Abstract
This paper presents an automatic classification segmentation tool for helping screening COVID-19 pneumonia using chest CT imaging. The segmented lesions can help to assess the severity of pneumonia and follow-up the patients. In this work, we propose a new multitask deep learning model to jointly identify COVID-19 patient and segment COVID-19 lesion from chest CT images. Three learning tasks: segmentation, classification and reconstruction are jointly performed with different datasets. Our motivation is on the one hand to leverage useful information contained in multiple related tasks to improve both segmentation and classification performances, and on the other hand to deal with the problems of small data because each task can have a relatively small dataset. Our architecture is composed of a common encoder for disentangled feature representation with three tasks, and two decoders and a multi-layer perceptron for reconstruction, segmentation and classification respectively. The proposed model is evaluated and compared with other image segmentation techniques using a dataset of 1369 patients including 449 patients with COVID-19, 425 normal ones, 98 with lung cancer and 397 of different kinds of pathology. The obtained results show very encouraging performance of our method with a dice coefficient higher than 0.88 for the segmentation and an area under the ROC curve higher than 97% for the classification.
Keywords: Deep learning, Multitask learning, Image classification, Image segmentation, Coronavirus (COVID-19), Computed tomography images
Highlights
-
•
Multitask deep learning based model can be used to detect COVID-19 lesions on CT scans.
-
•
The proposed model can improve state of the art U-NET by leveraging useful information contained in multiple related tasks.
-
•
Obtained a dice coefficient of 88% for image segmentation and an accuracy of 94.67 for multiclass classification.
-
•
The proposed model can be used as a support tool to assist physicians.
1. Introduction
The novel coronavirus disease (COVID-19) spread rapidly around the world, changing the daily lives of billions of people. The infection can lead to severe pneumonia that can causes death. Also, COVID-19 is highly contagious, which is why it must be detected quickly, in order to isolate the infected person very fast to limit the spread of the disease. Today, the gold standard for detecting COVID-19 is the Reverse Transcription Polymerase Chain Reaction (RT-PCR) [42], which consists of detecting viral RNA from sputum or nasopharyngeal swab. The limitation with the RT-PCR test is due to the time needed to get the results, the availability of the material which remains very limited in hospitals [42] and its relatively low sensitivity, which does not meet the major interest of rapidly detecting positive cases as soon as possible in order to isolate them [39]. An alternative solution for rapid screening could be the use of medical imaging such as chest X-ray images or computed tomography (CT) scanners [39].
Identifying COVID-19 at an early stage through imaging would indeed allow the isolation of the patient and therefore limit the spread of the disease [39]. However, physicians are very busy fighting this disease, hence the need to create decision support tools based on artificial intelligence to not only detect but also segment the infection at the lung level in the image [42]. Artificial intelligence has seen a major and rapid growth in recent years with deep neural networks [43] as a first tool to solve different problems such as object detection [29,37]speech recognition [37], drug interaction [27] and image classification [10]. More specifically, convolutional neural networks [22] showed astonishing results for image processing [21]. For image segmentation, several works have shown the power and robustness of these methods [5]. CNNs architectures for medical imaging also have been used with very good results [14], for both image classification [3,6] or image segmentation [19].
1.1. Related work
For the detection of COVID-19 and the segmentation of the infection at the lung level, several deep learning works on chest X-ray images and CT scans have emerged and reported in Ref. [34]. In Ref. [25] Ali Narin et al. created a deep convolutional neural network to automatically detect COVID-19 on X-ray images. To that end, they used transfer learning based approach with a very deep architectures such as ResNet50, InceptionV3 and Inception-ResNetV2. The algorithms were trained on the basis of 100 images (50 COVID vs 50 non-COVID) in 5 cross-validation. Authors claimed 97% of accuracy using InceptionV3 and 87% using Inception-ResNetV2. However, due to the very limited size of the dataset and the very deep models, overfitting would rise and could not be ruled-out, hence the need to validate those results in a larger database is necessary. Also in Ref. [15], Hemdan et al. created several deep learning models to classify X-ray images into COVID vs non-COVID classes reporting best results with an accuracy of 90% using VGG16. Again, the database was very limited with only 50 cases (25 COVID vs 25 non-COVID). A resembling study was conducted by wang and wang [38] where they trained a CNN on the ImageNET database [11] then fine-tuned on X-ray images to classify cases into one of four classes: normal, bacterial, non-COVID-19 viral and COVID-19 viral infection, with an overall performance of 83.5%. For CT images, Jinyu Zhao et al. [45] created a container for CT scans initially with 275 CT COVID-19 on which they also applied a transfer learning algorithm using chest-x-ray14 [40] with 169-layer DenseNet [17]. The performance of the model is 84.7% with an area under the ROC curve of 82.4%. As of today, the database contains 347 CT images for COVID-19 patients and 397 for non-COVID patients.
Instead of using CNNs, other works have used network capsules which were first proposed in Refs. [16] to solve the problems of CNNs architectures which need a large amount of data and many parameters. In Ref. [2] the authors opted for this method. They created a capsule network to identify COVID-19 cases in X-ray images. The results were encouraging with an accuracy of 95.7%, sensitivity at 90% and specificity at 95.8%. They compared their results with Sethy et al. [33] where they created a model based on resnets50 with SVMs and obtained a performance of 95.38%, a sensitivity of 97.29% and a specificity of 93.47%.
In [18], Jin et al. created and deployed an AI tool to analyze CT images of COVID-19 in 4 weeks. To do this, a multidisciplinary team of 30 people collaborated together using a database of 1136 images including 723 positive COVID-19 images from five hospitals, to achieve a sensitivity of 0.974 and a specificity of 0.922. The system was deployed in 16 hospitals and performed over 1300 screenings per day. They proposed a combined model for classification and segmentation showing lesion regions in addition to the screening results. The pipeline is divided into 2 steps: segmentation and classification. They used several models including 3D U-NET++, V-NET, FCN-8S for segmentation and InceptionV3, ResNet50 and others for classification. They were able to achieve a dice coefficient of 0.754 using 3D U-NET ++ trained on 732 cases. The combination of 3D U-NET ++ and ResNet50 resulted in an area under the OCR curve of 0.991 with a sensitivity of 0.974 and a specificity of 0.922. In practice, the model continued to improve by re-training. The model proved to be very useful to physicians by highlighting regions of lesions which improved the diagnosis. What should be noted here is that the two models are independent and therefore they cannot help each other to improve both classification and segmentation performances.
