern present in the vicinity of one of the maturation sites of the SARS-CoV-2 S protein contributes towards its enhanced pathogenicity and replication cycle [16]. (iii) Contrary to SARS-CoV S protein, SARS-CoV-2 S protein holds the ability to undergo a conformational change by decreasing its energy barrier enabled via a reduction in its thermostability [17]. Plausibly, these aforementioned factors contribute to the high transmission efficiency of SARS-CoV-2 with respect to SARS-CoV.
Fig. 1.
Structure of SARS-CoV-2.
Bequeathed with a robust and multifunctional nature, CQDs-based bioisosteres (bioisosteres are considered chemical groups having similar physical and chemical properties with broadly similar type of biological activity) can inhibit various interactions between the viruses and the host cells. This is underpinned by the large surface-to-volume ratio of CQDs that serve as a platform for binding of ligand molecules, adding to their ability to intervene in the viral-host cell attachment [18], thereby blocking the cellular entry. In an interesting study, the response of heteroatom doping in the carbon framework of CQDs was evaluated. Two groups of CQDs were synthesized via hydrothermal route using 4-aminophenylboronic acid and phenylboronic acid as precursors for inhibiting human coronavirus 229E (HCoV-229E). The underlying inhibitory mechanism of the viral entry was plausibly due to the interactions of CQDs and virus entry receptors. Surprisingly, substantial inhibition was also observed at the viral replication step [10].
Entry of human coronaviruses into the host cell
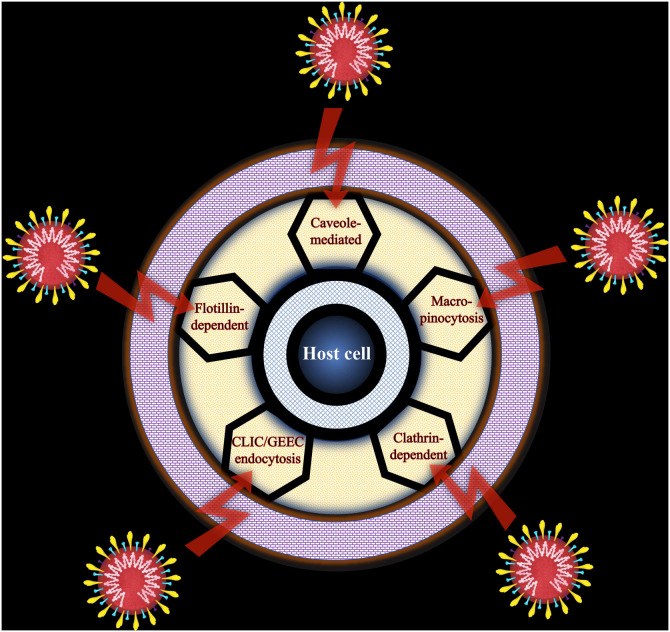
Upon internalization into the host cell, the virus releases its genetic content to specific sites for replication. The cargo transport in the cytosol occurs through major cytoskeletons i.e. actin filaments and microtubules. Moreover, the virus takes over cellular transport systems to target replication sites and protein-making machinery of the host cell. For instance, to gain entry into the host cells, spike (S) protein of the SARS-CoV-2 binds to its receptor hACE2 in the human cells via its RBD and is proteolytically triggered. Polar interactions mediate the contact between hACE2 and SARS-CoV-2 [13]. The RBD of S1 binds to the peptidase domain of the hACE2, while the membrane fusion occurs with the help of S2 [19,20]. Previous reports reveal that the primary cell entry mechanism of SARS-CoV-2 inside the human host cell plays a crucial role in eluding immune surveillance via strong hACE2 binding affinity of the RBD, hidden RBD in the S protein, and furin pre-activation of the S protein. The two types of coronaviruses, SARS-CoV and the human coronavirus NL63 (HCoV-NL63), are known to utilize hACE2 as a receptor for cellular entry. A question arises whether there can be other entry routes inside the host cell independent of hACE2? SARS-CoV-2 might also utilize receptor(s) other than hACE2 to unlock the doors for cellular entry through various possible mechanistic pathways. Reportedly, coronaviruses gain entry into the host cells primarily via the endosomal pathway (clathrin) apart from the non-endosomal pathway (non-clathrin), both depending upon hACE2 receptor [16]. The endocytic routes for SARS-CoV-2 entry include clathrin-dependent, caveolae-mediated, clathrin-independent carriers (CLIC)/glycosylphosphatidylinositol-anchored protein enriched compartments (GEEC), flotillin-dependent, and macropinocytosis [21] (Fig. 2 ) whereas caveolae/lipid-raft mediated, macropinocytosis, and clathrin-mediated are primary energy-dependent pathways for the cellular uptake of graphene-based nanomaterials. The cousin member of CQDs, i.e. GQDs, are considered to internalize through endocytosis. Recently, our group has explored the internalization of sulfur-doped graphene quantum dots (S-GQDs) in animal cell lines [22]. Interestingly, loss-of-function analysis via knockdown of the crucial regulatory proteins using various endocytic pathways can shed light on their relative contribution for SARS-CoV-2 infection [21].