Other works have also emerged recently with interesting results. In Ref. [26] T. Ozturk et al. proposed a deep neural network called DarkCovidNet to detect automatically COVID-19 cases on X-ray images. The model inspired from Darknet-19, is a classifier model that forms the basis of a real-time object detection system named YOLO (You only look once) [30]. They implemented a 17 convolutional layers network achieving a results of 98.08% for binary classes and 87.02% for multi-class cases. In Ref. [27], Y. Pathak et al. used a transfer learning approach to classify COVID-19 infected patients. They introduced a top-2 smooth loss function with cost-sensitive attributes to handle noise and imbalanced dataset. The model was trained on a public dataset of chest CT images, and then used to classify COVID-19 infected patients. The model achieved an accuracy of 0.93, a sensitivity of 0.91 and a specificity of 0.94. Other works such as in S. Dilbag et al. [35] tried a multi-objective differential evolution–based convolutional neural networks to classify COVID-19 patients from chest CT images.
1.2. Motivation
Multi-task learning (MTL) [8] is a type of learning algorithm whose goal is to combine several pieces of information from different tasks to improve the performance of the model and its ability to better generalize [44]. The basic idea of MTL is that different tasks can share a common features representation [44], and therefore, training them jointly. The use of different datasets of different tasks makes it possible to learn an effective feature representation that is common to all tasks, because all datasets are used to obtain it, even if each task has a small dataset, which leads to improve the performance of each task. Different approaches have been proposed in MTL such as hard parameter sharing [8] or soft parameter sharing [32]. Hard parameter sharing is the most commonly used approach to MTL in neural networks and greatly reduces the risk of overfitting [32], see Fig. 4 . It is generally applied by sharing the hidden layers between all tasks, while keeping several task-specific output layers. Soft parameter sharing defines a model for each task with its own parameters, and the distance between the parameters of the model is regularized in order to encourage the parameters to be similar.
Fig. 4.
Hard parameter sharing for multi-task learning in deep neural networks used in our proposed architecture.
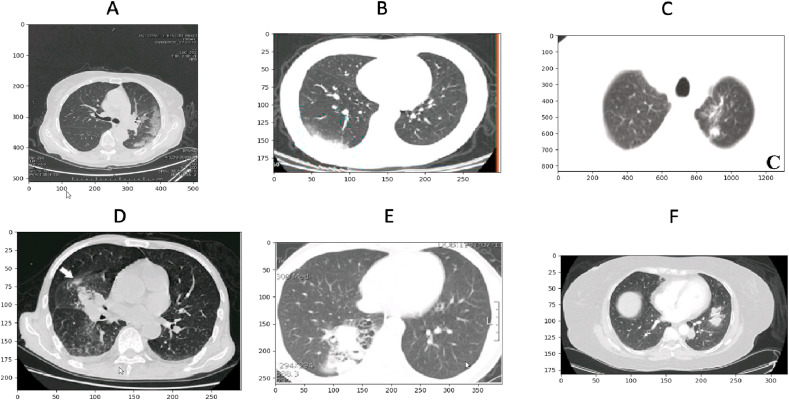
In this work, we propose a novel multi-task deep learning model for jointly detecting COVID-19 image and segmenting lesions. The main challenges of this work are: 1) the lack of data and annotated data, because the databases were collected from multiple sources with a huge variation in images and most of the images are noisy (see Fig. 3). 2) instead of expensive models like ResNet 50 or DenseNet, developing a multitasking approach to reduce overfitting and improve results. In Fig. 1 we can see an example of different image formats (png (B C E F), Nifti (D), DICOM (A), different scales of visualisation window (strong centering on lung (B E C), low (A,D, F), different image sizes, with/without annotations (with (C,D E), without (A B F). Facing these challenges, we proposed to train our neural network with three tasks: reconstruction, classification and segmentation, in order to both classify COVID/Non-COVID images and to segment the lesion regions. We add the reconstruction task often used in unsupervised learning, to better learn the disentangled feature representation with a single common encoder. Based on this features representation, three neural networks are designed to finally accomplish the three tasks.
Fig. 3.
The different databases used in this study.
Fig. 1.
An example of exams heterogeneities between different CT images for COVID (upper) and non-COVID (bottom). Patient images do not have the same resolution. Also, images show different image formats (Dicom(A), png (B C E F), Nifti (D).
The paper is organized as follows. In Section 2, we describe our multi-task model, which is mainly based on classification and segmentation tasks. Section 3 presents the experimental studies. In section 4, we describe the validation methodology used in this study. Section 5 is showing the results of our work. Section 6, 7 are for discussion and conclusion respectively.
1.3. Data
In this study, three datasets from different hospitals including one thousand three hundred and ninety-six CT images are used. The first one is a public available data set coming from Refs. [45] which includes 347 COVID-19 images and 397 non-COVID images with different types of pathologies. The database was pre-processed and stored in png format. The dimension varies from 153 to 1853 with an average of 491 for the height, while the width varies from 124 to 383 with an average of 1485. The second dataset coming from [http://medicalsegmentation.com/covid19/] in which 100 COVID-19 CT scan with ground truths lesions segmentation are available. The ground truth was defined by the physicians of different hospitals. Three lesion labels are provided: ground glass, consolidation and pleural effusion. As all lesion labels are not given in all images, for the purpose of this study, we merged the three labels into one lesion label (See Fig. 2). The third dataset coming from the Henri Becquerel Cancer Center (HBCC) in Rouen city of France includes 425 CT scans of normal patients and 98 of lung cancer. All the three image datasets were resized to have the same size 256 × 256 and the intensity normalized between 0 and 1 prior to analysis. Table 1 summarizes how to split the datasets for training, validation and test.