Fig. 2.
Endosomal pathways for the entry of SARS-CoV-2.
Additionally, host proteases such as transmembrane protease serine 2 (TMPRSS2) play a pivotal role in the non-endosomal entry of SARS-CoV-2 by cleaving the S protein at the S1/S2 cleavage site [23]. Furthermore, SARS-CoV-2 employs a broad range of host proteases such as cathepsin B and L, furin, elastase, factor X, trypsin, and TMPRSS2 for S protein priming, thereby enabling cell entry [15]. On the other hand, additional coronavirus receptors such as dipeptidyl peptidase 4 and aminopeptidase N were not used for SARS-CoV-2 entry [24]. Plausibly, the mechanism of viral entry is contextual dependent i.e. on the type of cell and virus involved [25]. Viruses make use of molecules such as heparan sulfate proteoglycans (HSPGs) on the cell membrane to invade the host cells which are perturbed by a glycoprotein, lactoferrin (LFN). HSPGs, virtually expressed on all cell types of the body, are linear polymers made up of alternate glucosamine and hexuronic acid moieties sulfated at several locations. They serve as a binding site for many viruses including the SARS-CoV invasion at the early attachment phase. Moreover, HCoV-NL63 also utilizes HSPGs for attaching and infecting the target cells, implying their function as adhesion molecules [26]. LFN can ameliorate the activity of natural killer cells and neutrophil aggregation using adhesion and regulate host defense mechanisms by binding to HSPGs, thereby blocking its interaction with the coronaviruses [27]. In this regime, the boronic acid (B-(OH)2) surface-functionalized bioisosteres, anticipated to interact with HSPGs on the host cell membrane, can help us to understand the viral inhibitory mechanism [28]. More recently, the interaction between the RBD of spike S1 protein (RBD-S1) of the SARS-CoV-2 and heparin were unraveled. The primary sequence analysis of the modeled RBD-S1 structure and expressed protein domain revealed few patches of basic Aa residues exposed on the surface of protein, comprising the potential heparin-binding regions. The RBD-S1 undergoes a conformational change, mainly related to the antiparallel and the helix content, indicating that the RBD-S1 can interact with an aqueous solution of heparin [29].
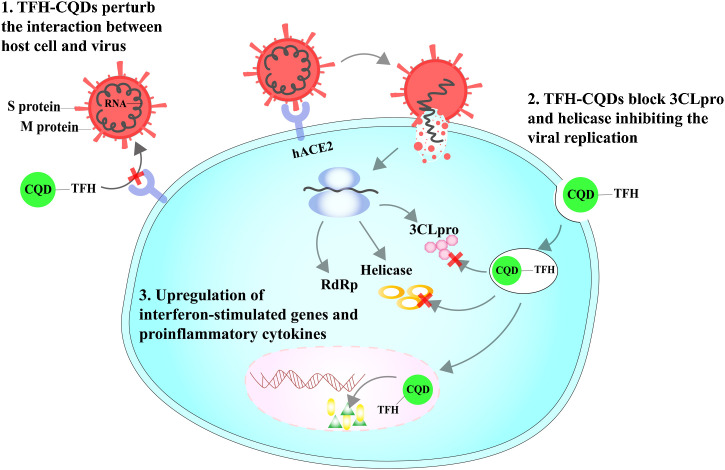
In the pursuit of finding an efficacious antiviral agent, the basic interplay and efficacy of type 1 interferon (IFN-1), one of the first cytokines to be formed in a viral response, against SARS-CoV-2 infection could accelerate the search for potential therapeutics. IFN-1 is secreted by several cell types when the pattern recognition receptors distinguish the viral components. The main mechanisms of action to fight a viral attack involve either the production of antiviral proteins or the activation of cellular immunity. The elevated levels of IFN-1 are linked to the phosphorylation of signal transducer and activator of transcription 1 (STAT1) and amplified expression of IFN-stimulated genes (ISGs) [30]. Compared to IFN-α, IFN-β1 is a more potent inhibitor of coronaviruses owing to the defending activity of IFN-β1 in the lungs [31]. Interestingly, human coronavirus SARS-CoV does not hold the ability to trigger the pathway for activation of IFN-α/β, which has a critical role in alleviating viral replication and dissemination at an early stage, along with an overall stimulatory effect on the host immune system [32]. In an effort to unleash their hidden potential, Du et al. demonstrated that CQDs can be used to activate IFN-1 levels, and consequently the expression of ISGs, both of which play an important role in inhibiting viral replication (Fig. 3 ). Contrary to control, the cells treated with CQDs displayed 5 folds higher mRNA expression levels of IFN-α along with an up-regulation in the expression of interferon-inducible protein (IP-10), interferon-stimulated gene 54 (ISG-54), and interferon-stimulated gene 15 (ISG-15). These results imply that the inhibitory effect of CQDs on viruses was probably due to the activation of IFN-α and production of ISGs. Additionally, ISG-54 can stimulate the expression of phosphorylated STAT1, which further plays a crucial part in the immune and inflammatory responses [33]. In conjunction, the up-regulation of pro-inflammatory cytokines and the production of ISGs might have a crucial role in the inhibitory effect of CQDs on viruses. This cross-talk related to the interactions of CQDs and IFNs for antiviral activity calls for unfolding additional mechanistic pathways to account for the explicit antiviral activity of CQDs. Herein, a myriad of unexplored possibilities can be posited to overcome the three crucial aspects of COVID-19: detection, treatment, and prevention. Thus, a new consortium stemming from multifarious research disciplines will offer us core capabilities to combat human coronaviruses with the potential approach of click chemistry.
Fig. 3.
Proposed mechanism of action of TFH-CQDs against SARS-CoV-2.
Inhibitory mechanism of human coronaviruses using TFH-CQDS
Understanding the therapeutic possibilities of human coronaviruses is important to circumvent their ramifications, and consequently, unravelling the mystery of inhibitory mechanisms using TFH-CQDs. Herein, we propose the potential development of a carbon-nanomaterial i.e. triazole-based CQDs template using a series of bioisosteres as antiviral agents to treat human coronavirus infection. Functionalization of nanomaterials is of keen importance and there are many ways to do it such as non-covalent modifications that can be done via hydrophobic interactions, electrostatic interactions, and π–π stacking, whereas the list of covalent functionalization strategies is even longer. CQDs have a lot of hydrophilic functional groups on the edges that make them suitable for diverse biomedical applications. The surface functionality of CQDs is vital to tune the level of interactions with the viruses [34]. Carbon dots functionalized with amine and boronic acid restrict the cellular entry of herpes simplex virus type 1 (HSV-1) infection [28]. Moreover, CQDs when conjugated with boronic acid moieties, could prevent the human immunodeficiency virus (HIV-1) infection by blocking its interaction with gp120 [35]. These investigations offer guidance for intervention strategies, targeting the receptor recognition of SARS-CoV-2. Furthermore, hydrophobic patches present in the vicinity of the active site of some enzymes may interact via π-π stacking owing to its aromatic nature. The anionic functionality may inhibit the enzymatic activity of serine proteases by electrostatically binding with the positively charged patches in the proximity of its active site [36]. The presence of functionalities on CQDs such as hydroxyl, carboxyl, carbonyl, epoxide, etc. can offer a platform for functionalization with diverse organic ligands including antiviral agents. The conjugated π structure ease the protein adsorption, facilitating the interactions between CQDs and the viral enzymes [37]. In this regard, click chemistry can pave a new horizon for the surface-functionalization of heteroatom co-doped CQDs where TFH-CQDs can add to the armory of antiviral materials.
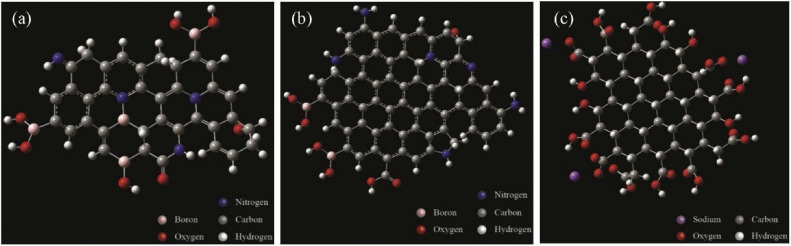
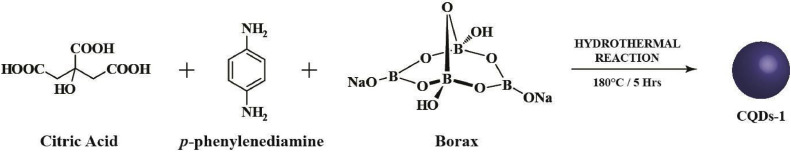
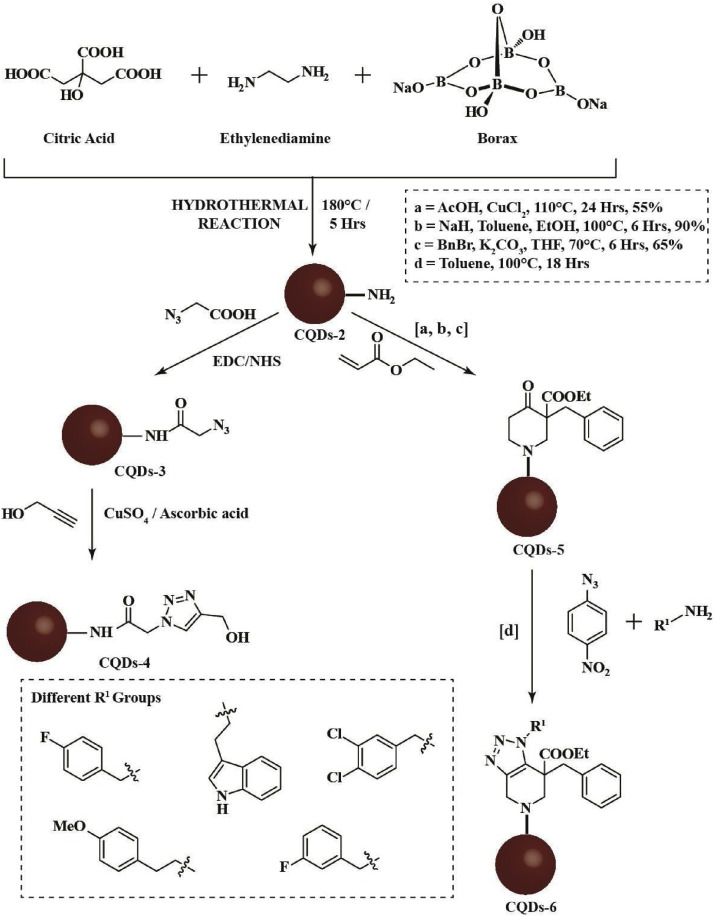
Although there are many ways to synthesize doped CQDs, one-pot hydrothermal synthesis method is widely used due to its ease and one-step process. N and B co-doped CQDs-1 (Fig. 4 a) can be prepared by hydrothermal carbonization of citric acid, p-phenylenediamine, and borax (Scheme 1 ). The -NH2 pendant groups on the CQDs-2 (Fig. 4b), synthesized from citric acid, ethylenediamine, and borax, can be used to form substituted triazoles with various R1 groups (Scheme 2 ). Intriguingly, various R1 groups exist in the literature for their activity against the enzyme 3CLpro of the human coronavirus [38]. Moreover, the -NH2 pendant groups on the edges can be functionalized to fabricate CQDs derivatives: CQDs-3 and hence, TFH-CQDs-4 with