Fig. 2.
Example of COVID-19 segmentation on CT scan for 2 patients: first column - original CT scanner; Second column - one label segmentation, Third column - 3 labels segmentation: ground glass (green), consolidation (blue) and pleural effusion (yellow).
Table 1.
Data split.
| Train | Validation | Test | Total | |
|---|---|---|---|---|
| Normal | 325 | 50 | 50 | 425 |
| COVID-19 | 349 | 50 | 50 | 449 |
| Other Infections | 395 | 50 | 50 | 495 |
| Total | 1069 | 150 | 150 | 1369 |
2. Method
2.1. Model description
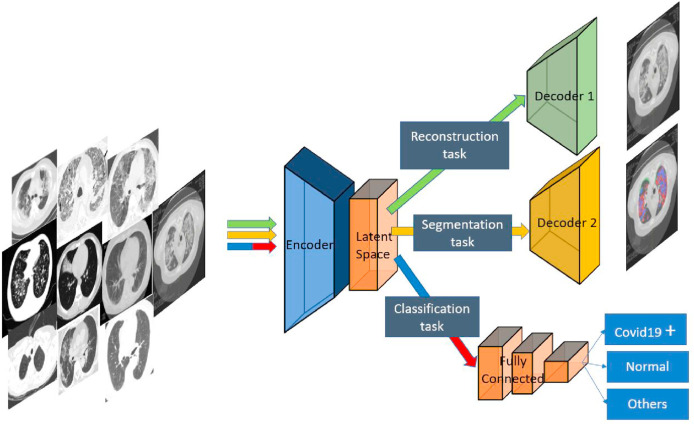
We propose a new MTL architecture based on 3 tasks: 1) COVID vs Normal vs Other Infections classification, 2) COVID lesion segmentation, 3) image reconstruction. The two first tasks are essential ones, while the third task is added to enhance the feature representation extracted. In this work, we choose to use a hard parameter sharing to share parameters between the different tasks (see Fig. 4). We create a common encoder for the three tasks which takes a CT scan as input, and its output is then used to the reconstruction of the image via a first decoder, to the segmentation via a second decoder, and to the classification of COVID vs Normal vs Other Infections classification image via a multi-layer perceptron.
Each convolutional layer, denoted C(m), consists of F(m) feature maps, where m is the layer number. For the first layer, C(1), each feature map is obtained by convolving the volume of interest with a weight matrix Wi (1) to which a bias term bi (1) is added, where i is the feature map number. Then, the output goes through a non linear function f(x), where x is the input to a neuron, such as:
| (1) |
Each element of a feature map, ci (1), is obtained by convolving the input x with a kernel. The F(1) weight matrices (one matrix per feature map) are learned by looking at different position of the input, leading to the extraction of the description of features. Thus, the weight parameters are shared for all lesion or infection input sites, so that the layer has an equivariance property and is invariant to the input lesion transformations (such as translation and rotation). It also results in a sparse weight, which means that the kernel can detect small, but meaningful features.
In order to extract high-level features from the low-level ones obtained in the initial layer, other layers are added. Each feature map in the other layers are obtained as follow:
| (2) |
2.1.1. The encoder-decoder
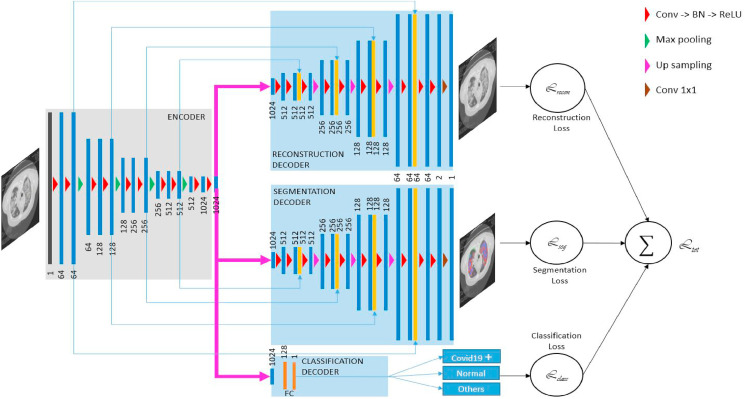
The encoder-decoder is based on the U-NET architecture [31] for both reconstruction and segmentation tasks (Fig. 5 ). The encoder is used to obtain the disentangled feature representation. It includes a convolutional block followed by skip connection. In order to maintain the spatial information, we use a convolution with stride = 2 to replace pooling operation. It's likely to require different receptive fields when segmenting different regions in an image. All convolutions are 3 × 3 and the number of filter is increased from 64 to 1024. Each decoder level begins with upsampling layer followed by a convolution to reduce the number of features by a factor of 2. Then the upsampled features are combined with the features from the corresponding level of the encoder part using concatenation.
Fig. 5.
Our proposed architecture, composed of an encoder and two decoders for image reconstruction and infection segmentation. A fully connected layers are added for classification (COVID vs Normal vs Other Infections classification).
2.1.2. The reconstruction task T1
We trained the model with a linear activation for the output and a mean squared error for the loss function (Lrecon) and used accuracy as the metric:
| (3) |
where y_true is the true label and y_predict is the predicted label.
2.1.3. The segmentation task T2
We used the same architecture as the reconstruction except for the activation function for the output, which is a sigmoid. The loss function is based on the dice coefficient loss (Lseg) which is considered as the metric:
| (4) |
| (5) |
where ε is the smoothing factor and used to avoid a division by zero.
2.1.4. The classification task T3
The resulting set of feature maps, encloses the entire spatial local information, as well as the hierarchical representation of the input. Then, each feature map is flattened out, and all the elements are collected into a single vector V of dimension K, providing the input for a fully connected hidden layer, called h, consisting of H units. The activation of the i(th) unit of the h hidden layer is given by:
| (6) |
In details, the output of the encoder is a tensor of mini_batch x 32 × 32 x 1024 to which we add a convolutional layer followed by a maxpooling, and then a flatten operation to convert the data to a mono-dimensional tensor to perform the classification. The multilayer perceptron consist of a two Dense layer with 128 and 64 neurons respectively, with a dropout of 0.5 and the activation function . The last layer is a Dense layer with three neurons for image classification using a sigmoid activation and a binary cross entropy is used as the loss function (Lclass):
| (7) |
which is a special case of the multinomial cross-entropy loss function for m = 2:
| (8) |
where n is the number of patients, y is the class label. The output layer consists of 3 neurons, each one output a binary value such as: ŷij (0,1): j ŷij = 1 i,j is the prediction of a COVID presence for the first neuron. The second neuron returns 1 if the patient is normal, 0 otherwise, and for the third neuron 1 if the patient has another infection and 0 otherwise. The patient is positive if he has the COVID-19. In our experiments, the Adam optimizer [20] algorithm was used with a mini batches of 4 and a learning rate of 0.0001. The global loss function (loss glob) for the 3 tasks is defined by:
| (9) |
Our model was trained for 500 epochs with an early stopping of 10.
2.2. Implementation
The implementation of our method was done using the keras library with tensorflow in backend. The model was performed on an nvidia p6000 quadro gpu with 24 GB, and 128 RAM.
3. Experiments
Three experiments are conducted to evaluate our model.
Experiment 1: The first experiment consisted of tuning the hyperparameters and add/remove a task to find the best model using only the training dataset. Several models were developed by combining the tasks 2 by 2 and the 3 tasks with different resolutions of images (512 × 512 and 256 × 256). The combination of the first task and the second one is only to evaluate segmentation results, since it is for image reconstruction and infection segmentation, while the peer T1 and T3 is for classification.
Experiment 2: The second experiment is to compare our model with state of the art method U-NET in order to evaluate the performance on the segmentation task. Two U-NET with different resolutions were trained: 512 × 512 and 256 × 256.
Experiment 3: Different state of the art models were compared to ours on the classification task. We use: Alexnet, VGG-16, VGG-19, ResNet50, 169-layer DenseNet, InceptionV3, Inception-ResNet v2 and Efficient-Net. We have also added an 8 layer deep neural network with 6 convolutional layers, where each one is followed by a Maxpooling and a Dropout regularization of 25% to prevent the model from overfitting. The feature maps go from 8 to 256 by a factor of 2 between each two layers. We used 3 × 3 filter for convolution and 2 × 2 for Maxpooling. Then a Flatten followed by two Dense layers with 128 neurons and 3 neurons respectively. A Dropout of 50% is also applied to the first layer to reduce and prevent overfitting. The activation function is for all layers except the last one which is a . The loss function is the binary cross-entropy and the metric is the accuracy, with the Adam optimizer. The CNN was optimized in order to ensure a fair comparison with our proposed model. The model was trained for 500 epochs with an early stopping of 10, in the same condition as our model.
To find the best hyperparameters, the influence of F, the number of feature maps (8–64), the number of neurons (128–4096) were evaluated, as well as different receptive field sizes (, ) and different sizes of mini-batch (2–16). Several expressions of f(x), the activation function, were also evaluated (relu, elu, selu and tanh), to choose finally .
4. Validation methodology
For the validating methodology, we split the data for training, validation and test as shown in Table 1. Among the 349 COVID cases in the training, the ground truth for the infection label (segmentation task) was available for 50 CT scans. Twenty others were in the validation and thirty in the test set. For normal patients, 50 were used in validation, 50 in test and 325 in training. For other infections, different kinds of pathology such as lung cancer or cases were selected randomly in training, validation and test. For a fair comparison, the other methods were trained, validated and tested on the same group of data. The performance of the models were evaluated using the dice coefficient for the segmentation task, area under the ROC curve (AUC) for the classification, and the accuracy (Acc), sensitivity (Sens), specificity (Spec) for both [1], such as:
| (10) |
where TP is the true positives, FN is the false negatives and TP + FN is the number of patients classified positively, or the segmented lesion infection.
| (11) |
where TN is the true negatives, FP is the false positives and TN + FP is the number of patients classified negatively, or the non segmented region.
| (12) |
For each curve, the definition of the thresholds was determined using the method proposed by Fawcett [13], and the optimal cut-off point was defined using Youden's index.
5. Results
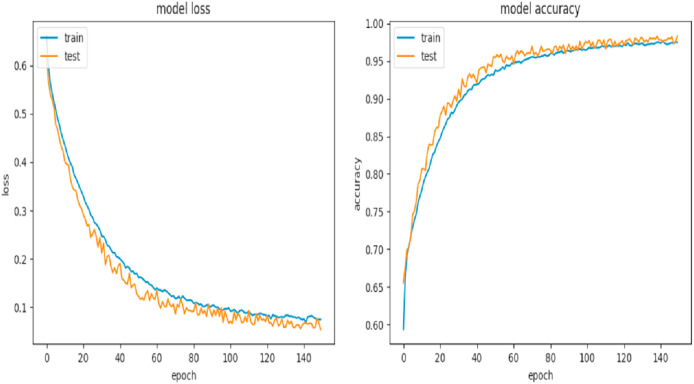
The main results of the three experiments are shown in Table 2, Table 3 . Metrics include: dice coefficient, accuracy, sensibility, specificity and the area under the ROC curve. The neural network was trained for 500 epochs with an early stopping of 10. In Fig. 6 , the learning curve and the loss curve obtained from the train and validation sets respectively show the stability of our model. From Fig. 6 we can observe that the training and validation losses decrease to a point of stability, and have a small gap between them. Early stopping of the training provides a robust mechanism to avoid overfitting, like the behavior of our model.
Table 2.
Segmentation results. Experiment 1: results with different tasks (T1: classification, T2: Segmentation, T3: reconstruction). Experiment 2: segmentation results comparing with U-NET.
| Method | Dice_coef | ACC | Sens | Spec |
|---|---|---|---|---|
| Experiment 1 | ||||
| T1 & T2 | 80.3% | 88.34% | 76.8% | 98.8% |
| T2 & T3 | 79.34% | 85.67% | 74.7% | 98.3% |
| Ours (T1&T2&T3 512 x 512) | 84.8% | 91.03% | 84.10% | 99.41% |
|
Ours 256 x 256 |
88.0% |
95.23% |
90.2% |
99.7% |
| Experiment 2 | ||||
| U-NET 512 × 512 | 76.03% | 81.14% | 69.51 | 97.34% |
| U-NET 256 × 256 | 77.69% | 83.40% | 73.10% | 98.24% |
| Ours 256 x 256 | 88.0% | 95.23% | 90.2% | 99.7% |
Table 3.