the help of Cu(I) catalyzed azide-alkyne cycloaddition conjugation reaction. Triazoles can offer an aromatic heterocyclic core for improved therapeutics owing to their higher thermodynamic stability. Additionally, in silico analysis have underscored the ability of substituted triazoles as helicase (proteins that are responsible for the separation of duplex oligonucleotide strands) inhibitors, thereby preventing viral replication. Interestingly, CQDs doped with N and B can be conjugated with substituted triazole derivatives and further explored for their ability to inhibit viral helicase [39]. TFH-CQDs-6 formed via intermediate CQDs-5 can serve as a propitious candidate for their synergistic antiviral effect (Scheme 2). Plausibly, aromatic compounds such as TFH-CQDs can be helpful in targeting the helicase of human coronaviruses with one of the following mechanisms: (i) interfering its binding with ATP and depriving it of the energy vital for unwinding and translocation, (ii) stabilizing the helicase in the low helicase activity state by an allosteric mechanism, and (iii) obstructing the binding of nucleic acids with helicases by sterically blocking its translocation [40].
Fig. 4.
3D ball and stick model of (a) CQDs-1 (b) CQDs-2 (c) CQDs-7.
Scheme 1.
The proposed hydrothermal route of heteroatom co-doped CQDs-1 from citric acid, p-phenylenediamine and borax as precursors.
Scheme 2.
The proposed hydrothermal route for heteroatom co-doped CQDs-2 from citric acid, ethylenediamine, and borax as precursors. CQDs-2 functionalized with triazole moiety via click chemistry giving rise to TFH-CQDs-4. Plausible pathway for the formation of CQDs-6 from CQDs-2 with a variable R1 group (as shown in the inset).
Owing to the similarity with the previously encountered strains, the SARS-CoV-2 genome is also found to encode for non-structural proteins including the 3CLpro [41]. This protease has a vital role in processing the polyproteins that are formed via translation of the viral RNA, making it an alluring target for coronavirus inhibition. To date, no human proteases with similar cleavage specificity are known, thereby inhibiting the activity of this enzyme is unlikely to create toxicity [42]. Intriguingly, triazole derivatives have been reported to play a crucial role in blocking the 3CLpro for exerting their antiviral response against HCoV-229E. Furthermore, an in silico study predicted that the molecular interactions between these compounds and 3CLpro is imperative for the inhibition of viral replication. The non-peptidic inhibitors of the 3CLpro rely on the presence of a benzotriazole group that can interact with the catalytic dyad (consisting of Cys145 and His41 of the 3CLpro active site). With the help of molecular modeling, some triazole compounds were observed to form two hydrogen bonds with the Glu166 and His163 wherein the positioning of the triazole ring in the vicinity of the catalytic dyad suggesting that this interaction pattern is crucial for the 3CLpro inhibition. The maintenance of these interactions throughout the course of molecular dynamics study further strengthens their bioactivities [38].