Classification results: Experiment 1 for optimizing hyperparameters and choosing the best combination of tasks. Experiment 3 for classification.
| Method | ACC | Sens | Spec | AUC | |
|---|---|---|---|---|---|
| Experiment 1 | T1 & T3 | 78.66 | 0.88 | 0.79 | 0.83 |
| T2 & T3 | 79.33 | 0.81 | 0.75 | 0.81 | |
| Ours (T1 & T2 & T3 512 × 512) | 91.13 | 0.94 | 0.85 | 0.94 | |
|
Ours(T1 & T2 & T3 256 x 256) |
94.67 |
0.96 |
0.92 |
0.97 |
|
| Experiment 3 | CNN 8-layers | 74.67 | 0.8 | 0.70 | 0.78 |
| Encoder-Dense | 70.04 | 0.75 | 0.61 | 0.72 | |
| AlexNET | 56.67 | 0.67 | 0.64 | 0.66 | |
| VGG-16 | 62.67 | 0.67 | 0.65 | 0.66 | |
| VGG-19 | 66.14 | 0.77 | 0.61 | 0.69 | |
| ResNet50 | 86.67 | 0.9 | 0.83 | 0.88 | |
| 169-layer DenseNet | 83.33 | 0.91 | 0.83 | 0.88 | |
| InceptionV3 | 82.67 | 0.88 | 0.78 | 0.82 | |
| Inception-ResNet V2 | 85.33 | 0.84 | 0.88 | 0.90 | |
| Efficient-Net | 90.67 | 0.91 | 0.85 | 0.93 | |
| Ours | 94.67 | 0.96 | 0.92 | 0.97 |
Fig. 6.
Learning curve of our proposed model. Left is the model loss and right is the model accuracy per epoch.
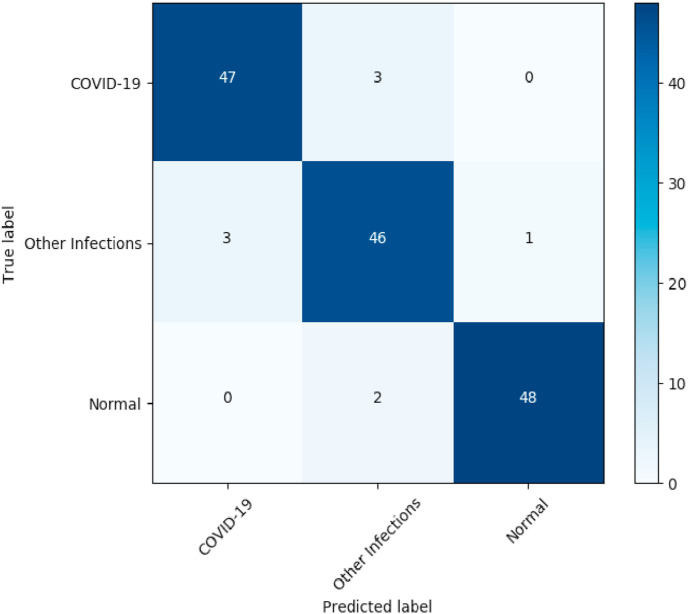
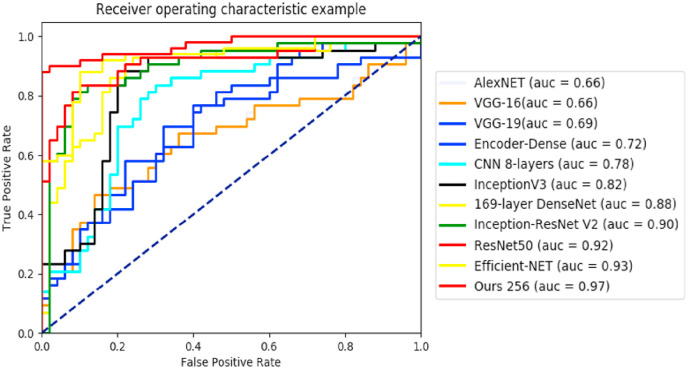
Experiment 1: As shown in Table 2 for the segmentation and Table 3 for the classification, the best dice coefficient = 88%, accuracy (ACC = 0.94.7) and area under the curve (AUC = 0.97) were obtained with the combination of the three tasks of image reconstruction, infection segmentation and image classification, with all images resized to 256 × 256. The results of 4 other experiments were also shown with multi-task learning but with a higher resolution of 512 × 512, and the combination 2 by 2 of the other tasks. The major differences between our best model and the model with higher resolution are in sensitivity (0.96 vs 0.94) and specificity (0.92 vs 0.85). Compared to the peer combination of T1 and T3 for segmentation our model proved to be more performant with an improvement of +9% of dice, and a higher AUC and accuracy compared to the peer T1 and T3 for classification only. On the ROC curve in Fig. 8 , the advantage of using all three tasks combined can be seen by obtaining a significantly better area under the curve than when two tasks are used. The same results were observed for the peer segmentation and classification without reconstruction. Those results confirm the usefulness of the reconstruction task to extract meaningful features and to help improve the results of the other two tasks. For multi-class classification, as we can observe from the confusion matrix of our model in Fig. 10 , 47 of 50 COVID cases were classified correctly, while only 3 were miss-classified as other infections. The same observation for normal patients, only 2 were misclassified as other infections, while no normal patient was misclassified as COVID.
Fig. 8.
ROC curve of Experiment 1 for COVID-19 classification.
Fig. 10.
Confusion matrix of our model.
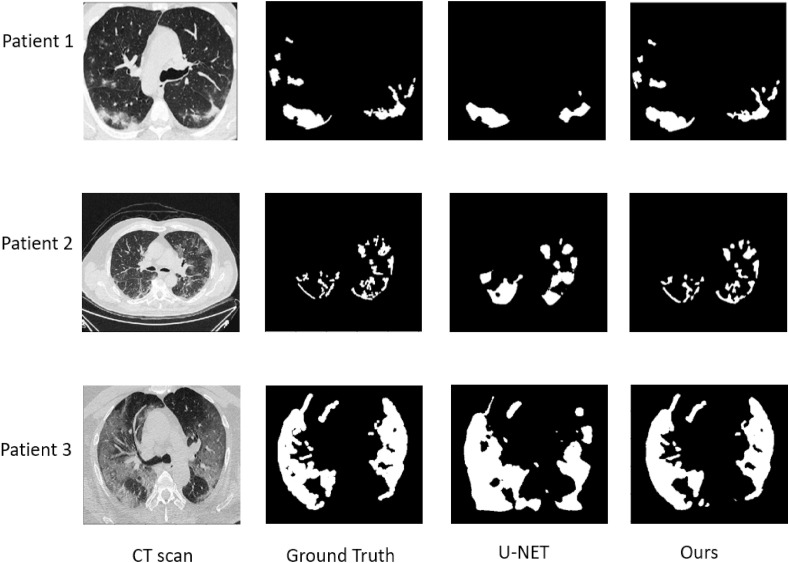
Experiment 2: In Table 2, the best results for image segmentation was obtained using our method with a dice_coef of 88% versus 77.69% and 76.69% using U-NET with 256 × 256 and 512 × 512 resolutions respectively. The combination of the reconstruction, segmentation and classification provide a higher accuracy to detect infection regions, compared to the use of the U-NET model alone. Fig. 7 shows a comparison between our model and U-NET for infection segmentation.
Fig. 7.
A comparison between our model and U-NET for infection segmentation. From left to right columns: input images, ground truth, results of U-NET, results of our method.
Experiment 3: The results of experiment 3 are also given in Table 3. We compared our multi-task deep learning model with multiple deep convolutional neural networks. The obtained results show that our model outperformed the CNN in both accuracy and AUC. The ROC curve for experiment 3 is shown in Fig. 9.
Fig. 9.
ROC curve of Experiment 3 for COVID-19 classification.
Finally, we compared our model with the state of the art methods on COVID-19 for classification and segmentation. Table 4 shows the results on the classification task. Results vary from an accuracy of 66.67% in Ref. [24] to 92.6% [38] in X-ray images, and from 84.7% to 90.8% for CT scans. Our model outperformed state of the art methods with an accuracy of 94.67%. Zhou et al. [47], who performed only the segmentation task achieved 61.0% using a modified U-NET and 69.1% using an attention mechanism. Other results reported in Ref. [9] reached a dice coefficient of 85.0%, which is less than our model with a dice coefficient of 88.0%. The results for the segmentation are shown in Table 5 .
Table 4.
A quantitative comparison between our model and state of the art for the classification task.
| Method | Modality | ACC | Sens | Spec |
|---|---|---|---|---|
| Alexnet Loey et al. (2020) [24] | X-ray | 66.67 | 66.67 | – |
| Resnet18 Loey et al. (2020) [24] | X-ray | 69.47 | 66.67 | – |
| ShuffleNet + SVM Sethy and Behera [33] | X-ray | 70.66 | 65.26 | – |
| Googlenet Loey et al. (2020) [24] | X-ray | 80.56 | 80.56 | – |
| CNN Zhao et al. (2020) [45] | CT | 84.7 | 76.2 | – |
| Ying et al. [36] | CT | 86 | – | – |
| Xu et al. [7] | CT | 86.7 | – | – |
| Ozturk et al. (multiclass) [26] | X-ray | 87.02 | – | – |
| Li and Zhu [23] | X-ray | 88.9 | – | – |
| Hemdan et al. [15] | X-ray | 90 | – | – |
| Zheng et al. [46] | CT | 90.8 | – | – |
| Wang et al. [38] | X-ray | 92.6 | – | – |
| Ours | CT | 94.67 | 96 | 92 |
Table 5.
A quantitative comparison between our proposed model and state of the art for the segmentation task.
| Method | Dice_coef |
|---|---|
| U-Net + DL Zhou et al. (2020) [47] | 61.0% |
| U-Net + FTL Zhou et al. (2020) [47] | 66.7% |
| AU-Net + DL Zhou et al. (2020) [47] | 68.5% |
| AU-Net + FTL Zhou et al. (2020) [47] | 69.1% |
| Backbone + PPD + RA + EA Fan et al. (2020) [12] | 73.9% |
| JCS Wu et al. (2020b) [41] | 77.5% |
| JCS′ Wu et al. (2020b) [41] | 78.3% |
| U-net Chen et al. (2020b) [9] | 82.0% |
| M − A Chen et al. (2020b) [9] | 85.0% |
| M − R Chen et al. (2020b) [9] | 84.0% |
| Ours | 88.0% |
6. Discussion
We have developed a new deep learning multi-task model to jointly detect COVID-19 CT images and segment the regions of infection. Our architecture is general, which means that it can be used for other segmentation-classification applications. We have also compared with several state of the art algorithms such as U-NET and CNNs. To show the performance of our method, we tested the different combinations of tasks 2 by 2 and all the 3 tasks simultaneously with different images resolutions. Our motivation was to leverage useful information contained in multiple related tasks to improve both segmentation and classification performances.
Multitasking can handle small data problems well, although each task can have a relatively small data set. In our study, we were able to increase the size of the database, in total 1044 images by the combination of several databases, to learn the disentangled representation. Although we have a database of 100 images for segmentation, thanks to the learned latent representation, we have obtained good segmentation results.