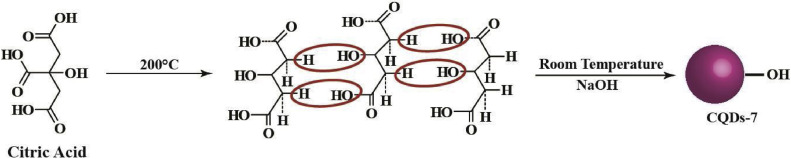
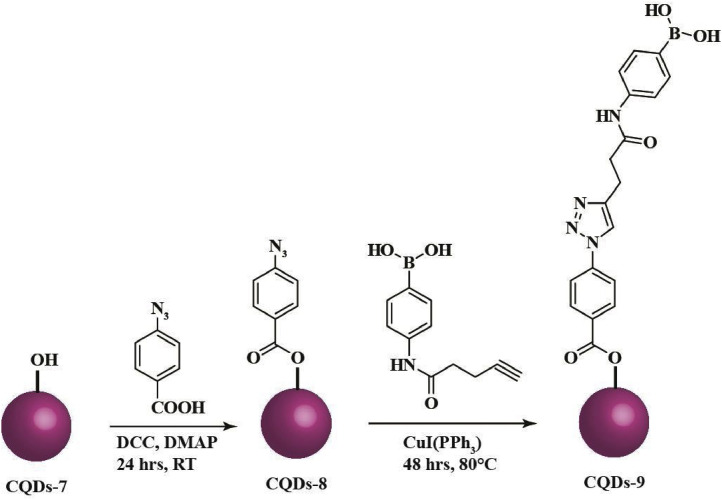
Addition of CQDs at different time frames and stages of the virus life cycle during the infection process exposed that CQDs can surmount the viral entry and also the replication process. Their ability to act either via signal transduction or directly with the cytosolic proteins, upon internalization, needs to be addressed further. Reportedly, the incubation of viral particles with CQDs could not impair the HCoV-229E infection [10], implying the emergence of a possible target for preventing viral entry. Recently, heteroatom doped CQDs have been vetted for their antiviral activity against the human coronaviruses. CQDs surface-functionalized with boronic acid (B-(OH)2) are anticipated to interact strongly with HSPGs on the host cell membrane, thereby inhibiting the viral attachment [28]. Boronic acid moieties (CQDs-9) help to form tetravalent boronate diester cyclic complexes reversibly and selectively with 1,2- or 1,3-cis-diols [43]. Hydroxyl derivatives of CQDs i.e. CQDs-7 (Fig. 4c) can be synthesized from citric acid via hydrothermal route (Scheme 3 ). CQDs-7 can be further used to synthesize CQDs-9, having boronic acid as a functional group, via click chemistry using CQDs-8 as intermediate (Scheme 4 ). Interestingly, CQDs conjugated with triazole moieties were found to display antiviral activity which was further enhanced with the addition of boronic acid as a functional group. These surface-functionalized CQDs undergo pseudolectin-type interactions with the S protein of the HCoV-229E. However, when the boronic acid functional groups were blocked, the remaining antiviral activity of these CQDs was ascribed to the triazole moiety. The above findings emphasize the potential of triazole moieties as antiviral agents. Moreover, heteroatom co-doped CQDs could exert a significantly higher antiviral response than surface-functionalized CQDs. Thus, based on the effective 50 % inhibition concentration (EC50) values, it is evident that N and B co-doped CQDs exhibit a more pronounced antiviral activity than triazole functionalized CQDs and CQDs that are surface-functionalized with boronic acid via click chemistry [10].
Scheme 3.
The proposed hydrothermal route for CQDs-7 using citric acid as precursor.
Scheme 4.
The proposed route for the formation of CQDs-9 with boronic acid as a functional group via click chemistry.
CQDs can act as a multi-site inhibitor by blocking the viral entry, RNA strand synthesis, and replication step. Furthermore, upon viral exposure, the accumulation of overexpressed reactive oxygen species in cells are also reduced with a concurrent upregulation in the expression of proinflammatory cytokines and ISGs [44]. Surprisingly, it was also reported that CQDs synthesized from glycyrrhizic acid exhibited an intensified antiviral behavior towards RNA viruses compared with DNA viruses [45]. We expect that TFH-CQDs or its functionalized analogues can be further exploited for their antiviral activity to inhibit human coronaviruses having RNA as the genetic material. However, the varying responsiveness of some viruses indicates that CQDs also differ in their antiviral strategies and further assessment is needed for a rational outcome.
Summary and outlook
Considering the inescapable exigency to depict new chemical entities for a treatment modality against human coronaviruses, herein, we proposed the conceptual insights into a series of bioisosteres derived from TFH-CQDs based on investigations and design of peptide inhibitors to combat human coronaviruses. These TFH-CQDs can act by several pathways. They may either block the viral entry by perturbing various interactions with the host cells or block the viral enzymes such as helicase and 3CLpro, important for viral replication. Therefore, we envisage that heteroatom co-doping (N and B) in the carbon framework of the CQDs surface-functionalized with triazole and its derivatives can synergistically exert an intensified antiviral response against human coronavirus infection. Complying with the study of CQDs, a special class of 0D carbon nano-crystalline materials, GQDs, are envisaged as standout choices for inhibiting various interactions of viruses with their host cells. Reason suggests that GQDs, with various surface functional groups painted on their canvas, in nano-sized and with excellent water solubility could be a winning candidate against the human coronaviruses. The toolbox of chemistries that TFH-CQDs embrace can help us disseminate our contributions and strategies for the benefit of mankind and with further research and exploration, they can be translated effectively from bench-to-bedside. From a broader perspective, insights into entry mode and viral itinerary warrant future studies, reshaping the currently available therapeutics and guiding new intervention strategies.