The state of the art U-NET has shown impressive results to deal with image segmentation in recent years, just like the classification with Alexnet, VGG-16, VGG-19, ResNet50, DensNet. However, these segmentation or classification methods usually require a large amount of annotated datasets to work efficiently. Due to the lack of annotated data in the medical imaging field, other mechanisms can be included to improve its generalisation ability. In this work, we propose to use a multitask learning approach that can jointly improve the U-NET model and classification models by enhancing its encoder. Indeed, by using a shared encoder for the classification and segmentation tasks, it is able to extract more meaningful information from the CT scan relating to the COVID-19 characteristics, which improves both tasks simultaneously with less annotated datasets. Furthermore, adding a third task for image reconstruction allows the encoder to refine the image characteristics to make a further improvement for both the classification and segmentation tasks. Thus, multitask learning can be used to improve U-NET and other classification models especially in the case where annotated data are limited.
In addition to the many advantages of using CT images to spot early COVID-19 patients and isolate them, deep learning methods using CT images can be used as a tool to assist physicians fighting this new spreading disease, as they can be used also to not only classify and segment images in the medical field, but also to predict the outcome of treatment [4,28].
7. Conclusion
In this paper, we proposed a multi-task learning approach to detect COVID-19 from CT images and segment the regions of interest simultaneously. Our method can improve segmentation results even if we don't have many segmentation ground truths. This thanks to the ground truths of classification which are relatively easy to obtain compared to those of segmentation. Our method shows very promising results. It outperformed the state of the art methods when used alone for image segmentation such as U-NET or image classification such as CNNs. We have shown that by combining jointly these two tasks, the method improves for both segmentation and classification performances. Moreover, adding a third task such as image reconstruction, the encoder can extract meaningful feature representation which help the other tasks (classification and segmentation) to improve even more their performances.
From experiment 2, we observe a neat improvement when using the multitask approach with a dice coefficient of 88% for segmentation, 10% higher than when using the state of the U-net alone. With a specificity of 99.7% and a sensitivity of 90.2%, the segmentation results outperformed other models without multitask learning approach and when combining a peer of tasks. For the classification results, with an AUC = 0.97 and an accuracy higher than 94%, our model shows a big improvement compared to other models with results ranging from 56% to 90%. Our method uses only CT images. Other information, like patient information, is not included in our architecture. In addition, the performance of our method was performed from a dateset of 150 patients. In future work, we will study new types of networks to take into account other useful information and test our method on a larger database to confirm its good performance.
Declaration of competing interest
None Declared.
Biographies

Amine Amyar, Publications:
1- Amyar, A. et al. “
3-D RPET-NET: development of a 3-D pet imaging convolutional neural network for radiomics analysis and outcome prediction.” IEEE Transactions on Radiation and Plasma Medical Sciences 3.2 (2019): 225-231.
2-Amyar, Amine, et al. “Radiomics-net: Convolutional neural networks on FDG PET images for predicting cancer treatment response.” Journal of Nuclear Medicine 59.supplement 1 (2018): 324-324
3- Amyar, Amine, et al. “Weakly supervised pet tumor detection usingclass response.” arXiv preprint arXiv:2003.08337 (2020).

Romain Modzelewski, Publications:
1- Belharbi, Soufiane, et al. “Spotting L3 slice in CT scans using deep convolutional network and transfer learning.” Computers in biology and medicine 87 (2017): 95-103.
2- Paul, Desbordes, et al. “Feature selection for outcome prediction in oesophageal cancer using genetic algorithm and random forest classifier.” Computerized Medical Imaging and Graphics 60 (2017): 42-49.
3- Lanic, Hélène, et al. “Sarcopenia is an independent prognostic factor in elderly patients with diffuse large B-cell lymphoma treated with immunochemotherapy.” Leukemia & lymphoma 55.4 (2014): 817–823.

Hua Li, Publications:
1- Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. KK Brock, S Mutic, TR McNutt, H Li, ML Kessler Medical physics 44 (7), e43-e76
2- Vessels as 4-D curves: Global minimal 4-D paths to extract 3-D tubular surfaces and centerlines H Li, A Yezzi IEEE transactions on medical imaging 26 (9), 1213-1223
3- Current status of Radiomics for cancer management: Challenges versus opportunities for clinical practice H Li, I El Naqa, Y Rong Journal of applied clinical medical physics 21 (7), 7

Su Ruan, Publications:
1- Nie, Dong, et al. “Medical image synthesis with context-aware generative adversarial networks.” International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2017.
2- Joint tumor segmentation in PET-CT images using co-clustering and fusion based on belief functions C Lian, S Ruan, T Denoeux, H Li, P Vera IEEE Transactions on Image Processing 28 (2), 755-766
3- Segmenting multi-source images using hidden markov fields with copula-based multivariate statistical distributions J Lapuyade-Lahorgue, JH Xue, S Ruan IEEE Transactions on Image Processing 26 (7), 3187-3195
References
- 1.Abbasian Ardakani A., Bitarafan-Rajabi A., Mohammadzadeh A., Mohammadi A., Riazi R., Abolghasemi J., Homayoun Jafari A., Bagher Shiran M. A hybrid multilayer filtering approach for thyroid nodule segmentation on ultrasound images. J. Ultrasound Med. 2019;38:629–640. doi: 10.1002/jum.14731. [DOI] [PubMed] [Google Scholar]
- 2.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. 2020. Covid-caps: A Capsule Network-Based Framework for Identification of Covid-19 Cases from X-Ray Images. arXiv preprint arXiv:2004.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amyar A., Ruan S., Gardin I., Chatelain C., Decazes P., Modzelewski R. 3-d rpet-net: development of a 3-d pet imaging convolutional neural network for radiomics analysis and outcome prediction. IEEE Transactions on Radiation and Plasma Medical Sciences. 2019;3:225–231. [Google Scholar]
- 4.Amyar A., Ruan S., Gardin I., Herault R., Clement C., Decazes P., Modzelewski R. Radiomics-net: convolutional neural networks on fdg pet images for predicting cancer treatment response. J. Nucl. Med. 2018;59 324–324. [Google Scholar]