Author statement
Piyush Garg, Sujata Sangam, and Monalisa Mukherjee have conceptualized and written the manuscript. Dakshi Kochhar has prepared the graphical abstract and contributed in the mechanistic insight of the perspective. Siddhartha Pahari has collected the literature along with Piyush Garg. Chirantan Kar has given the schematic representation of the series of TFH-CQDs.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
Sujata Sangam is grateful to the Council of Scientific & Industrial Research (CSIR) – University Grants Commission (UGC), India for providing financial support in the form of SRF. Monalisa Mukherjee thanks the Department of Science and Technology (DST), Science and Engineering Research Board (SERB) (EMR/2016/00561) for funding and Amity University Uttar Pradesh (AUUP, Noida) for providing research infrastructure and library facility.
Biographies

Piyush Garg is pursuing his B. Tech. + M. Tech. dual degree in Biotechnology from Amity University (Noida). Presently, he is a Research Trainee at the Molecular Science and Engineering Laboratory under the supervision of Professor Monalisa Mukherjee at Amity Institute of Click Chemistry Research and Studies, Amity University (Noida), U.P., India. His current research focuses on the biomedical applications of carbon-based nanomaterials and polymer nanocomposites.

Sujata Sangam received M.Sc. (Biotechnology) and M.Tech. (Environmental Engg. & Management) from SHUATS, Allahabad and IIT Kanpur, UP, respectively. Then she joined Amity Institute of Biotechnology, Amity University (Noida), U.P., India to pursue Ph.D. under the guidance of Prof. Monalisa Mukherjee. She is currently involved with Amity Institute of Click Chemistry Research and Studies. She is recipient of GATE and CSIR-UGC JRF Fellowship. Her current research focuses on the interfaces of molecular chemistry, nanotechnology, biotechnology, environmental engg. and studies based on biomimetic smart carbon-nanomaterials, hydrogels, nanocomposites and their biomedical applications.

Dakshi Kochhar received her B. Tech. in Biotechnology from Amity University (Noida), U.P., India and is starting her MS in Biomedical Engineering from Carnegie Mellon University, Pittsburgh, United States. During her under-graduation studies, she joined Amity Institute of Click Chemistry Research and Studies as a Research Intern where her focus is hydrogel nanocomposites and biomimetic exotic hydrogels for scarless burn wound healing and targeted drug delivery.

Siddhartha Pahari is a senior year high school student in Amity International School, Mayur Vihar, Delhi, India. During his internship at Amity Institute of Click Chemistry Research and Studies, Amity University (Noida), U.P., India, his primary focus was on nanoparticles and their relevance in biological applications such as antiviral properties and drug delivery.

Dr. Chirantan Kar obtained his Ph. D. in Chemistry from Indian Institute of Technology (Guwahati) in 2014. He did his first postdoctoral research work as a JSPS fellow in Keio University, Tokyo from 2015 to 2017. Later, he did his second postdoctoral research in Indian Institute of Chemical Biology from 2017 to 2018. He joined Amity University (Kolkata), India in 2018 and presently, he is working as an Assistant Professor in the Department of Chemistry. His current research interests include applied organic chemistry, materials chemistry and chemical biology.

Prof. Monalisa Mukherjee completed her Bachelors and Master degree from University of North Bengal, India. She received her Ph. D. from CBME, IIT (Delhi) in 2006. Presently, she is serving as Director at Amity Institute of Click Chemistry Research & Studies and Professor at Amity Institute of Biotechnology, Amity University (Noida), U.P., India. She is also the recipient of UK-India Distinguished Visiting Scientist Award. During the past 5 years, Dr. Mukherjee has made exemplary contributions to the field of Materials Science. Her research is not only reinforced by her remarkable publications and patents but also by its translational potential.
References
- 1.Alexander S., Armstrong J., Davenport A., Davies J., Faccenda E., Harding S., Levi-Schaffer F., Maguire J., Pawson A., Southan C., Spedding M. A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development. IUPHAR review 29. Br. J. Pharmacol. 2020 doi: 10.1111/bph.15094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmieri V., Papi M. Can graphene take part in the fight against COVID-19? Nano Today. 2020;33 doi: 10.1016/j.nantod.2020.100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tajik S., Dourandish Z., Zhang K., Beitollahi H., Van Le Q., Jang H.W., Shokouhimehr M. Carbon and graphene quantum dots: a review on syntheses, characterization, biological and sensing applications for neurotransmitter determination. RSC Adv. 2020;10:15406–15429. doi: 10.1039/d0ra00799d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L., Zhu S.J., Wang H.Y., Qu S.N., Zhang Y.L., Zhang J.H., Chen Q.D., Xu H.L., Han W., Yang B., Sun H.B. Common origin of green luminescence in carbon nanodots and graphene quantum dots. ACS Nano. 2014;8:2541–2547. doi: 10.1021/nn500368m. [DOI] [PubMed] [Google Scholar]
- 5.Zhang R., Ding Z. Recent advances in graphene quantum dots as bioimaging probes. J. Anal. Test. 2018;2:45–60. doi: 10.1007/s41664-018-0047-7. [DOI] [Google Scholar]
- 6.Cayuela A., Soriano M.L., Carrillo-Carrión C., Valcárcel M. Semiconductor and carbon-based fluorescent nanodots: the need for consistency. Chem. Commun. (Camb.) 2016;52:1311–1326. doi: 10.1039/c5cc07754k. [DOI] [PubMed] [Google Scholar]
- 7.Fan Z., Li S., Yuan F., Fan L. Fluorescent graphene quantum dots for biosensing and bioimaging. RSC Adv. 2015;5:19773–19789. doi: 10.1039/C4RA17131D. [DOI] [Google Scholar]
- 8.Li L., Wu G., Yang G., Peng J., Zhao J., Zhu J.J. Focusing on luminescent graphene quantum dots: current status and future perspectives. Nanoscale. 2013;5:4015–4039. doi: 10.1039/c3nr33849e. [DOI] [PubMed] [Google Scholar]
- 9.Xie R., Wang Z., Zhou W., Liu Y., Fan L., Li Y., Li X. Graphene quantum dots as smart probes for biosensing. Anal. Methods. 2016;8:4001–4006. doi: 10.1039/c6ay00289g. [DOI] [Google Scholar]
- 10.Łoczechin A., Séron K., Barras A., Giovanelli E., Belouzard S., Chen Y., Metzler-nolte N., Boukherroub R., Dubuisson J., Szunerits S. Functional carbon quantum dots as medical countermeasures to human coronavirus. ACS Appl. Mater. Interfaces. 2019;11:42964–42974. doi: 10.1021/acsami.9b15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao S., Liang K., Zhu J., Yang B., Zhao D., Kong B. Hetero-atom-doped carbon dots: doping strategies, properties and applications. Nano Today. 2020;33 doi: 10.1016/j.nantod.2020.100879. [DOI] [Google Scholar]
- 12.Cyranoski D. Profile of a killer: the complex biology powering the coronavirus pandemic. Nature. 2020;581:22–26. doi: 10.1038/d41586-020-01315-7. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan G., Madou M.J., Kalra S., Chopra V., Ghosh D., Martinez-Chapa S.O. Nanotechnology for COVID-19: therapeutics and vaccine research. ACS Nano. 2020 doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]
- 14.Weiss C., Carriere M., Fusco L., Capua I., Regla-Nava J.A., Pasquali M., Scott J.A., Vitale F., Unal M.A., Mattevi C., Bedognetti D., Merkoçi A., Tasciotti E., Yilmazer A., Gogotsi Y., Stellacci F., Delogu L.G. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano. 2020;14:6383–6406. doi: 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
- 15.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00502. https://doi.org/10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itani R., Tobaiqy M., Al Faraj A. Optimizing use of theranostic nanoparticles as a life-saving strategy for treating COVID-19 patients. Theranostics. 2020;10:5932–5942. doi: 10.7150/thno.46691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y., Král P. Computational design of ACE2-Based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020 doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sangam S., Gupta A., Shakeel A., Bhattacharya R., Sharma A.K., Suhag D., Chakrabarti S., Garg S.K., Chattopadhyay S., Basu B., Kumar V., Rajput S.K., Dutta M.K., Mukherjee M. Sustainable synthesis of single crystalline sulphur-doped graphene quantum dots for bioimaging and beyond. Green Chem. 2018;20:4245–4259. doi: 10.1039/c8gc01638k. [DOI] [Google Scholar]