- 5.Badrinarayanan V., Kendall A., Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39:2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 6.Basavegowda H.S., Dagnew G. Deep learning approach for microarray cancer data classification. CAAI Trans. Intell. Technol. 2020;5:22–33. [Google Scholar]
- 7.Butt C., Gill J., Chun D., Babu B.A. vol. 1. Applied Intelligence; 2020. (Deep Learning System to Screen Coronavirus Disease 2019 Pneumonia). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caruana R. Multitask learning. Mach. Learn. 1997;28:41–75. [Google Scholar]
- 9.Chen X., Yao L., Zhang Y. 2020. Residual Attention U-Net for Automated Multi-Class Segmentation of Covid-19 Chest Ct Images. arXiv preprint arXiv:2004.05645. [Google Scholar]
- 10.Ciregan D., Meier U., Schmidhuber J. 2012 IEEE Conference on Computer Vision and Pattern Recognition, IEEE. 2012. Multi-column deep neural networks for image classification; pp. 3642–3649. [Google Scholar]
- 11.Deng J., Dong W., Socher R., Li L.J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition, Ieee. 2009. Imagenet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 12.Fan D.P., Zhou T., Ji G.P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-net: automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imag. 2020;39(8) doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 13.Fawcett T. An introduction to roc analysis. Pattern Recogn. Lett. 2006;27:861–874. [Google Scholar]
- 14.Greenspan H., Van Ginneken B., Summers R.M. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans. Med. Imag. 2016;35:1153–1159. [Google Scholar]
- 15.Hemdan E.E.D., Shouman M.A., Karar M.E. 2020. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 16.Hinton G.E., Sabour S., Frosst N. 2018. Matrix Capsules with Em Routing. [Google Scholar]
- 17.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 18.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., Xu Z., Zheng Z. 2020. Ai-assisted Ct Imaging Analysis for Covid-19 Screening: Building and Deploying a Medical Ai System in Four Weeks. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayalibay B., Jensen G., van der Smagt P. 2017. Cnn-based Segmentation of Medical Imaging Data. arXiv preprint arXiv:1701.03056. [Google Scholar]
- 20.Kingma D.P., Ba J. 2014. Adam: A Method for Stochastic Optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- 21.Krizhevsky A., Sutskever I., Hinton G.E. Advances in Neural Information Processing Systems. 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 22.LeCun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE. 1998;86:2278–2324. [Google Scholar]
- 23.Li X., Zhu D. 2020. Covid-xpert: an Ai Powered Population Screening of Covid-19 Cases Using Chest Radiography Images. arXiv preprint arXiv:2004.03042. [Google Scholar]
- 24.Loey M., Smarandache F., M Khalifa N.E. Within the lack of chest covid-19 x-ray dataset: a novel detection model based on gan and deep transfer learning. Symmetry. 2020;12:651. [Google Scholar]
- 25.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. 103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S., Shukla P.K. IRBM; 2020. Deep Transfer Learning Based Classification Model for Covid-19 Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul D., Su R., Romain M., Sébastien V., Pierre V., Isabelle G. Feature selection for outcome prediction in oesophageal cancer using genetic algorithm and random forest classifier. Comput. Med. Imag. Graph. 2017;60:42–49. doi: 10.1016/j.compmedimag.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Qi G., Wang H., Haner M., Weng C., Chen S., Zhu Z. Convolutional neural network based detection and judgement of environmental obstacle in vehicle operation. CAAI Trans. Intell. Technol. 2019;4:80–91. [Google Scholar]
- 30.Redmon J., Divvala S., Girshick R., Farhadi A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. You only look once: unified, real-time object detection; pp. 779–788. [Google Scholar]
- 31.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 32.Ruder S. 2017. An Overview of Multi-Task Learning in Deep Neural Networks. arXiv preprint arXiv:1706.05098. [Google Scholar]
- 33.Sethy P.K., Behera S.K. 2020. Detection of Coronavirus Disease (Covid-19) Based on Deep Features. [Google Scholar]
- 34.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. 2020. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation and Diagnosis for Covid-19. arXiv preprint arXiv:2004.02731. [DOI] [PubMed] [Google Scholar]
- 35.Singh D., Kumar V., Kaur M. Classification of covid-19 patients from chest ct images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020:1–11. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Zhao H., Jie Y., Wang R. medRxiv; 2020. Deep Learning Enables Accurate Diagnosis of Novel Coronavirus (Covid-19) with Ct Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szegedy C., Toshev A., Erhan D. Advances in Neural Information Processing Systems. 2013. Deep neural networks for object detection; pp. 2553–2561. [Google Scholar]
- 38.Wang L., Wong A. 2020. Covid-net: A Tailored Deep Convolutional Neural Network Design for Detection of Covid-19 Cases from Chest Radiography Images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X. medRxiv; 2020. A Deep Learning Algorithm Using Ct Images to Screen for Corona Virus Disease (Covid-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 41.Wu Y.H., Gao S.H., Mei J., Xu J., Fan D.P., Zhao C.W., Cheng M.M. 2020. Jcs: an Explainable Covid-19 Diagnosis System by Joint Classification and Segmentation. arXiv preprint arXiv:2004.07054. [DOI] [PubMed] [Google Scholar]
- 42.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Chen Y., Su J., Lang G. 2020. Deep Learning System to Screen Coronavirus Disease 2019 Pneumonia. arXiv preprint arXiv:2002.09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yosinski J., Clune J., Bengio Y., Lipson H. Advances in Neural Information Processing Systems. 2014. How transferable are features in deep neural networks? pp. 3320–3328. [Google Scholar]
- 44.Zhang Y., Yang Q. 2017. A Survey on Multi-Task Learning. arXiv preprint arXiv:1707.08114. [Google Scholar]
- 45.Zhao J., Zhang Y., He X., Xie P. 2020. Covid-ct-dataset: A Ct Scan Dataset about Covid-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 46.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. medRxiv; 2020. Deep Learning-Based Detection for Covid-19 from Chest Ct Using Weak Label. [Google Scholar]
- 47.Zhou T., Canu S., Ruan S. 2020. An Automatic Covid-19 Ct Segmentation Based on U-Net with Attention Mechanism. arXiv preprint arXiv:2004.06673. [DOI] [PMC free article] [PubMed] [Google Scholar]