- 23.Kinaret P.A.S., del Giudice G., Greco D. Covid-19 acute responses and possible long term consequences: what nanotoxicology can teach us. Nano Today. 2020;35 doi: 10.1016/j.nantod.2020.100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., Guo H., Jiang R., Liu M., Chen Y., Shen X., Wang X., Zhan F., Wang Y., Xiao G., Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang N., Shen H.-M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milewska A., Zarebski M., Nowak P., Stozek K., Potempa J., Pyrc K. Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 2014;88:13221–13230. doi: 10.1128/JVI.02078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang J., Yang N., Deng J., Liu K., Yang P., Zhang G., Jiang C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barras A., Pagneux Q., Sane F., Wang Q., Boukherroub R., Hober D., Szunerits S. High efficiency of functional carbon nanodots as entry inhibitors of herpes simplex virus type 1. ACS Appl. Mater. Interfaces. 2016;8:9004–9013. doi: 10.1021/acsami.6b01681. [DOI] [PubMed] [Google Scholar]
- 29.Mycroft-West C., Su D., Elli S., Guimond S., Miller G., Turnbull J., Yates E., Guerrini M., Fernig D., Lima M., Skidmore M. The 2019 coronavirus (SARS-CoV-2) surface protein (Spike) S1 receptor binding domain undergoes conformational change upon heparin binding. BioRxiv. 2020 doi: 10.1101/2020.02.29.971093. [DOI] [Google Scholar]
- 30.Lokugamage K.G., Hage A., de Vries M., Valero-Jimenez A.M., Schindewolf C., Dittmann M., Rajsbaum R., Menachery V.D. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. BioRxiv. 2020 doi: 10.1101/2020.03.07.982264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sallard E., Lescure F., Yazdanpanah Y., Mentre F., Peiffer-Smadj N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florindo H.F., Kleiner R., Vaskovich-Koubi D., Acúrcio R.C., Carreira B., Yeini E., Tiram G., Liubomirski Y., Satchi-Fainaro R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du T., Liang J., Dong N., Liu L., Fang L., Xiao S., Han H. Carbon dots as inhibitors of virus by activation of type I interferon response. Carbon N. Y. 2016;110:278–285. doi: 10.1016/j.carbon.2016.09.032. [DOI] [Google Scholar]
- 34.Gholami M.F., Lauster D., Ludwig K., Storm J., Ziem B., Severin N., Böttcher C., Rabe J.P., Herrmann A., Adeli M., Haag R. Functionalized graphene as extracellular matrix mimics: toward well-defined 2D nanomaterials for multivalent virus interactions. Adv. Funct. Mater. 2017;27 doi: 10.1002/adfm.201606477. [DOI] [Google Scholar]
- 35.Fahmi M.Z., Sukmayani W., Khairunisa S.Q., Witaningrum A.M., Indriati D.W., Matondang M.Q.Y., Chang J.-Y., Kotaki T., Kameoka M. Design of boronic acid-attributed carbon dots on inhibits HIV-1 entry. RSC Adv. 2016;6:92996–93002. doi: 10.1039/C6RA21062G. [DOI] [Google Scholar]
- 36.De M., Chou S.S., Dravid V.P. Graphene oxide as an enzyme inhibitor: modulation of activity of α-chymotrypsin. J. Am. Chem. Soc. 2011;133:17524–17527. doi: 10.1021/ja208427j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B., Chen D., Nie M., Wang J., Li Y., Yang Y. Carbon dots/Cu2O composite with intrinsic high protease-like activity for hydrolysis of proteins under physiological conditions. Part. Part. Syst. Charact. 2018;35 doi: 10.1002/ppsc.201800277. [DOI] [Google Scholar]
- 38.Karypidou K., Ribone S.R., Quevedo M.A., Persoons L., Pannecouque C., Helsen C., Claessens F., Dehaen W. Synthesis, biological evaluation and molecular modeling of a novel series of fused 1, 2, 3-triazoles as potential anti-coronavirus agents. Bioorg. Med. Chem. Lett. 2018;28:3472–3476. doi: 10.1016/j.bmcl.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaher N.H., Mostafa M.I., Altaher A.Y. Design, synthesis and molecular docking of novel triazole derivatives as potential CoV helicase inhibitors. Acta Pharm. 2020;70:145–159. doi: 10.2478/acph-2020-0024. [DOI] [PubMed] [Google Scholar]
- 40.Briguglio I., Piras S., Corona P., Carta A. Inhibition of RNA helicases of ssRNA+ virus belonging to flaviviridae, coronaviridae and picornaviridae families. Int. J. Med. Chem. 2011 doi: 10.1155/2011/213135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khanal M., Vausselin T., Barras A., Bande O., Turcheniuk K., Benazza M., Zaitsev V., Teodorescu C.M., Boukherroub R., Siriwardena A., Dubuisson J., Szunerits S. Phenylboronic-acid-modified nanoparticles: potential antiviral therapeutics. ACS Appl. Mater. Interfaces. 2013;5:12488–12498. doi: 10.1021/am403770q. [DOI] [PubMed] [Google Scholar]
- 44.Du T., Dong N., Fang L., Lu J., Bi J., Xiao S., Han H. Multi-site inhibitors for enteric coronavirus: antiviral cationic carbon dots based on curcumin. ACS Appl. Nano Mater. 2018;1:5451–5459. doi: 10.1021/acsanm.8b00779. [DOI] [PubMed] [Google Scholar]
- 45.Tong T., Hu H., Zhou J., Deng S., Zhang X., Tang W., Fang L., Xiao S., Liang J. Glycyrrhizic‐acid‐based carbon dots with high antiviral activity by multisite inhibition mechanisms. Small. 2020;16 doi: 10.1002/smll.201906206. [DOI] [PMC free article] [PubMed] [Google Scholar]